Abstract
Neutrophils are professional phagocytes that are important for innate host defenses against pathogens and resolution of inflammation. Traditionally, the phagocytic capacity of neutrophils was quantified by enumeration of cells containing either internalized or bound bacteria or other cargo from a series of microscopic images. Here we describe an imaging flow cytometry-based protocol and analysis method for quantifying the binding and uptake of Neisseria gonorrhoeae by primary adherent human neutrophils. Imaging flow cytometry combines the capacity for quantitative, high-throughput analysis of tens of thousands of cells per condition, with the imaging power of fluorescence microscopy. Here, all bacteria are labeled with Tag-it Violet™ and bound bacteria are differentially stained with a DyLight™ 650-conjugated antibody. Images are analyzed using spot count and other algorithms. Outputs include the percent of neutrophils associated with bacteria, the percent of neutrophils with internalized bacteria, and the percent of internalized bacteria. This basic protocol can be adapted to a variety of particle types and can be used for multiplex analysis in combination with staining for different neutrophil surface and intracellular markers.
Keywords: imaging flow cytometry, neutrophils, bacteria, phagocytosis, binding, internalization, fluorescence
1. Introduction
Neutrophil phagocytic activity plays an important role in clearing microbial pathogens, resolving inflammation, and regulating immune responses (Elliott and Ravichandran 2010; Fond and Ravichandran 2016; Lim et al. 2017; Sarantis and Grinstein 2012). Phagocytosis of a target particle involves the coordinated steps of recognition, binding to phagocytic receptors, and internalization, which if successful leads to its elimination. Measurement of neutrophil phagocytic capacity has historically been performed in one of two ways. One, fluorescent target particles such as bacteria are incubated with neutrophils, and fluorescence-positive neutrophils are quantified by flow cytometry. However, this does not discriminate between bound and internalized targets (Hampton and Winterbourn 1999; Jersmann et al. 2003; Lehmann et al. 2000). Two, neutrophils are incubated with fluorescent targets, and surface-bound targets are recognized with an antibody or other reagent of a different fluorophore. Differentially labeled bacteria can be visualized by immunofluorescence microscopy and quantified, but this approach is laborious, time consuming, and low throughput (Agerer et al. 2004; Criss et al. 2009). To overcome the limitations of each approach, we and others have employed imaging flow cytometry to enable simultaneous fluorescent and morphological analysis of phagocytes and their targets in a large cell population (Haridas et al. 2017; Johansson et al. 2015; Phanse et al. 2012; Ploppa et al. 2011; Smirnov et al. 2015). Different methods have been used to quantify the binding and internalization of targets by phagocytes, including masks to delineate the outside of cells, bright detail similarity score between single- and dual-labeled targets, and spot counting (Johansson et al. 2015; Kuhn et al. 2019; Smirnov et al. 2015). We have found spot counting to be the most accurate approach for protrusive cells like neutrophils, especially in conditions where a sizable percentage of target particles are bound and not internalized (Smirnov et al. 2015).
Here we describe a protocol that was developed to quantify Neisseria gonorrhoeae bacteria that are bound to and/or internalized by adherent, interleukin 8 (IL-8)-treated, primary human neutrophils (Smirnov et al. 2017). Neutrophils purified from peripheral blood were infected with Tag-It Violet™ labeled bacteria. Neutrophils were fixed, lifted, and stained with an anti-Neisseria gonorrhoeae antibody coupled to DyLight™ 650. In the absence of permeabilization, intracellular bacteria are inaccessible to the antibody and remain single stained (Tag-it Violet™), while bound bacteria are double stained (Tag-it Violet™ and DyLight™ 650). Use of these fluorophores is an improvement over earlier versions of this protocol that used carboxyfluorescein succinimidyl ester, which reserves this green fluorophore for additional staining procedures if desired by the user (Smirnov et al. 2015; Smirnov et al. 2017). Fluorescent and imaging data were acquired using ImageStreamX Mk II with INSPIRE® acquisition software. Data analysis was performed using IDEAS® v. 6.2, and the spot count algorithm was applied to count the number of Tag-it Violet™ and DyLight™ 650 positive bacteria in individual neutrophils in a population of thousands of cells. These two measurements were used to calculate: 1) percent of neutrophils with cell-associated bacteria (either bound or internalized); 2) percent of cell-associated bacteria that are internalized; and 3) percent of neutrophils with internalized bacteria (neutrophils with at least one internalized bacterium).
2. Materials
All the solutions for working with primary human neutrophils are prepared in endotoxin free water using aseptic techniques.
2.1. Primary Human Neutrophils
Neutrophils are purified from peripheral human blood using dextran sedimentation of erythrocytes, followed by separation on a Ficoll-Paque PLUS gradient (GE Healthcare) and hypotonic lysis of residual erythrocytes (Boyum 1968; Stohl et al. 2005). Purified neutrophils are resuspended in Dulbecco’s modified PBS, pH 7.4 without calcium and magnesium (Thermo Scientific), kept on ice, and used within 2 h from purification (see Note 1).
2.2. Tissue Culture
25 mm tissue culture plastic coverslips (Sarstedt)
6-well tissue culture dish
Interleukin 8 (IL-8) (R&D Systems)
RPMI without glutamine (HyClone) supplemented with 10% of heat-inactivated fetal bovine serum (FBS) (Thermo Scientific)
2.3. Bacterial Culture
Neisseria gonorrheae culture grown to mid-logarithmic phase in liquid gonococcal medium base medium containing Kellogg’s supplements I and II (Kellogg et al. 1963) (see Note 2).
2.4. Bacteria Labeling
Tag-it Violet™ Proliferation and Cell Tracking Dye (TIV) (Biolegend).
5 mM MgSO4 in Phosphate Buffered Saline (PBS), pH 7.4
2.5. Fixation and Staining
16% paraformaldehyde (Electron Microscopy Sciences)
Blocking solution: 10% normal goat serum (Thermo Fisher Scientific) in PBS pH 7.4
Goat anti-Neisseria gonorrhoeae antibody (Biosource) labeled with DyLight™ 650 (DL650) according to the manufacturer’s protocol (Thermo).
2.6. Instrument and Software
ImageStreamX Mk II imaging flow cytometer with INSPIRE® and IDEAS® v. 6.2 Software packages (Amnis Luminex Corporation)
3. Methods
3.1. Short-term Culture of IL-8 Treated Primary Adherent Human Neutrophils
Add 25 mm tissue culture plastic coverslips to 6-well tissue culture dishes (see Note 3).
Dilute the neutrophil suspension into RPMI+10%FBS containing 10 nM human interleukin 8 (IL-8), to a final concentration of 2.5×106 neutrophils/ml (see Note 4).
Gently pipette 400 μl of the cell suspension onto the middle of the coverslip, allowing it to form “a dome”.
Incubate neutrophils for 30 min in a humidified incubator at 37°C, 5% CO2.
3.2. Labeling of Bacteria
Measure optical density of the bacterial culture. Using a standard curve of enumerated bacterial colony-forming units per optical density reading, set by the researcher’s laboratory, pellet 2×108 bacteria by centrifugation at 10,000 × g for 3 min at room temperature.
Resuspend bacteria in 1 ml of PBS containing 5 mM MgSO4.
Add 3 μl of TIV solution (50 μM TIV in DMSO).
Incubate for 20 min in a water bath at 37°C.
Pellet bacteria by centrifugation at 10,000 × g for 3 min at room temperature.
Remove and discard supernatant by aspiration.
Resuspend pellet in 1 ml PBS containing 5 mM MgSO4
Pellet bacteria by centrifugation at 10,000 × g for 3 min at room temperature.
Resuspend pellet in 0.5 ml RPMI+10% FBS.
Dilute the bacteria to a final concentration of 1×107 per ml in RPMI + 10% FBS (see Note 5).
3.3. Infection of Neutrophils
Remove 6-well plates with adherent neutrophils from the incubator and place them on ice or shipping packs that are prechilled at 4°C.
Add 700 μl of RPMI+10% FBS per well and incubate in the cold for 5 min.
Add 100 μl of the TIV-labeled bacterial suspension per well. Gently swirl the plate to mix.
Centrifuge the plates in a refrigerated tabletop centrifuge with swinging bucket rotor and microplate carriers at 600 × g for 4 min at 4°C.
Return the plates to the humidified 37 °C, 5% CO2 incubator and incubate for desired times (see Note 6).
3.4. Sample Fixation and Collection
Add 400 μl of a 16% aqueous solution of paraformaldehyde (final concentration of 4%) and incubate for 10 min at room temperature protected from light (see Note 7).
Remove neutrophils from the coverslips using a cell scraper, and collect them from replicate wells into one 15 ml conical tube per experimental condition.
Pellet neutrophils by centrifugation in a tabletop centrifuge with a swinging bucket rotor and conical tube carriers at 600 × g for 5 min at room temperature (see Note 8).
Remove and discard supernatant.
Resuspend neutrophils in 0.5 ml of PBS and pour through a 70 μm pore size cell strainer to remove any clumps of neutrophils.
Add 2.5 ml of PBS, vortex gently, and pellet neutrophils by centrifugation in a tabletop centrifuge with a swinging bucket rotor and conical tube carriers at 600 × g for 5 min at room temperature.
Remove and discard supernatant.
Add 3 ml PBS, vortex gently, and pellet neutrophils by centrifugation tabletop centrifuge with a swinging bucket rotor and conical tube carriers at 600 × g for 5 min at room temperature.
Remove and discard supernatant.
Repeat Steps 8–9.
3.5. Staining cell-bound bacteria
Resuspend pelleted neutrophils in 200 μl of 10% normal goat serum in PBS and vortex gently (see Note 9).
Incubate for 15 min at room temperature, protected from light.
Pellet neutrophils by centrifugation in a tabletop centrifuge with a swinging bucket rotor and conical tube carriers at 600 × g for 5 min at room temperature.
Resuspend neutrophils in 100 μl of goat DL650-conjugated anti-Neisseria gonorrhoeae antibody, diluted in PBS containing 10% normal goat serum to final concentration of 1 μg/ml (see Note 10).
Incubate for 1 hr at room temperature, protected from light.
Add 2 ml PBS, vortex gently, and pellet neutrophils by centrifugation in a tabletop centrifuge with a swinging bucket rotor and conical tube carriers at 600 × g for 5 min at room temperature.
Remove and discard supernatant.
Repeat Steps 6 and 7 (see Note 11).
Resuspend neutrophils in 40 μl of PBS and transfer to 1.5 ml microfuge tubes (see Note 12).
3.5. Data acquisition
Analyze samples on the ImageStreamX Mk II imaging flow cytometer with INSPIRE® software. Collect fluorescence data for each fluorophore and brightfield imaging data using the following parameters:
Magnification 60×
Channel 1 (Ch1) Camera 1 Brightfield 1; LED intensity 38.5 mW; emission collected with 420–480 nm filter
Channel 6 (Ch6) Camera 1; Darkfield (side scatter, SSC); excitation with 4mW 785 nm laser; emission collected with 740–880 nm filter
Channel 7 (Ch7) Camera 2; Excitation with 405 nm laser at 100mW; emission collected with 420–550 nm filter
Channel 11 (Ch11) Camera 2; excitation with 642 nm laser at 45mW; emission collected with 660–740 nm filter
Channel 9 (Ch9) Brightfield 2; Camera; 2 LED intensity 52.75 mW; emission collected with 570–595 nm filter
In order to define the compensation matrix, prepare and collect data from single color samples with Brightfield and Darkfield illumination turned off: (1) TIV single color control, containing neutrophils infected with TIV-labeled bacteria and no DL650-conjugated antibody; (2) DL650 single color control, containing neutrophils infected with unlabeled bacteria and stained with DL650-conjugated anti-Neisseria gonorrhoeae antibody.
3.6. Data analysis
For data analysis, use IDEAS® v6.2 software (see Note 13).
Create a new compensation matrix.
Open one data file and apply the compensation matrix to it.
-
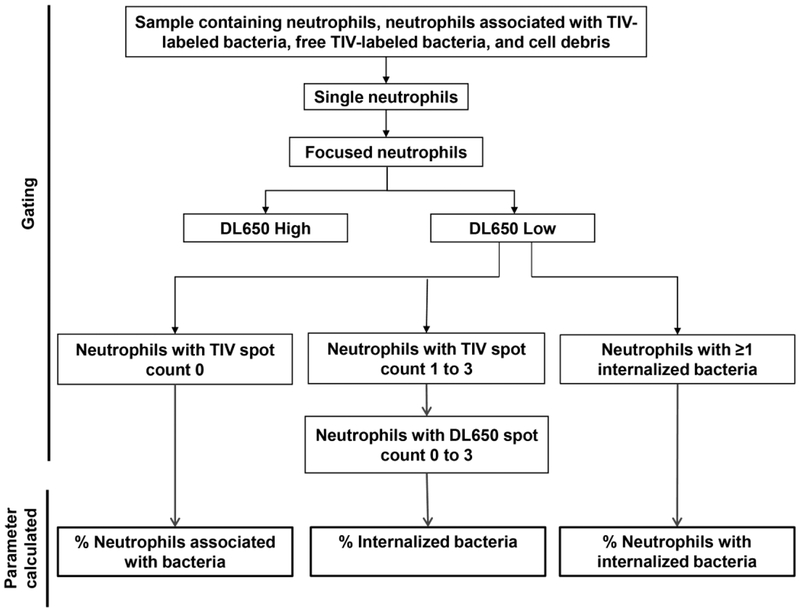
Create a data analysis file by defining populations as outlined in Fig. 1:
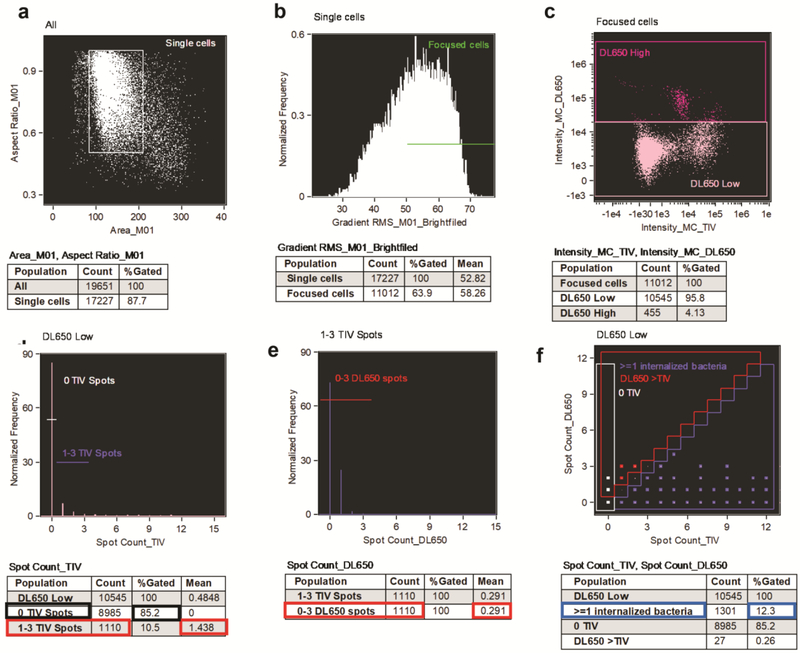
Singlet Gate: Generate a scatter plot with Area_M01 on the X-axis vs. Aspect Ratio_M01 on the Y-axis, and identify single neutrophils by gating on the population of neutrophils with Area_M01 between 85 and 270 and Aspect ratio_M01 between 0.5 and 1 (see Fig. 4a).
Focus gate: From the Singlet gate generate a histogram of Brightfield Gradient Root Mean Square (RMS). Identify the focused neutrophil population as neutrophils with RMS value ≥ 50 (see Note 14) (see Fig. 4b).
From the Singlet and Focus gated population generate a scatter plot Intensity of TIV on the X-axis vs. Intensity of DL650 on the Y-axis, and gate on the neutrophils with lower DL650 intensity, excluding cells with high DL650 intensity, which are not intact (see Note 15, Fig. 4c).
Use the Spot Count Wizard to create a TIV spot count feature (Spot_count_TIV) (see Note 16).
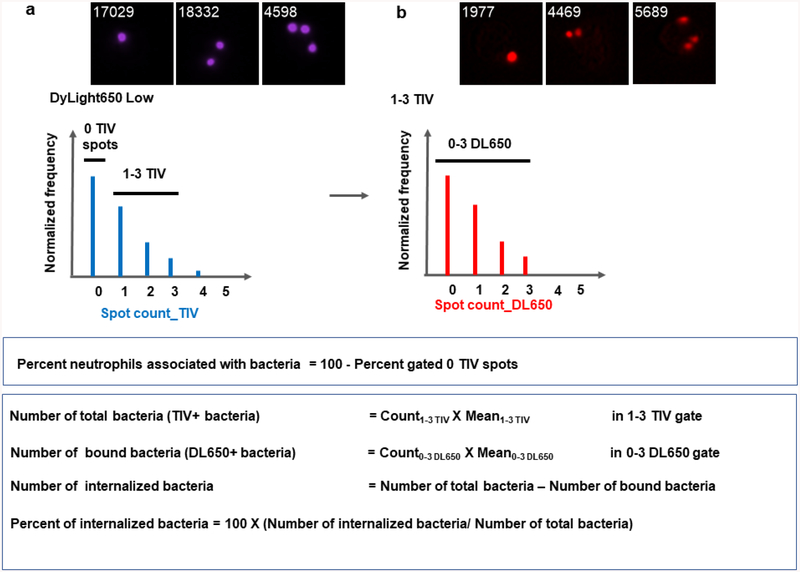
Create a histogram for the TIV spot count and gate on (1) the population of neutrophils with 0 TIV spots; (2) the population of neutrophils with 1 to 3 TIV spots (see Note 17) (Fig. 2a and 4d).
Use the Spot Count Wizard to create a DL650 spot count feature (Spot_count_DL650).
Create a histogram for the DL650 spot count for the population of neutrophils with 1–3 TIV spots and gate on the population of neutrophils with 0 to 3 DL650 spots (see Note 18) (Fig. 2b and 4e).
From the DL650 low neutrophil population (Step 3.6.4c), create an additional plot with Spot_count_TIV on the X axis and the spot_count_DL650 on the Y axis.
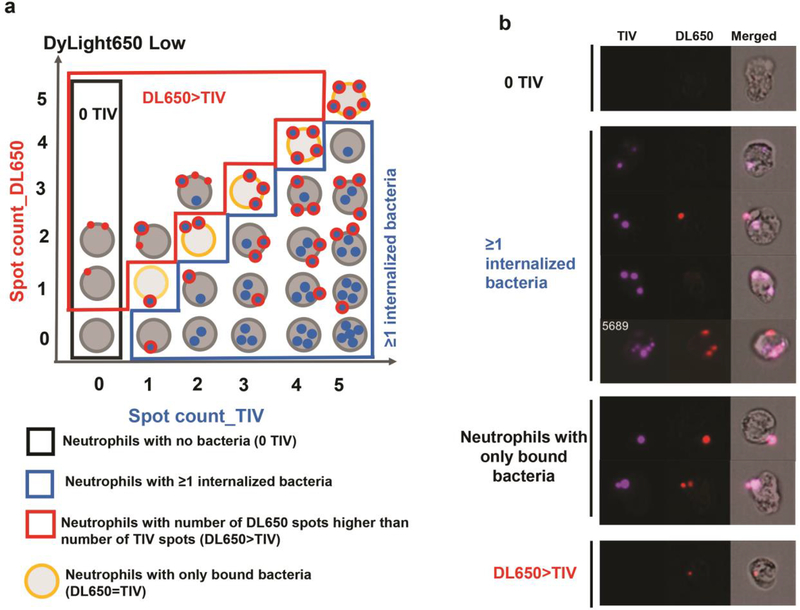
Identify the population of neutrophils with ≥1 internalized bacterium, the population of neutrophils with no TIV spots (0 TIV), and the population of neutrophils with number of DL650 spots higher than TIV spots using the coordinates indicated in Table 1 and Figs. 3a and 4f (see Note 19).
Save the file as an analysis template.
Apply compensation matrix and this analysis template to the rest of the files for the experiment.
Figure 1.
Gating strategy and data analysis workflow.
Figure 2.
Gating strategy, representative images, and calculation of the percent of neutrophils with associated bacteria and the percent of internalized bacteria.
Table 1.
Coordinates for gates described in Figure 3.
| X | −0.5 | 0.5 | −0.5 | 0.5 |
| Y | −0.5 | −0.5 | 5.5 | 5.5 |
| X | 0.5 | 0.5 | 1.5 | 1.5 | 2.5 | 2.5 | 3.5 | 3.5 | 4.5 | 4.5 | 5.5 | 5.5 |
| Y | −0.5 | 0.5 | 0.5 | 1.5 | 1.5 | 2.5 | 2.5 | 3.5 | 3.5 | 4.5 | 4.5 | −0.5 |
| X | −0.5 | 0.5 | 0.5 | 1.5 | 1.5 | 2.5 | 2.5 | 3.5 | 3.5 | 4.5 | 4.5 | −0.5 |
| Y | 0.5 | 0.5 | 1.5 | 1.5 | 2.5 | 2.5 | 3.5 | 3.5 | 4.5 | 4.5 | 5.5 | 5.5 |
Figure 3.
Gating strategy for calculating the percent of neutrophils with internalized bacteria. a) Schematic representatioon and b) representative images of cells in the four outlined populations: neutrophils with 0 TIV spots; neutrophils with ≥1 internalized bacteria; neutrophils with the number of TIV spots equal to the number of DL650 spots, and neutrophils with the number of DL650 spots higher than the number of TIV spots.
3.7. Parameter calculations
-
Calculate the percent of neutrophils associated with bacteria (this population includes neutrophils with both internalized and bound bacteria) (Fig. 2a and 4d outlined in black):
% neutrophils associated with bacteria = 100% - % Gated in “0 TIV” gate
-
Calculate the percent of cell-associated bacteria that are internalized within 1–3 TIV gate (Fig. 2 and Figs. 4d and 4e):
% internalized bacteria = number of internalized bacteria / number of total bacteria- Number of Internalized bacteria = Number of total bacteria – Number of bound bacteria
-
Calculate the percent of neutrophils with internalized bacteria in neutrophils within the DL650 Low population (Fig. 4f outlined in blue):
% neutrophils with internalized bacteria = % in the ≥1 internalized bacteria gate (see Note 20).
Figure 4.
Experimental data analysis. Data were collected from IL-8 primed, adherent primary human neutrophils that were infected with Tag-it Violet™-labled Nesseria gonorrhoeae for 1 hr, fixed, blocked and stained with anti-Neisseria gonorrhoeae specific antibody labeled with DyLight™ 650. Gating was performed as descibed in section 3.6. The percent of neutrophils with associated bacteria is 14.8% (d; data used for calculation outlined in black); the percent of internalized bacteria is 79.8% (d;e data used for calculation outlined in red); the percent of neutrophils with internalized bacteria is 12.3% (f; data used for calculation outlined in blue).
4. Notes
We purify neutrophils using the protocol as presented in Stohl et al (Stohl et al. 2005). The neutrophils purity is ≥ 90%. Purified neutrophils are resuspended in DPBS pH 7.4 and kept on ice to prevent activation. Any other protocol for purification of intact quiescent human neutrophils, including those published in this book, will be suitable for the procedure.
In this protocol we used Neisseria gonorrhoeae as a model organism. The bacteria were grown in rich liquid media with sequential dilution to ensure maximum bacterial viability at the time of infection, as described previously (Criss and Seifert 2008).
We observe better adherence of neutrophils to plastic coverslips compared to tissue culture plastic.
This system seeks to model the state of extravasated and transmigrated neutrophils in vivo, where they are exposed to the neutrophil chemoattractant and priming agent IL-8 (Stevens and Criss 2018). We found that adding 1 to 3 × 106 neutrophils per 25-mm diameter coverslip results in an optimal density of neutrophils. Neutrophils plated in higher density tend to clump during the course of the experiment. Normally 2 to 3 coverslips per condition are required to ensure there is a sufficient number of neutrophils for analysis (see Note 12).
The protocol for labeling bacteria with Tag-it™ Violet was adapted from the manufacturer’s protocol for labeling eukaryotic cells. Incubating Neisseria gonorrhoeae in PBS pH 7.4 with 5mM MgSO4 prevents bacterial autolysis. The labeling cannot be performed in RPMI +10% FBS. We did not test any other buffers or solutions besides those described in this protocol. To date multiple strategies for labeling bacteria and other particles have been used and validated for imaging flow cytometry (Haridas et al. 2017; Johansson et al. 2015; Phanse et al. 2012; Smirnov et al. 2015; Smirnov et al. 2017). The use of Tag-it Violet™ is advantageous for the following reasons: 1) It is excited with a violet laser at 405 nm and emits at 455 nm. This fluorescence profile provides flexibility in using other channels for multiplex analysis with additional markers; 2) unlike the other violet spectrum dye, DAPI, Tag-it Violet™ covalently binds to bacterial proteins and does not diffuse out of the bacteria, producing minimal background in host cells. Here we use a multiplicity of infection (MOI) equal to 1 Neisseria gonorrhoeae bacterium per neutrophil. The optimal MOI or particle-to-neutrophil ratio should be determined experimentally (see Note 17).
Prechilling neutrophils and the centrifugation step after addition of bacteria helps to synchronize the infection. We do not change the media following centrifugation since the neutrophils are loosely attached under these conditions. If the potential for loss of neutrophils is not a concern, investigators may choose to change the media after Step 4 to remove any bacteria that are not cell associated.
Adding paraformaldehyde to wells without removing the incubation medium is a particularly useful approach if lifting of neutrophils during the course of the experiment is a concern. Cell fixation prior to scraping also prevents dissociation of bound bacteria from the neutrophils during subsequent processing steps. The 16% paraformaldehyde solution should be aliquoted and stored at −20°C to avoid freeze-thaw.
The shape of the 15-ml conical tubes and spinning in the swing bucket rotor allows for maximum recovery of the neutrophils during washing and staining steps.
Normal goat serum is used to block nonspecific interaction of goat IgG with the human neutrophils to minimize background staining with the goat anti-Neisseria gonorrhoeae antibody used next. The choice of blocking solution depends on the investigator’s staining method.
Additional staining with antibodies for neutrophil surface markers can be performed at this step.
If staining with intracellular markers required neutrophils can be permeabilized and stained after step 3.5.8.
The recommended concentration of cells for imaging flow cytometry analysis is 2×107 cells per ml in a minimum volume of 15 μl, or a minimum of 3×105 cells. When planning the experiment, we account for some loss of cells during sample collection and processing. Normally, two 25-mm coverslips per condition as described in Note 4 yields a sufficient number of cells for analysis.
Data analysis manuals and tutorials can be found on the Amnis® Customer portal.
Gating on population based on the additional staining with neutrophil surface intracellular markers could be performed after this step (see Note 10 and Note 11).
Neutrophils with high cytoplasmic DL650 staining are assumed to not be intact and are excluded from downstream analysis by this gating strategy.
Spot_Count features are based on the masks defining the individual spots. The software calculates Spot_Count features based on a manually assigned truth population, containing neutrophils with high and low spot count for each fluorophore. The accuracy of the resulting mask should be verified by viewing populations of the neutrophils with different numbers of spots. If required, redefining of the truth population or manual adjustment of the spot count mask can be performed.
In our experience, the most accurate identification of individual spots was with 1 to 3 spots per neutrophil. The MOI should be determined experimentally to ensure that most neutrophils will fall into this range. The sensitivity and specificity of the antibody used to detect bound bacteria must also be determined experimentally. In our experiments, 99% of bacteria are recognized by the antibody.
Using an antibody with high specificity for the bacteria and at the lowest possible concentration for detection ensures that the DL650-positive spots are colocalized with TIV-positive spots, for accurate analysis of phagocytosis. If masking is incorrect, the number of DL650-positive spot will be greater than the number of TIV positive spots per cell. Neutrophils in the 0, 1, 2, 3…spot count bins should be visually inspected, and the mask should be corrected either manually or through redefining the truth population. Representative neutrophils for each population are shown in Fig. 2 and 3b.
Here, TIV and DL650 spot counts are performed on all intact single focused neutrophils to determine the percent of neutrophils with internalized bacteria. Fig. 3 shows a schematic of a gate that includes the population of neutrophils with up to 5 spots per cell. The gate could be extended to include all neutrophils, as shown in Fig. 4f. The percent of neutrophils where the spot count for DL650 exceeds the spot count for TIV will correspond to an analysis error. In our experience ≤ 5% of cells are in this category (see Note 16 and (Smirnov et al. 2015).
The analysis strategy should be validated in experimental conditions known to affect bacterial binding and/or internalization by neutrophils. Conditions may include infection of prefixed or dead neutrophils, treatment with inhibitors of phagocytosis, and infection at low temperature (Smirnov et al. 2015).
Acknowledgments
We thank Lacie Werner for technical assistance with the experiment presented in Figure 4
This work has been supported by R01 AI097312 to A.K.C. and NIH SIG-1S10RR031633 for the ImageStreamX Mk II (T. Bender).
References
- Agerer F, Waeckerle S, Hauck CR (2004) Microscopic quantification of bacterial invasion by a novel antibody-independent staining method J Microbiol Methods 59:23–32 doi: 10.1016/j.mimet.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Boyum A (1968) Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g Scand J Clin Lab Invest Suppl 97:77–89 [PubMed] [Google Scholar]
- Criss AK, Katz BZ, Seifert HS (2009) Resistance of Neisseria gonorrhoeae to non-oxidative killing by adherent human polymorphonuclear leucocytes Cell Microbiol 11:1074–1087 doi: 10.1111/j.1462-5822.2009.01308.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criss AK, Seifert HS (2008) Neisseria gonorrhoeae suppresses the oxidative burst of human polymorphonuclear leukocytes Cell Microbiol 10:2257–2270 doi: 10.1111/j.1462-5822.2008.01205.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS (2010) Clearance of apoptotic cells: implications in health and disease The Journal of Cell Biology 189:1059–1070 doi: 10.1083/jcb.201004096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fond AM, Ravichandran KS (2016) Clearance of Dying Cells by Phagocytes: Mechanisms and Implications for Disease Pathogenesis In: Gregory CD (ed) Apoptosis in Cancer Pathogenesis and Anti-cancer Therapy: New Perspectives and Opportunities. Springer International Publishing, Cham, pp 25–49. doi: 10.1007/978-3-319-39406-0_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton MB, Winterbourn CC (1999) Methods for quantifying phagocytosis and bacterial killing by human neutrophils J Immunol Methods 232:15–22 [DOI] [PubMed] [Google Scholar]
- Haridas V, Ranjbar S, Vorobjev IA, Goldfeld AE, Barteneva NS (2017) Imaging flow cytometry analysis of intracellular pathogens Methods 112:91–104 doi: 10.1016/j.ymeth.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jersmann HP, Ross KA, Vivers S, Brown SB, Haslett C, Dransfield I (2003) Phagocytosis of apoptotic cells by human macrophages: analysis by multiparameter flow cytometry Cytometry Part A : the journal of the International Society for Analytical Cytology 51:7–15 doi: 10.1002/cyto.a.10005 [DOI] [PubMed] [Google Scholar]
- Johansson J, Karlsson A, Bylund J, Welin A (2015) Phagocyte interactions with Mycobacterium tuberculosis--Simultaneous analysis of phagocytosis, phagosome maturation and intracellular replication by imaging flow cytometry J Immunol Methods 427:73–84 doi: 10.1016/j.jim.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Kellogg DS Jr., Peacock WL Jr., Deacon WE, Brown L, Pirkle DI (1963) Neisseria Gonorrhoeae. I. Virulence Genetically Linked to Clonal Variation J Bacteriol 85:1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J, Smirnov A, Criss AK, Columbus L (2019) Quantifying CEACAM targeted liposome delivery using imaging flow cytometry Mol Pharm doi: 10.1021/acs.molpharmaceut.8b01274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AK, Sornes S, Halstensen A (2000) Phagocytosis: measurement by flow cytometry J Immunol Methods 243:229–242 [DOI] [PubMed] [Google Scholar]
- Lim JJ, Grinstein S, Roth Z (2017) Diversity and Versatility of Phagocytosis: Roles in Innate Immunity, Tissue Remodeling, and Homeostasis Frontiers in cellular and infection microbiology 7:191–191 doi: 10.3389/fcimb.2017.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanse Y et al. (2012) Analyzing cellular internalization of nanoparticles and bacteria by multi-spectral imaging flow cytometry Journal of visualized experiments : JoVE:e3884 doi: 10.3791/3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploppa A, George TC, Unertl KE, Nohe B, Durieux ME (2011) ImageStream cytometry extends the analysis of phagocytosis and oxidative burst Scand J Clin Lab Invest 71:362–369 doi: 10.3109/00365513.2011.572182 [DOI] [PubMed] [Google Scholar]
- Sarantis H, Grinstein S (2012) Subversion of Phagocytosis for Pathogen Survival Cell Host Microbe 12:419–431 doi: 10.1016/j.chom.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Smirnov A, Solga MD, Lannigan J, Criss AK (2015) An improved method for differentiating cell-bound from internalized particles by imaging flow cytometry J Immunol Methods 423:60–69 doi: 10.1016/j.jim.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Solga MD, Lannigan J, Criss AK (2017) High-Throughput Particle Uptake Analysis by Imaging Flow Cytometry Current Protocols in Cytometry 80:11.22.11–11.22.17 doi: 10.1002/cpcy.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JS, Criss AK (2018) Pathogenesis of Neisseria gonorrhoeae in the female reproductive tract: neutrophilic host response, sustained infection, and clinical sequelae Curr Opin Hematol 25:13–21 doi: 10.1097/MOH.0000000000000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl EA, Criss AK, Seifert HS (2005) The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection Mol Microbiol 58:520–532 doi: 10.1111/j.1365-2958.2005.04839.x [DOI] [PMC free article] [PubMed] [Google Scholar]