Abstract
The main goal of this study was to test a rational combination of pre‐selected carbohydrate‐active enzymes (CAZymes) and sulphatases, individually or in combination, in order to evaluate its capacity to disrupt Arthrospira platensis cell wall, allowing the release of its valuable nutritional bioactive compounds. By the end, a two‐enzyme constituted mixture (Mix), composed by a lysozyme and a α‐amylase, was incubated with A. platensis suspension. The microalga cell wall disruption was evaluated through the amount of reducing sugars released from the cell wall complemented with the oligosaccharide profile by HPLC. An increase of the amount of reducing sugars up to 2.42 g/L in microalgae treated with the Mix relative to no treatment (p < .05), as well as a 7‐fold increase of oligosaccharides amount (p < .001), were obtained. With resort of fluorescence microscopy, a 36% reduction of fluorescence intensity (p < .001) was observed using Calcofluor White staining. In the supernatant, the Mix caused a 1.34‐fold increase in protein content (p = .018) relative to the control. Similarly, n‐6 polyunsaturated fatty acids (PUFA) (p = .007), in particular 18:2n‐6 (p = .016), monounsaturated fatty acids (MUFA) (p = .049) and chlorophyll a (p = .025) contents were higher in the supernatant of microalgae treated with the enzyme mixture in relation to the control. Taken together, these results point towards the disclosure of a novel two‐enzyme mixture able to partial degrade A. platensis cell wall, improving its nutrients bioavailability for monogastric diets with the cost‐effective advantage use of microalgae in animal feed industry.
Keywords: Arthrospira platensis, carbohydrate‐active enzymes, cell wall, fatty acids, reducing sugars, total proteins
1. INTRODUCTION
In recent years, the use of microalgae as a source of proteins, lipids, carbohydrates and other bioactive compounds has been the focus of intensive research (Chew et al., 2017), mainly directed to its use for biofuel, nutraceutical and pharmaceutical applications (Baudelet, Ricochonb, Lindera, & Munigliaa, 2017), as well as sustainable animal production (Lum, Kim, & Lei, 2013). The nutritional profile of microalgae is species‐specific but has, in general, contents of proteins, lipids, carbohydrates, vitamins, pigments and minerals that are comparable, if not superior, to conventional feedstuffs (Liu & Chen, 2016). Microalgae are highly rich in beneficial n‐3 long‐chain polyunsaturated fatty acids (n‐3 LCPUFA) (Madeira et al., 2017), turning microalgae into an untapped natural resource with well‐known health benefits for both animals and humans (Calder, 2012).
Arthrospira platensis is a filamentous microalga, classified as a blue–green alga (Cyanophyceae, also known as cyanobacteria) (Seyidoglu, Inan, & Aydin, 2017). The cell organization of A. platensis is typical of a prokaryote Gram‐negative bacterium, lacking membrane‐bound organelles. The cell wall constitutes an envelope composed by several layers, mostly of peptidoglycan and lipopolysaccharide nature. A. platensis grows naturally in alkaline lakes but is commercially produced in large outdoor or greenhouse ponds under controlled conditions (Sotiroudis & Sotiroudis, 2013; Van Eykelenburg, Fuchs, & Schmidt, 1980).
This specific microalga has been designated as a healthy food by the World Health Organization (WHO) (Seyidoglu et al., 2017) due to its content in bioactive substances (Ovando et al., 2018), in which stand out the highest protein content of any natural food (60%–70%), essential amino acids, fatty compounds, including the beneficial n‐3 LCPUFA, and carotenoids. A. platensis presents several applications, mainly in food, pharmaceutical, nutraceutical, cosmetics, wastewater treatments and animal feed industries (Holman & Malau‐Aduli, 2013; Seyidoglu et al., 2017; Soni, Sudhakar, & Rana, 2017). In fact, this microalga is responsible solely for 50% of worldwide production as feed supplement (Yamaguchi, 1997).
The majority of microalgae exhibit recalcitrant cell walls, largely indigestible by monogastric animals, preventing them from accessing their valuable nutritional compounds, such as proteins and lipids. For microalgae species, unlike macroalgae, the mechanical methods, such as hammer mills, are not commonly applied (Makkar et al., 2016). In turn, bead milling is a successfully, rising process in the food industry used to incorporate microalga cells as food additives. However, this mechanical process is laborious and expensive whereupon cells are massively destroyed. Therefore, it is imperative to find novel technologies, cheaper and under a strictly controlled process, to disrupt A. platensis cells to improve microalgal nutrient utilization, as proteins and lipids by monogastric animals (Austic, Mustafa, Jung, Gatrell, & Lei, 2013; Lum et al., 2013). Despite A. platensis presents a relatively less complex cell wall, it still remains a barrier in the use of its compounds, whereby its degradation will improve the accessibility to such compounds (Safi et al., 2014). This aspect is particularly relevant if microalgae are included at higher percentages in the diet, that is used as feed ingredient not as feed supplement (Madeira et al., 2017).
Exogenous carbohydrate‐active enzymes (CAZymes) are largely accepted as a class of feed additives for pigs and poultry diet formulations to surpass the negative effects of anti‐nutritional factors, and to improve the digestion of dietary components and, ultimately, animal's performance (Ravindran & Son, 2011). These enzymes are produced by micro‐organisms and are complex enzymes, in which the catalytic module(s) is (are) appended to one or more non‐catalytic carbohydrate binding modules (CBM) (Fontes & Gilbert, 2010). According to circumstances, the utilization of CAZymes for microalgae biomass might represent a good strategy to value the nutritional compounds of cereal‐based diets for monogastrics.
Taking into account these considerations, we hypothesized that the nutrients bioavailability of A. platensis could be greatly improved by using individually or combined CAZymes and sulphatases that can efficiently degrade the microalga cell wall and be used, in the long run, as feed catalysts for monogastric diets. The cell wall disruption was achieved by enzymatic treatment and assessed by optical and fluorescence microscopies, complemented with the amount of reducing sugars released and the oligosaccharide profile. The nutritional bioactive compounds were detailed by measuring proteins and pigments, as well as fatty acid profile in both supernatant and residue fractions, after incubation with the enzymatic mixture treatment.
2. MATERIALS AND METHODS
2.1. Microalgae cultivation
To cultivate A. platensis (LB 2342), axenic microalga cultures from the institutes algae banks were inoculated in an adapted Krauss medium (Vonshak, 1986) to stimulate A. platensis growth: NaNO3 (250 mg/L), KH2PO4 (105 mg/L), MgSO4 (75 mg/L), CaCl2 (25 mg/L), NaCl (25 mg/L), K2HPO4 (75 mg/L) and 3 ml of trace metal solution: FeCl3 (0.194 g/L), CoCl2 (0.16 g/L), MnCl2 (0.082 g/L), Na2MoO4 2H2O (0.008 g/L) and ZnCl2 (0.005 g/L). A. platensis was first grown in airlift bioreactors with 1 litre capacity and then scaled up to 25 L polyethylene bag bioreactors with bubbling filtered air, without carbon dioxide addition at low incident light conditions (150 µE m−2 s−1), and at 34°C, which is the optimal temperature for A. platensis. Once reached the stationary growth phase, the harvesting step was carried out without flocculation by removing agitation, followed by centrifugation in a continuous centrifuge LPX 40 (Alfa Laval) (25 L). Then, the concentrated biomass slurry was frozen and freeze‐dried (Powerdry LL 3000; Thermo), until analysis.
2.2. Recombinant enzymes: high‐throughput gene synthesis, cloning and protein expression/purification
One hundred and seventy‐eight CAZymes theoretically capable of disrupting A. platensis cell wall were selected from a vast library, comprising glycoside hydrolases (GH), pectate lyases (PL) and carbohydrate esterases (CE). Twenty‐two sulphatases likely involved in microalgae cell wall disruption were selected for screening, as well (Gerken, Donohoe, & Knoshaug, 2013). The coding genes for all of these enzymes were synthesized in vitro using NZYGene Synthesis kit (Nzytech). The protein sequence of each enzyme is presented as Supplementary Material (Table S1). Synthetic genes were codon optimized for expression in Escherichia coli using NZYTech´s codon optimization software ATGenium (Sequeira et al., 2017). All genes included the required 16 bp overhangs on both 5′ and 3′ ends for direct cloning into the bacterial expression vector pHTP1 (Nzytech), based on NZYEasy Cloning & Expression kit I (Nzytech) protocol. The generated recombinant plasmids were subjected to inducible T7 promoter control, while encoding the 200 enzymes fused to an N terminal His6 tag to allow purification using immobilized affinity chromatography (IMAC). The two hundred plasmids were sequenced to guarantee no mutations generated during gene synthesis and were used to transform E. coli BL21 (DE3) cells. The transformed cells were grown on solid media. The resulting colonies were used to inoculate 5 ml of NZY Auto‐Induction LB medium (Nzytech, Portugal) supplemented with kanamycin (50 μg/mL) at 37°C to early‐exponential phase (A600nm = 1.5–2.0). The recombinant protein was produced following a step of incubation at 25°C during 16 hr. All steps were performed in 24‐deep‐well plates (Sequeira et al., 2017). Cells were harvested by centrifugation at 75,000 g at 4°C during 15 min and lysed using the NZY Bacterial Cell Lysis buffer (NZYTech). The His6‐tagged recombinant enzymes were purified from cell‐free extracts by IMAC, based on an automated procedure that enables the purification of 96 proteins per day, as previously reported (Saez & Vincentelli, 2014). In short, the crude cell lysates were incubated with Sepharose chelating beads (200 μl with bound Ni2+) and transferred to 96‐well filter plates (Macherey‐Nagel). Then, wells were washed 2 × with buffer A (50 mM NaHepes, pH 7.5, 500 mM NaCl, 10 mM imidazole). The recombinant proteins were eluted from the column resin beads using 200 μL of elution buffer (50 mM NaHepes, pH 7.5, 500 mM NaCl, 300 mM imidazole) into 96‐deep‐well plates. All steps involved in protein purification were automated on a Tecan robot (Tecan) that contains a vacuum manifold. The homogeneity of purified proteins and the molecular mass of recombinant enzymes were evaluated by SDS‐PAGE in 14% (w/v) acrylamide gels. The protein concentration of enzymes was determined spectrophotometrically by the Bradford method (Bradford, 1976) and varied between from 0.5 to 20 g/L.
2.3. Preparation of microalga cell suspension
The concentration of A. platensis suspension was 20 g/L. The preparation of microalga cell suspension included a pre‐wash step with phosphate‐buffered saline (PBS), followed by centrifugation and re‐suspension of the microalgae pellet in PBS, as described by Coelho et al. (2019).
2.4. Enzymatic cell wall disruption
In order to disrupt A. platensis cell wall, the microalgae suspension was incubated with CAZymes, under strictly controlled conditions. The cell wall disruption assay was performed, according to Coelho et al. (2019).
2.5. Reducing sugars measurement
To quantify the amount of reducing sugars released, the 3,5‐dinitrosalicylic acid (DNSA) method (Miller, 1959) was used, as described by Coelho et al. (2019).
2.6. Thermostability and proteolysis experiments
Each enzyme composing the mixture (Mix; Provisional Patent number 20191000008190, INPI) was biochemically characterized, in particular for thermostability and proteolysis resistance. The thermostability analysis was performed, according to Coelho et al. (2019). As the temperature of incubation increased, the amount of protein in the supernatant reduced. This was validated by running 14% SDS‐PAGE gels in the supernatants and visualizing the intensity of the band. The resultant images were acquired with Bio‐Rad ChemiDoc XRS imaging system (Bio‐Rad). To evaluate the proteolysis resistance, each enzyme was incubated with porcine pancreatin (VWR Chemicals), as described by Coelho et al. (2019). The samples were then removed and analysed by 14% SDS‐PAGE gels. The proteolysis was confirmed by visualizing fragments with different molecular weights. The resultant images were once again acquired with Bio‐Rad ChemiDoc XRS imaging system (Bio‐Rad).
2.7. Determination of total oligosaccharides
After control and Mix treatments, the profile of mono‐ and oligosaccharides from the supernatants of A. platensis was analysed and quantified by high‐performance liquid chromatography (HPLC), following on a protocol developed by Coelho et al. (2019).
2.8. Optical and fluorescence microscopic observations
The residue fractions (pellets) from control and Mix treatments were analysed through optical and fluorescence microscopic observations. On the one hand, the optical microscopy enabled to count the number of cells in the microalgae suspension; on the other hand, the fluorescence microscopy, through fluorochrome Calcofluor White (Sigma‐Aldrich) staining that binds to the cell wall (Safi et al., 2014), enabled to quantify fluorescence intensity. The optical and fluorescence microscopic procedures are described in detail by Coelho et al. (2019).
2.9. Determination of protein content
After control and Mix treatments, the N content in lyophilized supernatant and residue fractions from A. platensis suspension, was quantified using the Kjeldahl method (984.13) (AOAC, 2000). Crude protein was calculated as 6.25 × N.
2.10. Pigment analysis
Chlorophyll a, chlorophyll b and total carotenoids were quantified in supernatant and residue fractions from A. platensis suspension, after control and Mix treatments, as reported by Hynstova et al. (2018) with slight modifications as described by Coelho et al. (2019).
2.11. Determination of fatty acid content and composition
The fatty acid profile and content of supernatant and residue fractions of A. platensis suspension after control and Mix treatments were determined, as described by Coelho et al. (2019).
2.12. Statistical analysis
Data were analysed using the generalized linear mixed (GLM) model of the sas software package (version 9.4; SAS Institute Inc.). All experiments were conducted in triplicate. Results are presented as mean and standard error of the mean (SEM) and considered significantly different when the p‐value was < .05.
3. RESULTS
3.1. Individual screening of enzymes in Arthrospira platensis cell wall disruption
Each one of CAZymes and sulphatases from our vast repertoire was incubated individually with the microalgae suspension to degrade A. platensis cell wall. The majority of the enzymes tested were unable to deconstruct the microalgae biomass, except 26 enzymes, as described in Table 1. The capacity to disrupt A. platensis cell wall was assessed by the amount of reducing sugars released through the DNSA method and applying the following qualitative scale (g/L): −, 0.00 < 0.005; +, 0.05 < 0.200; ++, 0.200 < 0.300; +++,>0.300. Among this set of 26 enzymes, the ones with ID 5, 14, 18, 37 to 42, 60 to 69, 78, 81, 85, to 104 and (2) 72 showed the highest amount of reducing sugars released from the biomass, whereas the others revealed a minimal or moderate capacity to attack the complex polysaccharides.
Table 1.
Screening of the selected individual CAZymes sulphatases and Mix in Arthrospira platensis cell wall disruption
| ID | Name | Category | E.C | Main Substrate | Reducing sugars released scale |
|---|---|---|---|---|---|
| 5 | Cellulose 1,4‐β‐cellobiosidase | Cellobiohydrolases | 3.2.1.91 | Phosphoric acid‐swollen cellulose, Avicel and others forms of insoluble cellulose | +++ |
| 10 | Laccase | Laccases | 1.3.3.5 | 2,20‐azinobis(3‐ethylbenzothiazoline−6‐sulphonic acid) (ABTS) | ++ |
| 14 | Laminarinase | 1,3‐β‐Glucanases | 3.2.1.39 | 1,3‐β‐glucans such as laminarin | +++ |
| 16 | Chitinase 1 | Chitinases & Chitosanases | 3.2.1.14 | Chitin and chitosan | ++ |
| 18 | Oligoalginate lyase | Alginate lyases | 4.2.2. | Low‐viscosity alginate | +++ |
| 25 | β−1,3–1,4‐glucanase P2 | 1,3–1,4‐β‐Glucanases | 3.2.1.73 | 1,3–1,4‐β‐glucans | + |
| 33 | β−1,3‐glucanase/ laminarinase | 1,3‐β‐Glucanases | 3.2.1.39 | Laminarin | ++ |
| 36 | Chitosanase | Chitinases & Chitosanases | 3.2.1.132 | Chitosan | + |
| 37 | Endo‐β−2,6‐fructanase | Fructanases | 3.2.1.65 | Levans | +++ |
| 38 | Cellobiohydrolase | Cellobiohydrolases | 3.2.1.91 | Amorphous and crystalline cellulose | +++ |
| 42 | Trans‐sialidase B | Sialidases | 3.2.1.18 | Sialic acids from complex carbohydrates and glycoprotein human alpha−1 (AGP) | +++ |
| 50 | α‐glucuronidase | Glucuronidases | 3.2.1.139 | Glucuronic acid from the xylan backbone | + |
| 60 | Exo‐β‐glucosaminidase | Glucosaminidases | 3.2.1.165 | The 1,4‐β‐glycosidic bond of cellooligosaccharides, also hydrolysis non‐reducing end of chitooligosaccharides (Glc‐PNP) | +++ |
| 66 | Alginate lyase | Alginate lyases | 4.2.2.3 | Polyguluronate and polymannuronate | +++ |
| 69 | α−1,3‐glucanase | α‐Glucosidases | 3.2.1.59 | 1,3‐α‐glucan | +++ |
| 73 | Exo‐β‐agarase D | Agarases | 3.2.1.81 | Agarose and neoagarooligosaccharides | + |
| 78 | Keratan sulphate hydrolase/ keratanase II | Acetylglucosaminidases | 3.2.1.103 | Cartilage keratan sulphate and cornea keratan sulphate | +++ |
| 81 | Exo‐β‐glucosaminidase | Glucosaminidases | 3.2.1.165 | Lactose, GlcNAc2, GlcNAc3, cellobiose and cellotriose, as well as colloidal chitin, cellulose, lichenan, laminarin and xylan | +++ |
| 82 | β−1,3‐glucanase B | Laminarinases | 3.2.1.39 | Insoluble 1,3‐β‐glucan | + |
| 85 | β‐galactosidase | β‐Galactosidases | 3.2.1.23 | β‐galactosides | +++ |
| 86 | Lytic transglycosylase | Peptidoglycan lytic exotransglycosylases | 4.2.2.n1 | 1,4‐β‐glycosidic bonds between N‐acetylmuramic acid and N‐acetylglucosamine residues in the cell wall peptidoglycan, producing 1,6‐anhydromuropeptides | +++ |
| 92 | Endo‐rhamnogalacturonan lyase | Rhamnogalacturonan lyases | 4.2.2.23 | Rhamnogalacturonan | +++ |
| 93 | Peptidoglycan N‐acetylmuramic acid deacetylase | Acetylglucosamine deacetylases | 3.5.1.104 | Peptidoglycan | +++ |
| 95 | Lysozyme | Lysozymes | 3.2.1.17 | Peptidoglycans | +++ |
| 104 | Lysozyme | Lysozymes | 3.2.1.17 | Peptidoglycans | +++ |
| (2)72 | α‐amylase | Amylases | 3.2.1.1 | Endohydrolysis of 1–4‐α‐D‐glucosidic linkages in polysaccharides containing three or more 1–4‐α‐linked D‐glucose units | +++ |
| Mix | Lysozyme 104 + α‐amylase (2) 72 | 2.42 g/L | |||
For each enzyme, is presented the ID, the name, the category, the E.C number, the main substrate and a qualitative scale of reducing sugars released. It is also presented the enzymatic constitution of the Mix as well as the value of the reducing sugars released in g/L. Qualitative scale on the amount of reducing sugars released (g/L): −, <0; +, 0.05 < 0.2; ++, 0.2 < 0.3; +++,>0.3.
3.2. Composition of a two‐enzyme constituted mix based on reducing sugars released
To disclose synergistic actions, the 26 enzymes presented in Table 1 were tested in combination for the capacity to release reducing sugars from the microalgae. From that point on, several mixtures were tested, in which enzymes were consecutively removed, according to results from DNSA method. By the end, a mixture (Mix) of two enzymes was found to be the most constrained mixture, showing the highest amount of reducing sugars released. This Mix was composed by a lysozyme (ID 104) and a α‐amylase (ID (2) 72) and is presented in Table 1. When this mixture was incubated with A. platensis suspension, a value of 2.42 g/L (p < .05) of reducing sugars released was obtained, representing a 1.24‐fold increase in relation to the highest value observed in the individual screening. The rates for released sugars were calculated as: for Mix versus control = 407.3%; for Mix versus lysozyme = 102%, and for Mix versus α‐amylase = 30.2%.
3.3. Thermostability and proteolysis assays
We next tested the thermostability of the two enzymes that constitute the Mix treatment, individually. The variation of protein concentration across the temperatures tested is shown in Figure 1. For the internal temperature of mammals and poultry which are, respectively, 37°C and 40°C, all enzymes maintained their stability. However, the stability of ID 104 decayed from 65°C upward, while ID (2) 72 remained stable up to 80°C.
Figure 1.
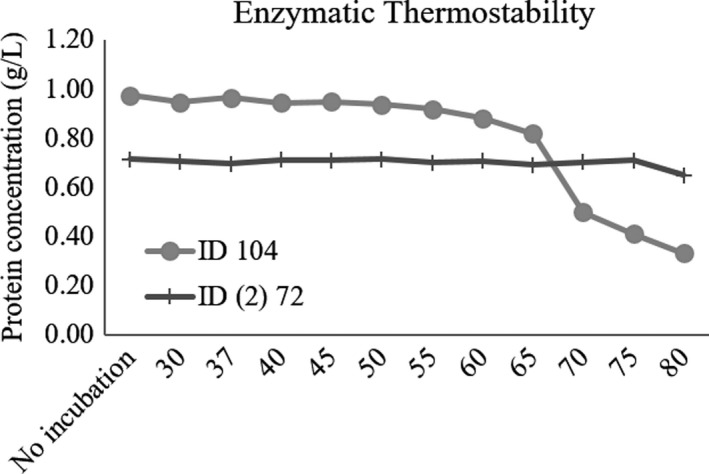
Thermostability characterization of the two enzymes constituting the Mix at different temperatures (30–80°C) and for the control without incubation
Next, the same enzymes were treated with pancreatin at 37°C to test their capacity to resist to proteolytic attack in the animal gastrointestinal tract. The proteolytic resistance scores of these enzymes are shown in Table 2. Enzyme ID (2) 72 displayed partial resistance along the entire assay; in contrast, ID 104 showed a complete degradation after 15 min of incubation (Figure 1).
Table 2.
Proteolysis resistance for each one of the two enzymes that constitute the Mix
| ID | Time | ||||
|---|---|---|---|---|---|
| 15 min | 30 min | 60 min | 90 min | 120 min | |
| 104 | − | − | − | − | − |
| (2) 72 | + | + | + | + | + |
Each enzyme, at a concentration of 1 g/L, was subjected to the proteolytic action of pancreatin, which was incubated at a final concentration of 2.5 g/L. The reactions were incubated at 37°C, at regular intervals of 15 min for 120 min. Results are presented at periods of 15, 30, 60, 90 and 120 min of incubation for each enzyme. Qualitative scale on proteolysis resistance: −, no resistant; +, partially resistant.
3.4. Effect of mix treatment on Arthrospira platensis cell number and cell wall integrity
The number of cells was kept unchanged between control and Mix (Figure 2a, p > .05) and was approximately 16,000 cells for both treatments (Figure 2b and c). When A. platensis was incubated with the Mix (Figure 2f), the fluorescence intensity was diminished by 36% (Figure 2d, p < .001), relative to the control (Figure 2e).
Figure 2.
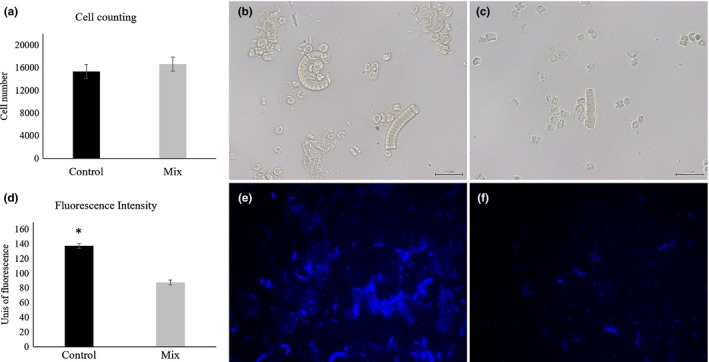
(a) Cell counting using a Neubauer chamber for control and Mix treatments. (b and c) light microscopy images (×400) of Arthrospira platensis suspension for control and Mix treatments, respectively (scale bar: 20 µm). (d) fluorescence intensity derived from Calcofluor White staining for control and Mix treatments. Asterisk denotes statistical difference at p < .001. (e and f) fluorescence images (×400) of A. platensis suspension stained with Calcofluor White for control and Mix treatments, respectively
3.5. Effect of mix treatment on the release of oligosaccharides from Arthrospira platensis cell wall
In the oligosaccharides region, three large peaks were detected in the Mix treatment chromatogram (Figure 3b), compared to the control (Figure 3a), which corresponds to a 7‐fold increase on the oligosaccharides content (p < .001; Figure 3c).
Figure 3.
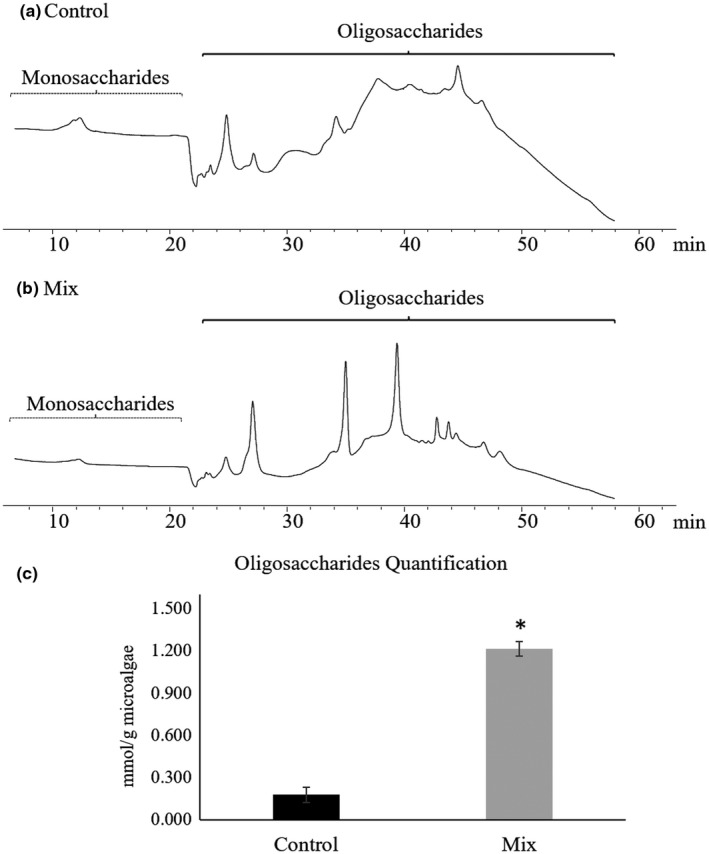
Illustrative chromatograms obtained by HPLC analysis of supernatants for the control (a) and the Mix (b) treatments. Monosaccharides and oligosaccharides regions are shown. The quantification of oligosaccharides are graphically displayed in C. Asterisk denotes statistical difference at p < .001
3.6. Effect of mix treatment on the release of proteins
In order to verify if the enzyme mixture favoured the release of proteins from A. platensis cells to the exterior, the amount of protein was quantified in supernatants and residues (Table 3). In the supernatant fraction, the Mix treatment led to a 1.34‐fold increase in protein content when compared to the control (p = .018). In the residue fraction, the Mix treatment caused a 1.14‐fold reduction when compared to the control (p = .003).
Table 3.
Content of protein, chlorophyll, carotenoids and fatty acids of the supernatant and residue fractions derived from incubation of Arthrospira platensis with control and Mix treatments
| Supernatant | Residue | |||||||
|---|---|---|---|---|---|---|---|---|
| Control | Mix | SEM | p‐value | Control | Mix | SEM | p‐value | |
| Total protein (mg/g microalgae) | 412 | 554 | 26.0 | .018 | 669 | 586 | 8.81 | .003 |
| Chlorophyll a (mg/g microalgae) | 0.4541 | 0.5201 | 0.016 | .025 | 6.102 | 7.602 | 0.324 | .017 |
| Chlorophyll b (mg/g microalgae) | 2.021 | 2.051 | 0.039 | .645 | 0.3552 | 0.5202 | 0.062 | .111 |
| Total chlorophylls (mg/g microalgae) | 2.481 | 2.571 | 0.054 | .274 | 6.462 | 8.712 | 0.725 | .071 |
| Total carotenoids (mg/g microalgae) | 0.1621 | 0.2091 | 0.017 | .102 | 3.042 | 2.732 | 0.164 | .218 |
| Total fatty acids (mg/g microalgae) | 4.27 | 3.63 | 0.136 | .016 | 46.7 | 41.8 | 0.910 | .009 |
| Fatty acid composition (% total fatty acids) | ||||||||
| 12:0 | 0.345 | 0.605 | 0.112 | .154 | 0.089 | 0.078 | 0.013 | .560 |
| 14:0 | 1.87 | 1.31 | 0.193 | .087 | 1.27 | 1.33 | 0.059 | .526 |
| 14:1c9 | nd | nd | ‐ | ‐ | 0.372 | 0.408 | 0.010 | .049 |
| 15:0 | 0.340 | 0.260 | 0.043 | .235 | 0.040 | 0.047 | 0.010 | .619 |
| 16:0 | 46.0 | 42.7 | 0.423 | .002 | 41.3 | 41.3 | 0.216 | .978 |
| 16:1c7 | 0.665 | 0.724 | 0.051 | .454 | 1.51 | 1.51 | 0.014 | .985 |
| 16:1c9 | 2.84 | 3.16 | 0.104 | .077 | 5.23 | 5.09 | 0.047 | .074 |
| 17:0 | 1.28 | 0.983 | 0.073 | .030 | 0.341 | 0.407 | 0.041 | .294 |
| 18:0 | 21.6 | 19.9 | 1.23 | .378 | 3.10 | 2.74 | 0.288 | .413 |
| 18:1c9 | 4.35 | 6.20 | 0.643 | .089 | 2.43 | 2.23 | 0.138 | .332 |
| 18:1c11 | 0.469 | 0.704 | 0.163 | .348 | 0.236 | 0.265 | 0.056 | .730 |
| 18:2n−6 | 9.06 | 10.4 | 0.284 | .016 | 18.4 | 18.8 | 0.086 | .013 |
| 18:3n−6 | 7.42 | 8.31 | 0.242 | .040 | 24.7 | 24.6 | 0.124 | .751 |
| 18:3n−3 | 0.141 | 0.345 | 0.062 | .058 | 0.090 | 0.106 | 0.004 | .032 |
| 20:0 | 1.18 | 1.18 | 0.087 | .964 | 0.202 | 0.224 | 0.040 | .709 |
| 22:0 | 1.58 | 1.81 | 0.105 | .182 | 0.181 | 0.209 | 0.011 | .111 |
| 22:2n−6 | 0.866 | 1.40 | 0.145 | .040 | 0.066 | 0.103 | 0.005 | .003 |
| Others | 0.068 | 0.062 | 0.065 | .952 | 0.436 | 0.479 | 0.037 | .439 |
| ∑ SFA | 74.1 | 68.7 | 1.06 | .011 | 46.5 | 46.4 | 0.283 | .669 |
| ∑ MUFA | 8.33 | 10.8 | 0.708 | .049 | 9.78 | 9.50 | 0.185 | .318 |
| ∑ PUFA | 17.5 | 20.4 | 0.484 | .005 | 43.3 | 43.7 | 0.187 | .163 |
| ∑ n−3 PUFA | 0.141 | 0.345 | 0.062 | .058 | 0.090 | 0.106 | 0.004 | .032 |
| ∑ n−6 PUFA | 17.3 | 20.1 | 0.491 | .007 | 43.2 | 43.6 | 0.188 | .179 |
Two mL of microalgae suspension was incubated with the two enzymes, which constitute the Mix, at a final concentration of 20 mg/L for each enzyme. The control treatment took the same amount of PBS. Incubations were done overnight at 37°C and 140 rpm. After incubations, supernatant and residue fractions were separated by centrifugation. Only fatty acids whose percentage was >0.25% are presented; nd, not detected.
Values measured in phosphate‐buffered saline (PBS).
Values measured after extraction with acetone.
3.7. Effect of mix treatment on the release of chlorophylls and carotenoids
Applying the same rationale as the previous point, the release of pigments from A. platensis cells to the exterior was quantified in supernatants and residues (Table 3). Chlorophyll a displayed a 1.15‐fold increment in the supernatant fraction of the Mix treatment relative to the control (p = .025), whereas in the residue fraction the Mix treatment led to a 1.24‐fold increase relative to the control (p = .017). Chlorophyll b, total chlorophylls and total carotenoids did not vary (p > .05).
3.8. Effect of mix treatment on the release of fatty acids
The fatty acid content and profile after incubation with the enzyme mixture was determined in supernatants and residues to verify if the Mix treatment promoted the beneficial release of fatty acids from A. platensis cells to the exterior (Table 3). The prevalent fatty acids were saturated fatty acids (SFA) > PUFA > n‐6 PUFA > monounsaturated fatty acids (MUFA) > n‐3 PUFA in both fractions. In the supernatant fraction, 16:0, SFA, total FA content and 17:0 were increased in the control relative to the Mix (p = .002, p = .011, p = .016, p = .030, respectively). In opposition, PUFA, n‐6 PUFA, 18:2n‐6, 18:3n‐6, 22:2n‐6 and MUFA increased in the Mix relative to the control (p = .005, p = .007, p = .016, p = .040, p = .040 and p = .049, respectively). In the residue fraction, the Mix treatment led to higher proportions of 22:2n‐6, 18:2n‐6, 18:3n‐3, n‐3 PUFA and 14:1c9 (p = .003, p = .013, p = .032, p = .032 and p = .049, respectively), and to a lower proportion of total fatty acids (p = .009) in comparison to the control.
4. DISCUSSION
In this work, a vast repertoire of 178 CAZymes and 22 sulphatases was created by recombinant expression in E. coli cells to assess the hypothesis that nutritional bioactive compounds availability of A. platensis may be enhanced after disruption of its cell wall. These 200 enzymes were chosen based on the composition of matrix polysaccharides of microalgae cell walls, which includes pectin, chitin agar, alginates and the aliphatic polymer algenan (Scholz et al., 2014), in addition to the cyanobacterium peptidoglycan (Palinska & Krumbein, 2000; Sotiroudis & Sotiroudis, 2013). The chosen enzymes were produced on a high‐throughput (HTP) system that includes several steps, from gene synthesis, gene cloning, protein expression to purification. These enzymes were screened, one by one, to disrupt A. platensis cell wall, by measuring the amount of reducing sugars released. In the following phase, the 26 recombinant enzymes able to partially or entirely disrupt A. platensis cell wall (Table 1) were combined and tested to achieve the maximum degradation of A. platensis cell wall. A two‐enzyme mixture (Mix) was found to be the most confined combination with the highest activity towards the disruption of A. platensis cell wall, and applied in subsequent steps. It was constituted by two well‐characterized recombinant glycosylases, a lysozyme (EC 3.2.1.17) and a α‐amylase (EC 3.2.1.1). Lysozyme belongs to the family 24 of glycoside hydrolases, according to the CAZy database (Cantarel et al., 2009). The enzyme‐coding gene was obtained from E. coli (Srividhya & Krishnaswamy, 2007) and has peptidoglycan, containing muramic acid δ‐lactam, as the main substrate (Babu, Arulandu, & Sankaran, 2018; Srividhya & Krishnaswamy, 2007). α‐amylase was firstly characterized by Liebl, Stemplinger, and Ruile, (1997) and, according to the CAZy database, belongs to the family 13, subfamily 36, of glycoside hydrolases (Cantarel et al., 2009). The enzyme‐coding gene was obtained from the aquatic hyperthermophilic Thermotoga maritima, and its main substrates comprise various α‐glucans, such as amylose, amylopectin and glycogen (Liebl et al., 1997).
It has been shown that cell walls of Gram‐negative bacteria, containing peptidoglycan, are susceptible to lysozyme, as appears to be the case of A. platensis (Sotiroudis & Sotiroudis, 2013; Van Eykelenburg et al., 1980). Mehta, Evitt, and Swartz, (2015) developed a complete lysis technique, which included the incorporation of a lysozyme using different cyanobacterial strains. Aikawa et al. (2013) observed that when lysozyme was added to the fermentation medium, the bioethanol production yield reached 86% of the theoretically expected amount, since lysozyme degraded A. platensis cell walls. Pyo, Kim, and Yoo, (2013) performed the extraction of bioethanol from two fresh water Gram‐negative cyanobacteria species, Microcystis aeruginosa and Anabaena variabilis with similar peptidoglycan cell wall layers (Sun, Jiang, Sato, Kawachi, & Lu, 2016; Thiel et al., 2014), which resemble A. platensis. In the same study, the authors used an enzyme mixture composed of three enzymes, including a α‐amylase to hydrolyse the cyanobacteria (Pyo et al., 2013). This finding was also supported by Carrillo‐Reyes, Barragán‐Trinidad, and Buitrón, (2016). The starch in cyanobacteria is a highly branched α‐1,4‐polyglucan, denominated as cyanophycean starch (Pulz & Gross, 2004), deeply located in the cyanobacterial cell wall with an irregular whitish spherical form (Lang, 1968; Pyo et al., 2013). In turn, A. platensis has a low content of internal energy storage, as starch due to high activities of α‐ and β‐amylase. The in vitro digestibility of A. platensis has been reported using an amylase (Pyne, Bhattacharjee, & Srivastav, 2017; Usharani, Saranraj, & Kanchana, 2012). Bearing those former observations in mind, that clearly establish a link between A. platensis cell wall composition and the degrading enzymes identified in this study, the enzymatic composition of the Mix described is in line with the cell wall composition of this microalga.
The two enzymes constituting the mixture were characterized individually for thermostability and proteolysis resistance. ID (2) 72 remained stable throughout the temperature range and resistant to the proteolytic attack of pancreatin. The tertiary structure of protein, which provides thermotolerance to enzymes, may provide inherent proteinase resistance, as reported by Fontes, Hall, Hirst, Hazlewood, and Gilbert, (1995). The high thermotolerance, which characterizes this enzyme, is due to the circumstance that it is isolated from Thermotoga maritima, one of the most thermophilic bacteria presently known, with maximum growth temperature at 90°C (Huber et al., 1986; Liebl et al., 1997; Singh, White, & Blum, 2017). In opposition, ID 104 was sensitive to temperature increase and proteolysis.
The Mix was proven capable at disrupting A. platensis cell wall through the increase of 1.24‐fold in reducing sugars relative to the highest individual value found, suggesting a synergistic action between these enzymes when combined, as demonstrated by Phong et al. (2018), when degrading diverse carbohydrate mixtures. Pyo et al. (2013) also selected the release of reducing sugars to assess the ability of different methods, including the enzymatic method, to hydrolyse the two species of Gram‐negative cyanobacteria above mentioned. Markou, Angelidaki, Nerantzis, and Georgakakis (2013) used different acids at different concentrations to hydrolyse A. platensis and quantified the outcome through the measurement of reducing sugars released. A higher amount of reducing sugars released corresponds to a higher hydrolysis yield (Markou et al., 2013; Pyo et al., 2013), which is in agreement with results obtained in our study.
The A. platensis cell number was not changed by the enzyme mixture. Contrarily, the fluorescence intensity reduced 36% after the Mix treatment, suggesting that the cell wall integrity was affected to a considerable extent. Safi et al. (2014) applied the same fluorochrome when testing various cell wall disruption methods (like, high pressure and ultrasonication). They concluded that after treatment a variation on fluorescence intensity was observed suggesting a clear change in cell wall structure, justifying the use of Calcofluor White staining. This evidence was reinforced by a 7‐fold augment of oligosaccharides content after the enzyme mixture treatment, as observed by Heo et al. (2017). These same authors, and contrarily to our findings, reported a large increase on glucose amount in a different species of microalga, Chlorella vulgaris, after osmotic shock suggesting the complete disruption of the cell wall. Contrarily, in our study, no complete degradation of carbohydrates from the cell wall was obtained, since a complex mixture of oligosaccharides rather than single sugars, was observed. In addition, Leal et al. (2017) observed that the implementation of an acidic method to hydrolyse microalgae/cyanobacteria cell walls led to a high release of oligosaccharides to obtain prebiotic oligosaccharides from A. platensis biomass via phosphoric acid hydrolysis.
The release of cytoplasmic (hydro‐)soluble proteins from A. platensis cell wall was observed after the enzymatic mixture treatment. This result was naturally followed by a decrease of protein content in the residue, which was expected. These findings concur with Safi et al. (2014), even if different mechanical and chemical cell wall disruption methods were applied. In addition, Lupatini, Colla, Canan, and Colla (2017) reported different methods of cell wall disruption, including enzymatic, as capable of promoting the extraction of microalgae proteins from A. platensis. The results obtained in our study assume particular relevance due to the high protein content of A. platensis (60%–70%) (Soni et al., 2017), re‐enforcing its great value as feed ingredient for animal production and human health (Holman & Malau‐Aduli, 2013; Lupatini et al., 2017).
Besides proteins, the enzymatic mixture treatment released chlorophyll a to the supernatant. A. platensis contains relevant amounts of chlorophylls, about 1%–1.5% (Jiménez, Cossío, Labella, & Xavier Niell, 2003; Leema, Kirubagaran, Vinithkumar, Dheenan, & Karthikayulu, 2010), which are located within thylakoid bundles circling the peripheral part of the cytoplasm with their associated structures, the phycobilisomes (containing the phycobiliproteins) on the surface of the thylakoids (Safi et al., 2014). In addition to phycocyanin (30%) (Cisneros & Rito‐Palomares, 2004; Leema et al., 2010), A. platensis also displays an appreciable content of other pigments, like carotenoids (0.2%–0.35%), including β‐carotene and lutein (Jiménez et al., 2003; Leema et al., 2010). Notwithstanding, no changes were detected for chlorophyll b and total carotenoids. Previous studies reported the successful disruption of the cell wall by several chemical and mechanical methods on the release of chlorophylls and carotenoids to the supernatant (Safi et al., 2015), and therefore concur with ours for chlorophyll a. We speculate that the cell wall degradation promoted by the Mix treatment favoured the release of chlorophyll a to the external aqueous medium due to the more hydrophilic nature of the chlorophyll molecule, which contains a hydrophilic part, compared with the hydrophobicity displayed by carotenoids (Schoefs, 2002).
The fatty acid content and detailed profile presented here for A. platensis cells are in agreement with other studies (Batista, Gouveia, Bandarra, Franco, & Raymundo, 2013; Bellou et al., 2014), despite the enzymatic treatment. Several authors applied different methodologies to extract and analyse the lipid fraction of A. platensis (Andrich, Zinnai, Nesti, & Venturi, 2006; Mendes, Reis, & Palavra, 2006). In none of the aforementioned studies, the authors took into account the presence of the cell wall. In parallel studies with C. vulgaris microalga, a considerable enzymatic cell wall degradation was reported using cellulases and β‐glucosidases (Cho, Oh, Park, Lee, & Park, 2013), or snailase, lysozyme and cellulose (Zheng et al., 2011) combined. In any case, the enzymatic treatment led to successful lipid extraction efficiency. Herein, our goal was not on whether the Mix led to improved lipid extraction efficiency, but instead, on the release of fatty acids from A. platensis to the extracellular space, through the partial or complete degradation of microalgae cell wall. Most differences were found for MUFA and some n‐6 PUFA, particularly 18:2n‐6 and 18:3n‐6, when A. platensis was incubated with the enzymatic mixture treatment, justifying a higher proportion of these fatty acids in the corresponding supernatant. The increased release of n‐6 PUFA, but not of SFA, in both residue and supernatant fractions, when applying this two‐enzyme constituted mixture, points out to future work due to their potential application in animal feed industry (Bellou et al., 2014).
5. CONCLUSIONS
Herein, we report the disclosure of a novel two‐CAZymes constituted mixture capable of efficiently degrade A. platensis cell wall under a strictly controlled process, thus allowing the release of trapped bioactive compounds with important nutritional value. Our findings set the opportunity to use feed catalysts for monogastric diets incorporated with microalgae, in particular with A. platensis, as feed ingredient.
Herein, we report the disclosure of a novel two‐CAZymes constituted mixture capable of efficiently degrade A. platensis microalga cell wall, thus allowing the release of trapped bioactive compounds with important nutritional value. Our findings set the opportunity to use feed catalysts for monogastric diets incorporated with microalgae, in particular with A. platensis, as feed ingredient. In addition, biotechnological applications, like those associated with biofuel, cosmetics and nutraceutical, are also envisaged. Animal trials are currently in progress to assess: (a) how essential really is α‐amylase, knowing that this enzyme is endogenously produced by monogastrics; (b) how capable this two‐enzyme constituted mixture actually is when using A. platensis microalga as feed ingredient (10%–15% of diet weight).
Supporting information
ACKNOWLEDGEMENTS
This work was supported by Fundação para a Ciência e a Tecnologia (FCT, Lisbon, Portugal; grant PTDC/CVT‐NUT/5931/2014), Portugal2020 (grant 08/SI/3399/2015), CIISA (project UID/CVT/00276/2019), a PhD fellowship to DC (SFRH/BD/126198/2016) and Post‐Doc fellowships to PAL (08/SI/3399/2015) and MSM (SFRH/BPD/97432/2013).
Coelho D, Lopes PA, Cardoso V, et al. A two‐enzyme constituted mixture to improve the degradation of Arthrospira platensis microalga cell wall for monogastric diets. J Anim Physiol Anim Nutr. 2020;104:310–321. 10.1111/jpn.13239
REFERENCES
- Aikawa, S. , Joseph, A. , Yamada, R. , Izumi, Y. , Yamagishi, T. , Matsuda, F. , … Kondo, A. (2013). Direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis processes. Energy and Environment Science, 6, 1844–1849. 10.1039/c3ee40305j [DOI] [Google Scholar]
- Andrich, G. , Zinnai, A. , Nesti, U. , & Venturi, F. (2006). Supercritical fluid extraction of oil from microalga Spirulina (Arthrospira) platensis . Acta Alimentaria, 35, 195–203. 10.1556/AAlim.35.2006.2.6 [DOI] [Google Scholar]
- AOAC (2000). Official methods of analysis, (17th ed .) Arlington, VA: Association of Official Analytical Chemists. [Google Scholar]
- Austic, R. , Mustafa, A. , Jung, B. , Gatrell, S. , & Lei, X. (2013). Potential and limitation of a new defatted diatom microalgal biomass in replacing soybean meal and corn in diets for broiler chickens. Journal of Agricultural and Food Chemistry, 31, 7341–7348. 10.1021/jf401957z [DOI] [PubMed] [Google Scholar]
- Babu, K. , Arulandu, A. , & Sankaran, K. (2018). The structure of DLP12 endolysin exhibiting alternate loop conformation and comparative analysis with other endolysins. Proteins: Structure, Function, and Bioinformatics, 86, 210–217. 10.1002/prot.25428 [DOI] [PubMed] [Google Scholar]
- Batista, A. P. , Gouveia, L. , Bandarra, N. M. , Franco, J. M. , & Raymundo, A. (2013). Comparison of microalgal biomass profiles as novel functional ingredient for food products. Algal Research, 2, 164–173. 10.1016/j.algal.2013.01.004 [DOI] [Google Scholar]
- Baudelet, P. H. , Ricochonb, G. , Lindera, M. , & Munigliaa, L. (2017). A new insight into cell walls of Chlorophyta. Algal Research, 25, 333–371. 10.1016/j.algal.2017.04.008 [DOI] [Google Scholar]
- Bellou, S. , Baeshen, M. N. , Elazzazy, A. M. , Aggeli, D. , Sayegh, F. , & Aggelis, G. (2014). Microalgal lipids biochemistry and biotechnological perspectives. Biotechnology Advances, 32, 1476–1493. 10.1016/j.biotechadv.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry, 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Calder, P. (2012). Mechanisms of action of (n‐3) fatty acids. The Journal of Nutrition, 142, 592–599. 10.3945/jn.111.155259 [DOI] [PubMed] [Google Scholar]
- Cantarel, B. L. , Coutinho, P. M. , Rancurel, C. , Bernard, T. , Lombard, V. , & Henrissat, B. (2009). The Carbohydrate‐Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Research, 37, 233–238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo‐Reyes, J. , Barragán‐Trinidad, M. , & Buitrón, G. (2016). Biological pretreatments of microalgal biomass for gaseous biofuel production and the potential use of rumen microorganisms: A review. Algal Research, 18, 341–351. 10.1016/j.algal.2016.07.004 [DOI] [Google Scholar]
- Chew, K. W. , Yap, J. Y. , Show, P. L. , Suan, N. H. , Juan, J. C. , Ling, T. C. , … Chang, J. S. (2017). Microalgae biorefinery: High value products perspectives. Bioresource Technology, 229, 53–62. 10.1016/j.biortech.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Cho, H. S. , Oh, Y. K. , Park, S. C. , Lee, J. W. , & Park, J. Y. (2013). Effects of enzymatic hydrolysis on lipid extraction from Chlorella vulgaris . Renewable Energy, 54, 156–160. 10.1016/j.renene.2012.08.031 [DOI] [Google Scholar]
- Cisneros, M. , & Rito‐Palomares, M. (2004). A simplified strategy for the release and primary recovery of c‐phycocyanin produced by Spirulina maxima . Chemical and Biochemical Engineering Quarterly, 18, 385–390. 10.15255/CABEQ.2014.550 [DOI] [Google Scholar]
- Coelho, D. , Lopes, P. A. , Cardoso, V. , Ponte, P. , Brás, J. , Madeira, M. S. , … Prates, J. A. M. (2019). Novel combination of feed enzymes to improve the degradation of Chlorella vulgaris recalcitrant cell wall. Scientific Reports, 9, 10.1038/s41598-019-41775-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes, C. M. G. A. , & Gilbert, H. J. (2010). Cellulosomes: Highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annual Review of Biochemistry, 79, 655–681. 10.1146/annurev-biochem-091208-085603 [DOI] [PubMed] [Google Scholar]
- Fontes, C. M. G. A. , Hall, J. , Hirst, B. H. , Hazlewood, G. P. , & Gilbert, H. J. (1995). The resistance of cellulases and xylanases to proteolytic inactivation. Applied Microbiology and Biotechnology, 43, 52–57. 10.1007/BF00170622 [DOI] [PubMed] [Google Scholar]
- Gerken, H. G. , Donohoe, B. , & Knoshaug, E. P. (2013). Enzymatic cell wall degradation of Chlorella vulgaris and other microalgae for biofuels production. Planta, 237, 239–253. 10.1007/s00425-012-1765-0 [DOI] [PubMed] [Google Scholar]
- Heo, Y. M. , Lee, H. , Lee, C. , Kang, J. , Ahn, J. W. , Lee, Y. M. , … Kim, J. J. (2017). An integrative process for obtaining lipids and glucose from Chlorella vulgaris biomass with a single treatment of cell disruption. Algal Research, 27, 286–294. 10.1016/j.algal.2017.09.022 [DOI] [Google Scholar]
- Holman, B. W. B. , & Malau‐Aduli, A. E. O. (2013). Spirulina as a livestock supplement and animal feed. Journal of Animal Physiology and Animal Nutrition, 97, 615–623. 10.1111/j.1439-0396.2012.01328.x [DOI] [PubMed] [Google Scholar]
- Huber, R. , Langworthy, T. A. , König, H. , Thomm, M. , Woese, C. R. , Sleytr, U. B. , & Stetter, K. O. (1986). Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 ºC. Archives of Microbiology, 144, 324–333. 10.1007/BF00409880 [DOI] [Google Scholar]
- Hynstova, V. , Sterbova, D. , Klejdus, B. , Hedbavny, J. , Huska, D. , & Adam, V. (2018). Separation, identification and quantification of carotenoids and chlorophylls in dietary supplements containing Chlorella vulgaris and Spirulina platensis using high performance thin layer chromatography. Journal of Pharmaceutical and Biomedical Analysis, 148, 108–118. 10.1016/j.jpba.2017.09.018 [DOI] [PubMed] [Google Scholar]
- Jiménez, C. , Cossío, B. R. , Labella, D. , & Xavier Niell, F. (2003). The feasibility of industrial production of Spirulina (Arthrospira) in Southern Spain. Aquaculture, 217, 179–190. 10.1016/S0044-8486(02)00118-7 [DOI] [Google Scholar]
- Lang, N. J. (1968). The fine structure of blue‐green algae. Annual Review of Microbiology, 22, 15–46. 10.1146/annurev.mi.22.100168.000311 [DOI] [PubMed] [Google Scholar]
- Leal, B. E. S. , Prado, M. R. , Grzybowski, A. , Tiboni, M. , Koop, H. S. , Scremin, L. B. , … Fontana, J. D. (2017). Potential prebiotic oligosaccharides from aqueous thermopressurized phosphoric acid hydrolysates of microalgae used in treatment of gaseous steakhouse waste. Algal Research, 24, 138–147. 10.1016/j.algal.2017.03.020 [DOI] [Google Scholar]
- Leema, J. M. , Kirubagaran, R. , Vinithkumar, N. V. , Dheenan, P. S. , & Karthikayulu, S. (2010). High value pigment production from Arthrospira (Spirulina) platensis cultured in seawater. Bioresource Technology, 101, 9221–9227. 10.1016/j.biortech.2010.06.120 [DOI] [PubMed] [Google Scholar]
- Liebl, W. , Stemplinger, I. , & Ruile, P. (1997). Properties and gene structure of the Thermotoga maritima alpha‐amylase AmyA, a putative lipoprotein of a hyperthermophilic bacterium. Journal of Bacteriology, 179, 941–948. 10.1128/jb.179.3.941-948.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , & Chen, F. (2016). Biology and Industrial Applications of Chlorella: Advances and Prospects. Advances in Biochemical Engineering and Biotechnology, 153, 1–35. 10.1007/10_2014_286 [DOI] [PubMed] [Google Scholar]
- Lum, K. , Kim, J. , & Lei, X. (2013). Dual potential of microalgae as a sustainable biofuel feedstock and animal feed. Journal of Animal Science Biotechnology, 4, 53 10.1186/2049-1891-4-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupatini, A. L. , Colla, L. M. , Canan, C. , & Colla, E. (2017). Potential application of microalga Spirulina platensis as a protein source. Journal of the Science of Food and Agriculture, 97, 724–732. 10.1002/jsfa.7987 [DOI] [PubMed] [Google Scholar]
- Madeira, M. S. , Cardoso, C. , Lopes, P. A. , Coelho, D. , Afonso, C. , Bandarra, N. M. , & Prates, J. A. M. (2017). Microalgae as feed ingredients for livestock production and meat quality: A review. Livestock Science, 205, 111–121. 10.1016/j.livsci.2017.09.020 [DOI] [Google Scholar]
- Makkar, H. P. S. , Tran, G. , Heuzé, V. , Giger‐Reverdin, S. , Lessire, M. , Lebas, F. , & Ankers, P. (2016). Seaweeds for livestock diets: A review. Animal Feed Science and Technology, 212, 1–17. 10.1016/j.anifeedsci.2015.09.018 [DOI] [Google Scholar]
- Markou, G. , Angelidaki, I. , Nerantzis, E. , & Georgakakis, D. (2013). Bioethanol production by carbohydrate‐enriched biomass of Arthrospira (Spirulina) platensis . Energies, 6, 3937–3950. 10.3390/en6083937 [DOI] [Google Scholar]
- Mehta, K. K. , Evitt, N. H. , & Swartz, J. R. (2015). Chemical lysis of cyanobacteria. Journal of Biological Engineering, 9, 10 10.1186/s13036-015-0007-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes, R. L. , Reis, A. D. , & Palavra, A. F. (2006). Supercritical CO2 extraction of γ‐linolenic acid and other lipids from Arthrospira (Spirulina) maxima: Comparison with organic solvent extraction. Food Chemistry, 99, 57–63. 10.1016/j.foodchem.2005.07.019 [DOI] [Google Scholar]
- Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428. 10.1021/ac60147a030 [DOI] [Google Scholar]
- Ovando, C. A. , de Carvalho, J. C. , Pereira, G. V. M. , Jacques, P. , Soccol, V. T. , & Soccol, C. R. (2018). Functional properties and health benefits of bioactive peptides derived from Spirulina: A review. Food Reviews International, 34, 34–51. 10.1080/87559129.2016.1210632 [DOI] [Google Scholar]
- Palinska, K. A. , & Krumbein, W. E. (2000). Perforation patterns in the peptidoglycan wall of filamentous cyanobacteria. Journal of Phycology, 36, 139–145. 10.1046/j.1529-8817.2000.99040.x [DOI] [Google Scholar]
- Phong, W. N. , Show, P. L. , Ling, T. C. , Juan, J. C. , Ng, E. P. , & Chang, J. S. (2018). Mild cell disruption methods for bio‐functional proteins recovery from microalgae ‐ recent developments and future perspectives. Algal Research, 31, 506–516. 10.1016/j.algal.2017.04.005 [DOI] [Google Scholar]
- Pulz, O. , & Gross, W. (2004). Valuable products from biotechnology of microalgae. Applied Microbiology and Biotechnology, 65, 635–648. 10.1007/s00253-004-1647-x [DOI] [PubMed] [Google Scholar]
- Pyne, S. K. , Bhattacharjee, P. , & Srivastav, P. K. (2017). Microalgae (Spirulina platensis) and its bioactive molecules: Review. Indian Journal of Nutrition, 4, 1–6. [Google Scholar]
- Pyo, D. , Kim, T. , & Yoo, J. (2013). Efficient extraction of bioethanol from freshwater cyanobacteria using supercritical fluid pretreatment. Bulletin of the Korean Chemical Society, 34, 379–383. 10.5012/bkcs.2013.34.2.379 [DOI] [Google Scholar]
- Ravindran, V. , & Son, J. (2011). Feed enzyme technology: Present status and future developments. Recent Patents on Food, Nutrition and Agriculture, 3, 102–109. 10.2174/2212798411103020102 [DOI] [PubMed] [Google Scholar]
- Saez, N. J. , & Vincentelli, R. (2014). High‐throughput expression screening and purification of recombinant proteins in E. coli . Methods in Molecular Biology, 1091, 33–53. 10.1007/978-1-62703-691-7_3 [DOI] [PubMed] [Google Scholar]
- Safi, C. , Frances, C. , Ursu, A. V. , Laroche, C. , Pouzet, C. , Vaca‐Garcia, C. , & Pontalier, P. Y. (2015). Understanding the effect of cell disruption methods on the diffusion of Chlorella vulgaris proteins and pigments in the aqueous phase. Algal Research, 8, 61–68. 10.1016/j.algal.2015.01.002 [DOI] [Google Scholar]
- Safi, C. , Ursu, A. V. , Laroche, C. , Zebib, B. , Merah, O. , Pontalier, P. Y. , & Vaca‐Garcia, C. (2014). Aqueous extraction of proteins from microalgae: Effect of different cell disruption methods. Algal Research, 3, 61–65. 10.1016/j.algal.2013.12.004 [DOI] [Google Scholar]
- Schoefs, B. (2002). Chlorophyll and carotenoid analysis in food products. Properties of the pigments and methods of analysis. Trends in Food Science and Technology, 13, 361–371. 10.1016/S0924-2244(02)00182-6 [DOI] [Google Scholar]
- Scholz, M. J. , Weiss, T. L. , Jinkerson, R. E. , Jing, J. , Roth, R. , Goodenough, U. , … Gerken, H. G. (2014). Ultrastructure and composition of the Nannochloropsis gaditana cell wall. Eukaryotic Cell, 13, 1450–1464. 10.1128/EC.00183-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira, A. F. , Turchetto, J. , Saez, N. J. , Peysson, F. , Ramond, L. , Duhoo, Y. , … Vincentelli, R. (2017). Gene design, fusion technology and TEV cleavage conditions influence the purification of oxidized disulphide‐rich venom peptides in Escherichia coli . Microbial Cell Factory, 16, 4 10.1186/s12934-016-0618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyidoglu, N. , Inan, S. , & Aydin, C. (2017).A prominent superfood: Spirulina platensis In Shiomi N., & Waisundara V. (Eds.), Superfood and functional food‐the development of superfoods and their roles as medicine (pp. 1–28). Rijeka, Croatia: IntechOpen. ISBN: 978‐953‐51‐2942‐4. [Google Scholar]
- Singh, R. , White, D. , & Blum, P. (2017). Identification of the ATPase subunit of the primary maltose transporter in the hyperthermophilic anaerobe Thermotoga maritima . Applied and Environmental Microbiology, 83, e00930–e1017. 10.1128/AEM.00930-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni, R. A. , Sudhakar, K. , & Rana, R. S. (2017). Spirulina‐From growth to nutritional product: A review. Trends in Food Science and Technology, 69, 157–171. 10.1016/j.tifs.2017.09.010 [DOI] [Google Scholar]
- Sotiroudis, T. G. , & Sotiroudis, G. T. (2013). Health aspects of Spirulina (Arthrospira) microalga food supplement. Journal of the Serbian Chemical Society, 78, 395–405. 10.2298/JSC121020152S [DOI] [Google Scholar]
- Srividhya, K. V. , & Krishnaswamy, S. (2007). Sub classification and targeted characterization of prophage‐encoded two‐component cell lysis cassette. Journal of Biosciences, 32, 979–990. 10.1007/s12038-007-0097-x [DOI] [PubMed] [Google Scholar]
- Sun, L. W. , Jiang, W. J. , Sato, H. , Kawachi, M. , & Lu, X. W. (2016). Rapid classification and identification of Microcystis aeruginosa strains using MALDI–TOF ID and polygenetic analysis. PLoS ONE, 11, e0156275 10.1371/journal.pone.0156275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, T. , Pratte, B. S. , Zhong, J. , Goodwin, L. , Copeland, A. , Lucas, S. , … Woyke, T. (2014). Complete genome sequence of Anabaena variabilis ATCC 29413. Standards in Genomic Sciences, 9, 562 10.4056/sigs.3899418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usharani, G. , Saranraj, P. , & Kanchana, D. (2012). Spirulina cultivation: A review. International Journal of Pharmaceutical and Biological Archive, 3, 1327–1341. [Google Scholar]
- Van Eykelenburg, C. , Fuchs, A. , & Schmidt, G. H. (1980). Some theoretical considerations on the in vitro shape of the cross‐walls in Spirulina spp. Journal of Theoretical Biology, 82, 271–282. 10.1016/0022-5193(80)90103-4 [DOI] [PubMed] [Google Scholar]
- Vonshak, A. (1986). Laboratory techniques for the cultivation of microalgae In Richmond A. (Eds.), Handbook of microalgal mass culture (pp. 117–143). Boca Raton, FL: CRC Press. [Google Scholar]
- Yamaguchi, K. (1997). Recent advances in microbial bioscience in Japan, with special reference to utilization of biomass and metabolites: A review. Journal of Applied Phycology, 8, 487–502. 10.1007/BF02186327 [DOI] [Google Scholar]
- Zheng, H. , Yin, J. , Gao, Z. , Huang, H. , Ji, X. , & Dou, C. (2011). Disruption of Chlorella vulgaris cells for the release of biodiesel‐producing lipids: A comparison of grinding, ultrasonication, bead milling, enzymatic lysis, and microwaves. Applied Biochemistry and Biotechnology, 164, 1215–1224. 10.1007/s12010-011-9207-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


