Abstract
UV resonance Raman (UVRR) spectroscopy is a powerful tool for investigating the structure of biological molecules, such as proteins. Numerous UVRR spectroscopic markers that provide information on the structure and environment of the protein backbone and of amino acid side chains have recently been discovered. Combining these UVRR markers with hydrogen-deuterium exchange and advanced statistics is a powerful tool for studying protein systems, including the structure and formation mechanism of protein aggregates and amyloid fibrils. These techniques allow crucial new insights into the structure and dynamics of proteins, such as polyglutamine peptides, which are associated with 10 different neurodegenerative diseases. Here we summarize the spectroscopic structural markers recently developed and the important insights they provide.
Keywords: Raman, UVRR, Fibrils, Polyglutamine
1.0. Introduction
Raman scattering is an inelastic light scattering phenomenon in which the Raman scattered light is frequency shifted from that of the incident light. The frequency shifts of the Raman scattered light correspond to the frequencies of molecular vibrations. Thus, Raman vibrational spectroscopy monitors the vibrations of a molecular species, which are sensitive to the conformations and environments of molecules.
In off-resonance Raman scattering, the incident excitation wavelength does not occur within an absorption band of the molecule. In contrast, resonance Raman spectroscopy uses an excitation wavelength within an analyte absorption band. Resonance Raman spectroscopy selectively enhances vibrations that couple to the resonant electronic transition, producing enhancement factors as high as 108-fold.
In non-resonance Raman spectroscopy all of a molecule’s vibrations contribute to the spectrum with comparable intensities. However, in resonance Raman spectroscopy, a small subset of vibrations localized on the resonant chromophore dominates the UVRR spectrum. Thus, the sensitivity and selectivity of UVRR is ideal for studying macromolecules that have crowded, complicated non-resonance Raman spectra that are difficult to analyze.
Protein folding is a topic of great interest because the final, folded structure of the protein dictates its biological activity [1]. An understanding of the rules of protein folding would allow for great insights into protein structure and function. This would lead to advances in many fields such as drug design, and for the treatment of diseases caused by protein misfolding. However, current spectroscopic methods used to investigate protein structure provide limited information on protein conformation and dynamics.
For example: UV absorption spectroscopy [2]and circular dichroism [3] (CD) are able to detect changes in protein conformation; however, these techniques provide only low-resolution structural information. X-ray crystallography allows for atomic level structural information of crystallized proteins [4]. However, the crystal structure of the protein can differ significantly from its native, solution functional structure. Also, X-ray crystallography generally does not provide insight into protein dynamics, which are important for proper protein function. NMR spectroscopy is a powerful tool for investigating protein structure and dynamics but requires expensive isotopic labeling, high protein concentrations, and long measurement times, which makes kinetic studies difficult [5]. Thus, there is a need for the development of additional methods that can obtain complementary information on protein structure and dynamics.
The selective enhancement of UVRR makes it a powerful tool for investigating protein structure and dynamics. Judiciously selecting the excitation wavelength allows for the enhancement of vibrations found on different chromophores of a protein [6, 7]. Excitation at ~200 nm is in resonance with the π→π* absorption band of amide groups and therefore selectively enhances vibrations of the backbone peptide bonds [6, 8] and of glutamine (Gln) and asparagine (Asn) side chains [9, 10]. In contrast, excitation at ~230 nm selectively enhances vibrations of the tyrosine (Tyr) and tryptophan (Trp) side chains [6, 11] while excitation at ~400 nm is in resonance with the Soret band of hemeproteins [12].
The frequencies and intensities of bands in the UVRR spectra are sensitive to the protein’s conformation and environment. If these spectral markers bands can be identified and quantitatively characterized, they can be used to obtain detailed information on the protein structure. Here we review UVRR protein structural markers that have been recently characterized and used to achieve important insights into protein structure. For a more detailed discussion on the biological applications of UVRR, review articles by Oladepo et al. [6] and Asher et al. [7] are recommended.
2.0. Discussion
2.1. UVRR Peptide Ψ Ramachandran angle marker
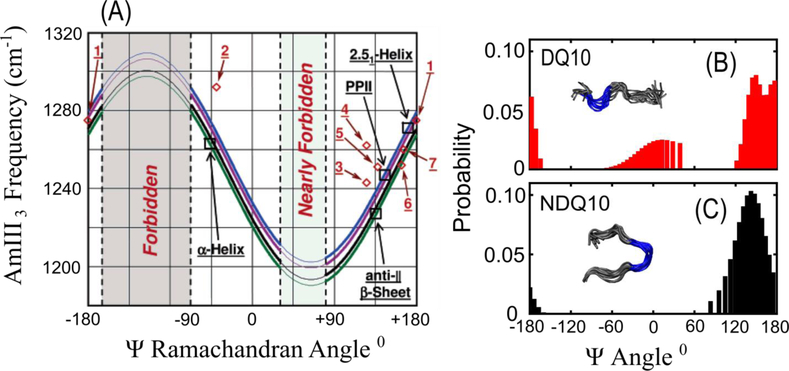
Asher et al. discovered a highly important UVRR spectral marker band that directly reports on the peptide backbone Ramachandran Ψ angle [13]. This band is called the amide III3 (AmIII3) or amide III3S (AmIII3S) band, where the superscript “S” denotes that the vibration derives from the secondary amide of the peptide backbone. The AmIII3S band consists predominantly of in phase C-N stretching and N-H bending motions and its frequency has a sinusoidal dependence on the Ψ Ramachandran angle of the peptide backbone (Figure 1a). This allows for detailed determination of a protein’s secondary structure. The sinusoidal dependence was found to derive from coupling of the N-H bending motion of the AmIII3 band with the CαH bending vibration. This coupling is strong when the N-H and CαH groups lie on the same side of the peptide bond (sort of in a cis-configuration), resulting in a downshift of the AmIII3 band frequency.
Figure 1:
(A) Sinusoidal dependence of the AmIII3S band frequency on the Ψ Ramachandran angle. The four curves show the correlation for peptide backbone in different environments: (blue) fully solvated, (purple) only C=O groups involved in peptide-peptide hydrogen bonding, (black) only N-H groups involved in peptide-peptide hydrogen bonding, and (green) N-H and C=O groups both involved in peptide-peptide hydrogen bonding. The red diamonds show experimental data points for small crystalline peptides: 1. Ala-Asp, 2. Gly-Ala-Leu, 3. Val-Glu, 4. Ala-Ser, 5. Val-Lys, 6. Ser-Ala, and 7. Ala-Ala. Figures B and C show Ψ angle distributions of the small monomeric polyQ peptides DQ10 (B) and NDQ10 (C) as derived from the AmIII3S-Ψ angle correlation. The insets show the structural models of the monomeric peptides produced from UVRR measurements and metadynamics simulations. Panel (A) was adapted with permission from reference 21. Copyright 2006 American Chemical Society. Panels B and C were adapted with permission from reference 39. Copyright 2017 American Chemical Society.
The frequency of the AmIII3S band is also dependent on the hydration of the peptide backbone and temperature of the sample. Mikhonin et al. derived a series of equations that allow for quantitative estimation of peptide Ψ angles for different sample temperatures and backbone solvation states (Figure 1a) [14].
The AmIII3S band is inhomogeneously broadened in the UVRR spectra of proteins due to the distribution of backbone conformations observed. Asher et al. showed that this inhomogeneous broadening can be used to estimate the protein peptide bond Ψ angle distribution. This is done by modeling the inhomogeneously broadened AmIII3 band as a sum of Lorentzian bands with a width equal to the homogenous linewidth of the AmIII3 band [15]. Using the set of equations derived by Mikhonin et al. [14], the Ψ angle for each Lorentzian band can be calculated, producing a Ψ angle distribution for the peptide (Figure 1b and c). Furthermore, we can calculate the Gibbs free energy landscape of the Ψ angle coordinate by applying the Boltzmann relation to the Ψ angle distribution [16, 17]. This methodology has been extensively used to investigate the secondary structure of peptides and proteins [6, 10, 18–22].
2.2. UVRR Side Chain Marker Bands
Numerous UVRR spectroscopic markers have been developed to monitor the structure and solvation environment of amino acid side chains. Aromatic amino acid side chains have strong absorption bands in the deep UV [11] and show selective resonance enhancement with ~230 nm excitation [11, 23]. Asher et al. found that the UVRR spectral intensities of resonance enhanced Tyr and Trp bands are dependent on their environment [24–26]. This is caused by an increase in the molar absorptivity and a redshift in the absorbance spectra of Tyr and Trp with decreasing solvation [26]. Because the resonance Raman intensity is proportional to the square of the molar absorptivity [27, 28], the UVRR band intensities of Trp and Tyr are dependent on side chain solvation. This effect has been used to investigate the structures and structural changes of various proteins including myoglobin [26, 29]. Also, Lednev et. al. have shown that the Raman cross section of the ~1000 cm−1 Phe Raman band, which derives from phenyl ring stretching, is dependent on water exposure of the Phe side chain. [30] Therefore, this band is a useful intrinsic UVRR indicator of local environment and protein tertiary structure.
The arginine (Arg) side chain is also resonance enhanced with deep UV excitation (~200 nm). The UVRR spectrum of arginine contains a band at ~1180 cm−1 that consists of C-N stretching and NH2 rocking of the guanadinium (Gdn) group and is sensitive to the hydration state of the Arg side chain [31]. Hong et al. found that the ~1180 cm−1 band of Arg downshifts to ~1174 cm−1 in acetonitrile [31]. This frequency shift is caused by changes in hydrogen bonding strength between the solvent and the NH2 groups of the Arg side chain. Stronger solvent-NH2 hydrogen bonding increases the NH2 bending force constant which increases the normal mode’s frequency. This UVRR band was used to investigate the solvation of Arg residues in small peptides [31].
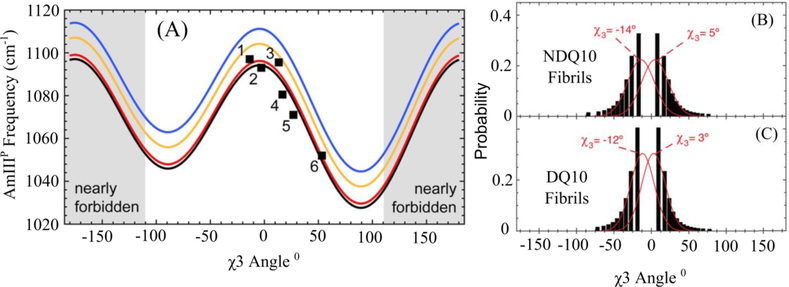
Glutamine (Gln) and asparagine (Asn) side chains show UV resonance enhancement (~200 nm excitation) from the π→π* transition of their primary amides. The UVRR spectra of Gln and Asn contain Raman bands sensitive to their amide conformations and hydration states. Punihaole et al. identified a UVRR spectral marker which reports on the χ3 (CβCγ-CδOε) torsion angle of the Gln side chain and the χ2 (CαCβ-CγOδ) torsion angle of the Asn side chain [9]. This band is called the AmIIIP band where the “P” superscript denotes that the vibration is located on the primary amide Gln or Asn side chain. The AmIIIP band consists of C-C stretching, NH2 rocking, and C-N stretching of the Gln (Asn) side chains and has a frequency that is sinusoidally dependent on the χ3 (χ2) torsion angle of Gln (Asn) (Figure 2a). This dependence originates from changes in hyperconjugation between the Cβ-Cγ (Cα-Cβ) σ and Cδ=Oε (Cγ=Oδ) π* orbitals as the χ3 (χ2) angle is rotated. When the torsion angle is at ~0° and ~180°, the σ and π* orbitals do not overlap. However, as the torsion angles approach ±90° the orbitals overlap causing electron density to shift from the σ to the π* orbital decreasing the Cβ-Cγ (Cα-Cβ) bond force constant and downshifting the AmIIIP band. Using the same procedure as that for the AmIII3-Ψ angle correlation, one can derive χ3 angle distributions from the inhomogeneous width of the AmIIIP band (Figure 2 b and c). This correlation was used to investigate the structure of the Gln side chain in small, monomeric polyglutamine (polyQ) peptides [9, 32] and in fibrils [22].
Figure 2:
(A) Dependence of the AmIII3P frequency on the Gln χ3 torsion angle. The black squares show experimental data for crystalline Gln-like compounds 1 L-Gln, 2 Gly-Gln, 3 D-Gln, 4 Gln t-butyl ester, 5 N-Acetyl-Gln, and 6 Ser-Asn. (B) and (C) show Gln side chain χ3 angle distributions for polyQ fibrils made from monomeric NDQ10 (B) and DQ10 (C) as derived from the AmIII3P-χ3 angle dependence. Panel (A) was adapted with permission from reference 15. Copyright 2015 American Chemical Society. Panels B and C were adapted with permission from reference 29. Copyright 2017 American Chemical Society.
Using UVRR, Punihaole et al. investigated the aqueous solvation dependence of the primary amide vibrations of propanamide, a small primary amide compound model for Gln and Asn side chains [33]. They discovered that almost all the bands in the UVRR spectrum increase in both intensity and frequency with increasing water solvation. In contrast, the AmI band of propanamide decreases in intensity and frequency with solvation. These trends are due to the changes in the geometry of the primary amide with solvation. As the amide group becomes increasingly hydrated, the primary amide polar resonance structure (−O-C=NH2+) becomes increasingly stable compared to the nonpolar resonance structure (O=C-NH2). As a result, the C=O bond force constant weakens while the C-N bond force constant strengthens with increasing aqueous solvation. UVRR enhancement of amide groups occurs selectively for bands that contain a large component of CN stretching [34]. Therefore, most UVRR enhanced bands will increase in frequency as the CN bond force constant increases with solvation. In contrast, the AmI band consists predominantly of C=O stretching motion whose frequency decreases as the C=O bond force constant decreases with aqueous solvation.
These structural changes also affect the intensity of propanamide’s UVRR bands. This is because the resonance Raman cross section is proportional to the square of the displacement of the enhanced vibrational normal coordinate (C-N bond) between the ground state and excited state equilibrium geometries [35]. In the primary amide excited state, the C-N and C-O bonds are elongated compared to the ground state. Therefore, most of propanamide’s UVRR bands show an increased intensity when solvated in water because the CN bond of the ground state contracts with aqueous solvation, which increases the CN bond displacement between the ground and excited state. However, the C=O bond elongates with aqueous solvation decreasing the C=O bond displacement between the ground and excited state. Thus, the AmI cross section decreases with aqueous solvation. These trends have been used to investigate the solvation of Gln side chains in monomeric polyQ peptides [9, 32] and in fibrils [22].
2.3. New insights into polyQ Peptide Structures
The structure of polyQ peptides is of significant interest because of their role in neurodegenerative diseases. Proteins containing expanded polyQ tracts are prone to aggregation, eventually forming well-ordered fibrils. This fibrillization is associated with at least 10 different neurodegenerative diseases including Huntington’s disease [36]. The Asher group used the UVRR spectral markers described above to investigate the structure of the small polyQ peptide D2Q10K2 (Q10) in the solution and fibril state.
2.3.1. Solution State Structure of Q10
Xiong et al. [10] and Punihaole et al. [32] studied the solution state structure of Q10 in two different monomeric conformations. Disaggregated Q10 (DQ10) is prepared by disaggregating the peptide using a well-established procedure [37]. Non-disaggregated Q10 (NDQ10) is made by simply dissolving the peptide in water. Using the correlation between the AmIII3S band and the Ψ Ramachandran angle [14], Punihaole et al. determined the Ψ angle distribution and secondary structure of NDQ10 and DQ10 (Figure 1 b and c) [32]. The UVRR results were quantitatively compared to Q10 structures predicted from Metadynamics simulations to obtain a molecular level structural model for NDQ10 and DQ10. They found that NDQ10 exists in a β-strand conformation that is collapsed due to intra-peptide hydrogen bonding and terminal residue electrostatic attractions. In contrast, DQ10 consists of short PPII helices interspersed with turn structures with the charged terminal residues in a 2.51-helix conformation. Punihaole et al. also qualitatively investigated the hydrogen bonding interactions of the Q10 monomer conformations and compared the results to structures found from Metadynamics simulations. This was done by examining the frequencies of the AmI bands of the peptide backbones and Gln side chains, which, as discussed above, are sensitive to the solvation state of the amide groups. They found that the Gln side chains and backbone of DQ10 are fully hydrated forming hydrogen bonds to water molecules, while NDQ10 contained a significant number of side chain-side chain and side chain-backbone hydrogen bonds.
Punihaole, et al., additionally investigated the effect of a low hydrogen bonding and dielectric environment on the structures of DQ10 and NDQ10 by adding acetonitrile to the solution. At ~50% acetonitrile, DQ10 contained a population of α-helix conformation while at higher acetonitrile concentrations DQ10 formed fibrils. Amazingly these fibrils re-dissolved in pure water; the re-dissolved peptide had the structure of the monomeric NDQ10 [32].
2.3.2. Fibril structures of Q10
Structural characterization of polyQ fibrils is difficult because of their non-crystalline and insoluble nature. UVRR is a promising new tool that can be used to gain insight into the structure of these types of systems. Punihaole et al. used UVRR spectroscopy and molecular dynamics (MD) simulations to investigate the structure of polyQ fibrils grown from the NDQ10 and DQ10 monomer states [22]. The fibrils were found in a dominantly antiparallel β-sheet conformation with an extended β-strand as the monomer unit. Using the correlation between the AmIII3P band and the Gln side chain χ3 angle [9] they determined that the χ3 angle of Gln side chains in NDQ10 and DQ10 fibrils occur at approximately −15° or 5° (Figure 2 b and c). This was consistent with their MD simulations showing that the side chains are approximately planar and extended, allowing for opposing β-sheets to form tightly interdigitated steric zippers.
2.4. Two-Dimensional Correlation Deep UV Resonance Raman Spectroscopy
Two-dimensional correlation spectroscopy (2DCoS) is a useful tool that can improve the resolution of individual bands and provide sequential and temporal information about structural and conformational changes, allowing for the determination of structural transitions, kinetics, and reaction mechanisms [38]. When combined with deep UVRR spectroscopy, 2DCoS can be used to study transient sates in complex biochemical processes, including nucleus formation in protein fibrillization [39]. Shashilov et al. used 2DCoS UVRR to investigate the early structural changes involved in hen egg white lysozyme (HEWL) fibrillization. They conclude that the fibrillization of HEWL begins with α-helix melting into disordered structures, followed by β-sheet formation, which is thought to nucleate fibrillization [40]. Additionally, Shashilov et al. performed kv correlation analysis on their 2DCoS UVRR data. This technique involves modeling the time-dependent concentration profiles as a set of exponential or sinusoidal reference functions, allowing one to characterize changes in spectral intensity as an effective rate constant. kv correlation analysis was used to determine the effective rate constants for the evolution of HEWL secondary and tertiary structures upon fibrillization [40].
One must be careful while interpreting the 2DCoS and kv analysis, as both can imply that the formation of disordered structures precedes α-helix melting. Rationally, this cannot be the case because melting of the α-helix must occur before the formation of disordered structures. This apparent anomalous order of events is explained by considering that the formation of an intermediate (i.e., random coil) means that there is a lag phase for formation of product, causing the contribution of the intermediate to appear ahead of the change to the α-helix [40].
The temporal insight provided by these 2D analysis techniques can be used to elucidate the mechanism of fibrillization. In the asynchronous 2D-Raman correlation data for lysozyme, the formation of random coil and β-sheet are not completely correlated, indicating the mechanism is a two-step process [39]. The Lednev group proved that, in the case of lysozyme denaturation, the transition of α-helix to partially unfolded structure is irreversible [41]. Combined with the negative correlation observed between the changes in α-helix and unordered structure, this suggests that the mechanism is as follows:
2.5. Hydrogen-Deuterium Exchange Combined with UVRR Spectroscopy for Probing the Structure of the Cross-β Fibril Core
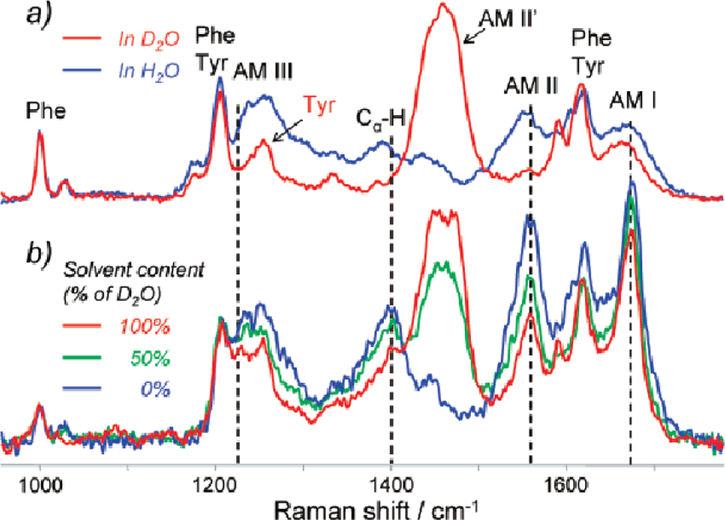
To investigate the secondary structure of the cross-β core of fibrils, one needs to isolate the UVRR signal from the fibril core from that of the rest of the fibril. To achieve this, Xu et al. used hydrogen-deuterium exchange (HX) combined with deep UVRR spectroscopy [42]. The labile protons exposed to solvent will exchange with deuterium, while those in the fibril core are protected from water exposure and cannot exchange. Upon HX of the backbone NH proton, the AmII and AmIII3 bands significantly downshift, therefore, only protonated amino acids contribute to the canonical AmII and AmIII3 band intensities [43, 44]. Figure 3 illustrates the frequency shifts that occur upon deuteration of native lysozyme. For lysozyme fibrils prepared in H2O and re-dispersed in D2O, the fibril core remains protonated because it is shielded from the solvent (Figure 3b, red curve).
Figure 3:
Deep UVRR spectra of (a) thermally denatured hen egg white lysozyme in H2O (blue) and D2O (red), and (b) lysozyme fibrils in H2O (blue), 50% D2O/H2O mixture (green), and 100% D2O (red). Spectra where collected using an excitation wavelength of 197 nm. Adapted with permission from reference [42]. Copyright 1982 American Chemical Society.
The complication of overlapping contributions of the deuterated and protonated UVRR spectra can be handled using a Bayesian approach [45]. Information about the known reference spectrum of the cross-β sheet and experimental shape of deuterated spectra are incorporated using Bayes’ theorem, which can be written as:
where P(Data|C, S, I) is the likelihood of measuring the quality of data fitting and P(C|I)∙P(S|I) are the prior probabilities for the individual component spectra and their concentrations, respectively.
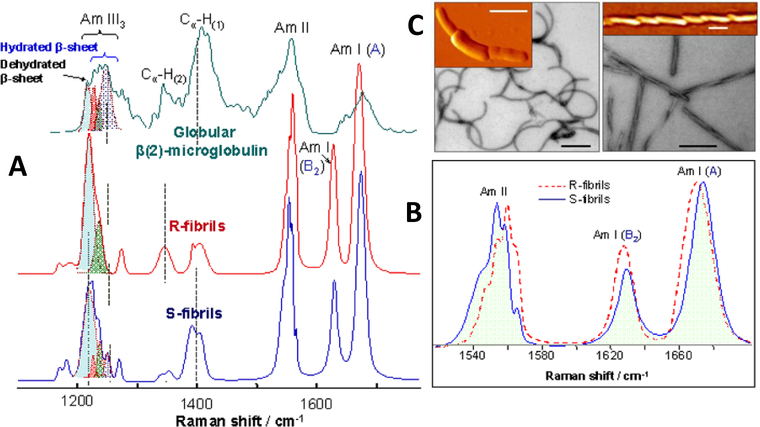
This method was used to characterize two different polymorphs (R- and S-fibrils) of amyloid fibrils formed from highly pure full-length recombinant prion protein with different morphologies (Figure 4a). For this study, globular β2-microglobulin (β2m), known to organize into two antiparallel β-sheets, was used for comparison against the R-fibrils and S-fibrils (Figure 4a). Figure 4b shows that the reconstructed Raman spectra of the cross-β-core of the R- and S-fibrils have different shifts in the AmI sub-bands and AmII bands, suggesting a difference in structural organization of the cores [46].
Figure 4:
(A) Deep UVRR spectra of antiparallel β-sheet-rich globular β2M (green) and cross-β-core of R-fibrils (red) and S-fibrils (blue). The cross-β-core spectra were extracted from the data sets obtained for partially deuterated fibrils using a Bayesian approach. (B) Amide I and amide II region of R- and S-fibril Raman spectra. (C) EM (black-and-white) and AFM (orange-yellow) images of S-fibrils (left) and R-fibrils (right). Scale bars correspond to 0.2 μm. Adapted with permission from reference [46]. Copyright 2016 American Chemical Society.
Popova et al. used UVRR and HX to probe the parallel and antiparallel fibril core structures of amyloid beta (Aβ) [47]. Raman spectra of the fibril core were obtained for fibrils prepared from a full length Aβ1–40 peptide and from Aβ32–42 fragments. The calculated Ψ dihedral angle distributions showed that Aβ1–40 peptide forms parallel β-sheet, and Aβ32–42 fragment forms antiparallel β-sheet [47]. The validity of this approach was verified by comparing the results to the solid-state NMR 3-dimensional structure of Aβ1–40 fibrils [48].
Using three genetically-engineered polypeptides of similar design, (GA)nGY(GA)nGE(GA)nGH(GA)nGK (n = 3, 4, or 5), it was directly shown that the structure of the cross-β core is sequence-dependent. Because the turn pattern of these three polypeptides is the same, pair-wise subtraction of the UVRR spectra resulted in the pure spectrum of the fibril β-sheet core. In addition, this work produced the first direct measurements of the Raman spectroscopic signature of β-turns [49]. The analysis of the AmIII3 region of UVRR spectra showed a narrow dihedral angular distribution, consistent with what was reported for lysozyme fibrils [42], suggesting that both fibrils contain a highly ordered β-sheet core. However, the experimentally determined β-sheet Ψ angle of the genetically-engineered polypeptides differs by ~15° from that of lysozyme fibrils providing the first direct evidence that the fibril structure is sequence dependent.
2.6. Spontaneous Refolding of Amyloid Fibrils
Raman spectroscopy is a convenient tool for checking the integrity of protein-based samples because little sample preparation is required, a wide range of concentrations can be probed, only small sample volumes are required, and the analysis is nondestructive. While monitoring the stability of apo-α-lactalbumin fibrils during sample preparation for atomic force microscopic characterization (desalting of the solution and cooling it from 37 °C to 25 °C), it was discovered that the protein undergoes β-sheet melting and refolding. This finding resulted in the discovery of a new protein folding-aggregation phenomenon: spontaneous refolding of protein fibrils from one polymorph to another [50]. During further investigation, HX-UVRR spectroscopy was used to characterize the different polymorphs, showing that the two polymorphs exhibit different AmIII3 peak patterns. This demonstrates a difference in the fibril β-sheet core structure of the two polymorphs [50]. Later, it was demonstrated that the spontaneous refolding of amyloid fibrils can also be induced in other ways, such as by changing sample pH [51, 52].
3.0. Conclusion
UVRR spectroscopy is a powerful tool for investigating the structure and dynamics of proteins. Careful selection of excitation wavelength allows for the selective enhancement of Raman bands of interest. For proteins, this can be used to selectively enhance the Raman bands of the peptide backbone and various side chains. The frequencies and intensities of certain UVRR bands are sensitive to the structure and solvation of the protein. Identification and characterization of these marker bands allows for insight into protein structure.
One important application of UVRR is its use in investigating protein fibrillization and its relation to disease. UVRR spectral markers allow for crucial structural insights into protein fibrils, which are difficult to study using classic biophysical techniques. By combining UVRR spectroscopy with hydrogen-deuterium exchange, one can isolate the spectral contributions of amino acids found in the core of a protein fibril, allowing for the direct interrogation of its structure. In addition, advanced statistical techniques, such as chemometrics and two-dimensional correlation spectroscopy, can by utilized to study, for example, the kinetics of fibril formation. The identification and characterization of new UVRR spectral markers and statistical techniques will enable additional probes into the structure of important protein systems such as disease causing protein fibrillization.
Acknowledgements
SAA, RSJ, and SEW acknowledges support from the University of Pittsburgh of the work described here. SEW gratefully acknowledges support through the NIH Molecular Biophysics and Structural Biology Training Grant (T32 GM088119). IKL and JH acknowledge support from the National Science Foundation Grant No. CHE-1152752.
References
- [1].Schulz GE, Schirmer RH, Principles of protein structure, Springer Science & Business Media 2013. [Google Scholar]
- [2].Rosenheck K, Doty P, The far ultraviolet absorption spectra of polypeptide and protein solutions and their dependence on conformation, Proceedings of the National Academy of Sciences of the United States of America, 47 (1961) 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fasman GD, Circular dichroism and the conformational analysis of biomolecules, Springer Science & Business Media 2013. [Google Scholar]
- [4].Strandberg B, Dickerson RE, Rossmann MG, 50 Years of Protein Structure Analysis, J. Mol. Biol, 392 (2009) 2–32.19712775 [Google Scholar]
- [5].Billeter M, Comparison of protein structures determined by NMR in solution and by X-ray diffraction in single crystals, Quarterly reviews of biophysics, 25 (1992) 325–377. [DOI] [PubMed] [Google Scholar]
- [6].Oladepo SA, Xiong K, Hong Z, Asher SA, Handen J, Lednev IK, UV resonance Raman investigations of peptide and protein structure and dynamics, Chemical reviews, 112 (2012) 2604–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Asher SA, UV resonance Raman spectroscopy for analytical, physical, and biophysical chemistry. Part 2, Anal Chem, 65 (1993) 201A–210A. [DOI] [PubMed] [Google Scholar]
- [8].Dudik JM, Johnson CR, Asher SA, Uv Resonance Raman Studies of Acetone, Acetamide, and N-Methylacetamide - Models for the Peptide-Bond, Journal of Physical Chemistry, 89 (1985) 3805–3814. [Google Scholar]
- [9].Punihaole D, Hong Z, Jakubek RS, Dahlburg EM, Geib S, Asher SA, Glutamine and Asparagine Side Chain Hyperconjugation-Induced Structurally Sensitive Vibrations, J Phys Chem B, 119 (2015) 13039–13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xiong K, Punihaole D, Asher SA, UV resonance Raman spectroscopy monitors polyglutamine backbone and side chain hydrogen bonding and fibrillization, Biochemistry, 51 (2012) 5822–5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Asher SA, Ludwig M, Johnson CR, Uv Resonance Raman Excitation Profiles of the Aromatic-Amino-Acids, J. Am. Chem. Soc, 108 (1986) 3186–3197. [Google Scholar]
- [12].Balakrishnan G, Hu Y, Oyerinde OF, Su J, Groves JT, Spiro TG, A conformational switch to β-sheet structure in cytochrome c leads to heme exposure. Implications for cardiolipin peroxidation and apoptosis, J. Am. Chem. Soc, 129 (2007) 504–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Asher SA, Ianoul A, Mix G, Boyden MN, Karnoup A, Diem M, Schweitzer-Stenner R, Dihedral psi angle dependence of the amide III vibration: A uniquely sensitive UV resonance Raman secondary structural probe, J. Am. Chem. Soc, 123 (2001) 11775–11781. [DOI] [PubMed] [Google Scholar]
- [14].Mikhonin AV, Bykov SV, Myshakina NS, Asher SA, Peptide secondary structure folding reaction coordinate: correlation between uv raman amide III frequency, Psi Ramachandran angle, and hydrogen bonding, J Phys Chem B, 110 (2006) 1928–1943. [DOI] [PubMed] [Google Scholar]
- [15].Asher SA, Mikhonin AV, Bykov S, UV Raman demonstrates that alpha-helical polyalanine peptides melt to polyproline II conformations, J. Am. Chem. Soc, 126 (2004) 8433–8440. [DOI] [PubMed] [Google Scholar]
- [16].Mikhonin AV, Myshakina NS, Bykov SV, Asher SA, UV resonance Raman determination of polyproline II, extended 2.5(1)-helix, and beta-sheet psi angle energy landscape in poly-L-lysine and poly-L-glutamic acid, J. Am. Chem. Soc, 127 (2005) 7712–7720. [DOI] [PubMed] [Google Scholar]
- [17].Ma L, Ahmed Z, Mikhonin AV, Asher SA, UV resonance Raman measurements of poly-L-lysine’s conformational energy landscapes: dependence on perchlorate concentration and temperature, J Phys Chem B, 111 (2007) 7675–7680. [DOI] [PubMed] [Google Scholar]
- [18].Xiong K, Ma L, Asher SA, Conformation of poly-L-glutamate is independent of ionic strength, Biophys Chem, 162 (2012) 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ma L, Hong Z, Sharma B, Asher S, UV resonance Raman studies of the NaClO4 dependence of poly-L-lysine conformation and hydrogen exchange kinetics, J Phys Chem B, 116 (2012) 1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hong Z, Damodaran K, Asher SA, Sodium dodecyl sulfate monomers induce XAO peptide polyproline II to alpha-helix transition, J Phys Chem B, 118 (2014) 10565–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xiong K, Asher SA, Impact of ion binding on poly-L-lysine (un)folding energy landscape and kinetics, J Phys Chem B, 116 (2012) 7102–7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Punihaole D, Workman RJ, Hong Z, Madura JD, Asher SA, Polyglutamine Fibrils: New Insights into Antiparallel beta-Sheet Conformational Preference and Side Chain Structure, J Phys Chem B, 120 (2016) 3012–3026. [DOI] [PubMed] [Google Scholar]
- [23].Ludwig M, Asher SA, Ultraviolet Resonance Raman Excitation Profiles of Tyrosine - Dependence of Raman Cross-Sections on Excited-State Intermediates, J. Am. Chem. Soc, 110 (1988) 1005–1011. [Google Scholar]
- [24].Chi ZH, Asher SA, UV resonance Raman determination of protein acid denaturation: Selective unfolding of helical segments of horse myoglobin, Biochemistry, 37 (1998) 2865–2872. [DOI] [PubMed] [Google Scholar]
- [25].Larkin PJ, Gustafson WG, Asher SA, A New Raman Cross-Section Measurement Technique Monitors the Tyrosine Environmental Dependence of the Electromagnetic-Field Strength, J Chem Phys, 94 (1991) 5324–5330. [Google Scholar]
- [26].Asher SA, Larkin PJ, Teraoka J, Ultraviolet resonance Raman and absorption difference spectroscopy of myoglobins: titration behavior of individual tyrosine residues, Biochemistry, 30 (1991) 5944–5954. [DOI] [PubMed] [Google Scholar]
- [27].Hong Z, Asher SA, Dependence of Raman and resonance Raman intensities on sample self-absorption, Appl Spectrosc, 69 (2015) 75–83. [DOI] [PubMed] [Google Scholar]
- [28].Albrecht AC, On the Theory of Raman Intensities, The Journal of Chemical Physics, 34 (1961) 1476–1484. [Google Scholar]
- [29].Chi Z, Asher SA, Ultraviolet resonance Raman examination of horse apomyoglobin acid unfolding intermediates, Biochemistry, 38 (1999) 8196–8203. [DOI] [PubMed] [Google Scholar]
- [30].Xu M, Ermolenkov VV, Uversky VN, Lednev IK, Hen egg white lysozyme fibrillation: a deep-UV resonance Raman spectroscopic study, Journal of Biophotonics, 1 (2008) 215–229. [DOI] [PubMed] [Google Scholar]
- [31].Hong Z, Wert J, Asher SA, UV resonance Raman and DFT studies of arginine side chains in peptides: insights into arginine hydration, J Phys Chem B, 117 (2013) 7145–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Punihaole D, Jakubek RS, Workman RJ, Marbella LE, Campbell P, Madura JD, Asher SA, Monomeric Polyglutamine Structures that Evolve into Fibrils, J Phys Chem B, (2017). [DOI] [PubMed] [Google Scholar]
- [33].Punihaole D, Jakubek RS, Dahlburg EM, Hong Z, Myshakina NS, Geib S, Asher SA, UV resonance Raman investigation of the aqueous solvation dependence of primary amide vibrations, J Phys Chem B, 119 (2015) 3931–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen XG, Asher SA, Schweitzer-Stenner R, Mirkin NG, Krimm S, Uv Raman Determination of the Pi-Pi* Excited-State Geometry of N-Methylacetamide - Vibrational Enhancement Pattern, J Am Chem Soc, 117 (1995) 2884–2895. [Google Scholar]
- [35].Heller EJ, Sundberg RL, Tannor D, Simple Aspects of Raman-Scattering, Journal of Physical Chemistry, 86 (1982) 1822–1833. [Google Scholar]
- [36].Orr HT, Zoghbi HY, Trinucleotide repeat disorders, Annu Rev Neurosci, 30 (2007) 575–621. [DOI] [PubMed] [Google Scholar]
- [37].Chen S, Wetzel R, Solubilization and disaggregation of polyglutamine peptides, Protein Sci, 10 (2001) 887–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shashilov VA, Lednev IK, Two-dimensional correlation Raman spectroscopy for characterizing protein structure and dynamics, J. Raman Spectrosc, 40 (2009) 1749–1758. [Google Scholar]
- [39].Shashilov V, Xu M, Ermolenkov VV, Fredriksen L, Lednev IK, Probing a Fibrillation Nucleus Directly by Deep Ultraviolet Raman Spectroscopy, J. Am. Chem. Soc, 129 (2007) 6972–6973. [DOI] [PubMed] [Google Scholar]
- [40].Shashilov VA, Lednev IK, 2D Correlation Deep UV Resonance Raman Spectroscopy of Early Events of Lysozyme Fibrillation: Kinetic Mechanism and Potential Interpretation Pitfalls, J. Am. Chem. Soc, 130 (2008) 309–317. [DOI] [PubMed] [Google Scholar]
- [41].Xu M, Shashilov VA, Ermolenkov VV, Fredriksen L, Zagorevski D, Lednev IK, The first step of hen egg white lysozyme fibrillation, irreversible partial unfolding, is a two-state transition, Protein Science, 16 (2007) 815–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Xu M, Shashilov V, Lednev IK, Probing the cross-beta core structure of amyloid fibrils by hydrogen-deuterium exchange deep ultraviolet resonance Raman Spectroscopy, J. Am. Chem. Soc, 129 (2007) 11002–+. [DOI] [PubMed] [Google Scholar]
- [43].Mikhonin AV, Asher SA, Uncoupled Peptide Bond Vibrations in α-Helical and Polyproline II Conformations of Polyalanine Peptides, The Journal of Physical Chemistry B, 109 (2005) 3047–3052. [DOI] [PubMed] [Google Scholar]
- [44].Kurouski D, Van Duyne RP, Lednev IK, Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: a review, Analyst, 140 (2015) 4967–4980. [DOI] [PubMed] [Google Scholar]
- [45].Shashilov VA, Sikirzhytski V, Popova LA, Lednev IK, Quantitative methods for structural characterization of proteins based on deep UV resonance Raman spectroscopy, Methods (San Diego, Calif, 52 (2010) 23–37. [DOI] [PubMed] [Google Scholar]
- [46].Shashilov V, Xu M, Makarava N, Savtchenko R, Baskakov IV, Lednev IK, Dissecting Structure of Prion Amyloid Fibrils by Hydrogen–Deuterium Exchange Ultraviolet Raman Spectroscopy, The Journal of Physical Chemistry B, 116 (2012) 7926–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Popova LA, Kodali R, Wetzel R, Lednev IK, Structural variations in the cross-beta core of amyloid beta fibrils revealed by deep UV resonance Raman spectroscopy, J Am Chem Soc, 132 (2010) 6324–6328. [DOI] [PubMed] [Google Scholar]
- [48].Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R, A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR, Proceedings of the National Academy of Sciences of the United States of America, 99 (2002) 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sikirzhytski V, Topilina NI, Higashiya S, Welch JT, Lednev IK, Genetic engineering combined with deep UV resonance Raman spectroscopy for structural characterization of amyloid-like fibrils, J Am Chem Soc, 130 (2008) 5852–5853. [DOI] [PubMed] [Google Scholar]
- [50].Kurouski D, Lauro W, Lednev IK, Amyloid fibrils are “alive”: spontaneous refolding from one polymorph to another, Chemical Communications, 46 (2010) 4249–4251. [DOI] [PubMed] [Google Scholar]
- [51].Kurouski D, Dukor RK, Lu X, Nafie LA, Lednev IK, Spontaneous inter-conversion of insulin fibril chirality, Chemical communications, 48 (2012) 2837–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Shanmugasundaram M, Kurouski D, Wan W, Stubbs G, Dukor RK, Nafie LA, Lednev IK, Rapid Filament Supramolecular Chirality Reversal of HET-s (218–289) Prion Fibrils Driven by pH Elevation, The journal of physical chemistry. B, 119 (2015) 8521–8525. [DOI] [PMC free article] [PubMed] [Google Scholar]