Abstract
Background
Debaryomyces hansenii exhibits a therapeutic effect on antibiotic-associated diarrhea (AAD) by promoting the growth of beneficial intestinal bacteria. Previous research has reported that AAD involves not only dysbacteriosis but also dysfunction of the activity of intestinal enzymes (such as lactase). Enzyme activities can be influenced by many other factors, such as gene expression. The present study showed that D. hansenii has a curative effect on AAD at the lactase gene level.
Material/Methods
The effect of D. hansenii on the lactase gene from intestinal bacteria in AAD mice was investigated. The diarrhea model was established with a gentamycin sulfate and cefradine capsule mixture. The antibiotic mixture (23.33 mL·kg−1·day−1) was intragastrically administered for 5 days. Subsequently, half of the diarrhea mice were treated with D. hansenii twice a day for 3 days while the other mice were intragastrically administered with the same volume of distilled water. Next, the intestinal contents were collected, and metagenomic DNA was extracted for high-throughput sequencing analysis.
Results
The Chao1 and Shannon indices decreased significantly following treatment with D. hansenii (P<0.01 and P<0.05, respectively). Moreover, the clusters in the D. hansenii group mice were quite different from those in the normal group mice and model group mice. Following treatment with D. hansenii, the quantity of lactase genes in Enterobacter sp. 638 and Modestobacter increased markedly, and the richness of intestinal bacterial lactase genes in Fretibacterium recovered.
Conclusions
D. hansenii altered the lactase-producing bacterial community structure and promoted the growth of several critical lactase-producing bacteria, such as Enterobacter sp. 638 and Modestobacter.
MeSH Keywords: Gastrula, Testolactone, Tubercidin
Background
Diarrhea is a common symptom that accompanies many diseases and occurs as 2 types: acute diarrhea and chronic diarrhea [1]. Antibiotic-associated diarrhea (AAD) has become a serious public health problem. AAD is an enteric dysbacteriosis disease caused by the overuse of antibiotics, which kills antibiotic-sensitive strains while allowing resistant strains to not only survive but also thrive [2,3]. As reported previously, the possible pathogenic features of AAD include interference with bacterial metabolism, disruption in the balance of intestinal bacteria, and reduction in disaccharidase (especially lactase) activity [4]. Our previous study determined that the lactase gene density of intestinal bacteria was reduced in AAD mice, and the ACE and Chao1 indices decreased. The abundance of some bacterial genera producing lactase was also decreased [5].
Intestinal lactase is mainly produced by microbes, such as Lactobacillus sp., Escherichia coli, Bacillus sp., Bifidobacterium sp., Enterobacter aerogenes, and Streptococcus thermophilus, although it is also produced by the intestinal mucosa, where it plays a critical role in modulating intestinal function [6–8]. Low activity and deficiency of lactase can induce diarrhea; in such cases, the symptoms can be improved if lactase is supplied [9,10]. Interestingly, lactase can be destroyed by antibiotics, especially β-lactam antibiotics such as cefradine [11,12]. Certain yeasts are probiotics that can confer a benefit to the host. These yeasts have the potential to treat diarrhea and dramatically alleviate symptoms [13]. The yeast D. hansenii can be isolated from food and the gut of experimental mice; it has important biotechnological potential in food and other industries [14,15]. D. hansenii has been shown to adjust the microecosystem balance in AAD mice [14,16]. Previous research on the impact of D. hansenii on AAD mainly focused on intestinal-cultured microbiota and various enzyme activities. A recent study reported that antibiotic-induced diarrhea was relieved by treatment with D. hansenii through adjustment of the richness and types of intestinal bacteria [17], although how D. hansenii affects bacterial functional genes is still unknown.
Because of its important role in human health, the gut microbiota has been a focus of research in recent years. Although 16S rRNA sequencing has been widely used for bacterial analysis, there have been few reports on the gut bacterial lactase gene or the sequencing of any other functional enzyme gene for bacterial analysis. As such, for the present study, a pair of special bacterial primers was designed and validated for amplifying the gut lactase gene [18]. The lactase gene sequencing results were used to probe the bacterial characteristics in the gut mucosa of AAD mice, and it was also used to explore the mechanism of Qiweibaizhu powder on AAD [19]. We previously found that the influence of Qiweibaizhu powder on bacteria in the intestinal mucosa of AAD mice was different from that on the intestinal contents of AAD mice [20]. Therefore, we aimed to investigate the therapeutic effect of D. hansenii on lactase from intestinal contents at the genetic level. The results suggest an experimental basis to explain the mechanism of D. hansenii on AAD. D. hansenii may be a novel microbioecologics.
Material and Methods
Reagents and medicine
D. hansenii was isolated from the mouse intestine and identified as described in a previous report [15]. We inoculated D. hansenii into potato sucrose liquid medium. Then, the D. hansenii was incubated for 36 h with shaking at 28°. After centrifugation at 2000 rpm for 4 min, D. hansenii cells were collected. The cells were repeatedly washed 2 times with sterile 0.85% NaCl solution to dislodge the medium component. Following this, the cells were counted by a hemocytometer and then diluted to 1010·mL−1 with an appropriate amount of sterile water. D. hansenii was stored at 4°C for use the next day [16].
Acetone, TE buffer, protease K, lysozyme, and phenol-chloroform-isoamyl alcohol (25: 24: 1/volume: volume: volume) were purchased from Dingguo Changsheng Biotechnology Company (Beijing, China). All other reagents for DNA extraction were prepared in our laboratory. Antibiotics were produced by Yichang Humanwell Pharmaceutical Company (Hubei, China) or Chung-Hwa Chemical & Pharmaceutical Industrial Company (Suzhou, Jiangsu, China).
PCR primers were 5′-TRRGCAACGAATACGGSTG-3′ (F) and 5′-ACCATGAARTTSGTGGTSARCGG-3′ (R), which were synthesized from Personal Biotechnology Company of Shanghai, China [18,19].
Animals and procedures
All mice (1 month old, weight 18–22 g) were specific pathogen-free (SPF). We purchased these mice from Hunan Slaccas Jingda Laboratory Animal Company. The sex ratio was male: female=1: 1 [SCXK (Xiang) 2013-0004]. All experiments involving animals were carried out following the relevant protocols and were approved by the university Animal Care and Use Committee. All tested mice were randomly divided into 3 groups: a model group (tlmn), a normal group (tlcn), and a D. hansenii group (tljn). There were 6 mice in each group (3 males and 3 females). The mice were kept in separate cages in a clean and quiet environment at 23–25°C and 50–70% humidity. To create the diarrhea model, the tlmn and tljn groups were intragastrically administered 0.35 mL 62.5 g/L (40 000 U/mL) cefradine capsule and gentamycin sulfate mixture, while the mice in the tlcn group were intragastrically administered 0.35 mL distilled water. All mice were intragastrically treated twice a day for 5 continuous days. After the AAD was established successfully, the model mice had symptoms of watery stool, erect coat, reduced food intake, and decreased activity. D. hansenii solution (0.35 mL, 1010·mL−1) [21] was administered to the tljn group mice twice a day for a total of 3 days. The tlcn and tlmn groups mice were intragastrically injected with 0.35 mL distilled water. Mice were then euthanized by cervical dislocation; then, the intestinal contents of the mice were collected for subsequent experiments [22]. Each sample contained mixed fecal material derived from 2 mice (1 male and 1 female).
Total DNA extraction from intestinal contents
The total DNA of the intestinal content was extracted following previously reported protocols [17,23]. The intestinal contents were washed twice with phosphate buffer solution (PBS) (0.1 mol·L−1). Subsequently, we transferred all the supernatants to fresh tubes, and the supernatants were centrifuged to collect the new sediment. The sediments were resuspended in TE buffer. The sediments were cleaned 1 time with PBS, 2 times with acetone, and 3 times with PBS. The sediments were incubated with proteinase K and lysozyme to release nucleic acids and other substances. Then, the DNA was dissolved into 50 μL TE buffer component for further analysis after being purified and extracted by CTAB/NaCl, phenol-chlororom-isoamyl alcohol (25: 24: 1/volume: volume: volume) saturated with Tris, chloroform-isoamyl alcohol (24: 1/volume: volume), absolute ethyl alcohol, and sodium acetate.
PCR amplification and sequencing
As in our previous report [18], bacterial lactase genes were PCR-amplified with our universal primers. The PCR mixture (25 μL volume) was composed of 5 μL 5×reaction buffer, 5 μL 5×high GC buffer, 0.5 μL 10 mmol/L dNTPs, 0.25 μL Q5 high-fidelity DNA polymerase, 1 μL 10 μmol/L each forward primer and reverse primer, 1 μL extracted DNA, and 11.25 μL sterilized ddH2O. The PCR was performed with initial denaturation at 98°C for 30 s, followed by denaturation at 98°C for 15 s, annealing at 46°C for 30 s, and extension at 72°C for 30 s, for a total of 32 cycles, a final extension cycle at 72°C for 5 min, and holding at 4°C. All the PCR products were determined by 2% agarose electrophoresis. Then, PCR products were sequenced by using the Illumina MiSeq method for bacterial lactase gene diversity. The sequencing service was provided by Personal Biotechnology Company (Shanghai).
Data analyses
Sequences were analyzed online using QIIME software [24] (http://qiime.org/) and Mothur software (http://www.mothur.org/). To analyze lactase gene diversity, 3 methods were used to determine the operational taxonomic units (OTUs): alpha diversity analysis (ADA), community composition analysis (CCA), and principal components analysis (PCA) [25–27]. Significant differences between different groups were determined using one-way ANOVA in SPSS 20.0. P<0.05 was regarded as a significant difference.
Results
Sample sequences and operational taxonomic units
The lactase gene sequencing data were analyzed by QIIME software (version 1.7.0, http://qiime.org/) to compare their homogeneity following initial filtering by Uchime software (version 1.35.1, http://www.mothur.org/) to remove the chimeric sequences. According to the sequencing results, there were 670 308 effective sequences and 658 355 high-quality sequences. In brief, the proportion of the bacterial lactase gene out of the total lactase gene was approximately 98% for each group.
According to gene homogeneity, the lactase gene sequences with over 97% similarity would be aligned and grouped into a single OTU. There were 1056, 1143, and 443 OTUs in the normal group, model group, and D. hansenii group, respectively. A total of 1348 OTUs were identified; with 122, 186, and 52 OTUs uniquely identified in these 3 groups, respectively (Figure 1). The results suggested that the variety of bacterial lactase genes decreased after treatment with D. hansenii.
Figure 1.

Bacterial lactase gene OTUs of intestinal contents. tlcn represents the normal group, tlmn represents the model group, and tljn represents the D. hansenii group.
Alpha diversity of lactase genes in AAD mice treated with D. hansenii
Rarefaction analysis showed that the OTUs of 9 samples gradually increased with the increasing number of measured sequences. Furthermore, the curve flattened, and the increasing trend decreased. These results indicated that the sequencing depth was sufficient to analyze the bacterial lactase gene characteristics. The diversity of the bacterial lactase gene in the D. hansenii treatment group was the lowest (Figure 2).
Figure 2.
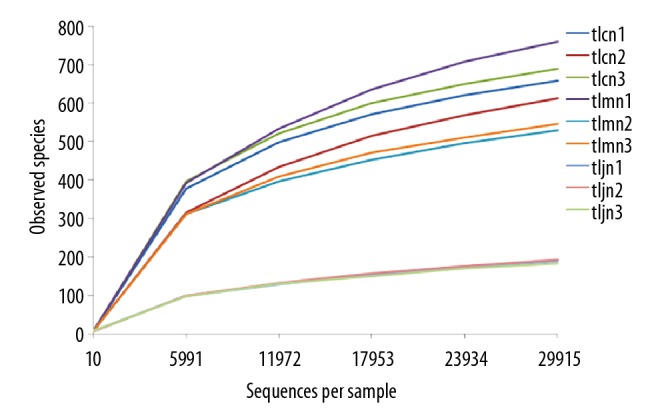
Rarefaction curve of gene from each sample. The abscissa represents the number of lactase gene sequences extracted randomly; the ordinate represents the number of observed OTUs of the lactase gene. The data indicate that the sequencing tended to be saturated and that increasing the amount of data would have no significant effect on obtaining a new OTU when the curve tended to be flat. tlcn1–3, tlmn1–3, and tljn1-3 represent normal groups 1–3, model groups 1–3, and D. hansenii groups 1–3, respectively.
The ACE, Chao1, and Shannon indices of lactase genes in the normal group and the model group were not significantly different, but the Chao1 and Shannon indices decreased significantly in the D. hansenii group compared with the other 2 groups (P<0.01). These results indicated that the community diversity of lactase-producing bacteria from intestinal contents of AAD mice had not recovered after being treated with D. hansenii (Table 1).
Table 1.
Alpha index of lactase genes in AAD mice treated with D. hansenii.
| Groups | Chao1 index | ACE index | Shannon index |
|---|---|---|---|
| tlcn | 532.00±48.14 | 570.22±77.44 | 5.30±0.24 |
| tlmn | 517.00±116.35 | 585.54±148.49 | 5.12±0.33 |
| tljn | 148.33±5.77*# | 184.19±24.44 | 3.42±0.51*# |
tlcn – normal group mice; tlmn – model group mice; tljn – D. hansenii group mice.
P<0.01 vs. the normal group;
P<0.01 vs. the model group.
Intestinal lactase gene beta diversity in AAD mice treated with D. hansenii
PCA analysis was conducted to measure differences in communities of lactase-producing bacteria at the genus level. The results show that the gene clusters of the D. hansenii group were quite different from those of the normal group and model group. This suggests that the bacterial community structure in the D. hansenii group was different from those in the normal group and model group. It also showed that there was little difference between the normal group and model group. The variation percentages of PC1 and PC2 were 99.22% and 0.73%, respectively (Figure 3).
Figure 3.
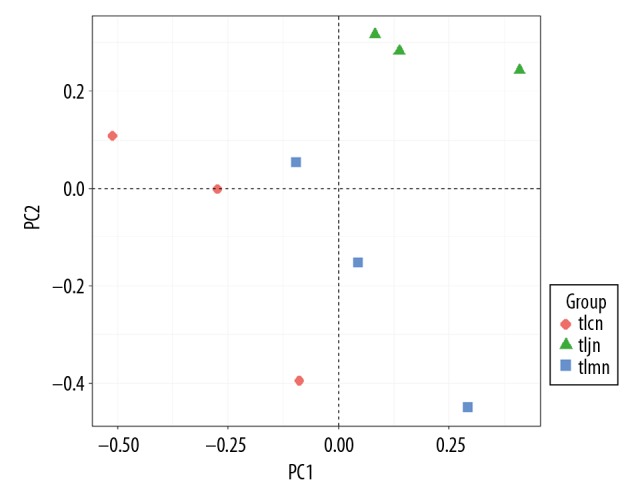
Community difference of bacterial lactase gene from intestinal contents. Different color points represent samples from different groups. A shorter distance between 2 points represents a smaller community difference of bacteria between the 2 groups. tlcn – normal group; tlmn – model group; tljn – D. hansenii group.
Intestinal lactase gene origination and abundance in AAD mice treated with D. hansenii
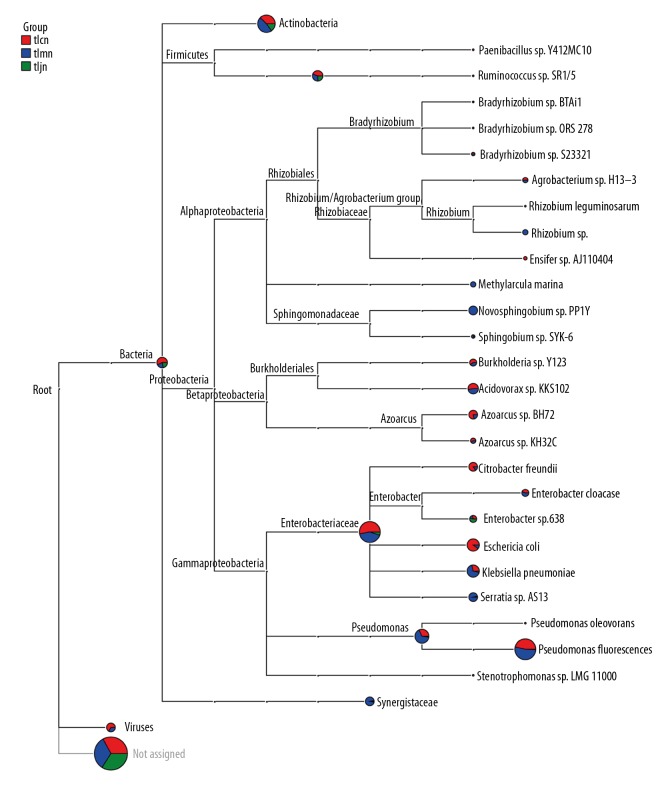
The results showed that intestinal lactase mainly originated from Actinobacteria, Firmicutes, and Proteobacteria (Figure 4), and Proteobacteria was the richest. Compared to the normal group and model group, we found that D. hansenii increased the richness of lactase genes originating from Enterobacter sp. 638.
Figure 4.
Lactase gene from intestinal contents of mice. The pie chart for each branch node indicated the abundance of this taxon in the sample; the larger the fan area, the higher the richness of lactase gene was. tlcn – normal group; tlmn – model group; tljn – D. hansenii group.
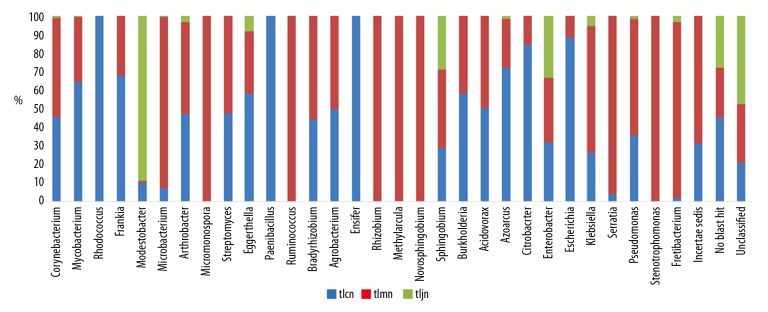
At the genus level, there were 24, 27, and 8 known sources in the normal group, model group, and D. hansenii group, respectively. Lactase-producing bacteria such as Paenibacillus, Rhodococcus, and Ensifer were exclusively detected in the normal group mice, while Micromonospora, Rhizobium, Methylarcula, Ruminococcus, Novosphingobium, and Stenotrophomonas were present only in the model group mice (Figure 5). In addition, most lactase-producing strains were unclassified bacteria or novel strains. After modeling, the quantity of the lactase gene originating from Fretibacterium was significantly increased, and it was recovered by D. hansenii. Similarly, after treatment with D. hansenii, the richness of detected genera in the D. hansenii group was lower than that in the other 2 groups, and, with the exception of Modestobacter, the richness was increased in the D. hansenii group.
Figure 5.
Relative abundance of the bacterial lactase gene at the genus level. tlcn – normal group mice; tlmn – model group mice; tljn – D. hansenii group mice.
Discussion
Approximately 1000 species of bacteria and approximately 1014 microbes inhabit the human intestinal tract [28]. It is well known that intestinal microbiota plays critical roles in metabolism, nutrient digestibility, and immunity [29]. Furthermore, intestinal microorganisms are a source of lactase [30]. Antibiotics save lives by combatting bacterial infections. Unfortunately, misuse of antibiotics frequently results in antibiotic-associated diarrhea in patients. The symptoms can be reduced or prevented using probiotics, such as D. hansenii [17].
Sequence quality was critical to the following sequencing and analysis. Our results showed that the high-quality sequences from each group were more than 97%, which ensured the accuracy and reliability of the subsequent analysis. In this study, all the bacterial lactase gene OTU numbers and Chao1 index and Shannon index values in intestinal contents were lowest in the D. hansenii group (P<0.01). These results showed that D. hansenii remarkably decreased the density and homogeneity of the lactase gene in comparison with the other 2 groups. PCA showed that genes in the D. hansenii group were clearly distinguishable from those in the normal group and model group, indicating that D. hansenii changed the community structure of lactase-producing bacteria.
The gut bacteria of the mice comprised Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, and Verrucomicrobia at the phylum level [17]. Lactase-producing genes in the intestine are mainly derived from Actinobacteria, Firmicutes, Proteobacteria, and other, unclassified, species. At the genus level, the number of lactase-producing species was lowest in the D. hansenii treatment group. Moreover, bacterial lactase genes from Rhodococcus, Paenibacillus, and Ensifer existed exclusively in the normal group, while those from Micromonospora, Ruminococcus, Rhizobium, Methylarcula, Novosphingobium, and Stenotrophomonas were detected only in the model group, which confirmed that the microbial communities had been changed by D. hansenii. Following treatment with D. hansenii, the abundance of lactase genes from Fretibacterium recovered significantly (P<0.01). It was also reported that cells of the strain of Fretibacterium were asaccharolytic [31]. Enterobacter sp. 638 was previously studied, and the genetic determinants required for sucrose metabolism are clustered on a genomic island [32]. In the present study, the results showed that treatment with D. hansenii increased the lactase gene from Enterobacter sp. 638. The abundance analysis also demonstrated that all bacterial lactase genes detected from intestinal contents, other than Modestobacter, were the lowest in the D. hansenii group. In conclusion, we speculate that Fretibacterium, Modestobacter, and Enterobacter sp. 638 are closely related to lactose metabolism. In addition, D. hansenii increased the number of unclassified lactase-producing bacteria.
Conclusions
In summary, the bacterial lactase gene diversity was decreased by D. hansenii. The improvement in lactase activity appears to be associated with the increasing abundance of several pivotal lactase-producing strains and the intestinal microflora balance reestablished by D. hansenii, thus treating AAD.
Footnotes
Source of support: The present study was supported by the National Natural Science Foundation of China (No. 81573951) and the Hunan Provincial Key Research and Development Program (No. 2018NK2025)
Conflict of interest
None.
References
- 1.Zong Y, Zhao HY, Liang XM, et al. Intestinal flora changes in patients with acute or chronic diarrhea. J Clin Int Med. 2006;23(2):89–90. [Google Scholar]
- 2.Lu LL. Probiotics in the prevention of antibiotics-associated diarrhea: A meta-analysis of Randomized Control Trails. J Clin Pediatr. 2010;28(11):1083–85. [Google Scholar]
- 3.Wu D, She KX. Antibiotic-associated diarrhea in critical patients. Chin J Nosocomiol. 2007;17(5):587–88. [Google Scholar]
- 4.Zhou XY. Pathogenesis of antibiotics-induced diarrhea. Chin J Microbiol. 2004;16(6):376–77. [Google Scholar]
- 5.Long CX, He L, Guo YF, et al. Diversity of bacterial lactase genes in intestinal contents of mice with antibiotics-induced diarrhea. World J Gastroenterol. 2017;23(42):7584–93. doi: 10.3748/wjg.v23.i42.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandre V, Even PC, Larue-Achagiotis C, et al. Lactose malabsorption and colonic fermentations alter host metabolism in rats. Brit J Nutr. 2013;110:625–31. doi: 10.1017/S0007114512005557. [DOI] [PubMed] [Google Scholar]
- 7.Juajun O, Nguyen TH, Maischberger T, et al. Cloning, purification, and characterization of β-galactosidase from Bacillus licheniformis DSM 13. Appl Microbiol Biot. 2011;89(3):645–54. doi: 10.1007/s00253-010-2862-2. [DOI] [PubMed] [Google Scholar]
- 8.Rhimi M, Aghajari N, Jaouadi B, et al. Exploring the acidotolerance of β-galactosidase from Lactobacillus delbrueckii subsp. bulgaricus: An attractive enzyme for lactose bioconversion. Res Microbiol. 2009;160(10):775–84. doi: 10.1016/j.resmic.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Lu R, Diao ZY, Xu XX. Efficacy of lactase preparations for treatment of secondary lactose intolerance caused by diarrhea in infants and young children. J Pedi Pharm. 2013;19(2):23–25. [Google Scholar]
- 10.Yang Y, Yang YP, Yao ML, et al. Clinical effect of lactase in the treatment of children with rotavirus enteritis deficiency. China Pract Med. 2014;9(15):24–25. [Google Scholar]
- 11.Hammer HF, Hammer J. Diarrhea caused by carbohydrate malabsorption. Gastroenterol Clin North Am. 2012;41(3):611–27. doi: 10.1016/j.gtc.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YY, Zhou LL, Yang SF. The basic research of primary lactase deficiency. Int J Pediatr. 2014;41(3):302–4. [Google Scholar]
- 13.Guandalini S. Are probiotics or prebiotics useful in pediatric irritable bowel syndrome or inflammatory bowel disease? Front Med (Lausanne) 2014;28(1):23. doi: 10.3389/fmed.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banjara N, Nickerson KW, Suhr MJ, et al. Killer toxin from several food-derived Debaryomyces hansenii strains effective against pathogenic Candida yeasts. Int J Food Microbiol. 2016;222:23–29. doi: 10.1016/j.ijfoodmicro.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Xiao XY, Liu YJ, Deng YL, et al. Isolation and identification of a yeast strain from intestine of mice. J Hunan Agric Univ (Nature Science) 2016;42(4):419–23. [Google Scholar]
- 16.Guo KX, Tan ZJ, Xie MZ, et al. The synergic effect of ultra-micro powder Qiweibaizhusan combined with yeast on dysbacteriotic diarrhea mice. Chin J Appl Environ Biol. 2015;21:61–67. [Google Scholar]
- 17.He L, Long CX, Liu YJ, et al. Effects of Debaryomyces hansenii treatment on intestinal microorganisms in mice with antibiotics-induced diarrhea. 3 Biotech. 2017;7(5):347. doi: 10.1007/s13205-017-0953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long CX, He L, Liu YW, et al. Universal primer for analysis of the diversity of intestinal bacterial lactase gene. Chin J Appl Environ Biol. 2017;23(4):758–63. [Google Scholar]
- 19.Long CX, Liu YW, He L, et al. Bacterial lactase genes diversity in intestinal mucosa of dysbacterial diarrhea mice treated with Qiweibaizhu powder. 3 Biotech. 2018;8(10):423. doi: 10.1007/s13205-018-1460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He L, Liu YW, Guo YF, et al. Diversity of intestinal bacterial lactases gene in antibiotics-induced diarrhea mice treated with Chinese herbs compound Qi Wei Bai Zhu San. 3 Biotech. 2018;8(1):4. doi: 10.1007/s13205-017-1024-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo KX. The Effects of ultra-micro powder Qiweibaizhusan collaborative with yeast on dysbacteriotic diarrheal mice. Hunan University of Chinese Medicine. 2014 [Google Scholar]
- 22.Zeng A, Zhang HL, Tan ZJ, et al. The construction of mice diarrhea model due to dysbacteriosis and curative effect of ultra-micro Qiweibaizhusan. Microbiol China. 2012;39(9):1341–48. [Google Scholar]
- 23.Wu H, Zhou SN, Guo C, et al. A Metagenome DNA extracting method of intestinal flora in mice for molecular diversity analysis based on PCR technology. Chin J Microecol. 2012;24(7):648–51. [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–36. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitta DW, Parmar N, Patel AK, et al. Bacterial diversity dynamics associated with different diets and different primer pairs in the rumen of Kankrej cattle. PloS one. 2014;9(11):e111710. doi: 10.1371/journal.pone.0111710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon CE. The mathematical theory of communication, 1963. MD Comput. 1997;14:306–17. [PubMed] [Google Scholar]
- 27.Mahaffee WF, Kloepper JW. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with Field-Grown Cucumber (Cucumis sativus L) Microbiol Ecol. 34(3):210–23. doi: 10.1007/s002489900050. 199. [DOI] [PubMed] [Google Scholar]
- 28.Thompson JA, Oliveira RA, Djukovic A, et al. Manipulation of the quorum sensing signal Al-2 effects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–71. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 29.Niu Q, Li P, Hao S, et al. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci Rep. 2015;5:9938. doi: 10.1038/srep09938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He L, Long CX, Liu YJ, et al. Research progress on microorganism lactase. Food Ferment Ind. 2017;43(6):268–73. [Google Scholar]
- 31.Vartoukian SR, Downes J, Palmer RM, et al. Fretibacterium fastidiosum gen. nov., sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2013;63(Pt 2):458–63. doi: 10.1099/ijs.0.041038-0. [DOI] [PubMed] [Google Scholar]
- 32.Taghavi S, van der Lelie D, Hoffman A, et al. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Gene. 2010;6(5):e1000943. doi: 10.1371/journal.pgen.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]