Abstract
Pleistocene climate fluctuations had profound influence on the biogeographical history of many biota. As large areas in high mountain ranges were covered by glaciers, biota were forced either to peripheral refugia (and possibly beyond to lowland refugia) or to interior refugia (nunataks). However, nunatak survival remains controversial as it relies solely on correlative genetic evidence. Here, we test hypotheses of glacial survival using two high alpine plant species (the insect‐pollinated Pedicularis asplenifolia and wind‐pollinated Carex fuliginosa) in the European Alps. Employing the iDDC (integrative Distributional, Demographic and Coalescent) approach, which couples species distribution modelling, spatial and temporal demographic simulation and Approximate Bayesian Computation, we explicitly test three hypotheses of glacial survival: (a) peripheral survival only, (b) nunatak survival only and (c) peripheral plus nunatak survival. In P. asplenifolia the peripheral plus nunatak survival hypothesis was supported by Bayes factors (BF> 100), whereas in C. fuliginosa the peripheral survival only hypothesis, although best supported, could not be unambiguously distinguished from the peripheral plus nunatak survival hypothesis (BF = 5.58). These results are consistent with current habitat preferences (P. asplenifolia extends to higher elevations) and the potential for genetic swamping (i.e., replacement of local genotypes via hybridization with immigrating genotypes [expected to be higher in the wind‐pollinated C. fuliginosa]). Although the persistence of plants on nunataks during glacial periods has been debated and studied over decades, this is one of the first studies to explicitly test the hypothesis instead of solely using correlative evidence.
Keywords: Alps, coalescent simulations, nunataks, peripheral refugia, Pleistocene glaciation
1. INTRODUCTION
Pleistocene climate fluctuations had profound influence on the biogeographical history of many biota (Hewitt, 1996, 2004). During the glacial periods, large areas in higher latitudes and in high mountain ranges were covered by ice sheets. It is of particular interest to identify where plants and animals occurring in formerly glaciated areas managed to survive during these glacial periods (Gabrielsen, Bachmann, Jakobsen, & Brochmann, 1997; Marr, Allen, & Hebda, 2008; Schönswetter, Stehlik, Holderegger, & Tribsch, 2005; Tremblay & Schoen, 1999; Wachter et al., 2012). In the case of mountain ranges, the focus of the present study, species might have retreated to unglaciated areas at their periphery (peripheral refugia: Holderegger & Thiel‐Egenter, 2009) and possibly beyond into lowlands (lowland refugia: Holderegger & Thiel‐Egenter, 2009), as supported by fossil data (Birks & Willis, 2008) and by molecular data (Comes & Kadereit, 1998; Cosacov, Sérsic, Sosa, Johnson, & Cocucci, 2010; Fulton, Norris, Graham, Semken, & Shapiro, 2013; Marr et al., 2008; Schönswetter et al., 2005; Stehlik, 2000; Tollefsrud, Bachmann, Jakobsen, & Brochmann, 1998) for many plant species. Alternatively, species may have survived within the ice shield on ice‐free mountain peaks, so‐called nunataks (nunatak refugia: Holderegger & Thiel‐Egenter, 2009), as suggested by the nunatak survival hypothesis (Schneeweiss & Schönswetter, 2011; Schönswetter et al., 2005; Stehlik, 2000). Due to a general lack of fossil evidence, nunatak survival is essentially inferred from molecular data only (Lohse, Nicholls, & Stone, 2011; Schönswetter & Schneeweiss, 2019; Stehlik, Blattner, Holderegger, & Bachmann, 2002; Stehlik, Schneller, & Bachmann, 2001; Westergaard et al., 2011). The incidence of nunatak survival may, however, be underestimated, especially in species with high gene flow, because during (re‐)colonization signals of nunatak survival can be genetically swamped by migrants from peripheral refugia (Gabrielsen et al., 1997; Schneeweiss & Schönswetter, 2011; Tollefsrud et al., 1998). The hypotheses of survival in peripheral versus nunatak refugia are, however, not mutually exclusive, and for some species both types of refugia have been inferred (Escobar García et al., 2012; Zhang, Zhu, Zhong, & Zhang, 2018).
The iDDC (integrative Distributional, Demographic and Coalescent modelling) approach provides a powerful framework allowing different glacial survival scenarios to be explicitly tested (Brown & Knowles, 2012; He, Edwards, & Knowles, 2013; He, Prado, & Knowles, 2017; Papadopoulou & Knowles, 2016). Briefly, using spatially explicit demographic models corresponding to the hypotheses to be tested (which may differ in, for instance, the carrying capacity landscapes or in migration rates), genetic patterns are simulated under the coalescent model. These models, which often are informed by species distribution modelling (SDM), are then evaluated by comparing them to the empirical genetic pattern using an Approximate Bayesian Computation (ABC) framework (for details, see Brown & Knowles, 2012: figure 4). In the context of glacial survival, demographic models differ with respect to whether species are allowed to persist in central glaciated and/or in peripheral unglaciated areas during the glacial period (Figure 1).
Figure 1.

Schematic of the three glacial survival scenarios used in the simulations. Suitability of cells derived from species distribution modelling (SDM) for the Last Glacial Maximum (22,000–10,000 years before present [YBP]) was modified to comply with the different glacial survival scenarios: (1) peripheral plus nunatak survival, (2) peripheral survival only and (3) nunatak survival only; suitabilities for the postglacial (10,000–0 YBP) were taken from the SDM for the present. Grey cells represent unsuitable areas of different altitude [Colour figure can be viewed at http://wileyonlinelibrary.com]
In the present study, glacial survival patterns of two plant species, Pedicularis asplenifolia and Carex fuliginosa, were investigated in the European Alps, a geographical model system to study Pleistocene range shifts (Escobar García et al., 2012; Lohse et al., 2011; Schönswetter et al., 2005). Both species are perennial herbs found exclusively in the alpine and, particularly P. asplenifolia, in the subnival belt. As species that can cope with cold harsh environments are likely to be able to survive in extreme habitats such as nunataks (Lohse et al., 2011; Stehlik et al., 2002), they are excellent candidates to test glacial survival hypotheses. In addition, their current distribution ranges encompass both areas situated in formerly glaciated regions, where they may have survived on nunataks, and areas outside the former ice‐sheet, where they may have survived in peripheral refugia (Figure 2). Using RAD‐seq (restriction site associated DNA sequencing) data analysed with the iDDC approach, we here test three glacial survival scenarios identified previously: peripheral survival only, nunatak survival only and peripheral plus nunatak survival (e.g., Escobar García et al., 2012; Schönswetter, Tribsch, Stehlik, & Niklfeld, 2004; Stehlik et al., 2002).
Figure 2.

(a) Genetic structure (K = 2) and (b) scaled number of private alleles of Pedicularis asplenifolia (circles) and Carex fuliginosa (squares). In (b), the size of the symbols are in proportion to the scaled number of nearly fixed private alleles (black symbols) and, shown nested therein, the scaled number of fixed private alleles (grey symbols). The dotted line indicates the permanent glacial snow line (i.e., altitude above which snow does not melt in climatically average years) during the Last Glacial Maximum (LGM), and the solid line indicates the maximum extent of the ice‐sheet during the LGM. The right insert shows the position of the study area [Colour figure can be viewed at http://wileyonlinelibrary.com]
2. MATERIALS AND METHODS
2.1. Study species
Pedicularis asplenifolia (Orobanchaceae) is endemic to the eastern Alps (from eastern Switzerland eastwards through the central Alps in Austria and adjacent parts of northern Italy; Meusel, Jäger, Rauschert, & Weinert, 1978). It is found in the alpine belt (i.e., the belt with closed swards) and the subnival belt (i.e., the belt with open vegetation), where it occurs in open swards and stabilized screes, often on base‐rich schists (Fischer, Oswald, & Adler, 2008). Based on flower morphology, P. asplenifolia is considered to be tightly adapted to bumblebee pollination (as in many Pedicularis species in the Alps: Heß, 2001), although in practice it appears to be essentially autogameous (Kreisch, 1993).
Carex fuliginosa (Cyperaceae) in its narrow circumscription is distributed in the eastern Alps and the Carpathians (Meusel, Jäger, & Weinert, 1965); occurrences in the western Alps, as indicated by Hultén & Fries (1986) for example, are probably erroneous (Schultze‐Motel, 1967–1980; Duhamel, 1994). C. fuliginosa is closely related to the circum‐Arctic C. misandra (Hultén & Fries, 1986), and both may be merged within one species either recognizing two geographically distinct subspecies or without such intraspecific taxa (Elven, Murray, Razzhivin, & Yurtsev, 2011). C. fuliginosa is found in the alpine belt, where it occurs in rocky, usually moist open swards, often on base‐rich schists (Fischer et al., 2008). As with most Carex species, C. fuliginosa is monoecious and wind‐pollinated (Schultze‐Motel, 1967–1980).
2.2. Molecular data generation
Leaf material of 28 P. asplenifolia and 46 C. fuliginosa individuals was collected from 10 and seven populations, respectively, across the species' entire distributional ranges in the eastern Alps (Table 1; Figure 2). Leaf material was stored in silica gel. DNA extractions were performed following Jang et al. (2013). Single enzyme‐ (PstI) digested RAD libraries (Baird et al., 2008) were constructed using the protocol described by Paun et al. (2015), and sequenced on the Illumina HiSeq2000 platform as single‐end 100‐bp reads at the Vienna Biocenter Core Facilities (https://www.vbcf.ac.at).
Table 1.
Collection information of the study species
| Species | Region | Coordinatesa | Population numberb | Number of individuals |
|---|---|---|---|---|
| Pedicularis asplenifolia | A, Rottenmanner und Wölzer Tauern | 47°26′/14°25′ | 83 | 4 |
| A, Schladminger Tauern | 47°16′/13°38′ | 107 | 4 | |
| A, Goldberggruppe | 46°57′/13°01′ | 66 | 4 | |
| A, Glocknergruppe | 47°04′/12°46′ | 113 | 4 | |
| A, Zillertaler Alpen | 47°05′/11°39′ | 286 | 4 | |
| A, Stubaier Alpen | 47°02′/11°05′ | 285 | 4 | |
| CH, Samnaungruppe | 46°54′/10°22′ | 287 | 4 | |
| Carex fuliginosa | A, Hochschwabgruppe | 47°36′/15°10′ | 279 | 5 |
| A, Rottenmanner und Wölzer Tauern | 47°26′/14°25′ | 82 | 5 | |
| A, Schladminger Tauern | 47°17′/13°36′ | 362 | 4 | |
| I, Julische Alpen | 46°22′/13°30′ | 284 | 4 | |
| A, Goldberggruppe | 46°57′/13°01′ | 71 | 5 | |
| A, Glocknergruppe | 47°04′/12°46′ | 114 | 5 | |
| A, Karnischer Hauptkamm | 46°38′/12°42′ | 283 | 4 | |
| A, Venedigergruppe | 46°59′/12°14′ | 282 | 5 | |
| A, Zillertaler Alpen | 47°00′/11°33′ | 280 | 4 | |
| I, Bergamasker Alpen | 46°03′/10°00′ | 281 | 5 |
Abbreviations: A, Austria; CH, Switzerland; I, Italy.
Latitude/longitude.
These correspond to collection numbers (“Schneeweiss XY”, where XY is the population number) as used on the vouchers, deposited in the herbarium of the University of Vienna (WU).
Raw reads were demultiplexed by allowing for a single mismatch at the barcodes using illumina2bam (https://github.com/gq1/illumina2bam) and stacks 2.3e (Catchen, Hohenlohe, Bassham, Amores, & Cresko, 2013). Reads with low quality scores (<10) were discarded. Single nucleotide polymorphism (SNP) calling was conducted employing the denovo_map.pl pipeline in stacks (Catchen et al., 2013) with default settings except that the minimum number of identical reads required to build stacks (−m) was set to 5. Loci containing missing data or more than 10 SNPs were filtered out; a cut‐off of 10 SNPs per locus was chosen to avoid a bias towards less variable loci. For final analyses, only one SNP per locus was retained to reduce linkage disequilibrium.
2.3. Inferring genetic structure
A Bayesian clustering method implemented in structure 2.3.4 (Pritchard, Stephens, & Donnelly, 2000) was used to identify population structure in the investigated species. Ten independent replicate runs were conducted for each number of populations, K, with K ranging from 1 to 8. For each run, we ran the admixture model with a burn‐in of 105 generations and sampling from the subsequent 106 generations. The best K was selected based on ΔK (Evanno, Regnaut, & Goudet, 2005) using structure harvester 0.6.94 (http://taylor0.biology.ucla.edu/structureHarvester/, Earl & vonHoldt, 2012). Structure results were plotted with distruct 1.1 (Rosenberg, 2004).
Relationships among individuals were visualized via a NeighbourNet (Bryant & Moulton, 2004), constructed using HKY85 (Hasegawa, Kishino, & Yano, 1985) distances in splitstree 4.14.2 (Huson & Bryant, 2006), and via a principal component analysis (PCA), conducted in the r package adegenet 2.1.1 (https://CRAN.R-project.org/package=adegenet). The number of fixed (present in all individuals of a population) and the number of nearly fixed (present in at least 75% of all individuals of a population) private alleles for each population were taken from the output of the “populations” function in stacks and corrected by nucleotide diversity as described by Westergaard et al. (2019).
2.4. iDDC approach
Species occurrence data were obtained from the GBIF database (https://www.gbif.org/), the project “Mapping the flora of Austria” (H. Niklfeld & L. Ehrendorfer, University of Vienna, unpubl. data) and the European Vegetation Archive (EVA; Chytrý et al., 2016). The accuracy of all georeferenced occurrence data was manually checked, and occurrences outside known distribution ranges were removed, resulting in 126 data points for P. asplenifolia and 206 data points for C. fuliginosa. Distributions of the two species were modelled for both the present and the last glacial maximum (LGM) period. Nineteen bioclimate variables representing current and past (LGM) climatic conditions were downloaded from the WorldClim database (http://www.worldclim.org/, Hijmans, Cameron, Parra, Jones, & Jarvis, 2005) at a resolution of 2.5 arc minutes. For both studied species, eight bioclimate variables were retained for further analyses (BIO1: annual mean temperature, BIO4: temperature seasonality, BIO8: mean temperature of wettest quarter, BIO9: mean temperature of driest quarter, BIO12: annual precipitation, BIO15: precipitation seasonality, BIO18: precipitation of warmest quarter, BIO19: precipitation of coldest quarter) after removing highly correlated (>0.7) variables. SDMs were calibrated by linking these climatic data to the species occurrence data using the ensemble modelling approach implemented in the package “biomod2” (Thuiller, Georges, Engler, & Breiner, 2016) of r (R Core Team, 2013). Thereby, we selected six modelling techniques: generalized linear model (GLM), generalized boosting model (GBM), generalized additive model (GAM), classification tree analysis (CTA), artificial neural network (ANN) and random forest (RF). To evaluate model quality for each species and modelling technique, the available occurrence data were randomly split into one part to calibrate the models (80%) and the remaining data to evaluate them (20%). To avoid random effects of splitting, we repeated this procedure 10 times. Only models with relative operating characteristic (ROC) values (Swets, 1988) >0.75 were used to subsequently generate ensemble projections of potential species distribution under current climate and under climatic conditions corresponding to the LGM. Ensemble predictions were defined as the means of projected occurrence probabilities of single models. Pseudo‐absence data were randomly generated (i.e., selected from the pool of unoccupied cells) with prevalence equal to 0.5 (i.e., absences have the same weight as presences) with 10 replicates.
Following the approach of Bemmels, Title, Ortego, and Knowles (2016), we up‐scaled the cell sizes of the SDMs from the original 2.5 × 2.5 arc minutes to 5 × 5 arc minutes (i.e., merging four cells resulting in a cell covering ~81 km2) using ArcGIS 9 (ESRI). Values for these larger cells were calculated as the mean value from the four smaller cells. Although a finer resolution for a highly heterogeneous landscape such as mountains would be desirable, this would have rendered computation time for the subsequent demographic modelling prohibitively long.
As precise delimitation of refugia is needed for the subsequent modelling, we specifically define interior refugia as areas within the permanent glacial snow line and peripheral refugia as areas outside the permanent glacial snow line, but still within the Alps (Schönswetter et al., 2005; Tribsch & Schönswetter, 2003); because there was only very limited evidence for lowland refugia from species distribution modelling (see Results), these were not considered. Our delimitations of interior and peripheral refugia are more specific than those of Holderegger and Thiel‐Egenter (2009), who explicitly do not consider the permanent glacial snow line to separate peripheral from interior (nunatak) refugia. Peaks protruding above the ice‐sheet were found in essentially all mountain ranges of the eastern Alps, but ice‐free areas were relatively small and isolated (Tribsch & Schönswetter, 2003); hence, studying survival in interior refugia at the level of single nunataks is not possible. Habitat suitabilities at the LGM were modified according to the three main scenarios (Figure 1). Specifically, in the peripheral survival only scenario (Peri), areas of interior refugia (i.e., within the permanent glacial snow line) were considered totally uninhabitable (i.e., suitability was set to zero). The location of the ancestral population (i.e., the geographical starting point for the demographic modelling) was either in the ice‐free eastern part (PeriEast scenario) or in the ice‐free southern part (PeriSouth scenario) of the Alps (corresponding to refugium IV and refugium III, respectively, of Schönswetter et al., 2005). In the nunatak survival only scenario (Nun), suitabilities in areas of interior refugia were reduced by 85% (as in Massatti & Knowles, 2016) and those in peripheral areas (i.e., outside the permanent glacial snow line) were set to zero; an ancestral population in the central glaciated area of the Alps was used. In the peripheral plus nunatak survival scenario (Peri + Nun), the suitabilities of cells of interior refugia was decreased by 85%, while those of peripheral refugia were left unchanged. The single ancestral population was located either in the ice‐free eastern part (PeriEast + Nun scenario), the ice‐free southern part (PeriSouth + Nun scenario) or the central glaciated part of the Alps (Peri + NunCentral scenario); using two ancestral populations was computationally not feasible with the available resources.
For each scenario, 106 demographic simulations were performed in splatche 3.0 (Currat, Arenas, Quilodran, Excoffier, & Ray, 2019). The generation times of both species were set to 40 years (resulting in 550 generations for the considered time period from 22,000 years before the present until the present), following a rescaling approach (the actual generation times are probably considerably shorter) similar to the one used by Massatti and Knowles (2016). The main reason for rescaling generation time is to make simulations computationally feasible, because increasing generation time results in fewer generations being simulated. The actual generation time will not affect model evaluations, but any interpretation of the estimated parameters would have to take the rescaling and possible differences in generation times between the two species into account (Massatti & Knowles, 2016). Demographic parameters, including migration rate m (the per‐generation probability of an individual moving out of a deme [i.e., grid cell] into neighbouring demes), maximum carrying capacity k, and population size of ancestral population Nanc, were drawn from uniform priors through abctoolbox 2.0 Beta (https://bitbucket.org/phaentu/abctoolbox-public/, Wegmann, Leuenberger, Neuenschwander, & Excoffier, 2010); these priors were sufficiently broad to prevent posteriors being unduly affected by narrow prior bounds. Specifically, the priors were m ~ U(0.001, 0.1), k ~ U(5 × 102, 2 × 104), Nanc ~ U(2 × 103, 5 × 106) for an ancestral population located in a peripheral area and Nanc ~ U(5 × 102, 5 × 104) for an ancestral population located in the glaciated central Alps, where populations are expected to have been smaller. The prior on the migration rate was set following the considerations of Bemmels et al. (2016). Specifically, the minimum value of the migration rate was chosen to allow the (climatically) suitable landscape to be fully colonized, and the maximum value was chosen to avoid colonization being too rapid, as this would eliminate any differences between models. However, the upper bound of the migration rate was set higher than in Bemmels et al. (2016) to compensate for scaled generation time (larger generation times result in fewer generations being simulated and thus may require higher migration rates). Cells with suitability <10% of the maximum were treated as totally unsuitable (i.e., their suitabilities were set to zero) to remove nonzero, although small, suitabilities mostly outside the mountain ranges (Figure 3). Subsequently, habitat suitabilities derived from the SDMs larger than zero were classified into 10 categories in increments of 10% of the maximum suitability found in the particular species using a modified python script from x‐origin (He et al., 2017). The carrying capacity of each cell was scaled according to its suitability. From generation 1 (22,000 years before the present) to 300 (10,000 years before the present), corresponding to the glacial period, demographic modelling used the modified SDM predictions for the LGM, as described above, whereas for generations 301–550 (present time) the modelling used the SDM predictions based on the current climate.
Figure 3.
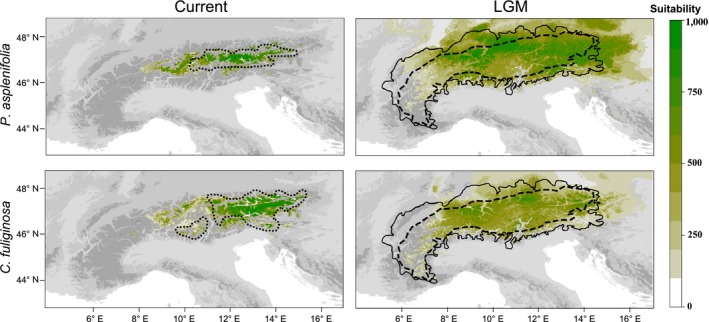
Projected suitabilities of Pedicularis asplenifolia and of Carex fuliginosa under current climate conditions (left panel) and under climate conditions at the Last Glacial Maximum (LGM; right panel). In the left panel, the dotted lines indicate the current distribution ranges of the two species; in the right panel, the dashed lines indicate the permanent glacial snow line during the LGM and the solid lines indicate the maximum extent of the ice‐sheet during the LGM [Colour figure can be viewed at http://wileyonlinelibrary.com]
Genetic data sets matching the dimensions of the empirical data set (i.e., having the same number of samples and SNPs) were generated on each of the 106 demographic simulations (i.e., using the stored demographic and migration histories of each grid cell at each generation) using splatche 3.0 (Currat et al., 2019). To this end, SNPs were considered unlinked and the minimum frequency of SNPs was set to zero; simulation of SNP data in splatche does not require a mutation rate to be specified (see manual available from http://www.splatche.com/splatche3). For both simulated and empirical data sets, summary statistics were calculated in arlequin 3.5 (Excoffier & Lischer, 2010), including mean number of alleles over loci for each population, mean number of alleles over loci and population, mean heterozygosity over loci for each population, mean heterozygosity over loci and population, mean total heterozygosity and pairwise population F ST. A total of 38 and 68 summary statistics were computed for P. asplenifolia and C. fuliginosa, respectively. These numbers differ because of the different number of populations analysed for the two species.
To identify the best supported scenario we employed ABC with abctoolbox 2.0 Beta (Wegmann et al., 2010). Instead of using all computed summary statistics directly, we converted summary statistics to partial least squares (PLS) components using the r package “pls” (Mevik & Wehrens, 2007) with Box–Cox treatment (Box & Cox, 1964). The number of PLS components to be used was determined based on the root mean squared error (RMSE) plots. For each scenario, 5,000 (0.5%) simulated genetic data sets that are closest to the empirical data set were retained for parameter estimation and model selection. A post‐sampling regression adjustment was applied using GLMs (Leuenberger & Wegmann, 2010). Marginal densities were used to evaluate models. For validation, p‐values were calculated to check if the models are able to generate the empirical data (Wegmann et al., 2010). Additionally, we checked whether parameter estimations are unbiased using 1,000 pseudo‐observations; a uniform distribution of posterior quantiles is expected if estimation of the parameter is unbiased (Wegmann et al., 2010).
3. RESULTS
3.1. RAD sequencing
After demultiplexing and quality filtering, more than 590,000 reads were obtained per individual (available under accession numbers SAMN13021464–SAMN13021537). After filtering of loci and retaining only one SNP per locus, the final data sets contained 2,185 SNPs for Pedicularis asplenifolia and 2,486 SNPs for Carex fuliginosa (Pan, Hülber, Willner, & Schneeweiss, 2019; available on Dryad at https://doi.org/10.5061/dryad.rg0134r).
3.2. Genetic structure
For both P. asplenifolia and C. fuliginosa, ΔK suggested K = 2 as the most likely number of groups (Figure S1). In P. asplenifolia these groups showed geographical structure and were longitudinally separated, whereas in C. fuliginosa, these groups showed no biologically easily interpretable structure, as the proportion of membership to the minor cluster in any individual was less than 0.4 (Figure 2a; Figure S2).
In the Neighbor‐Net of P. asplenifolia, three groups could be distinguished, corresponding to each of the two easternmost populations (pop. 83 and pop. 107, belonging to the same genetic cluster: Figure 2a) and to the remaining populations (Figure S3). The same three groups were identified in the PCA, where pops. 83 and 107 were separated from the other populations along the first axis (explaining 25.2% of the variation) and pops. Eighty‐three and 107 were separated from each other along the second axis (explaining 13.2% of the variation; Figure S4). In the Neighbor‐Net of C. fuliginosa, populations were clearly separated from each other, yet there was no major split suggesting any grouping among populations (Figure S3). In the PCA, pop. 284, supported by a long split in the Neighbor‐Net (Figure S3), was separated from the remaining populations along the first axis (explaining 14.3% of the variation), and the remaining populations were separated, mostly longitudinally, along the second axis (explaining 9.8% of the variation; Figure S4).
The numbers of (nearly) fixed private alleles were high in (nearly) peripheral populations both in P. asplenifolia (pops. 83 and 107) and in C. fuliginosa (especially pop. 284, less so in pops. 82, 279 and 281; Figure 2b, Table S1). The numbers of (nearly) fixed private alleles were constantly lower in each of the interior populations, but (except for the number of private alleles in pop. 113 of P. asplenifolia) were always larger than zero (Table S1) occasionally approaching levels of peripheral populations (e.g., interior pop. 114 vs. peripheral pop. 281 of C. fuliginosa; Figure 2b, Table S1).
3.3. Model evaluation
Based on current climate data, SDM predicted suitable areas for P. asplenifolia and C. fuliginosa that were mostly congruent with their current distribution ranges (Figure 3). According to the projections at LGM conditions, major parts of the Alps as well as peripheral areas (mostly adjacent midelevation mountain ranges) were suitable for P. asplenifolia and, to a lesser extent, also for C. fuliginosa (Figure 3).
Observed heterozygosity, H obs, and expected heterozygosity, H exp, ranged in P. asplenifolia from 0.1536 to 0.2368 and from 0.0958 to 0.1708, respectively; in C. fuliginosa, they ranged from 0.1372 to 0.1624 and from 0.0814 to 0.1196, respectively (Table S1). The mean heterozygosity over loci and population, H, ranged from 0.1094 to 0.1951 in P. asplenifolia and from 0.0930 to 0.1328 in C. fuliginosa (Table S1); accordingly, both the mean heterozygosity over loci and population, H mean, as well as the total heterozygosity, H total, were higher in P. asplenifolia than in C. fuliginosa (H mean: 0.1455 vs. 0.1112, respectively; H total: 0.1729 vs. 0.1413, respectively). The mean number of alleles over loci and population, K, ranged from 1.2485 to 1.4622 in P. asplenifolia and from 1.2071 to 1.3660 in C. fuliginosa (Table S1); accordingly, the mean number of alleles over loci and population, K mean, was higher in P. asplenifolia than in C. fuliginosa (1.3447 vs. 1.2885, respectively). Pairwise F ST values ranged from 0.008 to 0.332 in P. asplenifolia and from 0.087 to 0.422 in C. fuliginosa (Table S2).
Based on the RMSE plots (Figure S5), three to five PLS components were retained for calculating the distance between simulated and empirical data sets. In P. asplenifolia, the peripheral plus nunatak survival scenario with the ancestral population located in the eastern Alps (PeriEast + Nun) best explained the empirical genetic pattern (Table 2). All remaining scenarios were clearly rejected (Bayes factor [BF] support for the best model in all cases >100). In accordance, the PeriEast + Nun scenario better reproduced the empirical data (p = .736) compared to all alternative scenarios (p ≤ .001). In C. fuliginosa, the best supported model was the peripheral survival only scenario with the ancestral population located in the eastern Alps (PeriEast; Table 2), followed by the peripheral plus nunatak survival scenario with the ancestral population located in the eastern Alps (PeriEast + Nun; BF support for the best model = 5.58). For these models, p‐values were .993 and .992, respectively (Table 2). The remaining scenarios were clearly rejected (BF support for the best model >100) and had p‐values ≤ .001. In both species, prior distributions of parameter estimates in the best supported models were distinct from the posterior distributions (Figure S6), indicating that the data have power to estimate parameters. Parameter estimates were not unbiased, as posterior quantiles of all parameters departed from a uniform distribution (Kolmogorov–Smirnov test, Figure S7).
Table 2.
Comparison of Pleistocene survival scenarios of the study species
| Species | Modela | Marginal density | Bayes factorb | p |
|---|---|---|---|---|
| Pedicularis asplenifolia | Nun | 1.28 × 10−76 | >100 | <.001 |
| PeriEast + Nun | 2.57 × 10−5 | – | .736 | |
| PeriSouth + Nun | <1.00 × 10−100 | >100 | <.001 | |
| Peri + NunCentral | <1.00 × 10−100 | >100 | <.001 | |
| PeriEast | 2.48 × 10−13 | >100 | <.001 | |
| PeriSouth | <1.00 × 10−100 | >100 | <.001 | |
| Carex fuliginosa | Nun | 8.05 × 10−97 | >100 | <.001 |
| PeriEast + Nun | 1.51 × 10−4 | 5.58 | .992 | |
| PeriSouth + Nun | <1.00 × 10−100 | >100 | <.001 | |
| Peri + NunCentral | 2.00 × 10−96 | >100 | <.001 | |
| PeriEast | 8.43 × 10−4 | – | .993 | |
| PeriSouth | <1.00 × 10−100 | >100 | <.001 |
Nun, nunatak survival in interior refugia, the index indicating (where necessary) the location of the ancestral population (Central, central Alps); Peri, peripheral survival in peripheral refugia, the index indicating (where necessary) the location of the ancestral population (East, eastern Alps; South, southern Alps).
The ratio between marginal densities of the best model (i.e., the one with the highest marginal density) and of the alternative model: the higher the value, the higher the support for the best model.
4. DISCUSSION
Concerning the debate as to whether nunatak survival does matter, the answer may be species‐specific rather than universal (Gabrielsen et al., 1997; Tollefsrud et al., 1998; Wachter et al., 2016; Westergaard et al., 2011). Species traits affecting, for instance, dispersal capabilities shape current genetic patterns both through glacial survival per se (via, for instance, genetic bottlenecks; Schönswetter, Paun, Tribsch, & Niklfeld, 2003; Wachter et al., 2012) and through post‐glacial recolonization (via, for instance, gene flow or long‐distance dispersal; Paun, Schönswetter, Winkler, IntraBioDiv Consortium, & Tribsch, 2008; Schönswetter, Tribsch, Barfuss, & Niklfeld, 2002). As shown in this study, although Pedicularis asplenifolia and Carex fuliginosa have similar habitat preferences and current distribution ranges, unambiguous evidence for nunatak survival was only found in P. asplenifolia.
In P. asplenifolia, both peripheral and nunatak areas appear to have acted as refugia during the LGM. This is evident from the support for the peripheral plus nunatak survival scenario with the ancestral population located in the eastern Alps (PeriEast + Nun; Table 2), an area that acted as a glacial refugium also for other alpine plants (Schönswetter et al., 2005). Peripheral and nunatak survival were shown to jointly contribute to current genetic patterns in some high alpine plants (Escobar García et al., 2012; Schönswetter & Schneeweiss, 2019), and this appears also to be the case in P. asplenifolia.
In contrast, in C. fuliginosa, only peripheral areas in the easternmost Alps could be unambiguously confirmed as refugia, although based on BFs a nunatak survival cannot be ruled out (Table 2). A lack of nunatak survival in C. fuliginosa would agree with, compared with P. asplenifolia, a lower tolerance against harsh climate conditions expected to have occurred at Pleistocene nunataks. Such a lower tolerance is suggested by the current altitudinal distributions, as C. fuliginosa is restricted to the alpine zone whereas P. asplenifolia frequently extends into the subnival zone. Alternatively, however, the ambiguity with respect to nunatak survival might be because traces of nunatak survival may have been genetically swamped (Gabrielsen et al., 1997; Tollefsrud et al., 1998) after (re‐)colonization following deglaciation and subsequent gene flow between immigrants from peripheral refugia and in situ inhabitants in C. fuliginosa. The potential for genetic swamping due to gene flow is expected to be higher in wind‐pollinated species (Govindaraju, 1988) such as C. fuliginosa than in insect‐pollinated species such as P. asplenifolia. Genetic homogenization may also be responsible for the lack of geographical structure in the genetic data despite the presence of geographically distant populations in C. fuliginosa (Figure 2; Figures S2–S4). Wind‐pollination has been shown to mediate post‐glacial gene flow among refugia (Liepelt, Bialozyt, & Ziegenhagen, 2002). For taxa prone to genetic swamping, neither a correlative genetic approach nor a modelling approach as used here may allow nunatak survival to be detected, even if based on genomic data such as RAD‐seq data. In those cases, valuable information may be obtained by using mostly uniparentally inherited markers not prone to homogenization, as is the case for plastid or mitochondrial sequences (Schönswetter & Schneeweiss, 2019).
We acknowledge that our sampling is less intensive (four or five individuals per population) compared to other studies (e.g. Bemmels et al., 2016; Massatti & Knowles, 2016; Westergaard et al., 2019). This may, however, be compensated for by a larger number of SNPs (2,185 SNPs for P. asplenifolia and 2,486 SNPs for C. fuliginosa) when only retaining loci without any missing data as done here. Comfortingly, analyses of an earlier data set with fewer individuals (two individuals per population), but more SNPs (5,504 SNPs for P. asplenifolia and 4,976 SNPs for C. fuliginosa) resulted in the same ranks for the glacial survival scenarios as identified here, yet with reduced decisiveness (i.e., lower BFs; Table S3). This indicates that our inferences are robust with respect to sampling intensity.
As with any ABC approach, model validation is essential, because ABC will always produce posterior distributions independent of model quality (Bertorelle, Benazzo, & Mona, 2010; Wegmann et al., 2010). In our case, compared to alternative models, the most supported model had higher probability of generating data similar to the empirical one (high p‐values) than the alternative models, indicating that the model evaluation results are robust. The posterior quantiles from pseudo‐observations of all estimated parameters showed departure from a uniform distribution (Figure S7), suggesting that they are estimated inaccurately and that their biological interpretation should be avoided (Wegmann et al., 2010). However, in this study, we were not interested in the specific parameter values, as our primary objective was to distinguish alternative glacial survival scenarios.
Utilizing several summary statistics simultaneously, although not free from potential problems (e.g., the curse of dimensionality; Beaumont, 2010), is advantageous, as single summary statistics may not suffice to distinguish different evolutionary hypotheses (Hickerson, Dolman, & Moritz, 2006; Lin, Li, Schlötterer, & Futschik, 2011). This appears also to be the case for private alleles, which are expected to be high in populations that have experienced genetic drift in isolated refugia (Hewitt, 2004; Westergaard et al., 2019) and thus are good indicators for the position of refugia. In line with a hypothesis of peripheral refugia, the corrected numbers of (nearly) fixed private alleles were high in peripheral populations of both species (Figure 2b; Table S1). Evidence for or against nunatak survival from the distribution of (nearly) fixed private alleles was, however, ambiguous, because the corrected numbers of (nearly) fixed private alleles were nearly always above 0 (Table S1) and occasionally approached levels of peripheral populations. Only after taking the results from the iDDC approach into account, the elevated numbers of (nearly) fixed private alleles in pop. 286 of P. asplenifolia may be interpreted as indicating an interior refugium in the central Alps (as suggested previously: Escobar García et al., 2012; Schönswetter & Schneeweiss, 2019).
We acknowledge that testing more refined scenarios with respect to, for instance, geographical resolution of nunataks or model parameterization would be desirable, but there will be data‐imposed limits. Regardless, testing simple models does not compromise the biological insights from our study, which is that nunatak survival during periods of glaciation, at least potentially, contributed to the biogeography and evolution of alpine plant species.
5. CONCLUSION
In phylogeographical studies, multiple demographic histories may lead to similar genetic patterns. For example, high genetic diversity may be the outcome of either secondary contact or of temporally stable populations (Nettel, Dodd, Afzal‐Rafii, & Tovilla‐Hernández, 2008; Ursenbacher et al., 2008), or both geographical isolation and founder events might generate high genetic differentiation between populations (Gugerli et al., 2001; Schönswetter, Popp, & Brochmann, 2006). Using model‐based approaches as applied here allows the genetic pattern to be explicitly linked to the phylogeographical history of species (He et al., 2013; Massatti & Knowles, 2016). Thus, we could unambiguously demonstrate nunatak survival within the heavily glaciated central Alps in Pedicularis asplenifolia. Although the persistence of plants on nunataks during glacial periods has been debated and studied over decades (Gabrielsen et al., 1997; Schneeweiss & Schönswetter, 2011; Tollefsrud et al., 1998; Westergaard et al., 2011), this is one of the few studies (e.g., Westergaard et al., 2019) to explicitly test the hypothesis instead of solely using correlative evidence.
AUTHOR CONTRIBUTIONS
D.P. and G.S. conceived the study; D.P. and G.S. collected specimens; W.W. provided distribution data; K.H. advised on SDM methods; D.P. conducted laboratory work and data analyses; D.P., K.H., W.W. and G.S. wrote the manuscript.
Supporting information
ACKNOWLEDGEMENTS
We thank Ovidiu Paun (University of Vienna) for advice on the generation and analysis of RAD‐seq data. We thank Christian Parisod and four anonymous reviewers for their constructive criticisms, incorporation of which greatly improved the manuscript. Financial support from the China Scholarships Council programme (No. 201306860003) to D.P. is acknowledged. Computational work was performed on the Vienna Scientific Cluster (VSC; http://vsc.ac.at/) and the Life Science Computer Cluster at the University of Vienna (LiSC; http://cube.univie.ac.at/lisc).
Pan D, Hülber K, Willner W, Schneeweiss GM. An explicit test of Pleistocene survival in peripheral versus nunatak refugia in two high mountain plant species. Mol Ecol. 2020;29:172–183. 10.1111/mec.15316
DATA AVAILABILITY STATEMENT
Illumina sequence reads for Pedicularis asplenifolia and Carex fuliginosa are available under BioProject PRJNA577188, and genetic data (SNP data) are available at Dryad (https://doi.org/10.5061/dryad.rg0134r).
REFERENCES
- Baird, N. A. , Etter, P. D. , Atwood, T. S. , Currey, M. C. , Shiver, A. L. , Lewis, Z. A. , … Johnson, E. A. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE, 3(10), e3376 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont, M. A. (2010). Approximate Bayesian Computation in evolution and ecology. Annual Reviews in Ecology, Evolution, and Systematics, 41, 379–406. 10.1146/annurev-ecolsys-102209-144621 [DOI] [Google Scholar]
- Bemmels, J. B. , Title, P. O. , Ortego, J. , & Knowles, L. L. (2016). Tests of species‐specific models reveal the importance of drought in postglacial range shifts of a Mediterranean‐climate tree: Insights from integrative distributional, demographic and coalescent modelling and ABC model selection. Molecular Ecology, 25, 4889–4906. 10.1111/mec.13804 [DOI] [PubMed] [Google Scholar]
- Bertorelle, G. , Benazzo, A. , & Mona, S. (2010). ABC as a flexible framework to estimate demography over space and time: Some cons, many pros. Molecular Ecology, 19, 2609–2625. 10.1111/j.1365-294X.2010.04690.x [DOI] [PubMed] [Google Scholar]
- Birks, H. J. B. , & Willis, K. J. (2008). Alpines, trees, and refugia in Europe. Plant Ecology & Diversity, 1, 147–160. 10.1080/17550870802349146 [DOI] [Google Scholar]
- Box, G. E. , & Cox, D. R. (1964). An analysis of transformations. Journal of the Royal Statistical Society Series B (Methodological), 26, 211–252. 10.1111/j.2517-6161.1964.tb00553.x [DOI] [Google Scholar]
- Brown, J. L. , & Knowles, L. L. (2012). Spatially explicit models of dynamic histories: Examination of the genetic consequences of Pleistocene glaciation and recent climate change on the American Pika. Molecular Ecology, 21, 3757–3775. 10.1111/j.1365-294X.2012.05640.x [DOI] [PubMed] [Google Scholar]
- Bryant, D. , & Moulton, V. (2004). Neighbor‐Net: An agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution, 21, 255–265. 10.1093/molbev/msh018 [DOI] [PubMed] [Google Scholar]
- Catchen, J. , Hohenlohe, P. A. , Bassham, S. , Amores, A. , & Cresko, W. A. (2013). stacks: An analysis tool set for population genomics. Molecular Ecology, 22, 3124–3140. 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytrý, M. , Hennekens, S. M. , Jiménez‐Alfaro, B. , Knollová, I. , Dengler, J. , Jansen, F. , … Yamalov, S. (2016). European Vegetation Archive (EVA): An integrated database of European vegetation plots. Applied Vegetation Science, 19, 173–180. 10.1111/avsc.12191 [DOI] [Google Scholar]
- Comes, H. P. , & Kadereit, J. W. (1998). The effect of Quaternary climatic changes on plant distribution and evolution. Trends in Plant Science, 3, 432–438. 10.1016/S1360-1385(98)01327-2 [DOI] [Google Scholar]
- R Core Team . (2013). r: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; Retrieved from https://www.r-project.org/ [Google Scholar]
- Cosacov, A. , Sérsic, A. N. , Sosa, V. , Johnson, L. A. , & Cocucci, A. A. (2010). Multiple periglacial refugia in the Patagonian steppe and post‐glacial colonization of the Andes: The phylogeography of Calceolaria polyrhiza . Journal of Biogeography, 37, 1463–1477. 10.1111/j.1365-2699.2010.02307.x [DOI] [Google Scholar]
- Currat, M. , Arenas, M. , Quilodran, C. , Excoffier, L. , & Ray, N. (2019). splatche3: Simulation of serial genetic data under spatially explicit evolutionary scenarios including long‐distance dispersal. Bioinformatics, 35, 4480–4483. 10.1093/bioinformatics/btz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhamel, G. (1994). Le Carex fuliginosa Schkuhr existe‐t‐il en France? Acta Botanica Gallica, 141, 85–89. 10.1080/12538078.1994.10515139 [DOI] [Google Scholar]
- Earl, D. A. , & vonHoldt, B. M. (2012). structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Elven, R. , Murray, D. F. , Razzhivin, V. , & Yurtsev, B. A. (2011). Checklist of the Panarctic Flora (PAF): Vascular plants, draft version. Oslo: Natural History Museum, University of Oslo; Retrieved from http://panarcticflora.org/ assessed 17 September 2019. [Google Scholar]
- Escobar García, P. , Winkler, M. , Flatscher, R. , Sonnleitner, M. , Krejčíková, J. , Suda, J. , … Schönswetter, P. (2012). Extensive range persistence in peripheral and interior refugia characterizes Pleistocene range dynamics in a widespread Alpine plant species (Senecio carniolicus, Asteraceae). Molecular Ecology, 21, 1255–1270. 10.1111/j.1365-294X.2012.05456.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: A simulation study. Molecular Ecology, 14, 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , & Lischer, H. E. (2010). arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under linux and windows . Molecular Ecology Resources, 10, 564–567. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- Fischer, M. A. , Oswald, K. , & Adler, W. (2008). Exkursionsflora für Österreich, Liechtenstein und Südtirol. Linz: Biologiezentrum der Oberösterreichischen Landesmuseen. [Google Scholar]
- Fulton, T. L. , Norris, R. W. , Graham, R. W. , Semken Jr, H. A. , & Shapiro, B. (2013). Ancient DNA supports southern survival of Richardson's collared lemming (Dicrostonyx richardsoni) during the last glacial maximum. Molecular Ecology, 22, 2540–2548. 10.1111/mec.12267 [DOI] [PubMed] [Google Scholar]
- Gabrielsen, T. M. , Bachmann, K. , Jakobsen, K. S. , & Brochmann, C. (1997). Glacial survival does not matter: RAPD phylogeography of Nordic Saxifraga oppositifolia . Molecular Ecology, 6, 831–842. 10.1046/j.1365-294X.1997.d01-215.x [DOI] [Google Scholar]
- Govindaraju, D. R. (1988). Relationship between dispersal ability and levels of gene flow in plants. Oikos, 52, 31–35. 10.2307/3565978 [DOI] [Google Scholar]
- Gugerli, F. , Sperisen, C. , Büchler, U. , Magni, F. , Geburek, T. , Jeandroz, S. , & Senn, J. (2001). Haplotype variation in a mitochondrial tandem repeat of Norway spruce (Picea abies) populations suggests a serious founder effect during postglacial re‐colonization of the western Alps. Molecular Ecology, 10, 1255–1263. 10.1046/j.1365-294X.2001.01279.x [DOI] [PubMed] [Google Scholar]
- Hasegawa, M. , Kishino, H. , & Yano, T. (1985). Dating of human‐ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution, 22, 160–174. 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- He, Q. , Edwards, D. L. , & Knowles, L. L. (2013). Integrative testing of how environments from the past to the present shape genetic structure across landscapes. Evolution, 67, 3386–3402. 10.1111/evo.12159 [DOI] [PubMed] [Google Scholar]
- He, Q. , Prado, J. R. , & Knowles, L. L. (2017). Inferring the geographic origin of a range expansion: Latitudinal and longitudinal coordinates inferred from genomic data in an ABC framework with the program x‐origin. Molecular Ecology, 26, 6908–6920. 10.1111/mec.14380 [DOI] [PubMed] [Google Scholar]
- Heß, D. (2001). Alpenblumen: Erkennen, Verstehen, Schützen. Stuttgart: Eugen Ulmer. [Google Scholar]
- Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247–276. 10.1111/j.1095-8312.1996.tb01434.x [DOI] [Google Scholar]
- Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London B Biological Sciences, 359, 183–195. 10.1098/rstb.2003.1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickersen, M. J. , Dolman, G. , & Moritz, C. (2006). Comparative phylogeographic summary statistics for testing simultaneous vicariance. Molecular Ecology, 15, 209–223. 10.1111/j.1365-294X.2005.02718.x [DOI] [PubMed] [Google Scholar]
- Hijmans, R. J. , Cameron, S. E. , Parra, J. L. , Jones, P. G. , & Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965–1978. 10.1002/joc.1276 [DOI] [Google Scholar]
- Holderegger, R. , & Thiel‐Egenter, C. (2009). A discussion of different types of glacial refugia used in mountain biogeography and phylogeography. Journal of Biogeography, 36, 476–480. 10.1111/j.1365-2699.2008.02027.x [DOI] [Google Scholar]
- Hultén, E. , & Fries, M. (1986). Atlas of North European vascular plants: North of the Tropic of Cancer I‐III. Königstein: Koeltz Scientific Books. [Google Scholar]
- Huson, D. H. , & Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Jang, T. S. , Emadzade, K. , Parker, J. , Temsch, E. M. , Leitch, A. R. , Speta, F. , & Weiss‐Schneeweiss, H. (2013). Chromosomal diversification and karyotype evolution of diploids in the cytologically diverse genus Prospero (Hyacinthaceae). BMC Evolutionary Biology, 13, 136 10.1186/1471-2148-13-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisch, W. (1993). Zur Blühphänologie und Blütenbiologie der frühblühenden entemophilen Arten einer subnivalen Pflanzengemeinschaft am Brennkogel (Glocknergruppe). Wissenschaftliche Mitteilungen aus dem Nationalpark Hohe Tauern, 1, 72–83. [Google Scholar]
- Leuenberger, C. , & Wegmann, D. (2010). Bayesian computation and model selection without likelihoods. Genetics, 184, 243–252. 10.1534/genetics.109.109058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepelt, S. , Bialozyt, R. , & Ziegenhagen, B. (2002). Wind‐dispersed pollen mediates postglacial gene flow among refugia. Proceedings of the National Academy of Sciences USA, 99, 14590–14594. 10.1073/pnas.212285399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K. , Li, H. , Schlötterer, C. , & Futschik, A. (2011). Distinguishing positive selection from neutral evolution: Boosting the performance of summary statistics. Genetics, 187, 229–244. 10.1534/genetics.110.122614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse, K. , Nicholls, J. A. , & Stone, G. N. (2011). Inferring the colonization of a mountain range – refugia vs. nunatak survival in high alpine ground beetles. Molecular Ecology, 20, 394–408. 10.1111/j.1365-294X.2010.04929.x [DOI] [PubMed] [Google Scholar]
- Marr, K. L. , Allen, G. A. , & Hebda, R. J. (2008). Refugia in the Cordilleran ice sheet of western North America: Chloroplast DNA diversity in the Arctic–alpine plant Oxyria digyna . Journal of Biogeography, 35, 1323–1334. 10.1111/j.1365-2699.2007.01879.x [DOI] [Google Scholar]
- Massatti, R. , & Knowles, L. L. (2016). Contrasting support for alternative models of genomic variation based on microhabitat preference: Species‐specific effects of climate change in alpine sedges. Molecular Ecology, 25, 3974–3986. 10.1111/mec.13735 [DOI] [PubMed] [Google Scholar]
- Meusel, H. , Jäger, E. , Rauschert, S. , & Weinert, E. (1978). Vergleichende Chorologie der zentraleuropäischen Flora 2. Jena: Gustav Fischer. [Google Scholar]
- Meusel, H. , Jäger, E. , & Weinert, E. (1965). Vergleichende Chorologie der zentraleuropäischen Flora 1. Jena: Gustav Fischer. [Google Scholar]
- Mevik, B. H. , & Wehrens, R. (2007). The pls package: Principal component and partial least squares regression in r . Journal of Statistical Software, 18, 1–24. 10.18637/jss.v018.i02 [DOI] [Google Scholar]
- Nettel, A. , Dodd, R. S. , Afzal‐Rafii, Z. , & Tovilla‐Hernández, C. (2008). Genetic diversity enhanced by ancient introgression and secondary contact in East Pacific black mangroves. Molecular Ecology, 17, 2680–2690. 10.1111/j.1365-294X.2008.03766.x [DOI] [PubMed] [Google Scholar]
- Pan, D. , Hülber, K. , Willner, W. , & Schneeweiss, G. (2019). Data from: An explicit test of Pleistocene survival in peripheral versus nunatak refugia in two high mountain plant species with contrasting pollination syndromes. Dryad Digital Repository, 10.5061/dryad.rg0134r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou, A. , & Knowles, L. L. (2016). Toward a paradigm shift in comparative phylogeography driven by trait‐based hypotheses. Proceedings of the National Academy of Sciences, 113, 8018–8024. 10.1073/pnas.1601069113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun, O. , Turner, B. , Trucchi, E. , Munzinger, J. , Chase, M. W. , & Samuel, R. (2015). Processes driving the adaptive radiation of a tropical tree (Diospyros, Ebenaceae) in New Caledonia, a biodiversity hotspot. Systematic Biology, 65, 212–227. 10.1093/sysbio/syv076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun, O. , Schönswetter, P. Winkler, M. , IntraBioDiv Consortium , & Tribsch, A. (2008). Historical divergence vs. contemporary gene flow: Evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Molecular Ecology, 17, 4263–4275. 10.1111/j.1365-294X.2008.03908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A. (2004). distruct: A program for the graphical display of population structure. Molecular Ecology Notes, 4, 137–138. 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- Schneeweiss, G. M. , & Schönswetter, P. (2011). A re‐appraisal of nunatak survival in arctic‐alpine phylogeography. Molecular Ecology, 20, 190–192. 10.1111/j.1365-294X.2010.04927.x [DOI] [PubMed] [Google Scholar]
- Schönswetter, P. , Paun, O. , Tribsch, A. , & Niklfeld, H. (2003). Out of the Alps: Colonization of Northern Europe by East Alpine populations of the glacier buttercup Ranunculus glacialis L. (Ranunculaceae). Molecular Ecology, 12, 3373–3381. 10.1046/j.1365-294X.2003.01984.x [DOI] [PubMed] [Google Scholar]
- Schönswetter, P. , Popp, M. , & Brochmann, C. (2006). Central Asian origin of and strong genetic differentiation among populations of the rare and disjunct Carex atrofusca (Cyperaceae) in the Alps. Journal of Biogeography, 33, 948–956. 10.1111/j.1365-2699.2006.01462.x [DOI] [Google Scholar]
- Schönswetter, P. , & Schneeweiss, G. M. (2019). Is the incidence of survival in interior Pleistocene refugia (nunataks) underestimated? Phylogeography of the high mountain plant Androsace alpina (Primulaceae) in the European Alps revisited. Ecology and Evolution, 9, 4078–4086. 10.1002/ece3.5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönswetter, P. , Stehlik, I. , Holderegger, R. , & Tribsch, A. (2005). Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology, 14, 3547–3555. 10.1111/j.1365-294X.2005.02683.x [DOI] [PubMed] [Google Scholar]
- Schönswetter, P. , Tribsch, A. , Barfuss, M. , & Niklfeld, H. (2002). Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium Sternb. & Hoppe (Campanulaceae) in the European Alps. Molecular Ecology, 11, 2637–2647. 10.1046/j.1365-294X.2002.01651.x [DOI] [PubMed] [Google Scholar]
- Schönswetter, P. , Tribsch, A. , Stehlik, I. , & Niklfeld, H. (2004). Glacial history of high alpine Ranunculus glacialis (Ranunculaceae) in the European Alps in a comparative phylogeographical context. Biological Journal of the Linnean Society, 81, 183–195. 10.1111/j.1095-8312.2003.00289.x [DOI] [Google Scholar]
- Schultze‐Motel, W. (1967–1980). Cyperaceae In Conert H. J., Hamann U., Schultze‐Motel W., & Wagenitz G. (Eds.), Gustav Hegi: Illustrierte Flora von Mitteleuropa II/1, 3rd ed (pp. 2–274). Berlin: Paul Parey. [Google Scholar]
- Stehlik, I. (2000). Nunataks and peripheral refugia for alpine plants during Quaternary glaciation in the middle part of the Alps. Botanica Helvetica, 110, 25–30. [Google Scholar]
- Stehlik, I. , Blattner, F. R. , Holderegger, R. , & Bachmann, K. (2002). Nunatak survival of the high Alpine plant Eritrichium nanum (L.) Gaudin in the central Alps during the ice ages. Molecular Ecology, 11, 2027–2036. 10.1046/j.1365-294X.2002.01595.x [DOI] [PubMed] [Google Scholar]
- Stehlik, I. , Schneller, J. J. , & Bachmann, K. (2001). Resistance or emigration: Response of the high‐alpine plant Eritrichium nanum (L.) Gaudin to the ice age within the Central Alps. Molecular Ecology, 10, 357–370. 10.1046/j.1365-294X.2001.01179.x [DOI] [PubMed] [Google Scholar]
- Swets, J. A. (1988). Measuring the accuracy of diagnostic systems. Science, 240, 1285–1293. 10.1126/science.3287615 [DOI] [PubMed] [Google Scholar]
- Thuiller, W. , Georges, D. , Engler, R. , & Breiner, F. (2016). Package ‘biomod2’. Retrieved from https://cran.r-project.org/web/packages/biomod2/index.html
- Tollefsrud, M. M. , Bachmann, K. , Jakobsen, K. S. , & Brochmann, C. (1998). Glacial survival does not matter – II: RAPD phylogeography of Nordic Saxifraga cespitosa . Molecular Ecology, 7, 1217–1232. 10.1046/j.1365-294x.1998.00452.x [DOI] [Google Scholar]
- Tremblay, N. O. , & Schoen, D. J. (1999). Molecular phylogeography of Dryas integrifolia: Glacial refugia and postglacial recolonization. Molecular Ecology, 8, 1187–1198. 10.1046/j.1365-294x.1999.00680.x [DOI] [PubMed] [Google Scholar]
- Tribsch, A. , & Schönswetter, P. (2003). In search for Pleistocene refugia for mountain plants: Patterns of endemism and comparative phylogeography confirm palaeo‐environmental evidence in the Eastern European Alps. Taxon, 52, 477–497. 10.2307/3647447 [DOI] [Google Scholar]
- Ursenbacher, S. , Schweiger, S. , Tomović, L. , Crnobrnja‐Isailović, J. , Fumagalli, L. , & Mayer, W. (2008). Molecular phylogeography of the nose‐horned viper (Vipera ammodytes, Linnaeus (1758)): Evidence for high genetic diversity and multiple refugia in the Balkan Peninsula. Molecular Phylogenetics and Evolution, 46, 1116–1128. 10.1016/j.ympev.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Wachter, G. A. , Arthofer, W. , Dejaco, T. , Rinnhofer, L. J. , Steiner, F. M. , & Schlick‐Steiner, B. C. (2012). Pleistocene survival on central Alpine nunataks: Genetic evidence from the jumping bristletail Machilis pallida . Molecular Ecology, 21, 4983–4995. 10.1111/j.1365-294X.2012.05758.x [DOI] [PubMed] [Google Scholar]
- Wachter, G. A. , Papadopoulou, A. , Muster, C. , Arthofer, W. , Knowles, L. L. , Steiner, F. M. , & Schlick‐Steiner, B. C. (2016). Glacial refugia, recolonization patterns and diversification forces in Alpine‐endemic Megabunus harvestmen. Molecular Ecology, 25, 2904–2919. 10.1111/mec.13634 [DOI] [PubMed] [Google Scholar]
- Wegmann, D. , Leuenberger, C. , Neuenschwander, S. , & Excoffier, L. (2010). abctoolbox: A versatile toolkit for approximate Bayesian computations. BMC Bioinformatics, 11, 116 10.1186/1471-2105-11-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard, K. B. , Alsos, I. G. , Popp, M. , Engelskjon, T. , Flatberg, K. I. , & Brochmann, C. (2011). Glacial survival may matter after all: Nunatak signatures in the rare European populations of two west‐arctic species. Molecular Ecology, 20, 376–393. 10.1111/j.1365-294X.2010.04928.x [DOI] [PubMed] [Google Scholar]
- Westergaard, K. B. , Zemp, N. , Bruederle, L. P. , Stenøien, H. K. , Widmer, A. , & Fior, S. (2019). Population genomic evidence for plant glacial survival in Scandinavia. Molecular Ecology, 28, 818–832. 10.1111/mec.14994 [DOI] [PubMed] [Google Scholar]
- Zhang, Y.‐Z. , Zhu, R.‐W. , Zhong, D.‐L. , & Zhang, J.‐Q. (2018). Nunataks or massif de refuge? A phylogeographic study of Rhodiola crenulata (Crassulaceae) on the world's highest sky islands. BMC Evolutionary Biology, 18, 154 10.1186/s12862-018-1270-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina sequence reads for Pedicularis asplenifolia and Carex fuliginosa are available under BioProject PRJNA577188, and genetic data (SNP data) are available at Dryad (https://doi.org/10.5061/dryad.rg0134r).


