Summary
Seed vigour and early establishment are important factors determining the yield of crops. A wheat nitrate‐inducible NAC transcription factor, TaNAC2, plays a critical role in promoting crop growth and nitrogen use efficiency (NUE), and now its role in seed vigour is revealed. A TaNAC2 regulated gene was identified that is a NRT2‐type nitrate transporter TaNRT2.5 with a key role in seed vigour.
Overexpressing TaNAC2‐5A increases grain nitrate concentration and seed vigour by directly binding to the promoter of TaNRT2.5‐3B and positively regulating its expression. TaNRT2.5 is expressed in developing grain, particularly the embryo and husk.
In Xenopus oocyte assays TaNRT2.5 requires a partner protein TaNAR2.1 to give nitrate transport activity, and the transporter locates to the tonoplast in a tobacco leaf transient expression system. Furthermore, in the root TaNRT2.5 and TaNRT2.1 function in post‐anthesis acquisition of soil nitrate. Overexpression of TaNRT2.5‐3B increases seed vigour, grain nitrate concentration and yield, whereas RNA interference of TaNRT2.5 has the opposite effects.
The TaNAC2‐NRT2.5 module has a key role in regulating grain nitrate accumulation and seed vigour. Both genes can potentially be used to improve grain yield and NUE in wheat.
Keywords: NAC transcriptional factor, NRT2, NUE, seed vigour, wheat
Introduction
Seed vigour is critical for rapid and uniform crop establishment and, ultimately, yield performance. Improving vigour remains a primary objective of the agricultural industry and breeders (Rajjou et al., 2012; Finchsavage & Bassel, 2016). Speed and uniformity of germination are important for defining seed vigour (Finchsavage & Bassel, 2016), and are determined by the growth conditions of the mother plant, genotype and storage conditions (Rajjou et al., 2012; Finchsavage & Bassel, 2016). It has been shown that seeds from the mother plant grown under high nitrogen (N) supply had a faster and higher percentage germination when compared with those grown under low N conditions in Arabidopsis (Alboresi et al., 2010; He et al., 2016), wheat (Naylor, 2010) and rice (Hara & Toriyama, 1998). Exogenously supplied nitrate can stimulate seed germination in many plant species (Hendricks & Taylorson, 1974; Alboresi et al., 2010). The identification of seed‐specific nitrate transporters underlines the importance of nitrate provided by the mother plant to the developing seed for subsequent germination and vigour. Overexpression of a seed‐specific high‐affinity nitrate transporter AtNRT2.7 in Arabidopsis increased seed nitrate concentration and germination, whereas knockout of this gene has the opposite effect (Chopin et al., 2007). The low‐affinity nitrate transporter AtNRT1.6 is involved in delivering nitrate from maternal tissue to the developing embryo in Arabidopsis, and a mutation in the gene reduces nitrate accumulation in mature seeds and increases seed abortion rate (Almagro et al., 2008).
The two hormones, abscisic acid (ABA) and gibberellins (GAs), play important roles in controlling seed dormancy and germination, ABA acts as a positive regulator of dormancy induction and maintenance, but as a negative regulator of germination; whereas GA counteracts ABA effects (Kucera et al., 2005). Both exogenous and endogenous nitrate can reduce ABA concentrations during seed imbibition by upregulating the ABA degradation gene CYP707A2 (Matakiadis et al., 2009; Carrillo‐Barral et al., 2014; Yan et al., 2016), and increasing bioactive GA concentrations by upregulating the biosynthesis of bioactive GA gene GA3ox2, to release seed dormancy and stimulate seed germination (Carrillo‐Barral et al., 2014). Mutation in CYP707A2 impairs nitrate‐promoted germination (Matakiadis et al., 2009; Yan et al., 2016). In a study screening Arabidopsis for mutants defective in the response of germination to nitrate, the transcription factor NIN‐like protein 8 (NLP8) was found to be essential for nitrate‐promoted seed germination (Yan et al., 2016). NLP8 positively regulates CYP707A2 expression, and thus reduces ABA concentrations in seeds. Previous studies found that nitrate could produce nitric oxide (NO), which also stimulates CYP707A2 expression and seed germination (Bethke et al., 2007; Zheng et al., 2009; Arc et al., 2013). However, NLP8 functions in nitrate‐promoted seed germination even in a nitrate reductase (NR)‐deficient mutant background (Yan et al., 2016). As such, nitrate‐promoted germination is triggered directly by nitrate signaling, but not by nitrate‐assimilation and its products. In fact, it has been documented by many authors that nitrate is a major signal in regulating plant growth, and in breaking seed dormancy independent of its reduction by NR (Hilhorst & Karssen, 1989; Matakiadis et al., 2009; Alboresi et al., 2010; Bewley et al., 2013). However, little is known about the regulation pathway for seed nitrate storage that will later trigger germination in crops.
The present authors found previously that the nitrate‐inducible NAC transcription factor TaNAC2‐5A controls nitrate signaling in wheat. TaNAC2 positively regulates expression of nitrate transporters and root nitrate influx rate and is valuable in breeding wheat with improved yield and N use efficiency (He et al., 2015). It is reported herein that TaNAC2‐5A directly regulates expression of the grain nitrate transporter TaNRT2.5‐3B. Overexpression of either TaNAC2‐5A or TaNRT2.5‐3B increases both grain nitrate concentration and seed vigour, whereas knockdown of TaNRT2.5 had the opposite effect. Furthermore, overexpression of TaNRT2.5‐3B improved grain yield and N accumulation in wheat.
Materials and Methods
Plant materials and vector construction
Wheat variety Longchun 23 (LC23) was used as wild‐type (WT) in the present study. TaNAC2‐5A overexpression lines, such as TaOE1 and TaOE2 were reported previously (He et al., 2015). To generate TaNRT2.5‐3B overexpression lines and TaNRT2.5 RNAi lines, TaNRT2.5‐3B CDS was inserted into the pUbi‐163 vector, resulting in the construct pUbi::TaNRT2.5‐3B‐OE; selected consensus sequence of TaNRT2.5 was inserted into the pUbi‐RNAi vector, resulting in the construct pUbi::TaNRT2.5‐3B‐RNAi. The two constructs were transformed into immature embryos of LC23 via the particle bombardment method (Shan et al., 2014). For overexpression, OE102‐6 and OE103‐1 are two independent TaNRT2.5‐3B overexpression lines from five positive selected lines. For RNAi, R100‐1 and R109‐2 are two independent RNAi lines from four positive selected lines. The primers used for vector construction are listed in Supporting Information Table S1.
Wheat growth conditions
The WT, TaNAC2‐5A overexpression lines and TaNRT2.5 transgenic lines were used in the hydroponic culture, soil pot and field experiments. The nutrient solution composition, methods for seed sterilization and germination and growth conditions for the hydroponic experiments were described previously (He et al., 2015). The pot seed germination experiments used a calcareous soil that was collected from the experimental station of the Institute of Genetics and Developmental Biology in Beijing. The soil was wetted to 20% soil water content, and 100 seeds of the WT and the transgenic lines were sowed in each pot at 20°C. Photographs were taken and the shoot length of seedlings were measured at 7 d after sowing.
The field experiment was conducted at the experimental station of the Institute for Cereal and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences, Hebei Province, China. The experiment had a random block design with four replications. The fertilizer supply was 18 g m−2 nitrogen (N) in the form of urea, with 12 g m−2 applied before sowing and 6 g m−2 applied at the stem elongation stage, and 13.5 g m−2 phosphorus (calcium superphosphate) applied before sowing. Total biomass yield per plant, grain yield per plant, spike number per plant, and grain number of the primary spike were recorded for 30 representative plants for each sample group. The 1000‐grain weight was determined according to the DW of 500 dried grains.
Seed germination assays on Petri dishes
Seeds collected at the same time were used for germination assays. The germination assays were performed at least one month after the seeds were harvested. Four independent biological replicates were used, and each replicate included 100 seeds. Seeds were surface‐sterilized with 30% H2O2 for 10 min and washed five times with sterile water, then were sown on 10 × 10 cm Petri dish with filter paper containing 10 ml saturated CaSO4 solution at 20°C. Germination was scored at the indicated time points and scored as positive when the radicle protruded from the seed.
Controlled deterioration test (CDT) and tetrazolium assay
The CDT was performed with some modifications (see Zuo et al., 2018). Briefly, a closed container was used with saturated KCl solution to reach 82% relative humidity. Seeds were equilibrated for 4 d at 25°C in the dark. Thereafter the seeds were artificially aged for 2 d at 82% relative humidity and at 45°C in the dark. Seeds were dried at room temperature for 1 d before the tetrazolium assay.
Tetrazolium assay was performed in three biological repeats as described by Salvi et al. (2016). Wheat seeds were initially sterilized with sodium hypochlorite solution (v/v) and then washed with sterilized distilled water for five times. Sterilized seeds were then incubated in 1% tetrazolium solution in darkness at 30°C for 48 h to stain. The staining patterns were observed under the anatomical microscope. Wheat seed with embryos totally or mostly stained to red were defined as viable seeds. Seed vigour = viable seed number/total seed number × 100%.
Nitrate concentration measurements
Grain nitrate concentrations were determined as described previously (Chopin et al., 2007). The whole wheat seeds or razor‐dissected grains were homogenized into powder and then 1 ml 80% (w/v) ethanol was added into 250 mg wheat seed powder at 4°C and extracted for 2 h. Nitrate content of seeds was analyzed by high‐performance liquid chromatography on an ICS‐5000 analyzer (Thermo Fisher, Waltham, MA, USA).
The nitrate concentrations in leaves or roots of wheat seedlings were measured by the salicylic acid (SA) concentrated sulfuric acid colorimetry method. Samples of 1 g fresh wheat leaves or roots were homogenized, and 10 ml sterile water was added, and then nitrate was extracted for 30 min in a boiling water bath. 0.1 ml extracted solution, 0.4 ml 5% SA‐concentrated sulfuric acid (w/v) solution and 9.5 ml 8% NaOH (w/v) solution were well mixed and OD410 was measured when the solution was cooled to 25°C. the nitrate concentration of the extracted solution was calculated using a standard curve.
Quantitative real‐time (qRT‐)PCR
Total RNA from plant tissues was extracted with the Plant RNeasy Kit (Qiagen). First‐strand complementary DNA was synthesized from 2 μg DNase I‐treated total RNA using the PrimeScript RT Reagent Kit (TaKaRa, Basel, Switzerland) according to the manufacturer’s instructions. The qRT‐PCR analysis was performed with a LightCycler 480 engine (Roche) using Light‐Cycler480 SYBR Green I Master Mix (Roche). The primers used for qRT‐PCR are detailed in Table S2.
Chromatin immunoprecipitation (ChIP) assay
The LC23 wheat seedlings were used for ChIP assays performed according to methods described previously (Bowler et al., 2004). Anti‐TaNAC2 antibody was used to precipitate the DNA–TaNAC2 complexes. The DNA was released with proteinase K and then purified for further PCR analysis. The enrichment of DNA fragments was determined using qRT‐PCR, with the primers detailed in Table S2.
Fusion protein preparation and electrophoretic mobility shift assays (EMSAs)
The full‐length CDS of TaNAC2‐5A were cloned into the pGEX‐6p‐1 vector. The recombinant GST‐fusion proteins were expressed in Escherichia coli BL21 (Transseta) and purified to homogeneity using GE sepharose 4B beads (GE, Boston, MA, USA). Oligonucleotide probes were synthesized and labeled with biotin at their 5’ ends (Invitrogen). EMSAs were performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA, USA). The primers used for vector construction are listed in Table S1.
Assay of nitrate transporter activity in Xenopus laevis oocytes
The CDSs of TaNRT2.5‐3A/B/D, TaNAR2.1‐6B, TaNAR2.2‐5B and TaNAR2.3‐6B were amplified and cloned into the oocyte expression vector pT7Ts between the restriction sites SpeI and BglII and then linearized with EcoRI. Capped mRNA was synthesized in vitro using the mMESSAGE mMACHINE kit (Ambion, AM1340) according to the manufacturer’s protocol. Xenopus laevis oocytes at stage V–VI were injected with 50 ng cRNA per oocyte. After injection, oocytes were cultured in MBS medium for 48 h and then used for 15N‐NaNO3‐uptake assays; 500 µM 15N–NaNO3 was used for the uptake assays, as described previously (Tong et al., 2005). The primers used for vector construction are listed in Table S1.
Subcellular localization and Western blot
The CDSs of TaNRT2.5‐3B and TaNAR2.1‐6B were fused in frame with green and red fluorescent proteins (GFP and RFP) via cloning into the binary vector pMDC83‐CaMV35S‐GFP and pJAH2044‐CaMV35S‐RFP. The resulting vectors were transformed into Agrobacterium strain GV3101 as GV3101‐TaNRT2.5‐3B and GV3101‐TaNAR2.1‐6B. Then GV3101‐TaNRT2.5‐3B and GV3101‐TaNAR2.1‐6B were co‐infiltrated or separately infiltrated into tobacco leaf epidermal cell. The GFP and RFP image was observed with a confocal microscope (Zeiss LSM 710 NLO). The primers used for vector construction are listed in Table S1. Eight gram infiltrated tobacco leaves of different combined GV3101 were used for Western blot assays. The membrane protein fractionation and Western blot assay methods were as described previously (Ueno et al., 2010).
15N‐nitrate influx, uptake rate and accumulation after anthesis assay
15N‐nitrate influx and uptake rate were determined using a 15N‐KNO3 assay. The 15N content of wheat seedling root and shoot was determined after 15 min or 12 h of uptake of 15N‐KNO3. 15N‐nitrate influx and uptake rate were calculated as the amount of 15N taken up per unit weight of roots per unit time. 15N‐nitrate influx and uptake rate assays were performed using 200 µM and 2 mM 15N‐KNO3, respectively. Roots and shoots were collected and dried at 70°C. Samples were ground and the 15N content was determined using an isotope ratio mass spectrometer with an elemental analyzer (Isoprime 100‐EA, Manchester, UK). To determine the 15N‐nitrate accumulation after anthesis, the field experiment was fertilized with 15N‐KNO3. The 15N‐nitrate accumulation was calculated as the amount of 15N taken up per plant in grain and straw. The N accumulation after anthesis from fertilizer also was determined as the ratio of the amounts of 15N to total N in grain and whole plant, and the 15N content was determined as described above.
Statistical analysis of data
Statistically significant differences using Spss 17.0 for Windows (Spss, Armonk, NY, USA) were computed based on Student’s t‐tests.
Results
Overexpressing TaNAC2‐5A increases grain nitrate concentration and seed vigour
The present authors’ previous study showed that the NAC transcription factor TaNAC2‐5A is nitrate inducible and overexpression of TaNAC2‐5A increases root nitrate influx rate, N uptake and grain yield in wheat (He et al., 2015). It was observed herein that overexpression of TaNAC2‐5A increased the speed and rate of germination in Petri dishes and also the seed vigour in the CDT experiment (Figs 1a, S1a). When the seeds were germinated in soil, the overexpression lines had significantly fewer seedlings with < 2 cm shoots when compared with WT (Figs 1c, d, S1b), indicating the transgenic lines grew more rapidly and uniformly established seedlings. As seed nitrate is a signal promoting germination and TaNAC2‐5A is nitrate‐responsive, the nitrate concentrations were measured and compared in the grains of the TaNAC2‐5A overexpression lines and WT. Grain nitrate concentration was increased by c. 71% in TaNAC2‐5A overexpression lines compared with WT (Fig. 1b), but overexpression only slightly increased total N concentration (He et al., 2015).
Figure 1.
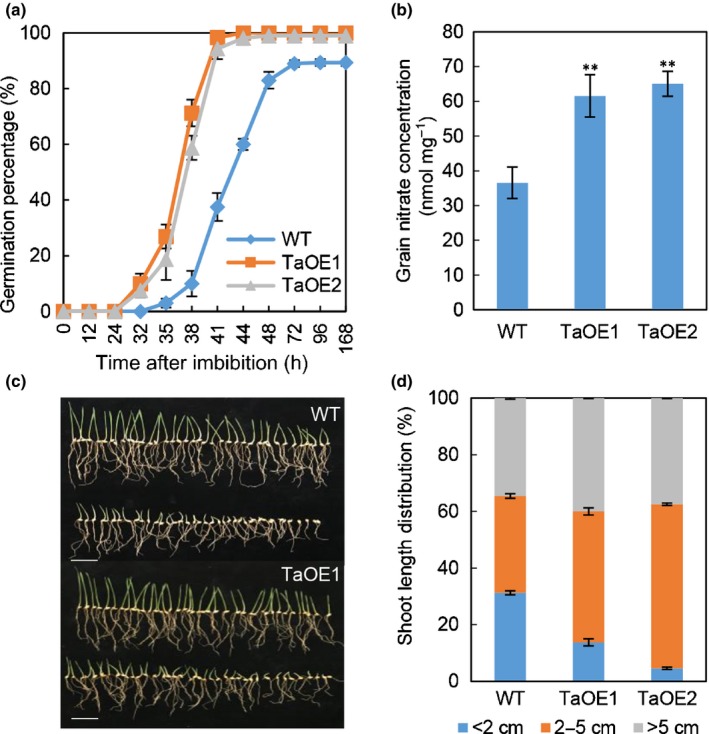
Overexpression of TaNAC2‐5A increases seed vigour and grain nitrate concentration. (a) Germination percentage of TaNAC2‐5A overexpression lines and wild‐type (WT) in Petri dishes. Values are means ± SE (n = 4). **, P < 0.01 (Student’s t‐test). (b) Grain nitrate concentration measured using the dry seeds. Values are means ± SE (n = 4). **, P < 0.01 (Student’s t‐test). (c) Seedling images of germination assay in pot experiment. Bars, 5 cm. (d) Shoot length (cm) distribution of (c). Values are means ± SE (n = 4).
Arabidopsis seeds from plants grown under high N supply had higher nitrate concentrations and germination rates than those from plants grown under low N conditions (Chopin et al., 2007; Rajjou et al., 2012). This also was observed in the wheat variety LC23 (WT for the transgenic lines) as the seeds harvested from high N treatment had a three‐fold higher nitrate concentration and 20% higher germination than those from low N treatment (Fig. S2a, b). Seed germination was assayed in the mini‐core collections of Chinese wheat varieties (Wang et al., 2012) and the three fastest germinating varieties were found to have much higher grain nitrate concentrations (Fig. S2c, d), and higher TaNAC2 expression in the germinating seeds when compared with the three slower ones (Fig. S2e).
Taken together, these results show that wheat grain nitrate concentration impacts on germination and overexpression of TaNAC2‐5A increased grain nitrate concentration resulting in rapid and uniform germination.
Identifying the downstream genes of TaNAC2
It is reported that TaNRT2.1‐6B, TaNPF7.1‐6D and TaGS2‐2A are directly and positively regulated by TaNAC2 (He et al., 2015), but these genes were not co‐expressed with TaNAC2 in the developing seeds during the grain‐filling stage herein (Fig. S3a). Other unknown downstream genes of TaNAC2 may be involved in delivering nitrate to the grain. To understand the mechanism of TaNAC2 control of grain nitrate concentration and germination, Chromatin Immunoprecipitation Sequencing (ChIP‐Seq) was performed using whole plants 2 d after germination. In total, ChIP‐Seq identified 61 candidate genes, including TaNRT2.5‐3B (Table S3). Further ChIP‐qPCR analysis showed that TaNAC2 could bind to the promoter of TaNRT2.5‐3B (Fig. 2a), TaLAX1‐3B and ‐1D, and TaARR12‐6A (Fig. S4). TaLAX1‐3B and ‐1D were predicted to encode auxin influx transporters, and TaARR12‐6B is a homologue of AtARR10/12 involving the cytokinin signaling. Among these newly identified downstream genes of TaNAC2‐5A, TaNRT2.5 was mainly expressed in the maturing seed (Fig. S3b–h) and was predicted to encode a putative high‐affinity NRT2‐type nitrate transporter. A more detailed analysis was then performed to determine whether TaNAC2 increased grain nitrate concentration and germination by regulating TaNRT2.5.
Figure 2.
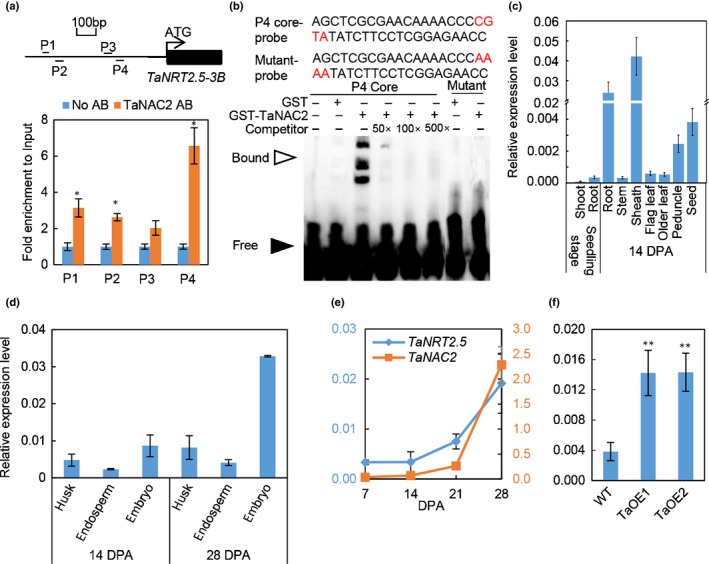
TaNAC2‐5A binds to the promoter of TaNRT2.5‐3B and regulates the expression. (a) Chromatin immunoprecipitation quantitative PCR (ChIP–qPCR) assay of TaNAC2‐5A binding to TaNRT2.5‐3B promoter. P1–P4 are fragments selected in TaNRT2.5‐3B promoter for ChIP–qPCR analysis. (b) Electrophoretic mobility shift assay (EMSA) of TaNAC2‐5A binding to P4 fragment from (a). (c) The expression pattern of TaNRT2.5 in different organs of wheat at seedling stage and 14 d post‐anthesis (DPA). (d) The expression pattern of TaNRT2.5 in different parts of wheat seeds in 14 and 28 DPA. (e) The expression pattern of TaNAC2 and TaNRT2.5 in developing seeds at 7, 14, 21 and 28 DPA. (f) The expression level of TaNRT2.5 in seeds of TaNAC2‐5A overexpression lines and wild‐type (WT). Significance for the difference between the means of the transgenic lines and WT: **, P < 0.01 (Student’s t‐test). TaActin was used as an internal reference. Data represented as means ± SE (n = 4).
TaNRT2.5 works downstream of TaNAC2
Phylogenetic analysis of NRT2s from wheat, rice, maize, barley, soybean and Arabidopsis showed that the NRT2 proteins appeared to be clustered into two main monophyletic groups, a larger Group A and a smaller Group B which contained TaNRT2.5s (Fig. S5).
A DNA EMSA was conducted to confirm the result in ChIP‐qPCR (Fig. 2a). As shown in Fig. 2(b), the GST‐TaNAC2 fusion proteins could bind biotin‐labeled DNA probes in TaNRT2.5‐3B promoter (proTaNRT2.5‐3B). Moreover, the addition of an excess amount of unlabeled proTaNRT2.5‐3B probe effectively reduced the binding of GST‐TaNAC2 to labeled DNA probe. The parallel experiment indicated that GST‐TaNAC2 was unable to bind a mutant form of the biotin‐labeled proTaNRT2.5‐3B probe (Fig. 2b). These results indicated that TaNAC2‐5A could bind to proTaNRT2.5‐3B in vivo and in vitro.
Next the tissue‐specific transcript of TaNRT2.5 was checked, revealing that there was higher expression in roots, leaf sheath, peduncle and developing seeds in wheat plants at 14 d post‐anthesis (DPA) (Fig. 2c). Measurements of the transcript level of TaNRT2.5 in dissected grains at 14 and 28 DPA showed that TaNRT2.5 was expressed mainly in the husk and embryo (Fig. 2d), which is similar to the nitrate distribution (Fig. S6a). The expression of both TaNAC2 and TaNRT2.5 in seeds also was found to be increased during grain filling (Fig. 2e), and overexpression of TaNAC2‐5A greatly upregulated TaNRT2.5 expression in seeds compared to WT (Fig. 2f). A hydroponic culture of seedlings was used to show that the root expression of TaNRT2.5 was nitrate inducible, and upregulated by low N availability (Fig. S7a, b), as has been shown for TaNAC2 (He et al., 2015). However, the response of TaNAC2 to nitrate induction was earlier than that of TaNRT2.5 (Fig. S7a). Taken together, TaNAC2 and TaNRT2.5 are co‐expressed in developing seeds, and TaNAC2 directly and positively regulates TaNRT2.5 expression. No significant differences in the expression patterns of TaNAC2 and TaNRT2.5 homeologs could be detected in the gene atlas (see Fig. S8).
TaNRT2.5‐TaNAR2.1 has nitrate transport activity and locates mainly on tonoplast
Sequence analysis predicts TaNRT2.5‐3B to be a high‐affinity nitrate transporter. As some NRT2 members require a partner protein NAR2, for nitrate transport at relatively low concentration ranges (Tong et al., 2005; Orsel et al., 2006; Feng et al., 2011), a search was carried out for TaNAR2s from the wheat genome database (http://plantSEnsembl.org/Triticum_aestivum/Info/Index) which were named according to their relationship to HvNAR2s in barley (Tong et al., 2005). Phylogenetic analysis showed that the NAR2 proteins clustered into three clades which included TaNAR2.1s, TaNAR2.2s and TaNAR2.3s, respectively (Fig. S7c).
The nitrate transport activity of TaNRT2.5 expressed in Xenopus laevis oocytes was tested by injecting oocytes with nuclease‐free water, TaNRT2.5‐3B cRNA alone and TaNRT2.5‐3B cRNA together with three different TaNAR2s cRNAs. Figure 3(a) shows that only oocytes co‐injected with TaNRT2.5‐3B and TaNAR2.1‐6B in combination accumulated significantly more 15N‐nitrate than the control oocytes. Further analysis revealed that all the homeolog genes of TaNRT2.5s (TaNRT2.5‐3A, ‐3B and ‐3D) required TaNAR2.1‐6B for nitrate transport activity. The NRT2 transport mechanism is generally thought to be cotransport with protons and significantly higher nitrate transport activity was measured in oocytes injected with TaNRT2.5‐3B/TaNAR2.1‐6B cRNAs incubated at pH 5.5 when compared with pH 7.5 (Fig. 3a). The predicted protein sequences of the three genes were very similar (Fig. S7e). Interestingly, among the TaNAR2s only the transcript of TaNAR2.1 was detected in seeds and mainly in the embryo (Fig. S7d).
Figure 3.
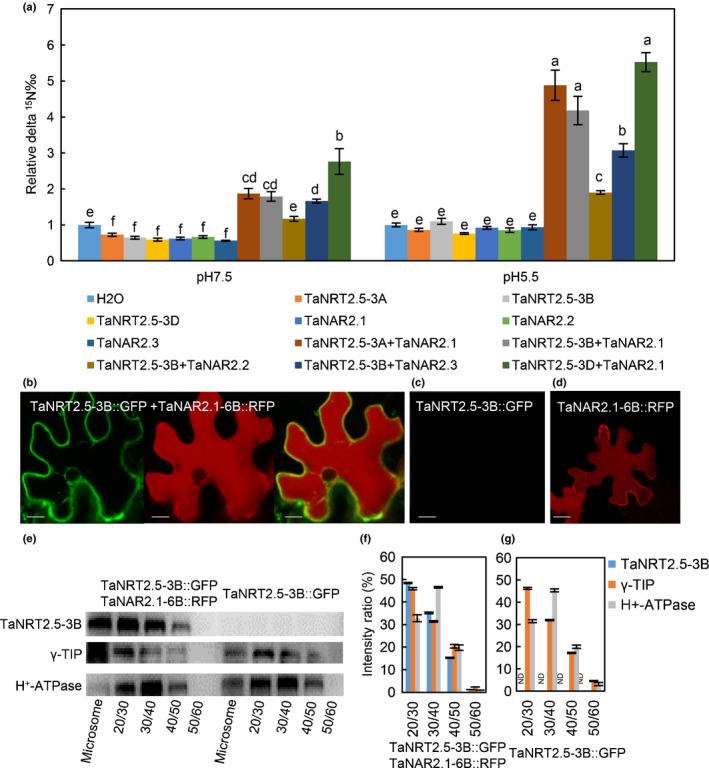
The nitrate transport activity and subcellular localization of TaNRT2.5. (a) The nitrate transport activity of different combinations of TaNRT2.5s and TaNAR2s in pH 7.5 and 5.5 relative to water‐injected controls. Uptake from 500 µM of 15N labelled nitrate in 6 h was used for Xenopus oocytes which were injected with water or different cRNA combinations. The delta‐15N values are shown as the mean ± SE for four oocytes. Different letters above the columns indicate statistically significant differences at the P < 0.05 level according to one‐way ANOVA. (b–d) Fluorescence of green fluorescent protein (GFP) in tobacco leaf epidermal cells expressing TaNRT2.5‐3B::GFP coupled with TaNAR2.1‐6B::RFP (b), TaNRT2.5‐3B::GFP alone (c) or TaNAR2.1‐6B::RFP (d) (Bars, 20 μm). (b) Left, TaNRT2.5‐3B::GFP signal; middle, TaNAR2.1‐6B::RFP signal; right, overlap of these two signals. (c) TaNRT2.5‐3B::GFP signal. (d) TaNAR2.1‐6B::RFP signal. (e–g) Microsome from whole leaf of expressed TaNRT2.5‐3B::GFP coupled with TaNAR2.1‐6B::RFP or TaNRT2.5‐3B::GFP alone were extracted and fractionated by sucrose‐density gradient centrifugation, then the different fractions were used for Western blot. Antibodies of anti‐GFP, anti‐γ‐TIP (tonoplast marker), and anti‐H+‐ATPase (plasma membrane marker) were used (e). Relative band intensity was measured using imagej software (NIH, Bethesda, MD, USA). The ratio of different band intensity to total intensity was calculated, and three independent experiments were performed. Data represented as means ± SE (n = 3) (f, g).
In order to investigate the subcellular localization of TaNRT2.5‐3B, the tobacco (Nicotiana benthamiana) leaf transient expression system was used. When TaNRT2.5‐3B::GFP and TaNAR2.1‐6B::mRFP were co‐infiltrated, a strong GFP signal was detected which seemed to localize to the tonoplast and on membranes around the nucleus (Fig. 3b). When TaNRT2.5‐3B::GFP was expressed alone, the GFP signal could not be detected under the same microscopy conditions (Fig. 3c). To confirm the tonoplast location of the TaNRT2.5‐3B and TaNAR2.1‐6B complex, membrane proteins were extracted and separated into different fractions using sucrose‐density gradient centrifugation. Western blot analysis showed that TaNRT2.5‐3B::GFP was present mainly in the same fraction as the anti‐γ‐TIP (a tonoplast marker), but in different fractions from the anti‐H+‐ATPase (a plasma membrane marker), when the tobacco leaves were co‐infiltrated with TaNRT2.5‐3B::GFP and TaNAR2.1‐6B::mRFP (Fig. 3e, f). By contrast, when TaNRT2.5‐3B‐GFP was expressed alone, the TaNRT2.5‐3B::GFP was not detected in any of the fractions (Fig. 3e, g). Taken together, these data suggest that TaNRT2.5‐3B was localized to the tonoplast when co‐expressed with TaNAR2.1‐6B, but TaNRT2.5‐3B alone seemed unstable.
TaNRT2.5 positively affects seed vigour and grain nitrate concentration
Because TaNAC2‐5A affected seed vigour and directly regulated TaNRT2.5, it was tested whether TaNRT2.5 could affect germination and grain nitrate concentration. Transcript analysis of TaNRT2.5 confirmed an increase in the TaNRT2.5‐3B overexpression lines and knock down in the TaNRT2.5 RNAi lines (Fig. 4d). Petri dish germination assays and the CDT both showed that the TaNRT2.5‐3B overexpression lines germinated earlier and finally achieved a higher germination percentage than WT seeds, whereas TaNRT2.5 RNAi lines had the opposite phenotype (Figs 4a, S9a, b). Overexpressing TaNRT2.5‐3B also increased germination, whereas knock‐down of TaNRT2.5 reduced seed vigour when germinated in soil (Fig. S9c–h). The nitrate concentration was measured in the seed used in the germination assays, revealing that the TaNRT2.5‐3B overexpression lines had significantly higher, and the RNAi lines lower grain nitrate concentrations compared with WT (Fig. 4b). The nitrate concentration was increased in the dissected parts of TaNRT2.5 transgenic lines by overexpression, but was decreased in the husk and embyro of the TaNRT2.5 RNAi lines (Fig. S6b), probably caused by the promoter expression patterns of TaNRT2.5. However, there was no significant difference in total grain N concentration between WT and the transgenic lines (Fig. 4c).
Figure 4.
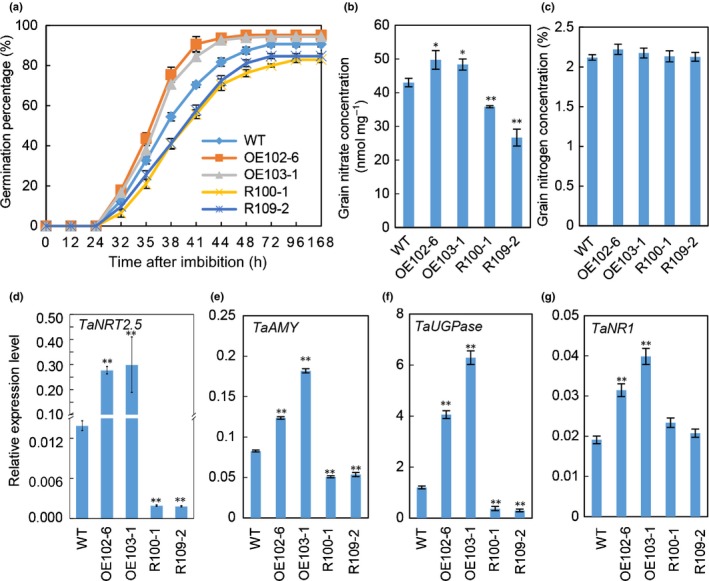
Overexpression of TaNRT2.5‐3B increases seed germination, grain nitrate concentration and modifies the expression of some associated genes. (a) Germination percentage of TaNRT2.5 transgenic lines and wild‐type (WT). (b) Grain nitrate concentration of TaNRT2.5 transgenic lines and WT. (c) Grain nitrogen concentration of TaNRT2.5 transgenic lines and WT. (d–g) Expression levels of TaNRT2.5 (d), TaAMY (e), TaUGPase (f) and TaNR1 (g) in the germinating seeds (60 h after sowing). OE102‐6 and OE103‐1, overexpression lines; R100‐1 and R109‐2, RNAi lines. TaActin was used as an internal reference. Data represented as means ± SE (n = 4). Significance for the difference between the means of the transgenic lines and WT: *, P < 0.05; **, P < 0.01 (Student’s t‐test).
In order to better understand how altering TaNRT2.5 expression affected germination, the expression of TaAMY encoding a starch‐degrading α‐amylase, TaUPGase encoding a UDP‐glucose pyrophosphorylase, and TaNR1 encoding a nitrate reductase in germinating seeds were measured 60 h after sowing. When compared with WT, overexpressing TaNRT2.5‐3B greatly increased expression of these three genes; however, knockdown of TaNRT2.5 dramatically reduced expression of TaAMY and TaUPGase, but did not significantly alter TaNR1 expression (Fig. 4d–g).
Overexpressing TaNRT2.5‐3B promotes seedling growth and N uptake under hydroponic conditions
As TaNRT2.5 mRNA was expressed at very low levels in wheat seedlings (Fig. 2c), only overexpression lines were used to investigate the effects of TaNRT2.5‐3B on N uptake and seedling growth (see Fig. 5a). Compared with WT, the overexpression lines had significantly higher root and shoot FW under both high and low N supply (Fig. 5b, c). The overexpression lines also displayed longer average primary root (PR) length under low N conditions (Fig. 5d), longer total lateral root (LR) length (Fig. 5e) under both high and low N conditions.
Figure 5.
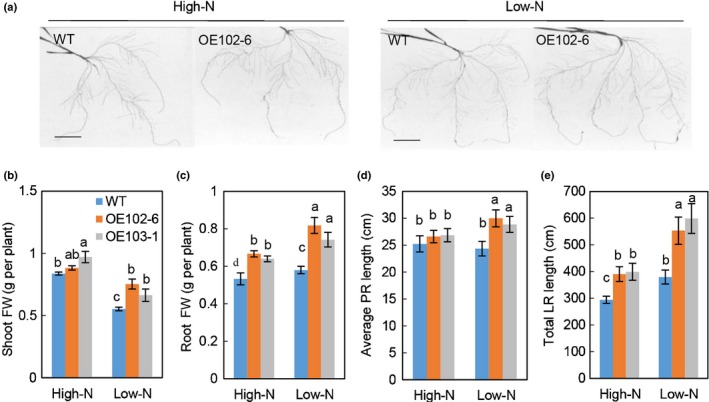
Overexpression of TaNRT2.5‐3B promotes wheat seedling growth. (a) Root images of TaNRT2.5‐3B overexpression lines (OE102‐6 and OE103‐1) and wild‐type (WT) grown under high and low nitrogen (N) supply conditions using a hydroponic culture system. Bars, 5 cm. (b) Shoot FW; (c) Root FW, (d) Average primary root (PR) length; (e) Total lateral root (LR) length. Data represented as means ± SE (n = 4). Different letters above the columns indicate statistically significant differences at the P < 0.05 level according to one‐way ANOVA.
Next the effects of overexpressing TaNRT2.5‐3B on nitrate influx were investigated, revelaing no difference in nitrate influx rate between TaNRT2.5‐3B overexpression lines and WT when nitrate influx rates were measured with 15N‐nitrate at 2 and 0.2 mM for 5 min (Fig. S10). But after the plants were exposed to 15N‐nitrate for 12 h, higher net nitrate uptake rates were detected in the TaNRT2.5‐3B overexpression lines relative to WT under both high and low nitrate supply (Fig. 6). These increases were more obvious under low nitrate supply (see Fig. 6a). Measurements of plant tissue nitrate revealed that the TaNRT2.5‐3B overexpression lines had significantly higher root nitrate concentrations under low nitrate supply and in shoots under high N supply (Fig. 6b, c). The overexpression lines had greater total N uptake under both high and low N conditions (Fig. 6f), but root and shoot N concentrations were not very different (Fig. 6d, e).
Figure 6.
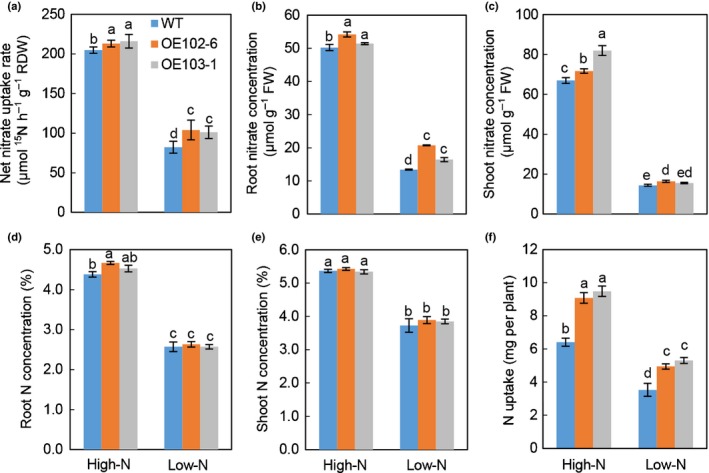
Overexpression of TaNRT2.5‐3B promotes nitrate uptake and accumulation at the seedling stage. The seedlings of TaNRT2.5‐3B overexpression lines (OE102‐6 and OE103‐1) and wild‐type (WT) grown under high and low nitrogen (N) supply conditions using a hydroponic culture system. (a) Net nitrate uptake rate. (b) Root nitrate concentration. (c) Shoot nitrate concentration. (d) Root N concentration. (e) Shoot N concentration. (f) N uptake. Data represented as means ± SE (n = 4). Different letters above the columns indicate statistically significant differences at the P < 0.05 level by one‐way ANOVA.
Overexpressing TaNRT2.5‐3B increases N uptake and yield under field conditions
A field experiment was carried out to investigate the effects of manipulating TaNRT2.5 expression on N use and yield performance. The data in Table 1 showed that TaNRT2.5‐3B overexpression lines had 16.9–17.9% (P < 0.05) higher biomass yield per plant, 19.4–21.3% (P < 0.05) higher grain yield per plant, 13.5–15.9% (P < 0.05) higher spike number per plant, 17.8–21.3% (P < 0.05) higher aerial N accumulation than WT, whereas the RNAi lines had the opposite phenotype. There were no significant differences in grain number per spike, 1000‐grain weight, grain and straw N concentrations between the transgenic lines and WT. These results indicated that manipulating TaNRT2.5 expression altered grain yield mainly by altering spike number and altered aerial N accumulation mainly by altering wheat growth, but not by changing N concentrations in the aerial parts.
Table 1.
Agronomic traits and nitrogen (N) accumulation of the TaNRT2.5 overexpression and RNAi lines and wild‐type (WT) in the field experiment.
| Trait | WT | OE102‐6 | OE103‐1 | R100‐1 | R109‐2 |
|---|---|---|---|---|---|
| Biomass (g per plant) | 36.58 ± 3.32 | 42.78 ± 5.49* | 43.11 ± 4.49* | 32.34 ± 3.75* | 32.65 ± 2.56* |
| Grain yield (g per plant) | 12.98 ± 1.66 | 15.75 ± 1.24* | 15.50 ± 1.02* | 10.94 ± 1.58* | 10.61 ± 1.71* |
| Spike number | 11.96 ± 1.58 | 13.58 ± 1.88* | 13.86 ± 2.17* | 9.90 ± 1.56* | 10.10 ± 2.46* |
| Grain number per main spike | 65.17 ± 1.41 | 64.58 ± 1.88 | 67.22 ± 1.92 | 58.50 ± 1.88* | 62.43 ± 1.41 |
| 1000‐grain weight (g) | 41.83 ± 0.38 | 42.95 ± 1.76 | 41.33 ± 0.89 | 41.04 ± 0.91 | 41.16 ± 0.66 |
| Grain N concentration (%) | 2.12 ± 0.04 | 2.22 ± 0.07 | 2.18 ± 0.06 | 2.13 ± 0.07 | 2.13 ± 0.06 |
| Straw N concentration (%) | 0.59 ± 0.09 | 0.60 ± 0.04 | 0.59 ± 0.03 | 0.58 ± 0.03 | 0.59 ± 0.03 |
| Aerial N accumulation (mg per plant) | 406.5 ± 41.0 | 492.9 ± 34.2* | 478.7 ± 33.5* | 347.3 ± 24.0* | 346.2 ± 18.2* |
OE102‐6 and OE103‐1, overexpression lines; R100‐1 and R109‐2, RNAi lines. Data were means ± SE of four biological replicates. Each replicate contained ≥ 10 plants. Asterisks indicate statistically significant difference between WT and transgenic line: *, P < 0.05 (Student’s t‐test).
It was observed that the expression of TaNRT2.5 in roots was much higher at the grain filling stage when compared with the seedling stage (Figs 2c, 7a), and TaNRT2.5 and the root‐specific TaNRT2.1 had comparable mRNA abundance in roots at the grain filling stage. Hence, it was questioned whether manipulating TaNRT2.5 expression affects N uptake and distribution after anthesis. After analyzing the 15N–N and total N accumulated in straw and grain of the physiologically matured plants which were treated with 15N‐nitrate in the soil at anthesis, the TaNRT2.5‐3B overexpression lines were detected to have significantly higher ratios of 15N–N over the total N accumulated in aerial parts relative to WT (Fig. 7b). Likewise, the 15N–N over total N accumulated in grains (Fig. 7c) compared with WT was significantly higher in overexpression plants, whereas the RNAi lines showed the opposite phenotype. These results suggest that TaNRT2.5 may be involved in late root N uptake and allocation to the grain after anthesis.
Figure 7.
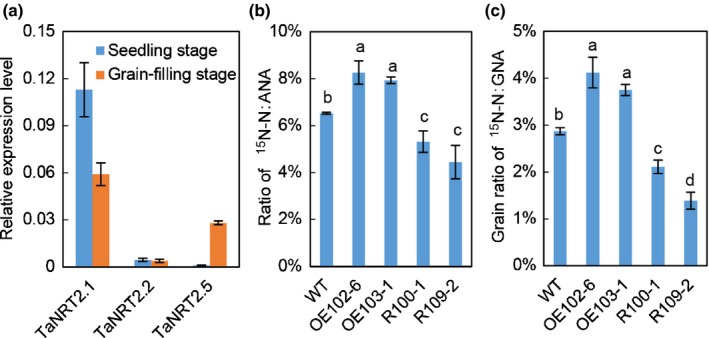
Overexpression of TaNRT2.5‐3B increases the contribution of post‐anthesis N uptake to N accumulated in aerial parts and grains. (a) The expression levels of different TaNRT2 family genes in roots of wild‐type (WT) seedling and the plants 14 d post‐anthesis (DPA). TaActin was used as an internal reference. (b, c) Ratio of 15N‐nitrogen (N) accumulation to ANA (b) and to GNA (c) of the TaNRT2.5 overexpression (OE102‐6 and OE103‐1) and RNAi (R100‐1 and R109‐2) lines and WT. ANA, aerial N accumulation; GNA, grain N accumulation. Data represented as means ± SE (n = 4). Different letters above the columns indicate statistically significant differences at the P < 0.05 level according to one‐way ANOVA.
Discussion
Good germination and subsequent rapid seedling establishment are key features for the successful propagation of plants. We showed that TaNAC2‐5A regulated grain nitrate concentration and germination. Furthermore, TaNAC2 mRNA levels showed a positive correlation (r 2 = 0.71) with grain nitrate concentration and germination in diverse wheat varieties (Fig. S2). In addition, overexpressing TaNAC2‐5A promotes seed germination, and increases grain nitrate concentration (Fig. 1). During germination, seedlings are particularly sensitive and may easily suffer during adverse environmental conditions. Overexpressing TaNAC2 in Arabidopsis has been shown to improve tolerance to drought and salt stress (Mao et al., 2012; Huang & Wang, 2016) and so enhancing TaNAC2 expression level by traditional breeding or transgenic methods may help wheat to resist these stresses and increase germination rate and improve yield. Nitrate may regulate seed germination by regulating the bioactive abscisic acid (ABA) and gibberellin (GA) content (Kucera et al., 2005; Matakiadis et al., 2009; Carrillo‐Barral et al., 2014). TaNAC2 is inducible by nitrate, as well as by ABA (Mao et al., 2012; Zhang et al., 2018) and the protein can bind to the promoters of downstream genes which were involved in nitrate transport and assimilation (He et al., 2015), phytohormone transport and some other transcription factors (TFs) (Fig. S4). TaNAC2 may be involved in the cross‐talk between nitrate and phytohormones in the processes of germination and early seedling establishment.
Multiple lines of molecular and biochemical evidence show that TaNAC2‐5A can bind the promoter of nitrate transporter TaNRT2.5‐3B and promote expression of TaNRT2.5 (Fig. 2). The biochemical data were nicely in agreement with genetic findings. Overexpressing TaNRT2.5‐3B promoted germination and increased grain nitrate concentration, but not the total seed nitrogen (N) (Fig. 4), and enhanced spikes and grain yield (Table 1). The NRT2 family size is different in each plant species (Miller et al., 2007; Pellizzaro et al., 2015; Bajgain et al., 2018; Fig. S5), and some NRT2s are capable of nitrate uptake only when co‐expressed with a partner NAR2 in Arabidopsis (Orsel et al., 2006; Kotur & Glass, 2015; O'Brien et al., 2016), barley (Tong et al., 2005), rice (Yan et al., 2011), wheat (Taulemesse et al., 2015) and chrysanthemum (Gu et al., 2016). TaNRT2.5 specifically interacted with TaNAR2.1 to transport nitrate in Xenopus oocytes, but not with two alternative TaNAR2s. In addition, like AtNRT2.5 (a closely related NRT2; see Fig. S5) TaNRT2.5 does not occur only in root membranes as a 150 kDa molecular complex with AtNAR2.1 in vivo (Kotur & Glass, 2015), the TaNRT2.5‐3B protein was unstable when expressed without TaNAR2.1 in the tobacco leaf system (Fig. 3), so TaNAR2.1 may be involved in TaNRT2.5 protein turnover. But unlike AtNRT2.5 (Lezhneva et al., 2015), overexpressing TaNRT2.5‐3B did not increase the 5‐min nitrate influx rate and TaNRT2.5‐3B seemed to locate to the tonoplast in the tobacco leaf system (Fig. 3). This shows that TaNRT2.5‐3B plays a role in nitrate transport to the vacuole and effects intracellular nitrate distribution, therefore having an indirect function in nitrate acquisition from the soil. Increasing the expression of TaNRT2.1 and TaNR1 may result in increased 12 h net nitrate uptake rate and nitrate concentration in seedlings of TaNRT2.5‐3B overexpression lines (Figs 6a, S10). Expression analysis of the TaNAR2s in these overexpressing lines showed that TaNAR2.1 and TaNAR2.2 had more transcript, but TaNAR2.3 did not (Fig. S11). This suggests that, perhaps like Arabidopsis AtNRT2.1, the TaNRT2.1 protein may be regulated by the levels of TaNAR2.1 and TaNAR2.2, but not TaNAR2.3. Additionally, overexpressing TaNRT2.5‐3B increases post‐anthesis nitrate uptake, resulting in more N accumulated in the aerial parts of the whole plant, including the grain (Fig. 7). In a wheat crop, the late root acquisition of N is known to be important for grain N concentration and quality. Although TaNRT2.1, another nitrate transporter, also seems to play a major role in nitrate uptake after anthesis (Taulemesse et al., 2015), the present work suggests that the expression of TaNRT2.1 in root was slightly enhanced in overexpression lines (Fig. S12a) in high N, and the expression of TaNRT2.5 in the root increased after anthesis (Fig. 7a). The transcript for nitrate reductase (TaNR1) only increased at high N in the overexpression lines (Fig. S12). These results suggested that TaNRT2.5 might cooperate with TaNRT2.1 to play a role in nitrate uptake after anthesis. The present authors’ previous research has already shown that TaNAC2 binds to the promoter of TaNRT2.1‐6B and TaNPF7.1‐6D and enhances expression of the latter two genes (He et al., 2015). TaNRT2.1 is root‐specifically expressed and may contribute to nitrate uptake by wheat plants (He et al., 2015). TaNPF7.1 has been suggested to participate in root nitrate xylem loading and nitrate translocation in shoots (Buchner & Hawkesford, 2014). Taking this information together, TaNAC2 controls nitrate transport from the soil to grain by regulating TaNRT2.1, TaNPF7.1 and TaNRT2.5.
Plants require a large amount of energy during seed germination, so carbohydrate metabolism is increased to power this process (Yu et al., 2014). Alpha‐amylase (α‐d‐1,4‐glucan‐4‐glucanohydrolases) is of critical importance for the breakdown of starch granules during seed germination (Bak‐Jensen et al., 2010; Ju et al., 2019). TaNRT2.5 transgenic lines have altered TaAMY expression and this may have influenced the seed germination rate (Fig. 4). In a previous study, UTP‐glucose pyrophosphorylase (UGPase) was associated with glycogenesis, the synthesis of UDP‐glucose from glucose‐1‐phosphate and UTP, which was upregulated throughout germination (Yu et al., 2014). In the present study, the expression level of TaUGPase also was found to increase in TaNRT2.5‐3B overexpression lines and decrease in RNAi lines.
Overexpressing TaNRT2.5‐3B increases grain nitrate concentration and germination, suggesting that the transporter may be a functional orthologue of AtNRT2.7 in Arabidopsis (Chopin et al., 2007). The two transporters both show seed‐specific expression pattern, tonoplast localization and nitrate transport activity, but the two genes have some divergent properties. For example, AtNRT2.7 does not require a NAR2 partner protein for the nitrate transport function. The NRT2 genes have functional differences in Arabidopsis and wheat although they may be performing the same function in storing nitrate in the seed. The key role of the TaNAC2‐NRT2.5 module in the grain nitrate signal linked to germination is now established in wheat. As in Arabidopsis (Yan et al., 2016), there is evidence that ABA might be involved as ABRE cis‐acting elements involved in ABA responses can be identified in the promoter regions of both AtNRT2.7 and TaNRT2.5 (://bioinformatics.psb.ugent.be/webtools/plantcare/html/). It may be important to separate grain N storage and the signaling role of nitrate for the TaNAC2‐NRT2.5 module. In wheat, increasing TaNRT2.5 expression by almost 30‐fold (Fig. 4d) did not result in similar fold‐increases in grain fill of N or nitrate. For the RNAi lines too, there was not a huge effect on grain storage of nitrate or N (Fig. 4b, c). Additional factors such as post‐translational regulation might be important, but TaNAC2‐5A is regulating the expression of other assimilatory genes that may interact and contribute to the balance of N storage forms in seed. Stored nitrate is important during the grain filling for subsequent germination and this has been summarized in a revised model that includes the novel information provided by the wheat research herein (Fig. 8). In conclusion, TaNRT2.5 has a specific role in seed nitrate accumulation, an important signal for seed vigour and crop establishment, and the TaNAC2‐NRT2.5 module potentially can be genetically engineered to improve germination rate, seed vigour and grain yield in wheat. TaNAC2 is the transcription factor regulating the ‘workhorse’ TaNRT2.5 transporter which drives nitrate accumulation and, therefore, seedling vigour.
Figure 8.
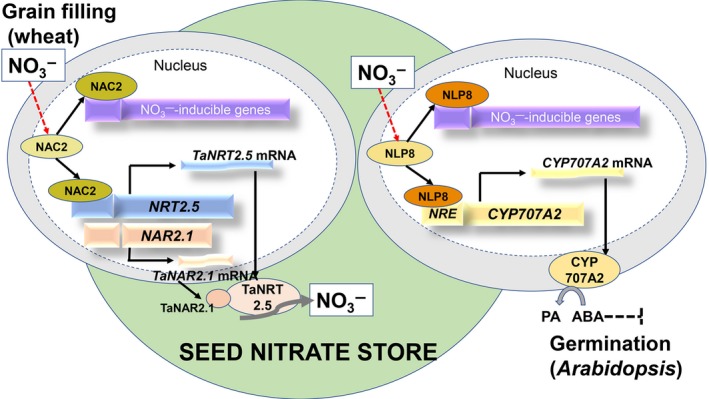
A proposed schematic model for NAC2/TaNRT2.5/TaNAR2.1 activity in nitrate grain filling and subsequent germination integrating data from Arabidopsis and wheat. Modified from Yan et al. (2016). Nitrate promotes TaNAC2 expression to activate the TaNRT2.5/TaNAR2.1 nitrate grain‐filling activity. Nitrate stored in the grain helps provide the signal promoting germination mediated by the abscisic acid (ABA) content controlled by NIN‐like protein 8 (NLP8) and CYP707A.
Author contributions
WL and XH designed and performed most of the experiments; YC contributed to the nitrate transporter activity in Xenopus laevis oocyte experiments; YJ, CS and JY performed experiments and interpreted the results; WT, XZ and WH provided technical assistance; MH and HL contributed to the field experiments; AJM and YT supervised the project and designed the experiments; and WL, XH, AJM and YT wrote the manuscript. WL and XH contributed equally to this work.
Supporting information
Fig. S1 Seed vigour after artificial aging treatment and seed germination in soil.
Fig. S2 Seed germination phenotypes of six Chinese wheat varieties.
Fig. S3 Expression levels of genes in different organs of wheat.
Fig. S4 Binding abilities of TaNAC2 to the promoter fragments of TaLAX1‐3B/1D, TaARR12‐6A and TaCLCc‐3A.
Fig. S5 Phylogenetic tree of NRT2 gene family in Arabidopsis, rice, maize, barley, soybean and wheat.
Fig. S6 Nitrate concentration in the dissected parts of wheat seed.
Fig. S7 Expression analysis and nitrate transport activity of TaNRT2.5s and TaNAR2s.
Fig. S8 Gene expression patterns for the TaNAC2‐5A/B/D and TaNRT2.5‐3A/B/D homeolog genes.
Fig. S9 TaNRT2.5 expression influences seed vigour.
Fig. S10 Nitrate influx rate of TaNRT2.5‐3B overexpression lines and WT at the seedling stage
Fig. S11 Expression levels of TaNAR2 genes at seedling stage in roots of TaNRT2.5‐3B overexpression lines and WT.
Fig. S12 Expression levels of nitrate transporter TaNRT2.1 and nitrate reductase TaNR1 at seedling stage in roots of TaNRT2.5‐3B overexpression lines and WT.
Table S1 Primers used in constructs.
Table S2 Primers used for qRT‐PCR.
Table S3 Gene information in the ChIP‐seq analysis of TaNAC2.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Caixia Gao’s laboratory (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for producing the transgenic wheat lines. This research was supported by the National Natural Science Foundation of China (31672214), the Centre of Excellence for Plant and Microbial Sciences (CEPAMS) established between the John Innes Centre and the Chinese Academy of Sciences, and funded by the UK Biotechnology and Biological Sciences Research Council, the Chinese Academy of Sciences, and the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision seed design and breeding).
Contributor Information
Anthony J. Miller, Email: tony.miller@jic.ac.uk.
Yiping Tong, Email: yptong@genetics.ac.cn.
References
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN. 2010. Nitrate, a signal relieving seed dormancy in Arabidopsis . Plant, Cell & Environment 28: 500–512. [DOI] [PubMed] [Google Scholar]
- Almagro A, Lin SH, Yi FT. 2008. Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arc E, Sechet J, Corbineau F, Rajjou L, Marion‐Poll A. 2013. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Frontiers in Plant Science 4: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgain P, Russell B, Mohammadi M. 2018. Phylogenetic analyses and in‐seedling expression of ammonium and nitrate transporters in wheat. Scientific Reports 8: 7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak‐Jensen KS, Sabrina L, Ole O, Christine F, Peter R, Birte S. 2010. Spatio‐temporal profiling and degradation of alpha‐amylase isozymes during barley seed germination. The FEBS Journal 274: 2552–2565. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Aoyama N, Chung YY, Still DW, Jones RL. 2007. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiology 143: 1173–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Bradford K, Hilhorst H, Nonogaki H, Bewley JD, Bradford K, Hilhorst H, Nonogaki H. 2013. Seeds: physiology of development, germination and dormancy. Seed Science Research 23: 289–289. [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. 2004. Chromatin techniques for plant cells. The Plant Journal 39: 776–789. [DOI] [PubMed] [Google Scholar]
- Buchner P, Hawkesford MJ. 2014. Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. Journal of Experimental Botany 65: 5697–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo‐Barral N, Matilla AJ, Rodríguez‐Gacio MC, Iglesias‐Fernández R. 2014. Nitrate affects sensu‐stricto germination of after‐ripened Sisymbrium officinale seeds by modifying expression of SoNCED5, SoCYP707A2 and SoGA3ox2 genes. Plant Science 217–218: 99–108. [DOI] [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe MF, Chardon F, Truong HN, Miller AJ, Krapp A, Daniel‐Vedele F. 2007. The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Fan X, Yan M, Liu X, Miller AJ, Xu G. 2011. Multiple roles of nitrate transport accessory protein NAR2 in plants. Plant Signaling & Behavior 6: 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finchsavage WE, Bassel GW. 2016. Seed vigour and crop establishment: extending performance beyond adaptation. Journal of Experimental Botany 67: 567–591. [DOI] [PubMed] [Google Scholar]
- Gu C, Song A, Zhang X, Wang H, Li T, Chen Y, Jiang J, Chen F, Chen S. 2016. Cloning of chrysanthemum high‐affinity nitrate transporter family (CmNRT2) and characterization of CmNRT2.1 . Scientific Reports 6: 23462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Toriyama K. 1998. Seed nitrogen accelerates the rates of germination, emergence, and establishment of rice plants. Soil Science and Plant Nutrition 44: 359–366. [Google Scholar]
- He H, Willems L, Batushansky A, Fait A, Hanson J, Nijveen H, Hilhorst HW, Bentsink L. 2016. Effects of parental temperature and nitrate on seed performance are reflected by partly overlapping genetic and metabolic pathways. Plant and Cell Physiology 57: 473–487. [DOI] [PubMed] [Google Scholar]
- He X, Qu BY, Li WJ, Zhao XQ, Teng W, Ma WY, Ren YZ, Li B, Li ZS, Tong YP. 2015. The nitrate‐inducible NAC transcription factor TaNAC2‐5A controls nitrate response and increases wheat yield. Plant Physiology 169: 1991–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks SB, Taylorson RB. 1974. Promotion of seed germination by nitrate, nitrite, hydroxylamine, and ammonium salts. Plant Physiology 54: 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HWM, Karssen CM. 1989. Nitrate reductase independent stimulation of seed germination in Sisymbrium officinale L. (hedge mustard) by light and nitrate. Ann Bot 63: 131–137. [Google Scholar]
- Huang Q, Wang Y. 2016. Overexpression of TaNAC2D displays opposite responses to abiotic stresses between seedling and mature stage of transgenic Arabidopsis . Frontiers in Plant Science 7: 1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Deng G, Liang J, Zhang H, Li Q, Pan Z, Yu M, Long H. 2019. Structural organization and functional divergence of high isoelectric point α‐amylase genes in bread wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.). BMC Genetics 20: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotur Z, Glass AD. 2015. A 150 kDa plasma membrane complex of AtNRT2.5 and AtNAR2.1 is the major contributor to constitutive high‐affinity nitrate influx in Arabidopsis thaliana . Plant, Cell & Environment 38: 1490–1502. [DOI] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubnermetzger G. 2005. Plant hormone interactions during seed dormancy release and germination. Seed Science Research 15: 281–307. [Google Scholar]
- Lezhneva L, Kiba T, Feria‐Bourrellier AB, Lafouge F, Boutet‐Mercey S, Zoufan P, Sakakibara H, Daniel‐Vedele F, Krapp A. 2015. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen–starved plants. The Plant Journal 80: 230–241. [DOI] [PubMed] [Google Scholar]
- Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R. 2012. TaNAC2, a NAC‐type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis . Journal of Experimental Botany 63: 2933–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou JP, Kamiya Y, Nambara E, Truong HN. 2009. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiology 149: 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A, Fan X, Shen Q, Smith S. 2007. Expression and functional analysis of rice NRT2 nitrate transporters. Comparative Biochemistry and Physiology Part A Molecular and Integrative Physiology 146: S241. [Google Scholar]
- Naylor REL. 2010. The effect of parent plant nutrition on seed size, viability and vigour and on germination of wheat and triticale at different temperatures. Annals of Applied Biology 123: 379–390. [Google Scholar]
- O'Brien J, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez R. 2016. Nitrate transport, sensing, and responses in plants. Molecular Plant 9: 837–856. [DOI] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel‐Vedele F, Miller AJ. 2006. Characterization of a two‐component high‐affinity nitrate uptake system in Arabidopsis. Physiology and protein–protein interaction. Plant Physiology 142: 1304–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzaro A, Clochard T, Planchet E, Limami AM, MorèreLe Paven MC. 2015. Identification and molecular characterization of Medicago truncatula NRT2 and NAR2 families. Physiologia Plantarum 154: 256–269. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D. 2012. Seed germination and vigor. Annual Review of Plant Biology 63: 507–533. [DOI] [PubMed] [Google Scholar]
- Salvi P, Saxena SC, Petla BP, Kamble NU, Kaur H, Verma P, Rao V, Ghosh S, Majee M. 2016. Differentially expressed galactinol synthase(s) in chickpea are implicated in seed vigor and longevity by limiting the age induced ROS accumulation. Scientific Reports 6: 35088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C. 2014. Genome editing in rice and wheat using the CRISPR/Cas system. Nature Protocols 9: 2395–2410. [DOI] [PubMed] [Google Scholar]
- Taulemesse F, Gouis JL, Gouache D, Gibon Y, Allard V. 2015. Post‐flowering nitrate uptake in wheat is controlled by N status at flowering, with a putative major role of root nitrate transporter NRT2.1 . PLoS ONE 10: e0120291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Zhou JJ, Li Z, Miller AJ. 2005. A two‐component high‐affinity nitrate uptake system in barley. The Plant Journal 41: 442–450. [DOI] [PubMed] [Google Scholar]
- Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF. 2010. Gene limiting cadmium accumulation in rice. Proceedings of the National Academy of Sciences, USA 107: 16500–16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ge H, Hao C, Dong Y, Zhang X. 2012. Identifying loci influencing 1,000‐kernel weight in wheat by microsatellite screening for evidence of selection during breeding. PLoS ONE 7: e29432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Vanathy E, Vivian C, Masanori O, Matthew I, Mitsuhiro K, Akira E, Ryoichi Y, Asher P, Gong Y. 2016. NIN‐like protein 8 is a master regulator of nitrate‐promoted seed germination in Arabidopsis . Nature Communications 7: 13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G. 2011. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant, Cell & Environment 34: 1360–1372. [DOI] [PubMed] [Google Scholar]
- Yu Y, Guo G, Lv D, Hu Y, Li J, Li X, Yan Y. 2014. Transcriptome analysis during seed germination of elite Chinese bread wheat cultivar Jimai 20. BMC Plant Biology 14: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Zhang Q, Pei CL, Li X, Huang XL, Chang CY, Wang XJ, Huang LL, Kang ZS. 2018. TaNAC2 is a negative regulator in the wheat‐stripe rust fungus interaction at the early stage. Physiological and Molecular Plant Pathology 102: 144–153. [Google Scholar]
- Zheng C, Jiang D, Liu F, Dai T, Liu W, Jing Q, Cao W. 2009. Exogenous nitric oxide improves seed germination in wheat against mitochondrial oxidative damage induced by high salinity. Environmental and Experimental Botany 67: 222–227. [Google Scholar]
- Zuo JH, Liu JD, Gao FM, Yin GH, Wang Z, Chen FY, Li XY, Xu JM, Chen TT, Li L et al. 2018. Genome‐wide linkage mapping reveals QTLs for seed vigor‐related traits under artificial aging in common wheat (Triticum aestivum). Frontiers in Plant Science 9: 1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Seed vigour after artificial aging treatment and seed germination in soil.
Fig. S2 Seed germination phenotypes of six Chinese wheat varieties.
Fig. S3 Expression levels of genes in different organs of wheat.
Fig. S4 Binding abilities of TaNAC2 to the promoter fragments of TaLAX1‐3B/1D, TaARR12‐6A and TaCLCc‐3A.
Fig. S5 Phylogenetic tree of NRT2 gene family in Arabidopsis, rice, maize, barley, soybean and wheat.
Fig. S6 Nitrate concentration in the dissected parts of wheat seed.
Fig. S7 Expression analysis and nitrate transport activity of TaNRT2.5s and TaNAR2s.
Fig. S8 Gene expression patterns for the TaNAC2‐5A/B/D and TaNRT2.5‐3A/B/D homeolog genes.
Fig. S9 TaNRT2.5 expression influences seed vigour.
Fig. S10 Nitrate influx rate of TaNRT2.5‐3B overexpression lines and WT at the seedling stage
Fig. S11 Expression levels of TaNAR2 genes at seedling stage in roots of TaNRT2.5‐3B overexpression lines and WT.
Fig. S12 Expression levels of nitrate transporter TaNRT2.1 and nitrate reductase TaNR1 at seedling stage in roots of TaNRT2.5‐3B overexpression lines and WT.
Table S1 Primers used in constructs.
Table S2 Primers used for qRT‐PCR.
Table S3 Gene information in the ChIP‐seq analysis of TaNAC2.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


