Summary
Abscission is a process in which plants shed their parts, and is mediated by a particular set of cells, the abscission zone (AZ). In grasses (Poaceae), the position of the AZ differs among species, raising the question of whether its anatomical structure and genetic control are conserved.
The ancestral position of the AZ was reconstructed. A combination of light microscopy, transmission electron microscopy, RNA‐Seq analyses and RNA in situ hybridisation were used to compare three species, two (weedy rice and Brachypodium distachyon) with the AZ in the ancestral position and one (Setaria viridis) with the AZ in a derived position below a cluster of flowers (spikelet).
Rice and Brachypodium are more similar anatomically than Setaria. However, the cell wall properties and the transcriptome of rice and Brachypodium are no more similar to each other than either is to Setaria. The set of genes expressed in the studied tissues is generally conserved across species, but the precise developmental and positional patterns of expression and gene networks are almost entirely different.
Transcriptional regulation of AZ development appears to be extensively rewired among the three species, leading to distinct anatomical and morphological outcomes.
Keywords: abscission zone, evo‐devo, gene co‐expression network, heterotopy, morphological diversity, Poaceae
Short abstract
See also the Commentary on this article by https://doi.org/10.1111/nph.16256.
Introduction
Plants have developed specific and sophisticated mechanisms for shedding parts, a process known as abscission. In general, the plant forms a specialised set of cells known as an abscission zone (AZ), marking the point at which the cells of the falling organ separate from those of the parent (Patterson, 2001). Abscission has received considerable attention in cereal grains because loss of abscission (retention of seeds on the parent plant, also called loss of shattering) was one of the earliest steps in plant domestication (Fuller & Allaby, 2009). Cereals are all members of the grass family (Poaceae), a monophyletic group of c. 12 000 species with global distribution (Kellogg, 2015; Soreng et al., 2015). In grasses, the AZ rarely forms below the fruit (caryopsis or grain), but rather in a position that allows the fruit to be dispersed with various floral and inflorescence parts. Grasses bear their tiny flowers in clusters known as spikelets, consisting of one to many flowers and associated surrounding bracts (glumes, lemmas and paleas). Spikelets are borne on stalks known as pedicels and, within the spikelet, flowers are borne on a central stalk known as the rachilla. The AZ may form below the flower in the rachilla, below sets of flowers in the pedicel or rachis, or even below the entire inflorescence (Fig. 1a) (Doust et al., 2014b; Yu & Kellogg, 2018).
Figure 1.
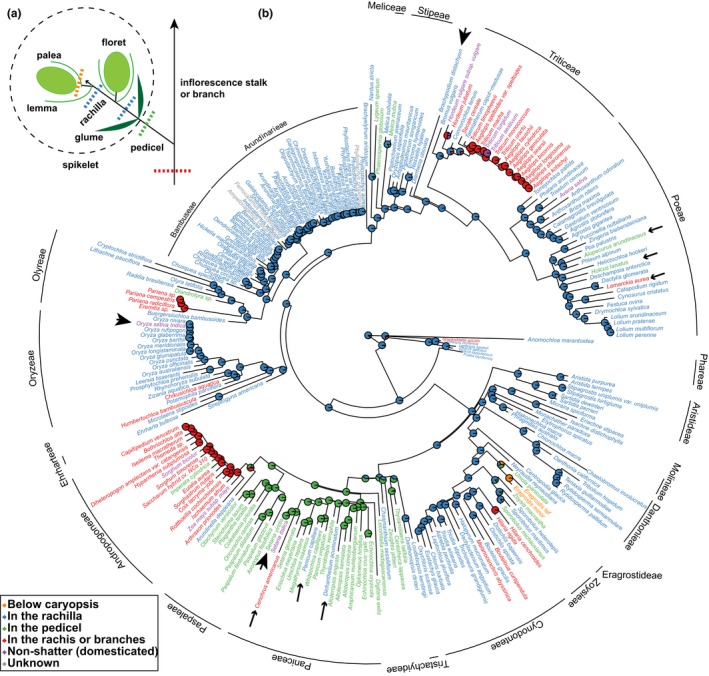
The abscission zone (AZ) position may shift from rachilla down to pedicel and rachis during evolution. (a) A diagram illustrating different AZ positions: below caryopsis and above lemma and palea (orange), in the rachilla below floret and above the glumes (blue), in the pedicel below the glumes (green), or in the rachis or inflorescence branches (red). A floret includes caryopsis and floral bracts (palea and lemma). A spikelet (marked by dotted circle) includes florets and the subtending pair of glumes. (b) Ancestral state reconstruction of the AZ positions in the grass family. Big arrowheads point to rice, Brachypodium and Setaria. Small arrowheads point to examples of species with AZ positions different from other species within the tribe.
AZs may differ anatomically. Often, the AZ is composed of one to a few layers of small, cytoplasmically dense and meristem‐like cells that are distinct from the surrounding cells (Sexton & Roberts, 1982; Patterson, 2001). For example, the AZ of rice (tribe Oryzeae) is located in the rachilla and is composed of small cells with thin nonlignified walls surrounded by larger lignified cells. By contrast, the AZs of Setaria viridis (tribe Paniceae) and wild barley (Hordeum vulgare subsp. spontaneum; tribe Triticeae) are located in the pedicel and inflorescence stalk (rachis), respectively, and are histologically indistinguishable from adjacent cells (Pourkheirandish et al., 2015; Hodge & Kellogg, 2016). Differences in the position and anatomy of the AZ raise the question of whether different grass species share the same underlying genetics and how molecular regulation generates morphological diversification.
Because evolution generally operates by modifying existing structures, we hypothesised that AZs in grasses would be developmentally and genetically similar despite their positional and anatomical differences. Genetic studies on the AZ have been most extensive in rice, and a number of genes have been identified that, when mutated, lead to loss of abscission. These include a myb transcription factor (TF) Shattering4 (SH4) (Li et al., 2006; Wu et al., 2017), BEL1‐like homeodomain TFs qSH1 (Konishi et al., 2006) and SH5 (Yoon et al., 2014), the KNOX TF OSH15 (Yoon et al., 2017), the APETALA2 (AP2) TF SHATTERING ABORTION1 (SHAT1) (Zhou et al., 2012), the YABBY TF YAB2/ObSH3 (Lv et al., 2018) and the carboxy‐terminal domain phosphatase‐like gene OsCPL1 (Ji et al., 2010). Data on species other than rice remain scarce. Only two genes are known to control AZ development in species with different AZ positions. YAB2/ObSH3 (also called Shattering1, SH1) leads to loss of shattering not only in rice (Lv et al., 2018), but also in Setaria (Doust et al., 2014a; Odonkor et al., 2018) and Sorghum (Lin et al., 2012), where AZs are located in the pedicel and rachis, respectively. Similarly, the wheat AZ forms in the rachis, which is a different position from rice, and yet orthologous AP2 (called SHAT1 in rice and Q in wheat) loci in both species play pleiotropic roles in inflorescence development, including AZ development (Simons et al., 2006; Zhou et al., 2012). OsAP2 is highly expressed in the AZ in rice and a null mutation completely abolishes the small and nonlignified AZ cells (Zhou et al., 2012), while the specific role of TaAP2 in wheat AZ development is unknown.
Outside Poaceae, evolution of the AZ has been studied extensively in fruits of Brassicaceae. Despite their diverse shapes, most studies support a conserved genetic regulatory pathway composed of several TFs identified in Arabidopsis thaliana, including the MADS‐box TFs FRUITFULL (FUL) (Ferrándiz et al., 2000), SHATTERPROOF1 (SHP1) and SHP2 (Liljegren et al., 2000), the basic helix‐loop‐helix (bHLH) TFs ALCATRAZ (Rajani & Sundaresan, 2001) and INDEHISCENT (IND) (Liljegren et al., 2004), and BEL1‐like homeodomain TF REPLUMLESS (RPL) (Roeder et al., 2003). Altered expression of these genes in other Brassicaceae species, including the naturally nonshattering Cakile lanceolata (Avino et al., 2012) and transgenic plants of shattering Lepidium campestre (Lenser & Theißen, 2013; Mühlhausen et al., 2013), changes abscission capability. The functions of FUL and SHP are also conserved in Solanaceae, a eudicot family distantly related to Brassicaceae. Overexpression of FUL and suppression of SHP in Nicotiana species reduces shattering as in Arabidopsis (Smykal et al., 2007; Fourquin & Ferrándiz, 2012). Furthermore, FUL2 is expressed in the AZ of Streptochaeta angustifolia (Preston et al., 2009), a lineage sister to the remainder of the grass family, suggesting deep conservation of FUL in AZ development, even though the organs where abscission occurs are not homologous between Brassicaceae and Poaceae: the former is between the carpels and the latter is along the inflorescence axis.
If the genetic pathway controlling AZ development is conserved and only shifts from one position to another, the differences in AZ position within Poaceae and even between Brassicaceae and Poaceae can be interpreted as heterotopy, an evolutionary change of position of a developmental process or program (Bateman, 1994; Zelditch & Fink, 1996). Activating a developmental process in a new position often leads to novel structures, and therefore heterotopy could be a means of evolutionary modification (Hall, 1999; Baum & Donoghue, 2002). Only a few examples of heterotopy have been demonstrated, however, including development of the jaw in gnathostomes (Shigetani et al., 2002), shell formation of turtles (Nagashima et al., 2009) and leaf‐borne inflorescences in some angiosperms (Baum & Donoghue, 2002).
To test whether variation in AZ position and anatomy fits the definition of heterotopy, we first need to understand the phylogenetic distribution of the different AZ positions and then to examine AZs in different positions to determine if they share similar anatomy, histology and underlying gene networks. In this study, we compared AZ development of three distantly related grass species, a shattering weedy rice (Oryza sativa, Os), purple false brome (Brachypodium distachyon, Bd) and green foxtail (Setaria viridis, Sv). Rice and Brachypodium are both members of one major clade of the grass family and have an AZ located in the rachilla, while Setaria belongs to the other major clade and its AZ is located in the pedicel. We show that the AZs of all three species differ in anatomy, cell wall properties and gene expression as shown by co‐expression network analyses and in situ hybridisation, suggesting divergence of regulatory mechanisms.
Materials and Methods
Ancestral state reconstruction
AZ position was assessed for 250 grass taxa by new observations of specimens at the herbarium of the Missouri Botanical Garden, supplemented with information from Barkworth et al. (2003, 2007) and Wu et al. (2006). AZ position was classified as either below the fruit, in the rachilla below the floret, in the pedicel below the spikelet, or in the rachis or branches (Fig. 1a). The ancestral AZ position was reconstructed on the grass phylogeny of Saarela et al. (2018) using phytools (Revell, 2012) in R (v.3.5.0) with an equal rates model.
Plant materials and growth conditions
De‐domesticated US weedy rice (Oryza sativa), straw‐hulled (Os‐SH) (Thurber et al., 2010), was provided by Dr Kenneth M. Olsen at Washington University at St Louis. Brachypodium distachyon (Bd21‐0) was provided by Dr Malia Gehan at Donald Danforth Plant Science Center. Setaria viridis (ME034V) was from seeds available in the Kellogg lab (Layton & Kellogg, 2014). Each species was grown under its own optimal growth conditions at the Danforth Center Plant Growth Facility. Rice was grown in Turface MVP soil (Turface Athletics, Buffalo Grove, IL, USA) in a glasshouse with 30–70% humidity and 15 h : 9 h, light : dark with temperatures of day : night, 24–28°C : 23–25°C. Brachypodium was grown in a growth chamber with 50% humidity, light : dark, 20 h : 4 h, with light intensity of 200 μmol m−2 s−1 and temperatures of day : night, 24°C : 18°C. Setaria viridis was grown in Metro‐Mix 360 (Sun Gro) in a growth chamber with 50% humidity, light : dark 12 h : 12 h, with light intensity of 250 μmol m−2 s−1 and temperatures of day : night, 31°C : 22°C.
Histology
At least three inflorescences per species were collected and dissected (Supporting Information Fig. S1). Post‐anthesis spikelets (seeds are half filled, corresponding to the ‘old stage’ described below) (Fig. S1d–i) were fixed in ice cold FAA (37% formaldehyde : ethanol : H2O : acetic acid = 10 : 50 : 35 : 5), and dehydrated in 50%, 70%, 85%, 95%, 100%, 100% and 100% ethanol and then 25%, 50%, 75%, 100%, 100% and 100% Histo‐Clear II (National Diagnostics, Atlanta, GA, USA) with ethanol as solvent, following Ruzin (1999). Paraplast® (Leica Biosystems, Wetzlar, Germany) was then added to each vial of samples and kept overnight, heated at 42°C, and placed in a 60°C oven. Solution was replaced with molten Paraplast® twice a day for 3 d. Samples were embedded in paraffin using a Leica EG1150 tissue embedder, sectioned into 10‐μm serial slices with a Leica RM2255 automated microtome, and mounted on microscope slides at 37°C on a Premiere XH‐2001 Slide Warmer overnight.
Sections were deparaffinised, rehydrated, stained with 1% (w/v) Safranin O for 3 h and 0.05% (w/v) Fast Green for 5–15 s following Ruzin (1999). Specimens were mounted with Permount, and images taken using a Leica DM750 LED Biological microscope with ICC50 camera module and leica acquire v.2.0 software (Leica Microsystems).
Transmission electron microscopy (TEM)
Old stage tissues from two biological samples per species were dissected and fixed in 2% glutaraldehyde in 0.1 M PIPES buffer for 90 min under vacuum. After three rinses in 0.1 M PIPES buffer for 7 min each, tissues were post‐fixed in 2% osmium tetroxide in 0.1 M PIPES for 90 min, followed by three rinses in water and dehydrated in 5%, 10%, 20%, 30%, 50%, 75%, 95% EtOH for 15 min, 100% EtOH for 30 min, 100% acetone for 15 min and a second 100% acetone for 45 min. Tissues were infiltrated in a series of Spurr's resin of 5% and 10% for 12 h, 25%, 50%, 75% and 100% for 24 h with acetone as solvent, and embedded in Spurr's resin at 60°C for 48 h. Specimens were sliced into 90‐nm sections using a Leica Ultracut UCT, mounted on a copper grid, stained in uranyl acetate for 12 min and lead salts for 3 min, and imaged using a LEO 912AB TEM.
RNA isolation, RNA‐Seq library construction, sequencing and mapping
Spikelets were sampled when anthers were at late sporogenesis and gametogenesis but the AZ lacked obvious secondary cell wall formation (the ‘young stage’), and also after anthesis, when seeds were approximately half filled (the ‘old stage’). The AZ (A) plus tissues immediately above (upper, U) and below (lower, L) were collected for transcriptomic analysis (Fig. S1). Each of the three biological replicates used spikelets from at least two plants per species per replicate at each developmental stage. Tissues were dissected in RNAlater™ Stabilisation Solution (ThermoFisher Scientific, Waltham, MA, USA) using a scalpel under a dissecting microscope. The upper tissues included the lowermost parts of the rachilla and bracts (lemma, palea, and/or glumes depending on species) but excluded reproductive organs, and lower tissues were rachilla or pedicel, again depending on species (Fig. S1). Dissected tissues were immediately transferred to the extraction buffer of PicoPure RNA Isolation Kit (Thermo Fisher Scientific), ground with pellet pestles, total RNA isolated using the kit with in‐column DNase I treatment following manufacturer's instructions, and quantified using a Qubit 3.0 Fluorometer (Life Technologies). Next, c. 300 ng total RNA was used for generating 3′ mRNA‐Seq libraries using QuantSeq 3′ mRNA‐Seq Library Prep Kit FWD for Illumina (Lexogen) (Moll et al., 2014). In total, 54 libraries were single‐end sequenced at 150 nt in two lanes on an Illumina HiSeq 4000 instrument at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana‐Champaign. Sequences were deposited in the Sequence Read Archive (SRR8635261‐SRR8635314).
Sequencing adaptors, polyA and low quality sequences were removed using Trimmomatic (v.0.35) (Bolger et al., 2014). Reads from rice, Brachypodium and Setaria were mapped to an indica rice genome Shuhui498 (R498) (Du et al., 2017), Brachypodium distachyon v.3.1 (International Brachypodium Initiative, 2010) and Setaria viridis ME034V v.1.0 (P. Huang et al. unpublished), respectively, using hisat2 software (v.2.0.6) (Pertea et al., 2016). Mapped read counts were calculated using htseq‐count (HTSeq v.0.9.1) (Anders et al., 2015). Raw counts were input into DEseq2 (v.1.22.2) in R (v.3.5.0) (Love et al., 2014). Genes with < 10 normalised counts in all of the libraries were filtered out before subsequent analyses.
Self‐organising map (SOM) clustering, weighted gene co‐expression network analysis (WGCNA) and gene co‐expression visualisation
Differentially expressed genes (DEG) were identified using a threshold of absolute fold change of 1.5 between A vs L or A vs U at young or old stage, and adjusted P‐value < 0.05, using DEseq2 (Love et al., 2014). Genes with at least one significant difference in the comparisons were used for SOM analysis using Multiple Experiment Viewer (v.4.8.1) with the bubble neighbourhood method (radius = 0.7).
Orthologues among rice, Brachypodium and Setaria were identified using OrthoFinder (Emms & Kelly, 2015). Normalised counts of one‐to‐one orthologues expressed in all three species were imported to WGCNA (v.1.66) in R for co‐expression network construction (Langfelder & Horvath, 2008). A minimal module size of 30 genes, cutHeight of 0.14, soft‐thresholding power of 16 and deepSplit of 2 were used for all three species. Degree distributions in each network followed the power law and satisfied the scale‐free topology criterion. Module preservation analysis used the R function module preservation in the wgcna package, and the composite statistic Zsummary value (average of module density and module connectivity) was calculated for evaluating module preservation between species (Langfelder et al., 2011). Co‐expressed genes were defined as a weight of interaction > 0.05, and those with weight > 0.1 were visualised using cytoscape v.3.7.1 with edge‐weighted spring embedded layout (Smoot et al., 2010).
In situ hybridisation
RNA probes were designed from the 3′ end of the transcript of each gene (primers and probe lengths in Table S1). Sequences were amplified from each species and cloned into the Zero Blunt™ TOPO™ PCR Cloning Kit (ThermoFisher). Digoxigenin‐UTP‐labeled sense and antisense probes were synthesised by in vitro transcription using T7 or SP6 RNA polymerase (Promega) and hydrolysed in carbonate buffer (pH 10.2) at 60°C into 150 nt.
In situ hybridisation followed published methods (Yang et al., 2018). Sections were deparaffinised with Histo‐Clear II (National Diagnostics) for 10 min twice, rehydrated in 100%, 100%, 90%, 70%, 50% and 30% ethanol series and DEPC treated water for 2 min each, followed by 5 μg ml−1 proteinase K (Roche) at 37°C for 20 min, refixation in 4% (w/v) formaldehyde for 10 min, and acetic anhydride for 10 min. Sections were then dehydrated with an ethanol series and kept under vacuum for 1–2 h. Hybridisation was performed at 50°C for 16–18 h in buffer composed of 0.5 ng ml−1 probe, 50% formamide, 1 mg ml−1 tRNA, 0.5 mg ml−1 polyA, 30 mM DTT, 0.3 M NaCl, 10 mM Tris‐HCl, 1 mM EDTA, 10% dextran sulfate, and 1 × Denhardt's solution. On the second day, slides were washed with 4 × SSC (0.6 M NaCl and 60 mM trisodium citrate) at 50°C for 15 min, treated with 30 μg ml−1 RNase A in RNase buffer (0.5 M NaCl and 0.01 M Tris‐HCl, pH 7.5) at 37°C for 30 min, washed with RNase buffer for 5 min three times, 0.5 × SSC for 20 min twice, and buffer1 (0.15 M NaCl and 0.1 M Tris‐HCl, pH 7.5) for 5 min twice, and incubated in blocking reagent (Roche) for 30 min. Sections were incubated with 1 : 1000 anti‐digoxigenin‐AP antibody (Roche) for 1 h at room temperature, washed with buffer 1 for 10 min three times and coloration buffer (0.1 M NaCl, 0.05 M MgCl2 and 0.1 M Tris‐HCl, pH 9.5) for 5 min. Alkaline phosphatase signal was visualised by incubating the slides in 5‐bromo‐4‐chloro‐3‐indolyl‐phosphate/nitro blue tetrazolium (Roche) in coloration buffer overnight at 37°C. Imaging used a Leica DM750 LED Biological microscope with ICC50 camera module and leica acquire v.2.0 software.
Results
AZ position shifts from the rachilla down to pedicel and rachis in evolutionary time
The ancestral position of the AZ is likely in the rachilla (below the floret), while the position shifts to the pedicel in tribes Paniceae and Paspaleae and to the rachis in tribes Andropogoneae and Triticeae (Fig. 1b). Additional shifts also occurred between closely related species in a number of tribes. For example, in the tribe Poeae, where most species have an AZ in the rachilla, the position shifted to the pedicel in Alopecurus arundinaceus and Holcus lanatus, and to the base of the inflorescence branches in Lamarckia aurea. Similarly, in the tribe Paniceae, with the AZ mostly located in the pedicel, the AZs of Panicum capillare and Dichanthelium acuminatum form in the rachilla, and in Cenchrus americanus at the base of the inflorescence branches (Fig. 1b; Table S2). The positional shift of the AZ in closely related species suggests that heterotopy of AZ development might be governed by simple genetics and few mutations.
AZs of rice, Brachypodium and Setaria differ in anatomical structure
We hypothesised that AZs in the same position should be anatomically similar and differ from those in different positions. To test this hypothesis, we compared the AZ anatomy of a shattering weedy rice and two wild shattering species, Brachypodium and Setaria. The AZ of rice and Brachypodium is located in the rachilla, although Brachypodium has multiple fertile florets and therefore multiple AZs within a spikelet while rice has only one. The AZ of Setaria is in the pedicel (Fig. 1b). Consistent with our hypothesis, the AZs of rice and Brachypodium are composed of one or two layers of cells that are much smaller than the adjacent ones (Fig. 2a,b), while the cells in the AZ of Setaria are not distinguishable from adjacent cells (Fig. 2c) (Hodge & Kellogg, 2016). The cells of the rice AZ lack the characteristic magenta colour of lignin stained with safranin O (Johansen, 1940; Ruzin, 1999) (Fig. 2a), whereas the AZ of Brachypodium is more strongly stained by safranin O than the surrounding cells (Fig. 2b), indicating different cell wall composition between the AZ of rice and Brachypodium despite their similar position and cell size. In Setaria, cells in the AZ lack lignin entirely except in the epidermis, confirming previous data (Fig. 2c) (Hodge & Kellogg, 2016).
Figure 2.
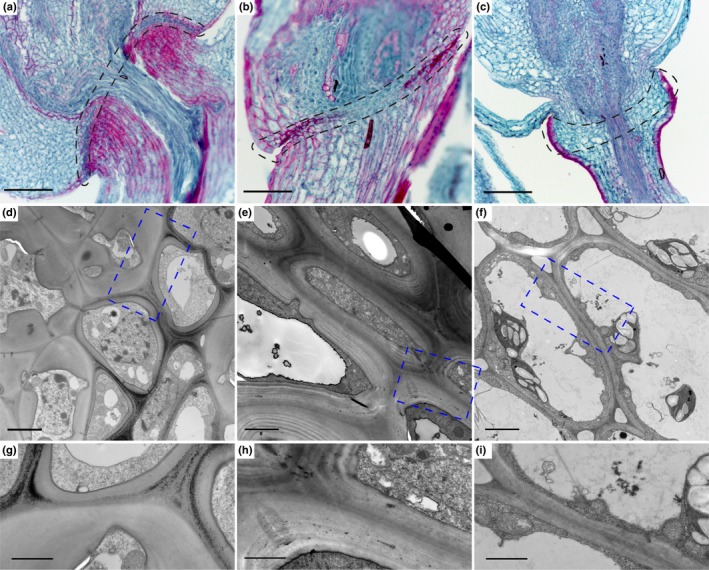
The abscission zones (AZ) of rice, Brachypodium and Setaria differ in histology and cell wall structures. (a–c) Fast green/Safranin O staining of mature spikelets in (a) rice, (b) Brachypodium and (c) Setaria. Safranin O stains secondary cell wall including lignin, suberin and cutin with a magenta colour, and fast green stains primary cell wall and cytoplasm with a green colour. The AZ is marked with black dotted lines. (d–f) Transmission electron microscopy images of the AZ and surrounding cells in (d) rice, (e) Brachypodium and (f) Setaria. (g–i) Zoomed in images of the blue dotted rectangles shown in (d‐f). Bars: (a ‐c) 100 µm; (d #x2010;f) 2 µm; (g #x2010;i) 1 µm.
TEM shows that cells in the rice AZ are thin‐walled and cytoplasmically dense, while the adjacent cells are thick‐walled, indicating secondary cell wall deposition (Fig. 2d,g). In Brachypodium, AZ cells also have dense cytoplasm but the wall is much thicker than that in rice, although thinner than that of the adjacent cells (Fig. 2e,h). Setaria exhibits only thin‐walled cells with large vacuoles and extensive intercellular spaces in the region of the AZ (Fig. 2f,i). In summary, the AZs of rice, Brachypodium and Setaria all differ in anatomy and cell wall structure, although rice and Brachypodium are more similar to each other than either is to Setaria.
Gene co‐expression patterns are conserved at similar developmental stages across three species
After filtering out genes with low expression, we identified 19 990, 20 408 and 20 381 genes expressed in at least one of AZ, upper or lower tissues in rice, Brachypodium and Setaria, respectively (Tables S3–S5).
Overall gene expression was similar among the three species. Among 12 951 one‐to‐one orthologues identified, 11 333, 11 235 and 11 456 genes are expressed in rice, Brachypodium and Setaria, respectively, and 10 273 genes are expressed in all three species (90.6% of Os, 91.4% of Bd and 89.7% of Sv expressed orthologues), suggesting substantial overlap and extensive conservation of gene expression in the examined tissues as a whole. The expressed one‐to‐one orthologues were then used for WGCNA in individual species. Overall, 15, 17 and 18 co‐expression modules were found in rice, Brachypodium and Setaria, respectively, including a module of unassigned genes (grey module) in each species (Figs S2–S4; Table S6). WGCNA designates modules by colours, which here apply only within individual species; they are not comparable among species.
To test whether any modules in one species were preserved in the other two, we calculated the preservation statistic Zsummary using the modules in one species as a reference network and the other two as test networks (Langfelder et al., 2011). Most modules were only weakly to moderately preserved (2 < Zsummary < 10), and only three to five modules were highly preserved (Zsummary > 10) (Fig. S5). For instance, when using rice as reference, the orange module in Brachypodium and the blue module in Setaria, preferentially expressed at young and old stages, respectively, were highly preserved (Fig. S5a,b). We observed the same pattern when using Brachypodium and Setaria as reference (Fig. S5c−f). Therefore, a partially conserved set of genes differentiates young and old stages of development in the study tissues.
We next calculated gene overlap between all pairs of modules (Fig. S6) and found significant overlap among all three species for modules upregulated at the young stage, again supporting conserved expression patterns for developmental stages (Fig. 3). In addition, modules highly expressed in the region below the AZ (L) at the old stage also overlap significantly among three species (Fig. S7a–f), and two pairs of modules with highly significant overlap are observed between Brachypodium and Setaria (Fig. S7g–l). Modules with highly significant overlap across species also have similar expression patterns (Figs 3, S7), suggesting that these genes may play fundamental roles in development of these tissue types.
Figure 3.
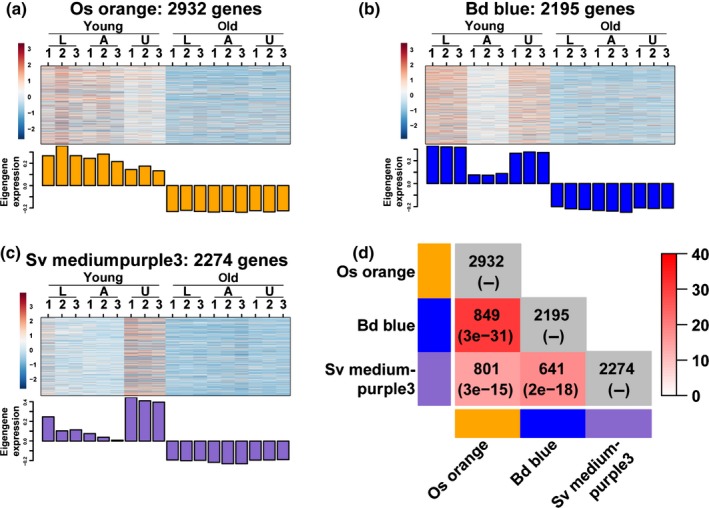
WGCNA modules with differential expression at the two developmental stages have substantial gene overlap among the three species. (a–c) Heatmap and eigengene bar graph of (a) rice orange module, (b) Brachypodium blue module and (c) Setaria mediumpurple3 module showing higher expression at the young stage than that at the old stage. (d) Pairwise comparisons of the number of overlapping genes between modules. The numbers in parentheses are P‐values from Fisher's exact test to test statistical significance between two modules. White to red colour key indicates –log10(P‐value). Module comparisons within species are not meaningful and indicated with grey colour.
AZ‐enriched co‐expression modules are largely nonoverlapping between species
Each species had AZ‐specific co‐expression modules. Rice had one AZ module (black, 962 genes) at the young stage and one (midnightblue, 300 genes) at the old stage (Fig. 4a,b). Brachypodium also had one at each stage (young: orange, 111 genes; old: darkred, 861 genes) (Fig. 4c,d). Setaria had two AZ modules at the young stage (purple and darkgrey, 298 and 327 genes, respectively) (Fig. 4e,f), but none at the old stage (given our module identification criteria). No module is upregulated in the AZ at both young and old stages in any species, suggesting different gene co‐expression networks controlling AZ development at the two stages (Fig. 4a–f).
Figure 4.
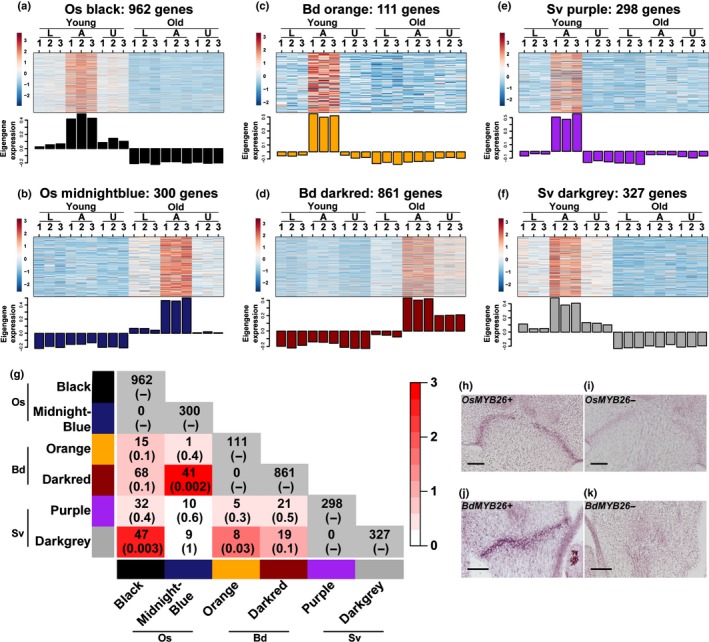
The number of overlapping genes between abscission zone (AZ) modules is small among the three species. (a–f) Heatmap and eigengene bar graph of (a) rice young AZ module (black), (b) rice old AZ module (midnightblue), (c) Brachypodium young AZ module (orange), (d) Brachypodium old AZ module (darkred) and (e, f) Setaria young AZ modules (purple and darkgrey). (g) Pairwise comparisons of the number of overlapping genes between modules. The numbers in parentheses are P‐values from Fisher's exact test to test statistical significance of overlap between two modules. White to red colour key indicates –log10(P‐value). Module comparisons within species are not meaningful and indicated with grey colour. (h–k) In situ hybridisation of MYB26 in (h, i) rice and (j, k) Brachypodium. (h, j) antisense probes; (i, k) sense probes. Bars, 50 μm.
We then compared shared genes between the AZ modules. By contrast with the conserved developmental stage modules (Fig. 3), AZ modules share many fewer genes (Fig. 4g; Table S6). Only three pairs of comparisons show significant overlap, including the young stage AZ modules Os black vs Sv darkgrey (Fisher's exact test, P = 0.003) and Bd orange vs Sv darkgrey (Fisher's exact test, P = 0.03), and the old stage modules Os midnightblue vs Bd darkred (Fisher's exact test, P = 0.002), with 47, 8 and 41 shared genes, respectively (Fig. 4g). Surprisingly, the set of genes preferentially expressed in the AZ was no more similar between rice and Brachypodium than between those species and Setaria, despite the similar position and anatomy of the AZ in the former two species (Figs 1, 2).
Only three genes are shared among the young stage AZ modules of all three species: MYB26, 4‐COUMARATE:CoA LIGASE 3 and a gene with a possible lysine decarboxylase domain (Table S7). The first two of these may regulate lignification or other aspects of secondary cell wall development (Yang et al., 2007; Li et al., 2015), while the role of the putative lysine decarboxylase is unknown.
We addressed the possibility that our result could be biased by the use of only one‐to‐one orthologues and WGCNA. As an alternative approach, we examined all of the expressed genes in each individual species and identified DEG in A vs L or A vs U comparisons. Rice, Brachypodium and Setaria had 2137, 2524 and 4072 DEG, respectively (Table S8). SOM clustering identified one AZ‐enriched cluster in each species at the young stage, with 318, 245 and 467 genes in rice, Brachypodium and Setaria, respectively (Fig. S8). At the old stage, rice had one AZ‐specific cluster of 121 genes and Brachypodium had two clusters of 252 and 301 genes each (Fig. S8). Consistent with our WGCNA results, no obvious AZ clusters are observed at the old stage in Setaria (Fig. S8c).
Comparing DEGs among AZ clusters again finds little overlap (Fig. S9; Table S9), suggesting that the AZ transcriptome is largely nonconserved across grass species regardless of the AZ position. However, three genes appear in the AZ clusters in all three species, two of which, MYB26 and the gene with a possible lysine decarboxylase domain, were also identified by WGCNA (Table S7). We validated expression of MYB26 by in situ hybridisation. The gene is indeed specifically expressed in the AZ of rice and Brachypodium (Fig. 4h–k), although expression in Setaria was not detectable (Fig. S10).
Orthologues of previously identified AZ genes show different co‐expression networks
To validate our RNA‐Seq analysis, we examined the co‐expression network of known AZ genes. In rice, the known AZ genes YAB2/ObSH3/SH1 (Lin et al., 2012; Lv et al., 2018), AP2/SHAT1/Q (Simons et al., 2006; Zhou et al., 2012), SH4 (Li et al., 2006), qSH1 (Konishi et al., 2006) and SH5 (Yoon et al., 2014) are all assigned to the AZ‐specific module at the young stage (black) of the WGCNA analysis (Fig. 5a; Table S6). AP2, SH4 and qSH1 also fall in AZ‐specific clusters in the SOM analysis (Table S8). In Setaria, YAB2/ObSH3/SH1 (Lin et al., 2012; Odonkor et al., 2018) is classified in the AZ module at the young stage in both WGCNA (purple module) and SOM analysis (Fig. 5a; Tables S6, S8). These results give credence to our RNA‐Seq co‐expression data. However, of the known AZ genes, only SH4 is found in an AZ module (orange) in Brachypodium. Other supposedly AZ‐specific genes show various co‐expression patterns (Fig. 5a), suggesting that their roles in AZ development may not be conserved across species.
Figure 5.
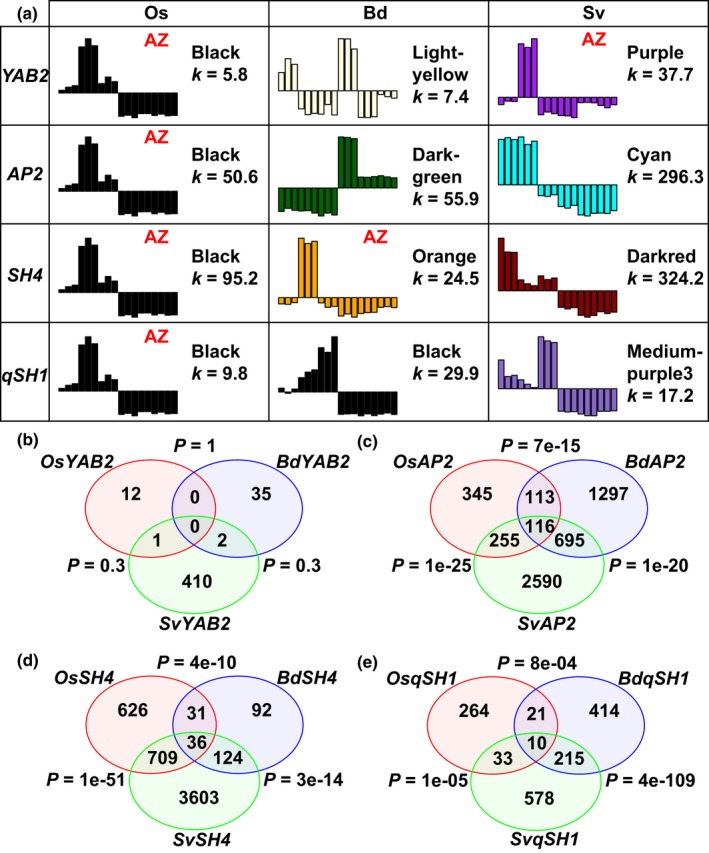
Previously identified abscission zone (AZ) genes are in different co‐expression modules in different species. (a) Modules in which YAB2, AP2, SH4 and qSH1 are classified. k stands for the total connectivity of a gene with all other expressed genes. (b–e) The number of overlapping co‐expressed genes of (b) YAB2, (c) AP2, (d) SH4 and (e) qSH1 in rice, Brachypodium and Setaria. P‐values are calculated from Fisher's exact test to test statistical significance of overlap between species.
We next compared the co‐expression networks of YAB2, AP2, SH4 and qSH1 in each species, using connectivity (sum of correlation strength with all other genes, k) as a measure of importance of a gene. The orthologues of these genes differ among species in connectivity and numbers of co‐expressed genes (Fig. 5; Table S10). For example, although both OsYAB2 and SvYAB2 are in AZ modules, OsYAB2 has a connectivity of 5.8 and 13 co‐expressed genes, whereas SvYAB2 has a connectivity of 37.7 and 413 co‐expressed genes (Fig. 5a,b), implying regulatory divergence of YAB2 in rice and Setaria.
We then hypothesised that the ‘AZ‐specific’ genes might share co‐expression networks, even if those networks are expressed at different positions in the plant. In other words, a subset of co‐expressed genes might shift their spatial expression patterns all together. Indeed, except YAB2, which has many fewer co‐expressed genes in rice and Brachypodium, networks for the other three genes all show highly significant overlap in all pairwise comparisons (Fisher's exact test, P < 0.001) (Fig. 5b–e), suggesting partially conserved co‐expression networks. AP2, SH4 and MYB26 are highly connected (i.e. co‐regulated) with each other in rice, and AP2 and SH4 are also connected in Setaria to a lesser degree, although not specifically enriched in the AZ (Fig. S11; Table S9).
Orthologues of known AZ genes exhibit different temporal and spatial expression patterns
Our RNA‐Seq and co‐expression analyses suggest that AZ transcriptomes at the same floral developmental stage differ substantially among the three species (Fig. 4). Even genes known to be critical for AZ development in one species do not have conserved expression patterns in the other two (Fig. 5). One possible explanation is that these genes might be expressed in the AZ but at times different from the two captured by our RNA‐Seq experiment. We tested this hypothesis with in situ hybridisation on YAB2, AP2, SH4 and qSH1, which are highly expressed in rice early in spikelet development at or before the young stage of our RNA‐Seq (Zhou et al., 2012; Lv et al., 2018). Therefore, we used in situ hybridisation at three stages of development in which the oldest (Stage 3, S3 hereafter) is the young stage of our RNA‐Seq experiment (Figs 6, 7, 8, 9). Of the YAB2 orthologues, only SvYAB2 is specifically enriched in the AZ compared with the adjacent tissues in all three tested stages, and expression in the AZ is even higher in stage 1 (S1) and stage 2 (S2) (Fig. 6i–l), consistent with WGCNA and SOM (Tables S6, S8). SvYAB2 is also expressed at the base of the upper floret (Fig. 6i–l), presumably the secondary AZ of Setaria, in which abscission occurs less frequently and later than at the pedicel AZ (Hodge & Kellogg, 2016). By contrast, OsYAB2 and BdYAB2 are expressed in the AZ as well as the bracts above (lemmas and paleas) so are not AZ specific, although expression in the AZ is more pronounced at stage 3 (S3) (Fig. 6a–h).
Figure 6.
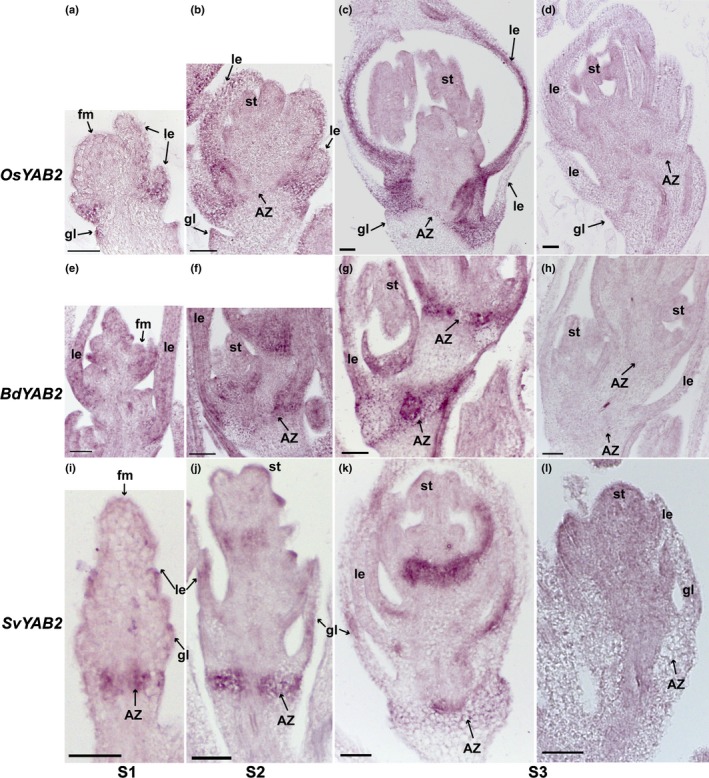
YAB2 is expressed in the abscission zone (AZ) and floral bracts by in situ hybridisation. In situ hybridisation of YAB2 in (a–d) rice, (e–h) Brachypodium and (i–l) Setaria was performed at (a, e, i) floret meristem stage (S1), (b, f, j) anther differentiation stage (S2) and (c, g, k) a stage when anthers were fully developed (S3). (d, h, l) are sense probe controls of YAB2 in (d) rice, (h) Brachypodium and (l) Setaria. fm, floral meristem; gl, glume; le, lemma; st, stamens. Bars, 50 μm.
Figure 7.
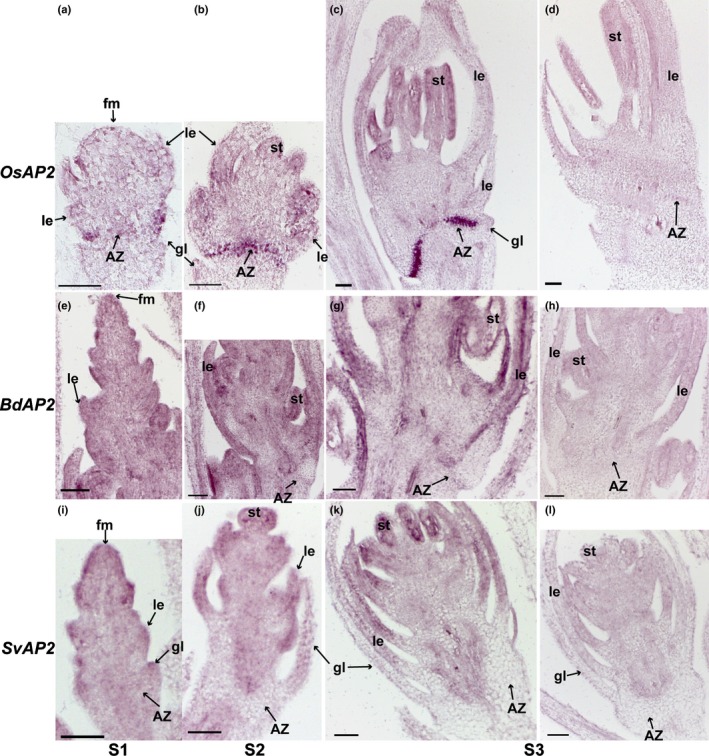
AP2 is highly enriched in the abscission zone (AZ) in rice but not in Brachypodium or Setaria. In situ hybridisation of AP2 in (a–d) rice, (e–h) Brachypodium and (i–l) Setaria was performed at (a, e, i) floret meristem stage (S1), (b, f, j) anther differentiation stage (S2) and (c, g, k) a stage when anthers were fully developed (S3). (d, h, l) are sense probe controls of AP2 in (d) rice, (h) Brachypodium and (l) Setaria. fm, floral meristem; gl, glume; le, lemma; st, stamens. Bars, 50 μm.
Figure 8.
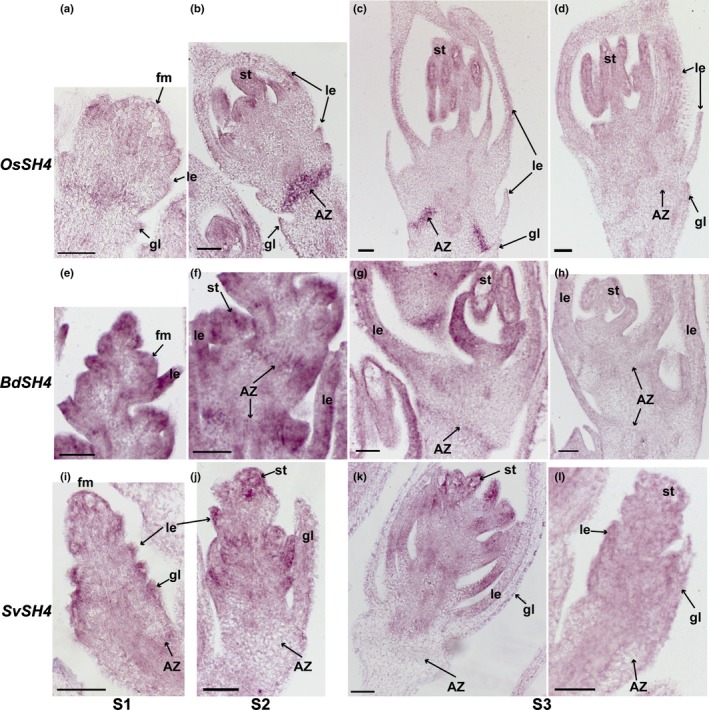
SH4 is expressed in the abscission zone (AZ) in rice and Brachypodium but not in Setaria. In situ hybridisation of SH4 in (a–d) rice, (e–h) Brachypodium and (i–l) Setaria was performed at (a, e, i) floret meristem stage (S1), (b, f, j) anther differentiation stage (S2) and (c, g, k) a stage when anthers were fully developed (S3). (d, h, l) are negative controls with sense probes of SH4 in (d) rice, (h) Brachypodium and (l) Setaria. fm, floral meristem; gl, glume; le, lemma; st, stamens. Bars, 50 μm.
Figure 9.
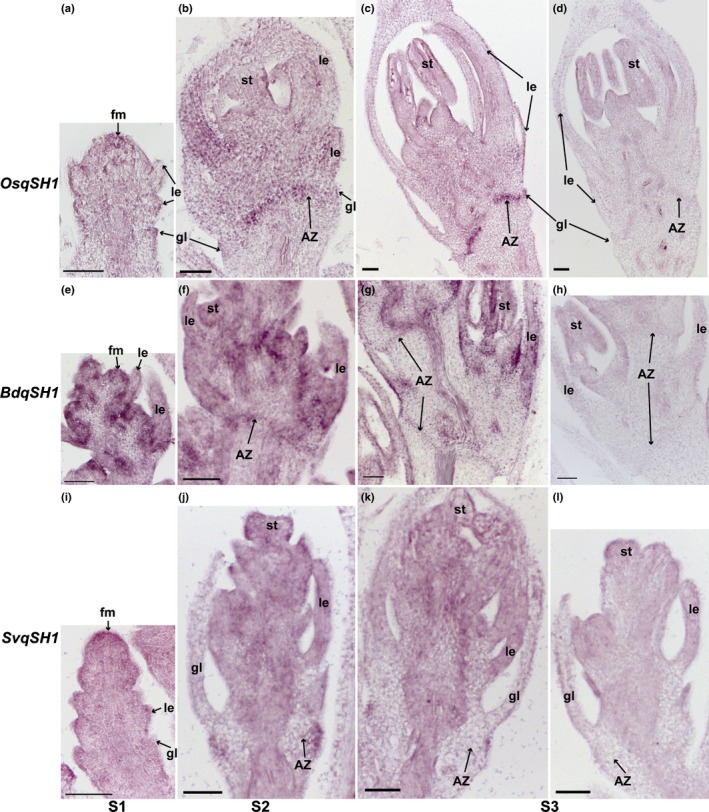
qSH1 is expressed in the abscission zone (AZ) in all three species with different expression patterns. In situ hybridisation of qSH1 in (a–d) rice, (e–h) Brachypodium and (i–l) Setaria was performed at (a, e, i) floret meristem stage (S1), (b, f, j) anther differentiation stage (S2) and (c, g, k) a stage when anthers were fully developed (S3). (d, h, l) are sense probe controls of qSH1 in (d) rice, (h) Brachypodium and (l) Setaria. fm, floral meristem; gl, glume; le, lemma; st, stamens. Bars, 50 μm.
In the case of AP2, the rice orthologue is strongly expressed in the AZ at all three stages, and relatively weakly expressed in floral bracts at S2 and stamens at S3 (Fig. 7a–d), while expression of BdAP2 and SvAP2 is more obvious in bracts and stamens. BdAP2 may be weakly expressed in the AZ at S2 and S3 (Fig. 7e–h), whereas SvAP2 is not detectable in the AZ (Fig. 7i–l).
For SH4, both OsSH4 and BdSH4 are preferentially expressed in the AZ compared with the adjacent tissues at all three stages (Fig. 8a–h), while no expression was observed in the AZ in Setaria (Fig. 8i–l), consistent with our RNA‐Seq results (Fig. 5a). Besides expression in the AZ, SH4 is also expressed in floral bracts and stamens in all three species, although the signal intensity varies (Fig. 8).
qSH1 is expressed in the AZ in all three species (Fig. 9). In rice, expression of qSH1 is specific to the AZ, especially at S3 (Fig. 9a–d), while BdqSH1 has strong expression in the upper tissues in addition to the AZ (Fig. 9e–h). SvqSH1 is expressed in the AZ at S2 and weakly at S3, and expression is restricted to the outer cell layers at the approximate position of the AZ (Fig. 9i–l).
In summary, all of these genes are involved in aspects of flower development and have somewhat conserved expression patterns. However, the intensity and spatial and temporal specificity of their expression in the AZ varies among species, implying relatively little conservation in genetic regulation of AZ development.
Discussion
Divergent gene co‐expression networks in a conserved background
Contrary to our initial hypothesis, we find that the AZs of rice, Brachypodium and Setaria differ in anatomy, cell wall structure and gene expression. No expression modules are completely conserved among species, and only a handful of genes are AZ specific in their expression in all cases (Figs 4, S9; Tables S6, S8). However, c. 90% of the expressed one‐to‐one orthologous genes are shared among all three species, demonstrating that the set of genes expressed in the broad region studied is largely conserved (Fig. S1). In addition, modules with differential expression between early and late development show high preservation among species (Figs 3, S5). Therefore, the set of AZ genes is a species‐specific subset of a generally conserved set of expressed genes, suggesting conservation of the overall developmental program through time but distinct spatial control, consistent with the observation of different position and anatomy of the AZ in rice, Brachypodium and Setaria (Figs 1, 2). We investigated specifically genes that had been identified in previous studies, generally in rice, as being required for shattering. We confirmed tissue specific expression with in situ hybridisation, and found, as with the RNA‐Seq experiments, that the details of when and where the genes are deployed differs among species (Figs 6, 7, 8, 9).
Genes controlling AZ development show both convergence and divergence at different phylogenetic scales
So far, YAB2/ObSH3/SH1 and AP2/SHAT1/Q are the only two genes demonstrated by genetic studies to play conserved roles in AZ development across different tribes of Poaceae (Simons et al., 2006; Lin et al., 2012; Zhou et al., 2012; Lv et al., 2018). Interestingly, in Arabidopsis thaliana, AtYAB1, AtYAB3 and AtAP2 also regulate fruit AZ development (Dinneny et al., 2005; Ripoll et al., 2011). Similarly, the orthologous Arabidopsis AtRPL (Roeder et al., 2003) and rice qSH1 (Konishi et al., 2006) are both involved in AZ development, although AtRPL is expressed in the pod replum adjacent to the AZ, while qSH1 is expressed in the AZ (Fig. 9). The Arabidopsis homologue of MYB26 regulates secondary cell wall thickening and anther dehiscence (Yang et al., 2007), and our results suggest that MYB26 also functions in AZ development in at least Brachypodium and rice (Figs 4h–k, S10). Together, these results suggest convergence or conservation of AZ genetic control with modification of expression pattern and molecular function of the genes.
Other evidence suggests mechanistic divergence. IND orthologues are confined to the Brassicaceae, indicating neofunctionalisation of IND in AZ development of that family (Pabón‐Mora et al., 2014). Btr1/Btr2, identified by their role in domestication of barley and wheat (Pourkheirandish et al., 2015; Avni et al., 2017), encode proteins with unknown functions. Their closest homologues in Brachypodium and rice only share limited sequence identity with barley and wheat, suggesting that the Btr1/2 genes may provide novel controls of AZ development in some Triticeae species (Pourkheirandish et al., 2015).
Previous studies mostly focus on one or a few genes in the genetic pathway controlling AZ development. Our transcriptomic study suggests high divergence in gene co‐expression patterns in grass AZ regulation, and genes indispensable for AZ function are few.
Differential lignification may not be required for abscission
Our results provide some hypotheses regarding AZ function. Studies on AZ anatomy in eudicot leaves showed that the cells in the AZ are often small and nonlignified (Sexton & Roberts, 1982). Genetic studies of the AZ in Arabidopsis and rice also suggest that cell size and differential lignification between the AZ and the adjacent cell layers are required for abscission, possibly creating tension between cell layers and therefore promoting cell separation. For example, in AZ development of Arabidopsis fruit, SHP1, SHP2, and IND regulate AZ differentiation and lignification of adjacent cells (Liljegren et al., 2000, 2004). When these genes are mutated, lignification is lost and dehiscence fails. In rice yab2/obsh3, sh4, qsh1 and ap2/shat1 mutants, lignin is deposited ectopically in the AZ and abscission is reduced or lost (Konishi et al., 2006; Li & Gill, 2006; Zhou et al., 2012; Lv et al., 2018), suggesting that the lack of lignin and smaller cell size of the AZ are essential for AZ function in rice. However, we show that this pattern does not apply to all grass species. In Brachypodium, lignin is deposited in both the AZ and the neighbouring cells (Fig. 2b), while in Setaria, lignin is only observed at the epidermal layer and no cell‐size difference was observed in the AZ (Fig. 2c). Similarly, lignification occurs throughout the AZ and the surrounding tissues of wild barley (Pourkheirandish et al., 2015), further supporting the hypothesis that lignification pattern is not a universal prerequisite for abscission.
The differences in AZ anatomy are consistent with different expression patterns of genes controlling the anatomy. For example, AP2 is highly enriched in the AZ of rice, and loss of AP2 function results in loss of nonlignified and small‐sized cells (Zhou et al., 2012). However, as Brachypodium has lignin in the AZ and the Setaria AZ cells are not smaller than surrounding cells (Fig. 2), AP2 may not be important for AZ development in those two species. Consistent with this interpretation, AP2 transcripts are not enriched in the AZ of Brachypodium or Setaria (Figs 5, 7).
Change in position of the AZ in evolutionary time
Heterotopy, expressing a developmental program in a different organismal position, has been hypothesized as a mechanism for morphological change. Haeckel (1866) used the term to interpret the origin of reproductive organs from different germ layers in different organisms during embryo development, although without the advantage of knowledge of genomes and molecular biology. In its modern sense, heterotopy can be interpreted as a change in the spatial expression pattern of genes essential for a developmental process, leading to positional change of the whole developmental program (Baum & Donoghue, 2002).
Both literature and our data support a much more limited version of heterotopy, applying to only a few key genes. For example, the functional conservation of FUL/SHP/ALC/IND pathway in fruit AZ development in the Brassicaceae regardless of AZ position (Ferrándiz et al., 2000; Avino et al., 2012; Lenser & Theißen, 2013) may be considered as heterotopy. Similarly, the expression of YAB2, qSH1 and MYB26 in the AZ in the rachilla in rice and Brachypodium but in the pedicel in Setaria may be another example (Figs 4h–k, 6, 9; Tables S7, S9). However, heterotopy is an insufficient explanation if applied to the whole transcriptomic network. The ancestral position of the AZ is conserved in rice and Brachypodium and is shared by most species in Poaceae (Fig. 1). Despite this positional conservation, the underlying development and genetic control differs between the two. The AZ of rice and Brachypodium differ in cell wall composition and thickness (Fig. 2). The number of shared genes enriched in the AZs of both Brachypodium and rice is as small as that between Brachypodium and Setaria or rice and Setaria (Figs 4, S9), suggesting that AZ regulatory networks are diverse regardless of position. Even in the case where genes in the same pathway shift together to a new position, such as co‐expression of SH4 and AP2 in both rice and Setaria (Fig. S11), their interactions with genes expressed in the new position differ, and therefore cause distinct co‐expression networks (Fig. 5).
The results presented here raise important questions about how gene regulatory networks change over time and the speed with which this change occurs. Our results show that similar AZ positions do not necessarily reflect similar regulatory networks. However, we do not know whether gene expression patterns of closely related species with different AZ positions are conserved. For example, both Setaria and sorghum belong to the subfamily Panicoideae, and their AZs are located in the pedicel and rachis, respectively (Fig. 1). Whether the underlying gene networks are shared is unknown. Likewise, we lack data comparing Elymus and Hordeum, two genera in the tribe Triticeae, although their histology and AZ positions differ markedly (Yu & Kellogg, 2018). Our data provide a framework for future studies on the rewiring of gene networks over evolutionary time.
Author contributions
AND and EAK designed the research and secured funding. AND, EAK and YY designed the experimental approach and YY performed the experiments. HH and YY analysed the RNA‐Seq data. EAK and YY drafted the manuscript, with input from AND and HH. All authors contributed to ideas, discussed the results and edited the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 The abscission zones (AZs) have different cell wall components at different developmental stages in rice, Brachypodium and Setaria.
Fig. S2 Weighted gene co‐expression network analysis (WGCNA) identified 15 co‐expression modules in rice.
Fig. S3 Weighted gene co‐expression network analysis (WGCNA) identified 17 co‐expression modules in Brachypodium.
Fig. S4 Weighted gene co‐expression network analysis (WGCNA) identified 18 co‐expression modules in Setaria.
Fig. S5 Weighted gene co‐expression network analysis (WGCNA) modules in one species are moderately preserved in the other two species.
Fig. S6 Numbers of overlapping genes in pairwise comparisons of modules between species.
Fig. S7 Modules with highly significant overlapping genes have similar expression patterns.
Fig. S8 Expression patterns of self‐organising map (SOM) clustering in rice, Brachypodium and Setaria.
Fig. S9 AZ clusters generated by self‐organising map (SOM) have a limited number of overlapping genes between species.
Fig. S10 MYB26 is expressed in the abscission zone in rice and Brachypodium.
Fig. S11 SH4 is co‐expressed with AP2 and/or MYB26.
Table S1 Probe primers used in in situ hybridization.
Table S2 Ancestral state reconstruction of the abscission zone positions in grasses.
Table S3 Deseq2 results of rice.
Table S4 Deseq2 results of Brachypodium.
Table S5 Deseq2 results of Setaria.
Table S6 Weighted gene co‐expression network analysis (WGCNA) modules of one‐to‐one orthologues in all three species.
Table S7 Pairwise comparisons of overlapping genes between the abscission zone modules by weighted gene co‐expression network analysis (WGCNA) in rice, Brachypodium and Setaria.
Table S8 Self‐organising map clusters of DEG in rice, Brachypodium and Setaria.
Table S9 Pairwise comparisons of overlapping genes between the abscission zone clusters by self‐organising map analysis in rice, Brachypodium and Setaria.
Table S10 Weights of all edges of YAB2, AP2, SH4 and qSH1 in rice, Brachypodium and Setaria.
Acknowledgements
We thank Dr Kenneth M. Olsen at Washington University at St. Louis and Dr Malia Gehan at Donald Danforth Plant Science Center for providing seed stocks. We thank Missouri Botanical Garden's Herbarium for providing herbarium specimens, and Rachel Tavares and Alexandria Pete for technical help. The study was funded by NSF‐IOS 1557633 and 1557640 to EAK and AND, respectively.
See also the Commentary on this article by https://doi.org/10.1111/nph.16256.
Contributor Information
Yunqing Yu, Email: yyu@danforthcenter.org.
Elizabeth A. Kellogg, Email: ekellogg@danforthcenter.org.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high‐throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avino M, Kramer EM, Donohue K, Hammel AJ, Hall JC. 2012. Understanding the basis of a novel fruit type in Brassicaceae: conservation and deviation in expression patterns of six genes. EvoDevo 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K. 2017. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. [DOI] [PubMed] [Google Scholar]
- Barkworth ME, Capels KM, Long S, Anderton LK, Piep MB, eds. 2007. Flora of North America North of Mexico vol. 24. Magnoliophyta: Commelinidae (in part): Poaceae, part 1. New York, NY, USA, Oxford, UK: Oxford University Press, 1–911. [Google Scholar]
- Barkworth ME, Capels KM, Long S, Piep MB, eds. 2003. Flora of North America North of Mexico vol. 25. Magnoliophyta: Commelinidae (in part): Poaceae, part 2. New York, NY, USA, Oxford, UK: Oxford University Press, 1–783. [Google Scholar]
- Bateman RM. 1994. Evolutionary‐developmental change in the growth architecture of fossil rhizomorphic lycopsids: scenarios constructed on cladistic foundations. Biological Reviews 69: 527–597. [Google Scholar]
- Baum DA, Donoghue MJ. 2002. Transference of function, heterotopy and the evolution of plant development In: Cronk QCB, Bateman RM, Hawkins JA, eds. Developmental genetics and plant evolution, 1st edn. London, UK: Systematics Association; Special Volume 65: 52–69. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF. 2005. A genetic framework for fruit patterning in Arabidopsis thaliana . Development 132: 4687–4696. [DOI] [PubMed] [Google Scholar]
- Doust AN, Lukens L, Olsen KM, Mauro‐Herrera M, Meyer A, Rogers K. 2014a. Beyond the single gene: How epistasis and gene‐by‐environment effects influence crop domestication. Proceedings of the National Academy of Sciences, USA 111: 6178–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Mauro‐Herrera M, Francis AD, Shand LC. 2014b. Morphological diversity and genetic regulation of inflorescence abscission zones in grasses. American Journal of Botany 101: 1759–1769. [DOI] [PubMed] [Google Scholar]
- Du H, Yu Y, Ma Y, Gao Q, Cao Y, Chen Z, Ma B, Qi M, Li Y, Zhao X. 2017. Sequencing and de novo assembly of a near complete indica rice genome. Nature Communications 8: 15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C, Liljegren SJ, Yanofsky MF. 2000. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289: 436–438. [DOI] [PubMed] [Google Scholar]
- Fourquin C, Ferrándiz C. 2012. Functional analyses of AGAMOUS family members in Nicotiana benthamiana clarify the evolution of early and late roles of C‐function genes in eudicots. The Plant Journal 71: 990–1001. [DOI] [PubMed] [Google Scholar]
- Fuller DQ, Allaby R. 2009. Seed dispersal and crop domestication: shattering, germination and seasonality in evolution under cultivation In: Østergaard L, eds. Annual plant reviews volume 38: fruit development and seed dispersal. Hoboken, NJ, USA: John Wiley & Sons, 238–295. [Google Scholar]
- Haeckel E. 1866. Generelle Morphologie der Organismen: Allgemeine Grundzuge der organischen Formen‐Wissenschaft, mechanisch begründet durch die von Charles Darwin reformirte Descendenz‐Theorie, vol. 2. Berlin, Germany: Verlag von Georg Reimer. [Google Scholar]
- Hall BK. 1999. Chapter 24. Time and place in evolution: heterochrony and heterotopy In: Hall BK, ed. Evolutionary developmental biology, 2 nd edn. Berlin, Germany: Springer, 375–391. [Google Scholar]
- Hodge JG, Kellogg EA. 2016. Abscission zone development in Setaria viridis and its domesticated relative, Setaria italica . American Journal of Botany 103: 998–1005. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative . 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463: 763. [DOI] [PubMed] [Google Scholar]
- Ji H, Kim SR, Kim YH, Kim H, Eun MY, Jin ID, Cha YS, Yun DW, Ahn BO, Lee MC. 2010. Inactivation of the CTD phosphatase‐like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. The Plant Journal 61: 96–106. [DOI] [PubMed] [Google Scholar]
- Johansen DA. 1940. Plant microtechnique. New York, NY, USA: McGraw‐Hill, 1–523. [Google Scholar]
- Kellogg EA. 2015. Poaceae: In: Kubitzki K, ed. Flowering plants. Monocots, 1 st edn. Berlin, Germany: Springer. [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M. 2006. An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. 2011. Is my network module preserved and reproducible? PLoS Computational Biology 7: e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton DJ, Kellogg EA. 2014. Morphological, phylogenetic, and ecological diversity of the new model species Setaria viridis (Poaceae: Paniceae) and its close relatives. American Journal of Botany 101: 539–557. [DOI] [PubMed] [Google Scholar]
- Lenser T, Theißen G. 2013. Conservation of fruit dehiscence pathways between Lepidium campestre and Arabidopsis thaliana sheds light on the regulation of INDEHISCENT . The Plant Journal 76: 545–556. [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. 2006. Rice domestication by reducing shattering. Science 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Li W, Gill BS. 2006. Multiple genetic pathways for seed shattering in the grasses. Functional & Integrative Genomics 6: 300–309. [DOI] [PubMed] [Google Scholar]
- Li Y, Im Kim J, Pysh L, Chapple C. 2015. Four isoforms of Arabidopsis 4‐coumarate: CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiology 169: 2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. 2000. SHATTERPROOF MADS‐box genes control seed dispersal in Arabidopsis . Nature 404: 766–770. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. 2004. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853. [DOI] [PubMed] [Google Scholar]
- Lin Z, Li X, Shannon LM, Yeh C‐T, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE. 2012. Parallel domestication of the Shattering1 genes in cereals. Nature Genetics 44: 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Wu W, Wang M, Meyer RS, Ndjiondjop M‐N, Tan L, Zhou H, Zhang J, Fu Y, Cai H. 2018. Genetic control of seed shattering during African rice domestication. Nature Plants 4: 331. [DOI] [PubMed] [Google Scholar]
- Moll P, Ante M, Seitz A, Reda T. 2014. QuantSeq 3′mRNA sequencing for RNA quantification. Nature Methods 11: 972.25317449 [Google Scholar]
- Mühlhausen A, Lenser T, Mummenhoff K, Theißen G. 2013. Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. The Plant Journal 73: 824–835. [DOI] [PubMed] [Google Scholar]
- Nagashima H, Sugahara F, Takechi M, Ericsson R, Kawashima‐Ohya Y, Narita Y, Kuratani S. 2009. Evolution of the turtle body plan by the folding and creation of new muscle connections. Science 325: 193–196. [DOI] [PubMed] [Google Scholar]
- Odonkor S, Choi S, Chakraborty D, Martinez‐Bello L, Wang X, Bahri BA, Tenaillon MI, Panaud O, Devos KM. 2018. QTL mapping combined with comparative analyses identified candidate genes for reduced shattering in Setaria italica . Frontiers in Plant Science 9: 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón‐Mora N, Wong GK‐S, Ambrose BA. 2014. Evolution of fruit development genes in flowering plants. Frontiers in Plant Science 5: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE. 2001. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiology 126: 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript‐level expression analysis of RNA‐seq experiments with HISAT, StringTie and Ballgown. Nature Protocols 11: 1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkheirandish M, Hensel G, Kilian B, Senthil N, Chen G, Sameri M, Azhaguvel P, Sakuma S, Dhanagond S, Sharma R. 2015. Evolution of the grain dispersal system in barley. Cell 162: 527–539. [DOI] [PubMed] [Google Scholar]
- Preston JC, Christensen A, Malcomber ST, Kellogg EA. 2009. MADS‐box gene expression and implications for developmental origins of the grass spikelet. American Journal of Botany 96: 1419–1429. [DOI] [PubMed] [Google Scholar]
- Rajani S, Sundaresan V. 2001. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Current Biology 11: 1914–1922. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Ripoll JJ, Roeder AH, Ditta GS, Yanofsky MF. 2011. A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138: 5167–5176. [DOI] [PubMed] [Google Scholar]
- Roeder AH, Ferrándiz C, Yanofsky MF. 2003. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Current Biology 13: 1630–1635. [DOI] [PubMed] [Google Scholar]
- Ruzin SE. 1999. Plant microtechnique and microscopy, vol.198. New York, NY, USA: Oxford University Press, 87–94. [Google Scholar]
- Saarela JM, Burke SV, Wysocki WP, Barrett MD, Clark LG, Craine JM, Peterson PM, Soreng RJ, Vorontsova MS, Duvall MR. 2018. A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions. PeerJ 6: e4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton R, Roberts JA. 1982. Cell biology of abscission. Annual Review of Plant Physiology 33: 133–162. [Google Scholar]
- Shigetani Y, Sugahara F, Kawakami Y, Murakami Y, Hirano S, Kuratani S. 2002. Heterotopic shift of epithelial‐mesenchymal interactions in vertebrate jaw evolution. Science 296: 1316–1319. [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai Y‐S, Gill BS, Faris JD. 2006. Molecular characterization of the major wheat domestication gene Q . Genetics 172: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P‐L, Ideker T. 2010. Cytoscape 2.8: new features for data integration and network visualisation. Bioinformatics 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal P, Gennen J, De Bodt S, Ranganath V, Melzer S. 2007. Flowering of strict photoperiodic Nicotiana varieties in non‐inductive conditions by transgenic approaches. Plant Molecular Biology 65: 233–242. [DOI] [PubMed] [Google Scholar]
- Soreng RJ, Peterson PM, Romaschenko K, Davidse G, Zuloaga FO, Judziewicz EJ, Filgueiras TS, Davis JI, Morrone O. 2015. A worldwide phylogenetic classification of the Poaceae (Gramineae). Journal of Systematics and Evolution 53: 117–137. [Google Scholar]
- Thurber CS, Reagon M, Gross BL, Olsen KM, Jia Y, Caicedo AL. 2010. Molecular evolution of shattering loci in US weedy rice. Molecular Ecology 19: 3271–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Liu X, Wang M, Meyer RS, Luo X, Ndjiondjop M‐N, Tan L, Zhang J, Wu J, Cai H. 2017. A single‐nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nature Plants 3: 17064. [DOI] [PubMed] [Google Scholar]
- Wu Z, Raven PH, Hong D. 2006. Flora of China. Poaceae. Beijing, China: Science Press; & St Louis, MO, USA: Missouri Botanical Garden Press. [Google Scholar]
- Yang C, Xu Z, Song J, Conner K, Barrena GV, Wilson ZA. 2007. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19: 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Thames S, Best NB, Jiang H, Huang P, Dilkes BP, Eveland AL. 2018. Brassinosteroids modulate meristem fate and differentiation of unique inflorescence morphology in Setaria viridis . Plant Cell 30: 48–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Cho L‐H, Antt HW, Koh H‐J, An G. 2017. KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiology 174: 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Cho LH, Kim SL, Choi H, Koh HJ, An G. 2014. The BEL1‐type homeobox gene SH5 induces seed shattering by enhancing abscission‐zone development and inhibiting lignin biosynthesis. The Plant Journal 79: 717–728. [DOI] [PubMed] [Google Scholar]
- Yu Y, Kellogg EA. 2018. Inflorescence abscission zones in grasses: diversity and genetic regulation. Annual Plant Reviews Online 2: 1–35. [Google Scholar]
- Zelditch ML, Fink WL. 1996. Heterochrony and heterotopy: stability and innovation in the evolution of form. Paleobiology 22: 241–254. [Google Scholar]
- Zhou Y, Lu D, Li C, Luo J, Zhu B‐F, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B. 2012. Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1 . Plant Cell 24: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 The abscission zones (AZs) have different cell wall components at different developmental stages in rice, Brachypodium and Setaria.
Fig. S2 Weighted gene co‐expression network analysis (WGCNA) identified 15 co‐expression modules in rice.
Fig. S3 Weighted gene co‐expression network analysis (WGCNA) identified 17 co‐expression modules in Brachypodium.
Fig. S4 Weighted gene co‐expression network analysis (WGCNA) identified 18 co‐expression modules in Setaria.
Fig. S5 Weighted gene co‐expression network analysis (WGCNA) modules in one species are moderately preserved in the other two species.
Fig. S6 Numbers of overlapping genes in pairwise comparisons of modules between species.
Fig. S7 Modules with highly significant overlapping genes have similar expression patterns.
Fig. S8 Expression patterns of self‐organising map (SOM) clustering in rice, Brachypodium and Setaria.
Fig. S9 AZ clusters generated by self‐organising map (SOM) have a limited number of overlapping genes between species.
Fig. S10 MYB26 is expressed in the abscission zone in rice and Brachypodium.
Fig. S11 SH4 is co‐expressed with AP2 and/or MYB26.
Table S1 Probe primers used in in situ hybridization.
Table S2 Ancestral state reconstruction of the abscission zone positions in grasses.
Table S3 Deseq2 results of rice.
Table S4 Deseq2 results of Brachypodium.
Table S5 Deseq2 results of Setaria.
Table S6 Weighted gene co‐expression network analysis (WGCNA) modules of one‐to‐one orthologues in all three species.
Table S7 Pairwise comparisons of overlapping genes between the abscission zone modules by weighted gene co‐expression network analysis (WGCNA) in rice, Brachypodium and Setaria.
Table S8 Self‐organising map clusters of DEG in rice, Brachypodium and Setaria.
Table S9 Pairwise comparisons of overlapping genes between the abscission zone clusters by self‐organising map analysis in rice, Brachypodium and Setaria.
Table S10 Weights of all edges of YAB2, AP2, SH4 and qSH1 in rice, Brachypodium and Setaria.


