Abstract
Gene set enrichment (GSE) testing enhances the biological interpretation of ChIP-seq data and other large sets of genomic regions. Our group has previously introduced two GSE methods for genomic regions: ChIP-Enrich for narrow regions and Broad-Enrich for broad regions. Here, we introduce Poly-Enrich, which has wider applicability, additional capabilities and models the number of peaks assigned to a gene using a generalized additive model with a negative binomial family to determine gene set enrichment, while adjusting for gene locus length. As opposed to ChIP-Enrich, Poly-Enrich works well even when nearly all genes have a peak, illustrated by using Poly-Enrich to characterize pathways and types of genic regions enriched with different families of repetitive elements. By comparing Poly-Enrich and ChIP-Enrich results with ENCODE ChIP-seq data, we found that the optimal test depends more on the pathway being regulated than on properties of the transcription factors. Using known transcription factor functions, we discovered clusters of related biological processes consistently better modeled with Poly-Enrich. This suggests that the regulation of certain processes may be modified by multiple binding events, better modeled by a count-based method. Our new hybrid method automatically uses the optimal method for each gene set, with correct FDR-adjustment.
INTRODUCTION
Regulatory genomics experiments help us understand how cells use more than their genetic sequence to carry out a vast repertoire of cellular programs. Common regulatory genomics methods include chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) and ATAC-seq, which identify transcription factor (TF) binding sites and open chromatin regions, respectively, across the genome. Other types of data, such as DNA methylation assays, copy number alterations, repetitive element families and groups of SNPs, also lead to large sets of genomic regions that potentially play a specific role in regulatory genomics, with each type having notably different properties in terms of the number, size and location of genomic regions.
Proteins that bind near a gene can regulate it in ways such as improving structural properties or physically blocking other proteins, often positively or negatively regulating the gene’s expression, respectively. Additionally, some proteins bind DNA several times in a clustered region (1) or in distant enhancer regions that interact with the same or distinct proteins bound in promoter regions (2). Binding sites also differ in strength; a protein may bind in only a portion of cells in a sample at the time of immunoprecipitation, either due to weak binding or due to varying chromatin accessibility among the cell types in the sample. These binding sites along the genome are interpreted as peaks of varying strengths, depending on the signal-to-noise ratio or significance level of the peak. In general, interpreting each peak’s target gene(s) and effects remains an active area of research, which over time may improve results on downstream tests such as gene set enrichment.
Gene Set Enrichment (GSE) is an approach to test for over (or under) representation of genes in a set of genes with similar functionalities. Gene Ontology (3), Reactome (4), KEGG pathways (5) and MsigDB (6) are widely used gene set databases. Although originally developed for gene expression data, GSE testing is now often used to help interpret ChIP-seq peak sets and other sets of genomic regions. Most of these tests are competitive, meaning that when testing gene sets, the null hypothesis is that genes in a gene set have no more signal (i.e. associated genomic regions) than genes not in the gene set, opposed to self-contained, which has the null hypothesis that the genomic regions in the gene set are not more significant than none (7). Existing methods for general GSE tests include Fisher’s exact test, random sets, logistic regression (e.g. LRPath (8)) and GSEA-type tests (9). GSE methods specific for ChIP-seq data include Genomic Regions Enrichment of Annotations Tool (GREAT) (10), ChIP-Enrich (11) and Broad-Enrich (12).
With so many different tests, one may wonder which test is optimal for their data, but there is no single recommendation across data types. Different tests are needed for different types of genomic regions as properties such as peak widths, number of peaks and location relative to genes all make a difference. Thus, GSE testing for genomic regions should not be a one-size-fits-all test; some methods work better than others in specific scenarios. For example, Cavalcante et al. showed that Broad-Enrich is more powerful than ChIP-Enrich for broad regions, but lacks power for narrow regions (12). As another example, GREAT does not account for variability among genes, so it is best used in situations where the probability of a peak is constant across genomic space (e.g. per kb), as opposed to clustered near transcription start sites or displaying variability among gene loci.
Our previous method, ChIP-Enrich, uses a binary score to classify a gene as having at least one peak. We saw that ChIP-Enrich tends to underperform when nearly all genes have at least one associated genomic region; in this case, ChIP-Enrich will not yield meaningful results. We hypothesized that a count-based, competitive enrichment method that captures the frequency of binding would perform better in those situations. In this paper, we introduce such a method, Poly-Enrich, to expand our available methods to be suitable for any set of narrow genomic regions including those that tend to saturate genes. The flexible structure of the Poly-Enrich test also allows additional capabilities, such as accounting for the strength of each ChIP-seq peak. Whereas ChIP-Enrich has the hypothesis that a single binding site is sufficient for regulation, Poly-Enrich allows for regulation that is incremental, i.e. more genomic regions correspond to stronger or more likely regulation. To identify under which situations one is more appropriate than the other, we performed a comparison of Poly-Enrich and ChIP-Enrich using a set of 90 transcription factor (TF) ChIP-seq datasets from the Encyclopedia of DNA Elements (ENCODE) (13). We also introduce a hybrid test that combines information from both ChIP-Enrich and Poly-Enrich.
To illustrate the usage of Poly-Enrich, we apply it to sets of repetitive elements in the human Alu and LINE1 families, revealing for the first time a comprehensive view of the processes and functions enriched or depleted with these repetitive elements in the human genome. Finally, we describe several updates to our ChIP-Enrich website and chipenrich Bioconductor package, including additional methods for assigning genomic regions to target genes, new gene set databases, and more supported species.
MATERIALS AND METHODS
Datasets
All ChIP-Seq data were obtained from Encyclopedia of DNA Elements (ENCODE) at University of Califonia, Santa Cruz (13). We used a total of 90 experiments over the three Tier 1 cell lines (Gm12878, H1-hESC and K562), and all 35 transcription factors that had available ChIP-seq data for at least two of the three Tier 1 cell lines (Supplementary Table S1).
The gene sets used were from Gene Ontology: Biological Processes (GOBP) ver. 3.4.2 (3). We filtered out gene sets with <15 genes or >2000 genes as gene sets with too few genes generally have insufficient power and may not satisfy the assumptions of the statistical model, and gene sets with too many genes are too vague to be biologically informative. In total, there were 5015 gene sets used.
Assigning regions to genes
The UCSC knownGene database for hg19 was used to define the transcription start sites across the genome (14). Each gene locus definition (e.g. nearest TSS, <5 kb etc.) was generated as a table containing the columns: chromosome, Start, End and gene ID.
Poly-Enrich model: a generalized linear model with a negative binomial family
We model the number of genomic regions assigned to each gene using a generalized linear model (GLM) with a negative binomial (NB) family. The model is:
 |
where for each gene  , GS is an indicator for whether the gene is in the gene set of interest or not (= 1 if in the gene set; 0 otherwise), μ is the mean of the negative binomial distribution for the number of genomic regions assigned to each gene, and the overdispersion parameter
, GS is an indicator for whether the gene is in the gene set of interest or not (= 1 if in the gene set; 0 otherwise), μ is the mean of the negative binomial distribution for the number of genomic regions assigned to each gene, and the overdispersion parameter 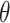 is simultaneously estimated so that
is simultaneously estimated so that 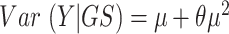 , where Y is the number of genomic regions for the gene. The function
, where Y is the number of genomic regions for the gene. The function  is a cubic smoothing spline that adjusts for the gene’s locus length and optionally adjusts for m, the mappability of the gene’s locus. Details about how we adjust for mappability can be found in the ChIP-Enrich manuscript (11). We use the gam function in the mgcv R package to fit the model, which uses a penalized likelihood maximization, and the smoothing spline penalty is a squared second derivative penalty (15). Use of a cubic smoothing spline to adjust for the genes’ locus lengths was first introduced in ChIP-Enrich, and has been shown to be a powerful, flexible way to model this relationship (11).
is a cubic smoothing spline that adjusts for the gene’s locus length and optionally adjusts for m, the mappability of the gene’s locus. Details about how we adjust for mappability can be found in the ChIP-Enrich manuscript (11). We use the gam function in the mgcv R package to fit the model, which uses a penalized likelihood maximization, and the smoothing spline penalty is a squared second derivative penalty (15). Use of a cubic smoothing spline to adjust for the genes’ locus lengths was first introduced in ChIP-Enrich, and has been shown to be a powerful, flexible way to model this relationship (11).
A likelihood ratio test (LRT) on the coefficient for the gene set is used to test for enrichment (or depletion) of each gene set: the test statistic is defined as  , where L follows a
, where L follows a  distribution under the null hypothesis that there is no association between gene set membership and number of genomic regions (i.e.
distribution under the null hypothesis that there is no association between gene set membership and number of genomic regions (i.e. 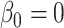 ), and
), and  are the maximum log likelihoods under the null and alternative hypotheses, respectively. We use the LRT instead of the Wald test, because the LRT was shown to perform significantly better than the Wald test with generalized linear models using a negative binomial family (16). We then look at the sign and significance of
are the maximum log likelihoods under the null and alternative hypotheses, respectively. We use the LRT instead of the Wald test, because the LRT was shown to perform significantly better than the Wald test with generalized linear models using a negative binomial family (16). We then look at the sign and significance of 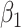 to test for enrichment, where a positive
to test for enrichment, where a positive 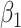 indicates enrichment, and a negative value indicates depletion (fewer regions than expected at random). For each gene set of interest, we estimate a different set of model parameters, and correct for multiple testing afterwards.
indicates enrichment, and a negative value indicates depletion (fewer regions than expected at random). For each gene set of interest, we estimate a different set of model parameters, and correct for multiple testing afterwards.
Poly-Enrich with weighting based on genomic region scores
In certain cases, each genomic region in a dataset may be associated with a numeric score. For example, ChIP-seq peak finding results often include a value denoting the strength of a peak, (e.g. signalValue in ENCODE dataset results or -10*log10(P-value) in MACS2 results). Poly-Enrich weights based on these scores by giving each genomic region a weight proportional to its signal value (or other score) and normalizing such that the mean of all weights is equal to 1. For every genomic region assigned to a gene, we sum all weights and substitute the weighted sum in place of the original count. The same model can still be used on non-whole number data as calculations are equivalent while using the Gamma function instead of a factorial.
Comparing P-values between methods
To compare P-values between methods, we use a scatterplot, plotting a signed -log10P-value per gene set. If a gene set is enriched, the sign is positive, and if the gene is depleted, the sign is negative. This allows us to detect if there are any cases where two methods may contradict each other’s conclusions.
Spline approximation for Poly-Enrich and ChIP-Enrich
With a library of over 20 000 genes and most gene sets being <1000 genes, the cubic smoothing spline estimate changes very little between gene sets. Thus, we have confirmed we can reasonably assume that the spline is approximately equal for any gene set of interest, including the spline with no gene set (Supplementary Figure S1A,B).
We first run the same model except without the gene set (GS) term:  . We then extract the fitted spline using the predict function with type = ’terms’ from the mgcv R package to obtain a spline-adjusted locus length for each gene. This new value is then input as a covariate in the model for every gene set, which allows us to fit a spline only once instead of once for each gene set. This saves a significant amount of time when testing a large number of gene sets (∼75% time saved when testing 4000 gene sets). Compared to the original model, we find that the -log P-values from the spline approximation model are nearly identical (Supplementary Figure S1C, D).
. We then extract the fitted spline using the predict function with type = ’terms’ from the mgcv R package to obtain a spline-adjusted locus length for each gene. This new value is then input as a covariate in the model for every gene set, which allows us to fit a spline only once instead of once for each gene set. This saves a significant amount of time when testing a large number of gene sets (∼75% time saved when testing 4000 gene sets). Compared to the original model, we find that the -log P-values from the spline approximation model are nearly identical (Supplementary Figure S1C, D).
Testing Type-I error
The null hypothesis of Poly-Enrich is that there is no true biological enrichment. To test the Type-I error, we randomly permuted genes to simulate scenarios where there is no association between genes and the number of peaks. However, to ensure that results are not biased by gene locus length or gene location, we performed two additional permutations: one permutes genes within bins of similar locus length, while the other permutes within bins of chromosomal locations. In both cases, genes are sorted by the variable of interest (locus length or location), and then assigned to consecutive bins of 100 genes each. These randomization tests are identical to those used in the Broad-Enrich manuscript (11).
For each of the 90 TF peak datasets chosen, after assigning the peaks to genes, we permuted the gene IDs using the randomization of interest, and then performed enrichment tests against GO biological processes. We ran a total of 10 trials and took the median P-value per gene set as the randomization P-value. Then, the proportion of P-values less than a defined confidence level was determined per experiment to calculate the overall Type-I error. We then plotted all 90 overall Type-I errors for each experiment in a box plot to convey overall Type-I error.
Testing power
To test statistical power, we chose three TF peak datasets of varying size (4194, 11129 and 40052 peaks) and two gene sets of varying size (42 and 471 genes) as our base scenarios. To illustrate how Poly-Enrich can detect enrichment for datasets with very large numbers of peaks (beyond what ChIP-Enrich can handle), we included two larger datasets: an ATAC-seq dataset with 99 478 genomic regions, and an Alu repetitive elements dataset with 1 094 736. After assigning the genomic regions to genes, we randomized the genes in bins of locus length to remove all true gene set enrichment signal while keeping locus length association, and then randomly added peaks into the gene set to simulate enrichment. We chose three scenarios of enrichment, each with varying levels (x% = 5, 10, 20 or 30) of enrichment:
CEbias: Enriched to closely satisfy the assumptions of the binary (ChIP-Enrich) model. We added peaks to x% of the remaining genes in the gene set without a peak. This increases the proportion of genes with a peak, without causing a large increase in the mean number of peaks per gene.
PEbias: Enriched to closely satisfy the assumption of the count-based (Poly-Enrich) model. We added a number of peaks, equal to x% of the number of peaks in the gene set, to a fraction of the genes in the gene set. This increases the mean number of peaks per gene, with little effect on the proportion of genes with a peak.
Balanced: We added a number of peaks, equal to x% of the number of peaks in the gene set, into the gene set weighted by gene locus length. This increases both the proportion of genes with a peak and the mean number of peaks per gene by a similar degree.
Defining the true positive transcription factor-GO term pairs
For each transcription factor, we identified the gene that codes for it from R package GO.db (17), and then identified every GO biological process that gene is assigned to. Pol2 is excluded as its functions are too widespread, resulting in 25 transcription factors, each of which is assigned to at minimum 50 GO BP terms. This set of GO terms, along with its parents and grandparents, is what we use as the true positive set. We also define a true negative set of GO BP terms as every other GO term, except ancestors, siblings and offspring of the true positive set, and terms with ≥2000 or ≤10 genes. Using the true positive and negative sets, we calculated empirical false positive rates (FPRs) for Poly-Enrich, GREAT and ChIP-Enrich. This estimated FPR serves as an upper bound for the true FPR as it is not a perfect gold standard (i.e. some negative GO BP terms may actually be novel true findings, since some functions of a TF may be unknown).
Hybrid test
The hybrid method introduced by Zhang et al. (16), which we employ, was shown to be especially beneficial when there is no one optimal test in all cases. Given n tests that test for the same hypothesis, the same Type-I error rate, and converted to P-values  , the Hybrid P-value is computed as:
, the Hybrid P-value is computed as: 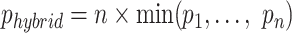 . This hybrid test will have at most the same Type-I error rate as the n tests, and if at least one test is consistent (power converges to 1 as sample size reaches infinity), the hybrid test will also be consistent. Proofs and simulations of the test in general were done by Zhang et al. (18). Here, we have implemented the hybrid test for users to use two methods (n = 2): ChIP-Enrich and Poly-Enrich. Users may also choose any two results files and run a hybrid test based on those.
. This hybrid test will have at most the same Type-I error rate as the n tests, and if at least one test is consistent (power converges to 1 as sample size reaches infinity), the hybrid test will also be consistent. Proofs and simulations of the test in general were done by Zhang et al. (18). Here, we have implemented the hybrid test for users to use two methods (n = 2): ChIP-Enrich and Poly-Enrich. Users may also choose any two results files and run a hybrid test based on those.
Clustering and heat maps
For every GO term, we calculated the difference in –log10P-value for each of the 90 experiments between ChIP-Enrich and Poly-Enrich, with positive values indicating a more significant result for Poly-Enrich. We then focused on GO terms where >10% of the experiments had an absolute log10P-value difference >2. Clustering was performed using uncentered correlation as the similarity metric and average linkage as the clustering method. Using Java TreeView, we extracted specific groups of GO terms that contain certain strings such as ‘cell cycle’ or ‘positive regul.’
Repetitive elements
Data were obtained from the UCSC Table Browser with RepeatMasker 3.0 on the hg19 genome. We chose the two most abundant families in the dataset: Alu and L1, as well as four methods of peak-to-gene assignments: Intron, Nearest TSS, >5 kb, and <5 kb. Poly-Enrich was then used to perform gene set enrichment. Before clustering for the heat map, we filtered out GO terms where there were 2 or fewer significant FDR values among the 8 categories. The clustering method was the same as mentioned in the previous section.
Website and bioconductor updates
The Chip-Enrich website (http://chip-enrich.med.umich.edu) was updated from the chipenrich package version 1.7.2 to version 2.5.0. (from https://github.com/sartorlab/chipenrich, on 8 August 2018). We have added the following reference genomes: human (hg38), rat (rn5, rn6), Drosophilla melanogaster (dm6) and zebrafish (danRer10). We also added the following databases from MSigDB (Version 6.0): Hallmark, Immunologic, MicroRNA, Transcription Factors and Oncogenic (6,19), and sets of genes that are known to be affected by particular environmental toxins from the Comparative Toxicogenomics Database (CTD) (20). We also provide direction in the vignette for how to use gene sets from other R packages, such as EGSEAdata (21).
In addition to the previous locus definitions (‘nearest TSS’, ‘nearest gene’, ‘≤1 kb from TSS’ and ‘≤5 kb from TSS’), we also now support gene locus definitions for regions <10 kb from a TSS and gene distal regions (>10 kb upstream of a TSS).
RESULTS
Motivation for development of Poly-Enrich
The motivation for our new methods comes from situations observed with real sets of genomic regions, often with ChIP-seq peak datasets, but also from other sources, such as families of repetitive elements or large sets of DNA polymorphisms such as those different between closely related species or sub-species. Although our original method, ChIP-Enrich, performs extremely well for most transcription factor (TF) ChIP-seq datasets (Figure 1A), because it uses a simple binary score for each gene, there are some scenarios where this simplification has a significant loss of information. For example, ChIP-Enrich models a gene with many peaks the same as a gene with only one peak, even though gene regulation may be affected by additional peaks (Figure 1B). Alternatively, if nearly every gene is assigned at least one peak, ChIP-Enrich would be unable to distinguish among them and thus unable to detect any gene set enrichment (Figure 1C).
Figure 1.

Three scenarios of ChIP-seq peak distributions illustrating how ChIP-Enrich and Poly-Enrich perform. Each color represents a different gene locus; the left three are in a gene set and the right three are not. (A) Peaks are relatively evenly distributed, with a small number across a subset of genes. Given this situation, ChIP-Enrich evaluates 2/3 versus 1/3 while Poly-Enrich evaluates {0,2,1} versus {0,1,0}; both methods perform well. (B) Some genes contain significantly more peaks than others, such that information is to be gained from the number per gene. ChIP-Enrich evaluates 2/3 versus 1/3, Poly-Enrich evaluates {0,2,3} versus {0,1,0}; ChIP-Enrich performs adequately, but Poly-Enrich is optimal. (C) Nearly all genes have at least one peak, with some having significantly more than others. ChIP-Enrich evaluates 3/3 versus 3/3, Poly-Enrich evaluates {1,2,3} versus {1,1,1}; ChIP-Enrich would not detect any enrichment, while Poly-Enrich can still detect gene sets enriched with more peaks.
Although the alternative current approach, GREAT, is also a count-based gene set enrichment method, Poly-Enrich differs significantly from it in two respects. First, whereas GREAT counts the number of peaks in an entire gene set, Poly-Enrich counts them per gene. By separating counts per gene, we are able to adjust for each gene’s locus length and the variability in peak count across genes, which we previously showed was an important adjustment to control for Type-I error (11). Second, the binomial model used by GREAT assumes that the background probability of a peak is constant across the genome. Poly-Enrich uses a more flexible, empirical approach to this that provides for a range of different assumptions about peak distribution. As previously shown, consequences are that GREAT does not provide accurate significance estimates (the resulting P-values are more significant than they ought to be), and it tends to rank gene sets with shorter genes more highly than those with longer genes (11). We therefore developed Poly-Enrich as a count-based competitive method that addresses all of the above-mentioned shortcomings of ChIP-Enrich and GREAT.
ChIP-Enrich, GREAT, and Poly-Enrich all use a region’s midpoint to define its location. These genomic regions can then be assigned to genes in different ways so that regulation from different types of regions (e.g. promoters, introns or regions distal to TSSs) can be studied. We define a gene’s locus definition as the region on the genome such that peaks in that region are assigned to the gene. These loci are defined using properties of the gene, such as within 5 kb of a gene’s transcription start site (TSS), or simply by assigning each region to the nearest TSS (Figure 2). In the new version of our GSE website and chipenrich Bioconductor package, we offer several additional choices, including exons, introns and distal regions only (>10 kb upstream from a TSS). Users can also upload their own custom locus definition, such as open chromatin regions for a specific cell type, or known enhancers and their target genes.
Figure 2.

Overview of peak-to-gene assignments given gene locus definitions. Given the gene locations and definitions for a genome, several different methods for assigning genomic regions to genes can be defined, referred to as gene locus definitions. Examples shown are: Exons—only peaks in any exon of a gene are assigned to that gene; <5 kb—peaks within 5 kb of a gene’s TSS are assigned to that gene; and nearest TSS—peaks are assigned to the gene with the closest TSS. A gene’s locus length is defined by the number of base pairs that could be assigned to the gene. In this toy example, peak 1 would be assigned to the blue gene for all three example gene locus definitions, peak 2 would only be assigned to the blue gene for the nearest TSS locus definition, and peak 3 would be assigned to the orange gene only for the <5 kb and nearest TSS locus definitions.
Testing Type-1 error and power
We tested the Type-I error rate of the count-based method under the null hypothesis of no enrichment signal. By permuting the genes in the peak-to-gene assignment pairs and breaking the peak–gene relationships, we mimicked three scenarios of no enrichment: (i) the ‘complete’ randomization was done by shuffling the gene IDs in the whole dataset; (ii) the ‘bylength’ randomization was performed to verify that our method adequately adjusts for locus length, by first grouping genes into bins of similar locus length to preserve the locus length relationship; (iii) the ‘bylocation’ randomization was performed to verify that the method adequately adjusts for relationships among genes in close proximity to each other, by grouping genes by their physical location to preserve relationships along the chromosomes (see ‘Materials and Methods’ section for more detail). We ran the randomizations on our 90 selected ChIP-Seq datasets from ENCODE (see ‘Materials and Methods’ section), and the proportion of P-values <0.05 and <0.001 for each dataset were plotted (Supplementary Figure S2A and S2B). We see that the test is properly controlled at an acceptable level for Type-1 error in all cases. That is, approximately 5% had P-values < 0.05 and ∼0.1% had P-values < 0.001 as expected. We observed a slight inflation in the ‘bylocation’ randomization, which upon examination, we found to be caused by certain large clusters of functionally related genes that are located near each other, for instance a cluster of histone genes that affected the results for Gm12878 ETS and H1hesc TBP (Supplementary Table S2). We previously showed that GREAT has an inflated Type-1 error under the ‘complete’ and ‘bylength’ randomizations, also using ENCODE ChIP-seq data (10).
To characterize the statistical power of Poly-Enrich under different situations, we permutated data while simulating enrichment of a gene set, and compared results with those from ChIP-Enrich. We used three datasets with a small, medium, and large number of peaks, and two GO terms with a small and large number of genes. Three types of enrichment were simulated: one that adds peaks mainly according to the regulatory assumptions of ChIP-Enrich (CEBias), one that adds peaks mainly according to the assumptions of Poly-Enrich (PEBias), and one that is balanced. For each type of enrichment, we simulated four levels of enrichment 0.05, 0.1, 0.2 and 0.3, which indicate the proportion of additional peaks added to the gene set (see ‘Materials and Methods’ section for more detail). Finally, we chose two different levels of significance: 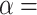 0.05 and 0.001, as our cutoffs.
0.05 and 0.001, as our cutoffs.
As expected, higher simulated enrichment resulted in higher power, since adding more signal increases the ability of a test to detect significance. Also, larger gene sets have higher power due to an increased confidence in the estimated mean number of peaks. Overall, we see that Poly-Enrich has more power in simulations that enrich a gene set by increasing the number of peaks per gene, while ChIP-Enrich has more power in simulations that enrich a gene set by adding peaks to genes without any previous peaks. Finally, the Balanced simulation results in the two methods having similar power in most cases (Supplementary Figure S3A and S3B).
With the two largest datasets, we tested power for the Balanced simulated gene sets to illustrate that Poly-Enrich is able to detect signal even when ChIP-Enrich fails. We see that ChIP-Enrich is can still perform reasonably well compared to Poly-Enrich with around 100 k peaks, but starts being unable to detect any enrichment in the dataset with over 1 million peaks where 81% of genes are assigned a peak in the small gene set, and actually loses power when more signal is added in the large gene set (Supplementary Figure S3C).
Validation with true positives
To complement our permutations and simulations, we compared Poly-Enrich, ChIP-Enrich and GREAT’s ability to find true positives while avoiding false positives with real ChIP-seq data. To do this, we first created a set of true positives comprised of GO term-TF pairs by using the GO term biological process (BP) assignments for the gene encoding the transcription factor (e.g. the gene encoding for JunD is assigned to the GO term, ‘cell death’). This makes the reasonable assumption that TFs tend to regulate genes in the same biological processes in which they are active. Out of the 25 TFs with at least 50 assigned GO terms, we found that GREAT had a larger empirical false positive rate (FPR) than both ChIP-Enrich or Poly-Enrich for 22 TFs (Supplementary Figure S4). Estimated FPRs were similar between Poly-Enrich and ChIP-Enrich, with 13 (52%) experiments being higher for ChIP-Enrich. The overall high FPR (compared to the expected 5%) can be attributed to the true positives being imperfect (see ‘Materials and Methods’ section).
Poly-Enrich with weighted genomic regions
The height and confidence of peaks in a ChIP-seq experiment can vary dramatically, thus we reasoned that incorporating this additional information would improve the ability to pinpoint the truly enriched pathways. Although the most apparent motivation for weighting genomic regions is to account for ChIP-seq peak strength, other situations exist where each peak or genomic region may be assigned a unique score (e.g. confidence or quality score). Due to the flexible nature of the Poly-Enrich model, we were able to easily add the option to weight regions by peak strength (using peak signal value; see ‘Materials and Methods’ section for details), and examined the extent to which adjusting for peak strength improves enrichment results using 90 ENCODE ChIP-seq datasets by comparing the –log10P-values per gene set.
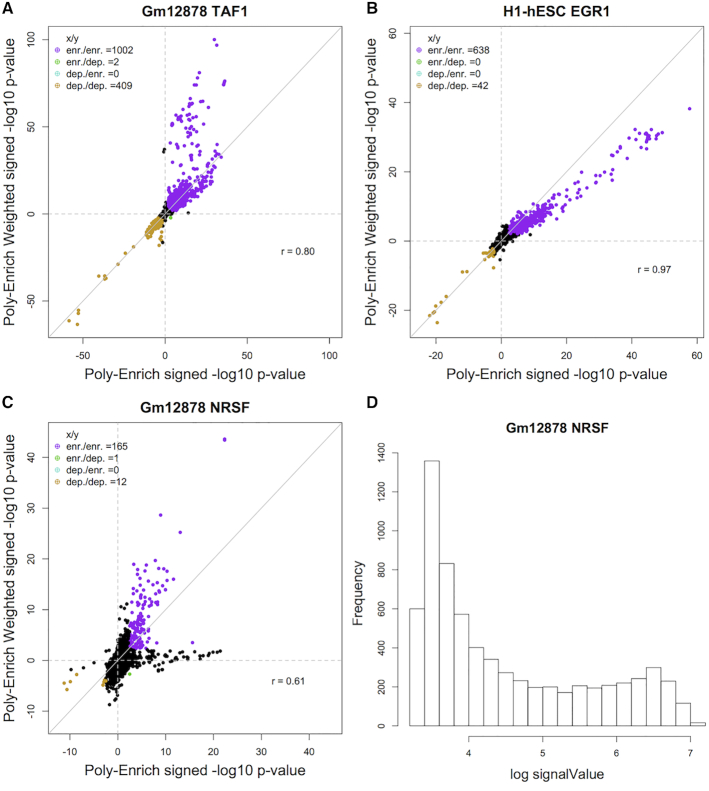
We noticed for 25% of the experiments, most enriched gene sets were more significant with weighting, thus as we hypothesized, binding events near genes in enriched GO terms were stronger than those near other genes (Figure 3A,B). In another 20% of experiments, the enrichment P-values were split between the two methods (Figure 3C). Interestingly, the distribution of log signal values for these experiments showed a bimodal pattern (Figure 3D). This suggests that some gene sets tend to have genes with significantly stronger binding peaks than others, and that both sets may be biologically interesting. For the remaining 55% of experiments tested, weighting made little difference on the results.
Figure 3.
Comparison of GO term enrichment results between standard Poly-Enrich and its weighted version using signal values as weights. Each point is a GO term’s –log10P-value of the two methods, signed positive for enriched, negative for depleted. (A) Using weighting results in more significant enrichment in many GO terms in the Gm12878 TAF1 ChIP-Seq experiment. (B) Using weighting results in slightly less significant enrichment in many GO terms in the H1-hESC EGR1 ChIP-Seq experiment. (C) Using weighting on the Gm12878 NRSF experiment results in several more significant GO terms as well as several less significant ones. (D) The histogram of log signal values from the NRSF experiment shows a bimodal pattern in the weights, suggesting that GO terms that are more significant with weighting than without may have genes that tend to have stronger bound peaks or vice versa.
Comparison of the count-based (Poly-Enrich) versus binary (ChIP-Enrich) model of enrichment
We next compared results from Poly-Enrich versus ChIP-Enrich on the same set of 90 ENCODE ChIP-seq datasets. Our initial hypothesis was that some experiments would be clearly modeled better by one method or the other (i.e. dependent on the transcription factor). However, our results strongly suggest that the optimal model for TF binding is more dependent on the gene set tested than the TF. This is visualized by a bifurcation in the significance levels of GO terms between the binary and count-based methods (Figure 4A), and suggests that a single transcription factor may regulate genes differently depending on the function of the gene. Thus, we sought to understand this further.
Figure 4.

Comparisons of Poly-Enrich with ChIP-Enrich. (A) Comparison of GO term significance levels between ChIP-Enrich and Poly-Enrich. Each point is the –log10P-value of a GO term from the two methods, signed positive for enriched or negative for depleted. Several gene sets are much more significant using Poly-Enrich and several are much more significant using ChIP-Enrich. This split pattern is representative of 32% of the tested datasets. (B) Heat map of –log10P-value differences between Poly-Enrich and ChIP-Enrich for GO terms and ChIP-seq experiments, where each row is a GO term and each column is a ChIP-seq experiment. Shown are GO terms where >15% of the experiments had a –log10P-value difference of 2 or larger. Red indicates Poly-Enrich was more significant, and blue indicates ChIP-Enrich was more significant. Light gray indicates the transcription factor used in the experiment was not assigned to the GO term and is omitted in the clustering. Representative GO terms are shown for each cluster. (C) GO terms containing ‘positive regulation of metabolism/biosynthesis’ are mostly red, indicating that a count score provides a more appropriate model. (D) GO terms related to cell cycle are mostly blue, indicating that a binary score provides a more appropriate model.
The binary model used by ChIP-Enrich assumes that a single binding event (i.e. a single genomic region) is sufficient for regulation, while the Poly-Enrich count-based model assumes that strength of regulation is incremental with the number of binding sites. Based on results above, we asked what kinds of genes were more consistent with either of those assumptions. We use the true positive set of known TF-GO combinations mentioned earlier in the validation section. Observing the enrichment results using the 5 kb locus definition for these true positive GO term–TF pairs, we used clustering to identify patterns of TFs and GO terms that are optimal with one of the methods. We found that the method that worked better was most often determined by the GO term (Figure 4B). For example, GO terms involving positive regulation of metabolic or biosynthetic processes tended to do better with Poly-Enrich except for those involving cell cycle, implying that related genes are regulated such that more binding sites increase regulation (Figure 4C,D). Conversely, GO terms related to ‘cell cycle’ clustered together and displayed greater power with ChIP-Enrich, implying that related genes are possibly regulated with only one binding site and having more have little additional effect. Parallel results using the Nearest TSS locus definition were similar (Supplementary Figure S5).
Poly-Enrich is recommended for experiments with a large number (>40 k) of peaks, as we showed that ChIP-Enrich starts losing power at around 100 ks of peaks (Supplementary Figure S3C). However, in many cases, the gene set, rather than the transcription factor, was a stronger determinant of the more appropriate method, we are not always able to recommend either Poly-Enrich or ChIP-Enrich for an entire experiment. We therefore developed a hybrid test that uses information from both ChIP-Enrich and Poly-Enrich.
Hybrid test
To obtain the best results across all types of GO terms and datasets, we developed a hybrid test that incorporates both the binary and count-based models. After performing the two models, the hybrid P-value of the two tests is defined as: 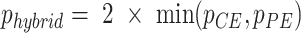 , where
, where  and
and  are the P-values given by ChIP-Enrich and Poly-Enrich, respectively (18). This is essentially a Bonferroni-adjusted P-value for two tests. This hybrid has been shown to be beneficial if the two tests are sufficiently different, but loses power and is conservative if the tests are identical or nearly identical (18). While the hybrid test is not as powerful as the better method between ChIP-Enrich and Poly-Enrich, it is dramatically more powerful than using the worse method, making it the optimal method to use across all GO terms (Figure 5). While this hybrid test currently only accommodates ChIP and Poly-Enrich, it can be extended to accommodate several additional gene set enrichment tests.
are the P-values given by ChIP-Enrich and Poly-Enrich, respectively (18). This is essentially a Bonferroni-adjusted P-value for two tests. This hybrid has been shown to be beneficial if the two tests are sufficiently different, but loses power and is conservative if the tests are identical or nearly identical (18). While the hybrid test is not as powerful as the better method between ChIP-Enrich and Poly-Enrich, it is dramatically more powerful than using the worse method, making it the optimal method to use across all GO terms (Figure 5). While this hybrid test currently only accommodates ChIP and Poly-Enrich, it can be extended to accommodate several additional gene set enrichment tests.
Figure 5.
Statistical power comparisons for Poly-Enrich (red), ChIP-Enrich (blue) and the hybrid test (gold). We compared datasets of three different sizes (i.e. number of peaks: small, medium and large) and two gene set sizes (small and large GS), under two significance levels: α = 0.05 (A) and 0.001 (B), and three different methods of simulated enrichment (CEbias: add peaks according to the regulatory assumptions of ChIP-Enrich, PEbias: add peaks mainly according to the assumptions of Poly-Enrich, Balanced: add peaks proportional to each gene’s locus length). The values on the X-axis indicate the percent of extra peaks added to simulate enrichment; a higher value simulates stronger enrichment. The hybrid test is shown to have much more power than the wrong method, and only slightly less power than the correct method.
Identifying biological processes enriched with or depleted in repetitive element families using Poly-Enrich
ChIP-Enrich is unable to identify enriched gene sets in cases where nearly all genes have at least one assigned genomic region (Figure 1C). Thus, to further illustrate the utility of Poly-Enrich, we used it to test large families of repetitive element regions. We asked whether we could identify gene sets that are either enriched or depleted for certain types of repetitive elements. Significant enrichment of repetitive elements in the promoter regions of genes, for example, can sequester the transcription factors that inhibit activities at another transcription factor binding site or other regulatory motif (22). Some of these mobile elements remain active with new insertions having neutral, detrimental or beneficial effects. Although repetitive element families have been well studied for over 30 years, little is yet known about the biological processes that they have adapted to help regulate or that they can easily disrupt and thus are negatively selected against (23). Using the database of human repetitive elements from the UCSC Table Browser (RepeatMasker 3.0) (24), we performed GSE testing on repetitive element families. Certain families of repetitive elements have over a million occurrences across the human genome, and thus virtually all genes have at least one nearby instance, making this an example where ChIP-Enrich performs poorly. Thus, in this situation, modeling the number of insertions per gene is critical to identify differences.
We examined two of the most abundant types of repetitive elements: the Alu and LINE1 (L1) elements, which make up an estimated 11% and 17% of the human genome, respectively (25,26). We also chose four gene locus definitions: Nearest TSS, <5 kb (promoter regions), >5 kb (distal regions) and Intron. We tested GO Biological Processes, and used clustering to identify related groups of biological processes enriched with or depleted of the repetitive elements (Figure 6). We found that both Alu and L1 elements are enriched in centrosome-related GO terms, which validate that our approach identifies known associations (27), and is only made possible with recent advancements in genome mapping near the centromeres (28). For Alu elements, we also found strong enrichment in GO terms describing metabolic processes, most significantly ‘ATP metabolic process’ and ‘rRNA metabolic process’, especially in promoter regions, which is consistent with an analysis of Alu distribution in chromosomes 21 and 22 that showed Alu elements on these chromosomes were enriched in or near metabolism and signaling genes (29). Conversely, Alu elements were sharply depleted in the promoter regions of many development and morphogenesis processes, with the strongest depletions in cell fate commitment and connective tissue development. Interestingly, depletions were also seen in the introns of genes in these gene sets, but not in regions >5 kb upstream, suggesting the negative selection is limited to the regions that are more commonly regulatory.
Figure 6.

Gene Ontology terms enriched or depleted with common repetitive element families. Shown are enrichment results using Poly-Enrich for the Alu (first four columns) and L1 (last four columns) repetitive element families using four different peak-to-gene assignments. Shown are signed –log10 FDR, where positive values (red) indicate enrichment and negative values (blue) indicate depletion. Only GO terms that were significant for at least three columns at the FDR = 0.05 level are displayed. We identified nine clusters of GO terms with similar enrichment patterns. Representative GO terms are used to label each cluster.
Novel insertions of L1 elements into or near key genes are known to be associated with neurological diseases (30). Consistent with this, we found that all neuro-related GO terms in Figure 6 were depleted for L1 (but not for all of Alu) (Supplementary Figure S6), which suggests that L1’s evolutionarily have been selected against occurring in the regulatory regions of neurological genes; when they are inserted into the introns or promoters of these genes, the inserted elements may have an unacceptably high risk of causing disease.
In general, we observed that the significance of the distal upstream regions (>5 kb locus definition) was lower than the other three locus definitions (with the exception of some enrichments for Alu elements) (Supplementary Table S3), implying that most repetitive element negative (or positive) selection has occurred in the promoter regions or introns of genes. Alternatively, the gene distal enriched and depleted regions may be limited to a specific set of enhancer regions, the signal from which could have been diluted in our analysis. Interesting additional findings are that L1 elements are enriched in chemical stimulus detection processes such as detection of chemical stimulus and sensory perception of chemical stimulus, while Alu elements are depleted in the genes in these processes. Finally, both Alu and L1 elements are significantly depleted in genes involved in many processes related to development and morphogenesis.
Availability, usage and updates
Poly-Enrich is available in the chipenrich Bioconductor package and as a web interface at http://chip-enrich.med.umich.edu. Several additional gene set databases and gene locus definitions (see ‘Materials and Methods’ section for details) have been added since our original publication (see http://chip-enrich.med.umich.edu/data/ChipenrichMethods.pdf).
To perform GSE analysis with either our Bioconductor package or web version, the user first needs a file of genomic regions, which may be a narrowPeak, BED, or text file with chromosome, start and end positions for each region. The user then selects a species, one or more gene set databases, a gene locus definition and the test method (ChIP-Enrich, Poly-Enrich, Hybrid or Fisher’s exact test). Optionally, the user can upload a custom/user-defined list of gene sets and/or gene locus definition. For narrow genomic regions (≤2–3 kb), we recommend using the Poly-Enrich method for sets of >100 000 regions, and the Hybrid method for sets of regions with fewer than this. For broad genomic regions (>2–3 kb), we still recommend the Broad-Enrich method (Supplementary Figure S7). The user can then also choose to weight the genomic regions based on a score of their choice, and apply a number of other options, such as adjustment for read mappability (recommended for read lengths <50 bp).
The enrichment function outputs five files:
opts: The options that the user input into the function.
peaks: A peak-level summary showing the peak-to-gene assignment for each peak.
peaks-per-gene: A gene-level summary showing gene locus lengths and the number of peaks assigned to each gene.
results: The results of the GSE tests. Lists the tested gene sets along with their descriptions, the test effect, odds ratio, enrichment status, P-value and FDR. Also included is the list of Entrez gene IDs with contributing signal for each enrichment test.
qcplot: A diagnostic plot of the gene locus lengths with a fitted smoothing spline.
The R code used to generate analysis and figures can be found at https://github.com/sartorlab/polyenrich.
DISCUSSION
Gene set enrichment testing methods for genomic regions should take into account the differing properties of the input datasets, including the widths and number of genomic regions, and where they tend to occur relative to genes. However, no single method is appropriate for all types, and therefore no single GSE method should be recommended for all sets of genomic regions. Although our previously developed ChIP-Enrich method for gene set enrichment with genomic regions performs well for most transcription factor ChIP-seq datasets (11), above we described common situations where it does not. Such cases include when nearly all genes are assigned at least one genomic region, and when the strength or likelihood of regulation increases incrementally with the number of genomic regions. As an example, the transcription factor NF-kappaB is known to regulate the gene NFKBIA by binding to a few or even many motif positions in the promoter (31), with gene expression correlated with the number of bound factors. Thus, motivated by specific examples of regulatory mechanisms, we developed Poly-Enrich, a method that models the number of regions per gene, empirically adjusts for each gene’s locus length, and takes into account variability among genes in each gene set. Poly-Enrich is also flexible, in that it easily allows for weighting of each genomic region by any score of interest. We used the example of weighting by peak strength, but other examples include weighting by SNP significance in a GWAS analysis, by the inverse distance to a gene, or by the probability that the region is in an open chromatin region in a particular cell type.
We showed that our count-based method, Poly-Enrich, is optimal when almost all genes are assigned a peak. In comparing when each test is most appropriate for typically sized ChIP-seq datasets, we discovered that the optimal test is mostly dependent on the gene set rather than the transcription factor being studied. Because in many cases we could not recommend a single best method to test all gene sets for an experiment, we developed and implemented a hybrid test that uses information from both methods and performs better than either test across GO terms for most datasets. However, as noted in the ‘Results’ section, specific situations exist when one particular method is optimal, and we therefore have provided specific recommendations to our users in choosing the most appropriate method.
When applying Poly-Enrich to repetitive element families, we both reconfirmed known associations and also identified novel findings. Poly-Enrich confirmed that Alu elements are over-represented in the promoters of metabolism genes and signaling by finding enrichment for related GO terms. Additionally, we know that L1 insertions into or near certain neurological-related genes are associated with neurological diseases (32). Indeed, we found that L1 is depleted in neuro-related process genes, implying there is natural selection against L1 elements inserting in the regulatory regions of these genes. We also found that there is little enrichment or depletion in the distal regulatory regions of genes, suggesting that repetitive elements may not have as large of an effect there due to mitigated regulatory activity at larger distances from transcription start sites. We also detected novel associations between repetitive element families and biological pathways. Both Alu and L1 elements were significantly depleted in development and morphogenesis-related gene sets, such as connective tissue development and skeletal system morphogenesis, suggesting that it is critical to have developmental regulatory regions for several different development systems free from potentially disruptive repetitive elements during early growth.
One shortcoming of our current methods (as well as current alternatives) is that they rely on associating each genomic region with the nearest gene(s). However, it is estimated that 79–95% of DNAse I hypersensitive sites, markers for enhancer regions, actually regulate a different, distal target gene (24,25). We are currently developing a set of enhancer locus definitions that identify and assign enhancer regions to their appropriate target genes, as was recently introduced by Chicco et al. (33), so peaks in enhancer regions will be correctly assigned and false positive peaks in nonfunctional intergenic regions will be filtered out. We believe this will improve all future gene enrichment analyses.
DATA AVAILABILITY
Poly-Enrich is available in the chipenrich Bioconductor package and as a web interface at http://chip-enrich.med.umich.edu.
The R code used to generate analysis and figures can be found at: https://github.com/sartorlab/polyenrich
Supplementary Material
Notes
Present address: Raymond G. Cavalcante, Epigenomics BRCF Core, University of Michigan, USA.
Present address: Chee Lee, LA Care Health Plan, Los Angeles, CA, USA.
Present address: Zing T. Y. Tsai, Illumina Inc, San Diego, CA, USA.
Contributor Information
Christopher T Lee, Biostatistics Department, University of Michigan, Ann Arbor, MI 48109, USA.
Raymond G Cavalcante, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Chee Lee, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Tingting Qin, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Snehal Patil, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Shuze Wang, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Zing T Y Tsai, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Alan P Boyle, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Maureen A Sartor, Biostatistics Department, University of Michigan, Ann Arbor, MI 48109, USA; Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Institutes of Health [R01 CA158286]; Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) Center is funded by the National Institute of Environmental Health Sciences (NIEHS) [P30 ES017885].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gotea V., Visel A., Westlund J.M., Nobrega M.A., Pennacchio L.A., Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010; 20:565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pennacchio L.A., Bickmore W., Dean A., Nobrega M.A., Bejerano G. Enhancers: five essential questions. Nat. Rev. Genet. 2013; 14:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B.et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018; 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011; 27:1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goeman J.J., Bühlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007; 23:980–987. [DOI] [PubMed] [Google Scholar]
- 8. Sartor M.A., Leikauf G.D., Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009; 25:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Welch R.P., Lee C., Imbriano P.M., Patil S., Weymouth T.E., Smith R.A., Scott L.J., Sartor M.A. ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res. 2014; 42:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cavalcante R.G., Lee C., Welch R.P., Patil S., Weymouth T., Scott L.J., Sartor M.A. Broad-Enrich: functional interpretation of large sets of broad genomic regions. Bioinformatics. 2014; 30:i393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosenbloom K.R., Dreszer T.R., Pheasant M., Barber G.P., Meyer L.R., Pohl A., Raney B.J., Wang T., Hinrichs A.S., Zweig A.S.et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010; 38:D620–D625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D. The UCSC Known Genes. Bioinformatics. 2006; 22:1036–1046. [DOI] [PubMed] [Google Scholar]
- 15. Wood S.N., Goude Y., Shaw S. Generalized additive models for large data sets. J. Roy. Statist. Soc. Ser. A. 2015; 64:139–155. [Google Scholar]
- 16. Robinson M.D., Smyth G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008; 9:321–332. [DOI] [PubMed] [Google Scholar]
- 17. Carlson M., Maintainer B.P. 2015; TxDb.Hsapiens.UCSC.hg19.knownGene: Annotation package for TxDb object(s). R package version 3.2.2.
- 18. Zhang S., Okhrin O., Zhou Q.M., Song P.X. Goodness-of-fit test for specification of semiparametric copula dependence models. J. Econometrics. 2016; 193:215–233. [Google Scholar]
- 19. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015; 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 2017; 45:D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alhamdoosh M., Law C.W., Tian L., Sheridan J.M., Ng M., Ritchie M.E. Easy and efficient ensemble gene set testing with EGSEA [version 1; peer review: 1 approved, 3 approved with reservations]. F1000Res. 2017; 6:2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu X., Wu B., Szary J., Kofoed E.M., Schaufele F. Functional sequestration of transcription factor activity by repetitive DNA. J. Biol. Chem. 2007; 282:20868–20876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brunner A.M., Schimenti J.C., Duncan C.H. Dual evolutionary modes in the bovine globin locus. Biochemistry. 1986; 25:5028–5035. [DOI] [PubMed] [Google Scholar]
- 24. Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2009; 25:4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- 25. Roy-Engel A.M., Carroll M.L., Vogel E., Garber R.K., Nguyen S.V., Salem A.H., Batzer M.A., Deininger P.L. Alu insertion polymorphisms for the study of human genomic diversity. Genetics. 2001; 159:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 27. de Sotero-Caio C.G., Cabral-de-Mello D.C., Calixto M.D.S., Valente G.T., Martins C., Loreto V., de Souza M.J., Santos N. Centromeric enrichment of LINE-1 retrotransposons and its significance for the chromosome evolution of Phyllostomid bats. Chromosome Res. 2017; 25:313–325. [DOI] [PubMed] [Google Scholar]
- 28. Aldrup-Macdonald M.E., Sullivan B.A. The past, present, and future of human centromere genomics. Genes (Basel). 2014; 5:33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wanichnopparat W., Suwanwongse K., Pin-On P., Aporntewan C., Mutirangura A. Genes associated with the cis-regulatory functions of intragenic LINE-1 elements. BMC Genomics. 2013; 14:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Solyom S., Kazazian H.H. Mobile elements in the human genome: implications for disease. Genome Med. 2012; 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giorgetti L., Siggers T., Tiana G., Caprara G., Notarbartolo S., Corona T., Pasparakis M., Milani P., Bulyk M.L., Natoli G. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol. Cell. 2010; 37:418–428. [DOI] [PubMed] [Google Scholar]
- 32. Thomas C.A., Paquola A.C., Muotri A.R. LINE-1 retrotransposition in the nervous system. Annu. Rev. Cell Dev. Biol. 2012; 28:555–573. [DOI] [PubMed] [Google Scholar]
- 33. Chicco D., Bi H.S., Reimand J., Hoffman M.M. BEHST: genomic set enrichment analysis enhanced through integration of chromatin long-range interactions. 2019; bioRxiv doi:15 January 2019, preprint: not peer reviewed 10.1101/168427. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Poly-Enrich is available in the chipenrich Bioconductor package and as a web interface at http://chip-enrich.med.umich.edu.
The R code used to generate analysis and figures can be found at: https://github.com/sartorlab/polyenrich