Abstract
Mutations in the BRCA1 and BRCA2 genes predispose afflicted individuals to breast, ovarian, and other cancers. The BRCA-encoded products form complexes with other tumor suppressor proteins and with the recombinase enzyme RAD51 to mediate chromosome damage repair by homologous recombination, and also to protect stressed DNA replication forks against spurious nucleolytic attrition. Understanding how the BRCA tumor suppressor network executes its biological functions would provide the foundation for developing targeted cancer therapeutics, but progress in this area has been greatly hampered by the challenge of obtaining purified BRCA complexes for mechanistic studies. In this article, we review how recent effort begins to overcome this technical challenge, leading to functional and structural insights into the biochemical attributes of these complexes and the multi-faceted roles that they fulfill in genome maintenance. We also highlight the major mechanistic questions that remain.
Keywords: Homologous recombination, DNA damage repair, replication fork repair, replication fork protection, genome maintenance, BRCA1, BARD1, BRCA2, tumor suppression
1. INTRODUCTION
The integrity of our genome is challenged continuously by environmental agents, such as high-energy radiations, and by reactive metabolites, e.g. aldehydes and free radicals, that damage DNA (1–3). Accordingly, a complex network of DNA damage checkpoints and repair pathways has evolved to minimize the harm that may be caused by these genotoxic agents. As such, a defect in any of these detoxification mechanisms leads to genome destabilization and disease, cancer in particular (2, 4). Studies on chromosome damage repair have taken center stage in cancer biology in recent years, and knowledge garnered from these endeavors is being translated into cancer therapeutics (5).
DNA damage checkpoints:
signal transduction cascades that result in cell cycle arrest and activation of DNA damage repair.
Below, we review chromosome damage repair that is mediated by homologous recombination (HR) genes of the RAD52 epistasis group (see Section 3.1 for description). This genome maintenance pathway helps eliminate DNA double-strand breaks (DSBs) and interstrand crosslinks (ICLs), and also repairs damaged DNA replication forks and protects stressed forks against nucleolytic digestion. We will specifically focus on the functional and structural insights derived from recent studies on the tumor suppressors BRCA1-BARD1 and BRCA2 that are integral components of the HR machinery.
2. BIOLOGICAL ROLES OF HOMOLOGOUS RECOMBINATION
2.1. Function as a DNA Double-Strand Break Repair Tool
HR and non-homologous DNA end-joining (NHEJ) are the major, mechanistically distinct DSB repair pathways in eukaryotic cells. HR primarily operates in the S and G2 phases of the cell cycle when the sister chromatid is available to template repair, whereas NHEJ functions throughout the cell cycle (6–8). HR also mediates the pairing of homologous chromosomes in meiosis to help ensure their proper disjunction in the first meiotic division (9).
Meiosis:
encompasses two cell divisions to generate haploid gametes. Pairing of meiotic chromosomes is triggered by DSBs and mediated by HR.
For HR to occur, a DNA break end must first undergo nucleolytic resection of the 5´-terminated strand to generate 3´-tailed single-stranded (ss) DNA to serve as the template for assembling the HR machinery (see Figure 1 and later sections). Under specific circumstances, e.g. when HR is inactivated by a genetic mutation, the ssDNA tails derived from DSB processing may be utilized by two other repair pathways, namely, single-strand annealing (SSA) and alternative DNA end joining (alt-EJ), to yield deletion type products. Details on the choice and regulation of DSB repair pathways can be found in several recent reviews (6–8, 10).
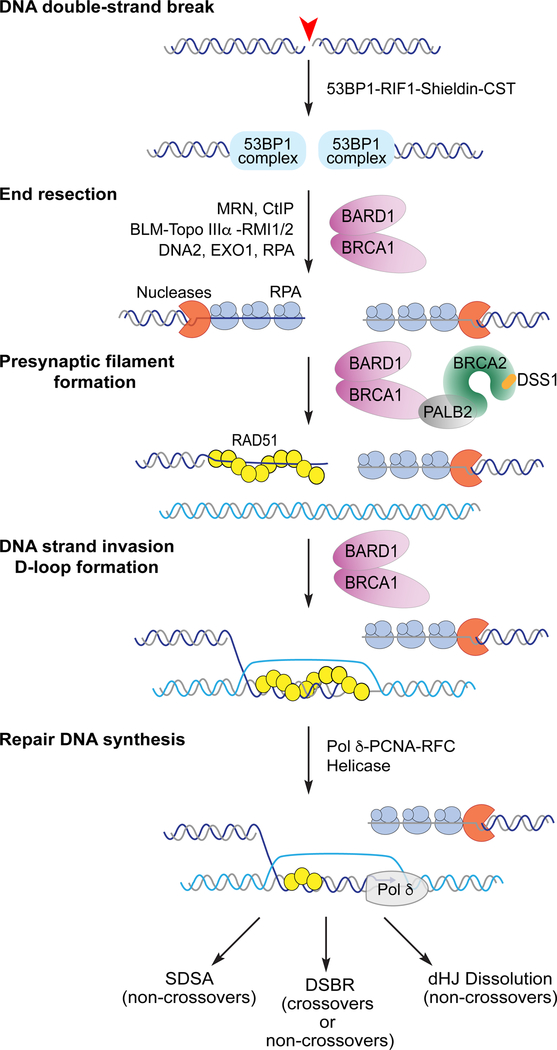
Figure 1.
DNA double-strand break repair by homologous recombination.
Emphasis is placed on the roles of BRCA1-BARD1 in DNA end resection and strand invasion and on the BRCA mediator complex (DSS1-BRCA2-PALB2-BRCA1-BARD1) in presynaptic filament formation. Note that many other factors that function in different stages of the recombination reaction are not shown (see Table 1 for the list of uncited factors). The three pathways of D-loop resolution are briefly discussed in Section 4.4 and in detail in recent reviews (8, 93). The four subunits of Shieldin are REV7 (MAD2L2), SHLD1 (C20orf196), SHLD2 (FAM35A), and SHLD3 (CTC534A2.2). The three subunits of the CST complex are CTC1, SNT1, and TEN1.
2.2. Involvement in Replication Fork Repair and Protection
DNA replication fork stalling induced by stress, such as upon nucleotide depletion or encountering a DNA lesion, triggers the DNA replication checkpoint for fork stabilization (11). One major means to circumvent fork stalling is the activation of a nearby, normally dormant replication origin (11). Alternatively, DNA lesions may be bypassed by one of several specialized DNA polymerases in a mechanism called translesion synthesis (12). In addition, the stalled fork can be converted by an ATP-dependent nucleic acid motor protein into a “chicken-foot” structure, in which the free DNA end is engaged by HR to mediate fork restart (11). Importantly, recent studies have provided evidence that a subset of HR factors, including the recombinase RAD51 and the tumor suppressors BRCA1 and BRCA2, help protect stressed replication forks from being degraded by nucleases such as MRE11 and DNA2 (13–17).
Translesion synthesis:
employs a specialized DNA polymerase to synthesize DNA across a lesion in the template strand.
Chicken foot structure:
also called a regressed fork, has four double-stranded arms, of which one harbors a free end.
2.3. Role in Interstrand DNA Crosslink Removal
An ICL arises when a covalent bond is formed between bases in the complementary strands of duplex DNA. This lesion impedes transcription and DNA replication, and the mechanism for its removal is exceedingly complex and normally coupled to DNA replication (18–21). The prevailing model for DNA replication-coupled ICL repair posits that lesion removal is triggered by two replication forks converging from opposite sides of the crosslink (19, 20). Then, lesion “unhooking” via endonucleolytic strand scissions 5´ and 3´ to the ICL generates two DNA molecules, with one harboring a DSB and the other a DNA gap with the unhooked crosslink appended as a mono-adduct (19, 20). Upon gap filling by DNA polymerase ζ (a translesion synthesis polymerase) and ligation of the resulting DNA nick, a duplex template is created to enable elimination of the DSB via HR (18, 20, 21). Accordingly, DSBs accumulate in HR-defective cells upon treatment with agents that induce ICLs (18, 20, 21). Ultimately, the unhooked DNA crosslink is removed by nucleotide excision repair (22). Aside from components of the HR pathway, the genes responsible for Fanconi Anemia (FA) are also critically important for replication-dependent ICL repair (18, 20, 21). We refer interested readers to recent reviews and research highlights on this topic (18–21, 23, 24).
Nucleotide excision repair:
removes bulky DNA lesions that distort the conformation of the DNA double helix.
The FANCONI ANEMIA DNA DAMAGE RESPONSE PATHWAY
Fanconi anemia (FA) is an autosomal recessive disorder characterized by bone marrow failure, developmental abnormalities, and a strong cancer predisposition in children and young adults. Cells from FA patients are hypersensitive to DNA damaging agents, in particular those that induce ICLs, and to reactive aldehydes that stem from cellular metabolism (18, 20, 21, 25). FA is genetically complex, comprising at least 22 complementation groups. Importantly, biallelic mutations in a number of HR genes can result in FA. This provides compelling evidence that HR-mediated DNA repair constitutes an integral component of the FA DNA damage response pathway (18, 20, 21, 25).
3. GENETICS OF HOMOLOGUS RECOMBINATION
3.1. Genes of the RAD52 Epistasis Group
The budding yeast S. cerevisiae has been a valuable model for dissecting the genetic complexity and mechanistic intricacy of HR. In S. cerevisiae, the majority of DSBs are resolved via HR, and the initial attempts at isolating mutants based on their hypersensitivity to X-rays found eight genetic loci designated RAD50 to RAD57 (26, 27). Later efforts utilizing genetic screens and bioinformatics led to the isolation of other HR genes (28, 29). For instance, SHU1 and SHU2 were identified as suppressors of the slow growth phenotype and DNA damage sensitivity of cells lacking DNA topoisomerase 3 (30). Collectively, these HR genes constitute the RAD52 epistasis group, and the majority of them possess an ortholog in metazoans ((31, 32); Table 1). Generally speaking, mutants of the RAD52 group are hypersensitive to agents that induce DSBs and replication stress, and are also defective in meiotic chromosome segregation (31). As will be discussed in the next section, metazoans rely on the tumor suppressors BRCA1, BARD1, BRCA2, and PALB2 for HR execution (4). Notably, S. cerevisiae does not harbor a structural homolog of any of these tumor suppressors.
Table 1.
Proteins and their functions in homologous recombination
| Function | H. sapiens | S. cerevisiae |
|---|---|---|
| DNA end resection | ||
| Initiation and short-range | MRE11-RAD50-NBS1 (MRN), CtIP BRCA1-BARD1 |
Mre11-Rad50-Xrs2 (MRX), Sae2 |
| Long-range | EXO1 | Exo1 |
| DNA2, BLM or WRN, TOPOIIIα-RMI1-RMI2 | Dna2, Sgs1-Top3-Rmi1 | |
| RPA | RPA | |
| SMARCAD1 | Fun30 | |
| Possibly BRCA1-BARD1 | ||
| Inhibitors of DNA resection | 53BP1-RIF1-Shieldin (REV7-SHLD1-SHLD2-SHLD3)-CST(CTC1-STN1-TEN1) | Rad9 |
| PTIP, Artemis | ||
| Presynaptic filament assembly and dynamics | ||
| Recombinase | RAD51 | Rad51 |
| Mediators | BRCA2-DSS1, Possibly BRCA1-BARD1, PALB2 |
Rad52 |
| Rad51 paralog complexes and partners | RAD51B-RAD51C-RAD51D-XRCC2, RAD51C-XRCC3, SWS1-SWSAP1 | Rad55-Rad57, Psy3-Csm2-Shu1-Shu2 |
| Recombinase stimulatory factors | RAD54, RAD54B SWI5-SFR1 |
Rad54, Rdh54 *Mei5-Sae3 |
| MMS22L-TONSL | ||
| Anti-recombinases | FBH1 | Srs2 |
| RECQ5 | ||
| Synaptic complex formation | ||
| BRCA1-BARD1 | ||
| PALB2 | ||
| RAD51AP1-UAF1 | ||
| HOP2-MND1 | *Hop2-Mnd1 | |
| DNA synthesis | ||
| DNA polymerase complex | Pol δ, PCNA, RFC | Pol δ, PCNA, RFC |
| DNA helicase | To be identified | Pif1 |
| DNA ligase | Ligase I | Ligase I |
Protein factors, including DNA helicases and resolvases, that help determine HR pathway choice are not listed above. Readers are referred to recent reviews on this topic (8, 32, 93). Blanks in the S. cerevisiae column indicate that no ortholog of the human factors exits. S. cerevisiae factors denoted by the asterisk (*) are expressed only during meiosis and function specifically with the RecA/Rad51-related recombinase Dmc1 (8, 69).
Epistasis group:
encompasses genes functioning in the same pathway, such that double mutants exhibit no more severe a phenotype than single mutants.
THE PALB2 GENE AND PROTEIN
PALB2 (Partner and Localizer of BRCA2) protein is stably associated with BRCA2 and required for the localization of BRCA2 to DNA damage (33). Through its interaction with BRCA1, PALB2 nucleates the assembly of the BRCA Mediator Complex (see Section 5.6b). Pathogenic mutations in PALB2 predispose afflicted individuals to breast, pancreatic, prostate, and other cancers (34, 35). Importantly, point mutations that disrupt the interaction of PALB2 with either BRCA1 or BRCA2 engender DNA damage sensitivity, HR defects, and cancer susceptibility (34, 35). PALB2 binds DNA and physically interacts with RAD51, and it also enhances the recombinase activity of RAD51 via promotion of synaptic complex formation (36, 37).
3.2. Dependence of HR on Tumor Suppressors in Metazoans
BRCA1, BARD1, BRCA2, and PALB2 are needed for the suppression of a wide variety of tumors, and they do so by enhancing the DSB repair capacity of cells. Efforts by many research groups have shown that these tumor suppressors form distinct complexes among themselves and with other HR proteins. These protein complexes play key roles in different stages of HR, DSB and replication fork repair, the protection of stressed replication forks, and ICL removal ((4); see Section 2). Given the important roles that HR fulfills during DNA replication, it is no surprise that deletion of these tumor suppressors engenders embryonic lethality at an early stage of development (38–43). Interestingly, biallelic mutations in BRCA1, BRCA2, and PALB2 can cause FA (44–46) and, hence, these genes are also known as FANCS, FANCD1 and FANCN, respectively (21). No FA-specific mutations have been described for BARD1 yet. It should be noted that cancer causative mutations have been identified in other HR genes such as those that encode the RAD51 paralogs (47) and FANCJ (48), a DNA helicase that is mutated in a subset of FA patients.
THE RAD51 PARALOGS IN HR AND CANCER
RAD51 paralogs are conserved HR factors that exhibit sequence similarity with and are structurally related to the recombinase RAD51 (4). Six such paralogs, namely, RAD51B, RAD51C, RAD51D, XRCC2, XRCC3 and SWS1AP1 (Table 1), have been described in humans, and they form complexes among themselves and with other factors (4, 49). However, the RAD51 paralogs are devoid of recombinase activity. Rather, they regulate RAD51 presynaptic filament assembly and appear to enhance the catalytic potential of the presynaptic filament (50, 51). Existing evidence suggests that the RAD51 paralogs function together with BRCA2 in presynaptic filament assembly (51, 52). Mutations in the RAD51 paralogs have been linked to breast and ovarian cancers and to Fanconi anemia (47).
4. CRITICAL STAGES IN HOMOLOGOUS RECOMBINATION
4.1. DNA End Resection
As touched upon earlier (see Section 2.1), the repair of DSBs or replication forks by HR is initiated via the resection of the 5´-terminated DNA strand at the break site, which generates a 3´-tailed ssDNA substrate for the assembly of the HR machinery that harbors the RAD51 recombinase and its ancillary factors (53). Studies, first done in S. cerevisiae and later in metazoans, have shown that DNA end resection occurs in two distinct stages (53). At DNA ends that are occluded by the NHEJ factor Ku, Sae2 (CtIP in humans; Table 1) activates the Mre11-Rad50-Xrs2 complex (MRE11-RAD50-NBS1 in humans) to incise the 5´-terminated DNA strand endonucleolytically (Figure 1). Subsequent long-range resection is mediated by two apparently redundant mechanisms that are dependent on either the 5´→ 3´ exonuclease Exo1 or the ensemble comprising the DNA helicase/endonuclease Dna2. Both of these long-range resection nucleases are dependent on other protein factors for optimal activity (54, 55). Specifically, Dna2 functions within the context of an obligate ensemble with RPA, the RECQ family helicase Sgs1 (BLM in metazoans), and other cofactors (Table 1), while the activity of Exo1 is enhanced by RPA and, in the case of the human enzyme, by BLM as well (53, 55, 56). Interestingly, WRN, another RECQ family helicase, can substitute for BLM in DNA2-dependent resection (57, 58). It should be noted that nucleolytic resection of the “chicken foot” structure stemming from the regression of stressed replication forks appears to proceed via mechanisms different than what occurs at DSBs (59, 60).
Notably, yeast Rad9 and its mammalian counterpart 53BP1 (p53-binding protein 1) suppress DNA end resection in the G1 phase of the cell cycle. The restriction on resection imposed by Rad9/53BP1 effectively renders NHEJ the default DSB pathway in G1 cells. The inhibitory effect of these proteins is relieved in the S cell cycle phase (6, 61). In metazoans, relaxation of the 53BP1-dependent resection block requires BRCA1 (61, 62). A great deal of effort has been expended to identify protein factors that act with 53BP1. The current model posits that the recruitment of 53BP1 occurs via its interaction with modified histones H2A-K15ub and H4-K20Me2, which then nucleates the formation of a higher order ensemble that encompasses RIF1, the CST complex and Shieldin ((6, 63–67); Figure 1).
4.2. Presynaptic Filament Assembly
4.2a. Recombinase presynaptic filament.
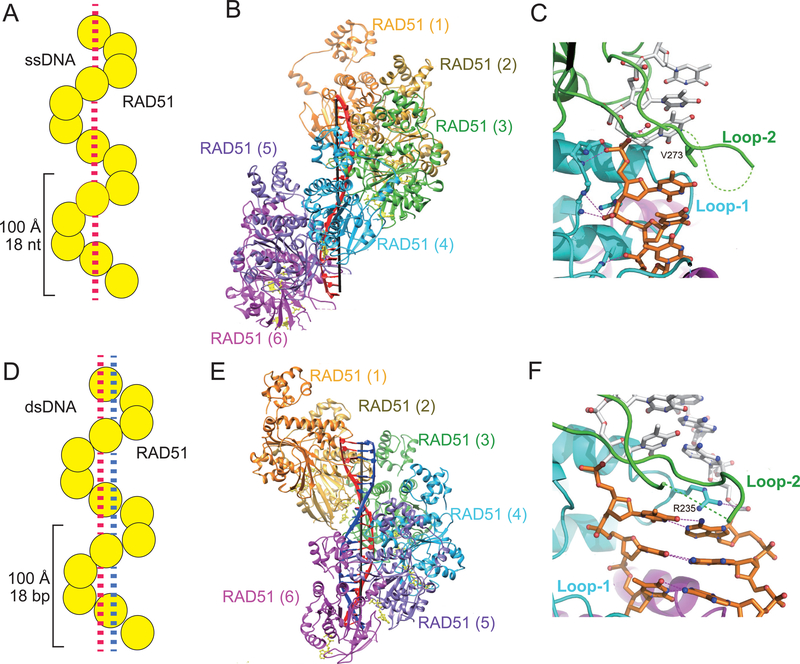
Rad51 belongs to the RecA/Rad51 family of recombinases that catalyze the pairing of homologous ssDNA and dsDNA and the exchange of DNA strands between the synapsed DNA molecules (68–70). Homologous DNA pairing and strand exchange occur within the confines of a right-handed, filamentous recombinase polymer assembled on the 3´-tailed ssDNA derived from DNA end resection ((53, 69, 70); Figure 1). Within the HR community, the Rad51-ssDNA nucleoprotein filament is referred to as the “presynaptic filament” or “presynaptic complex” (69, 70). The presynaptic filament of Rad51 has been examined by negative stain electron microscopy (71, 72) and, more recently, by cryo-electron microscopy (cryo-EM; (73, 74)). As revealed in earlier work (71, 72, 75) and highlighted in a near-atomic cryo-EM structure (73, 74), the presynaptic filament of human RAD51 comprises ~6 protein protomers and ~18 bases of DNA per turn with a 100 Å pitch ((74); Figure 2, A and B). Importantly, the ssDNA is extended to ~1.5 times the length of B-form DNA, a conserved feature of presynaptic filaments of RecA/Rad51 family members (74, 75). The majority of RAD51 protomer-protomer interactions in a functional presynaptic filament occur upon ATP being sandwiched between two adjacent protomers. Within the filament, RAD51 protomers primarily interact with the phosphate backbone of the DNA, with key amino acid residues in the L1 and L2 DNA binding loops of three contiguous protomers organizing the DNA ligand in base triplets (Figure 2C). In this configuration, the base triplets point towards the axial direction of the presynaptic filament and are poised to undergo pairing and strand exchange with a homologous duplex target ((74); Figure 2C).
Figure 2.
Structure of the RAD51-ssDNA presynaptic filament and RAD51-dsDNA post-synaptic complex.
(A, D) Cartoon illustrations of the presynaptic filament (A) and post-synaptic complex (D); RAD51 is shown in yellow and DNA is depicted as dashed lines. (B, E) Atomic structure of the presynaptic filament (B) and post-synaptic complex (E), the invading ssDNA is shown in red and complementary strand in blue. (C, F) RAD51-DNA interactions in the presynaptic filament (C) and post-synaptic complex (F). Base triplets interacting with V273 in RAD51 DNA binding Loop 2 in the presynaptic filament (C), and base pair triplets with R235 in DNA binding Loop 1 in the post-synaptic complex (F) are highlighted. See reference 74 for details.
4.2b. Mediators of presynaptic filament assembly and maintenance.
Owing its abundance and high affinity for ssDNA, RPA promptly occupies the 3´-tailed ssDNA derived from DNA end resection (69). Even though RPA protects the DNA tail from nucleolytic attack and removes secondary structure therein, it presents a strong impediment to the initial nucleation of RAD51 (76). Facilitators of RPA-Rad51 exchange have been found in different organisms, including S. cerevisiae Rad52 and the tumor suppressor BRCA2 in metazoans (69, 76). These HR “mediators” interact with RAD51 and bind ssDNA. Interestingly, while Rad52 associates with RPA, the small, acidic protein DSS1 provides the RPA-interacting interface in its stoichiometric complex with BRCA2 ((77); see Section 6.5).
The yeast Rad55-Rad57 and human RAD51B-RAD51C complexes enhance presynaptic filament assembly on RPA-coated ssDNA. As such, these complexes of Rad51 paralogs are HR mediators in their own right (78, 79). Moreover, Rad55-Rad57 has been shown to attenuate the ability of the Srs2 helicase, one of several anti-recombinases that have been identified, to dismantle the Rad51 presynaptic filament (8, 80, 81). Interestingly, the yeast Rad51 paralogs associate and synergize with Rad52 to facilitate Rad51 presynaptic filament assembly (51). Whether the metazoan RAD51 paralogs (76, 82) similarly interact and function with BRCA2 remains to be determined, but it should be noted that some of the human RAD51 paralogs show an epistatic relationship with BRCA2 (52).
Anti-recombinases:
ATP-dependent motor proteins that either dismantle the presynaptic filament or HR intermediates, or inhibit repair DNA synthesis.
4.3. Homologous DNA Pairing and Strand Exchange
4.3a. Assembly of the synaptic complex:
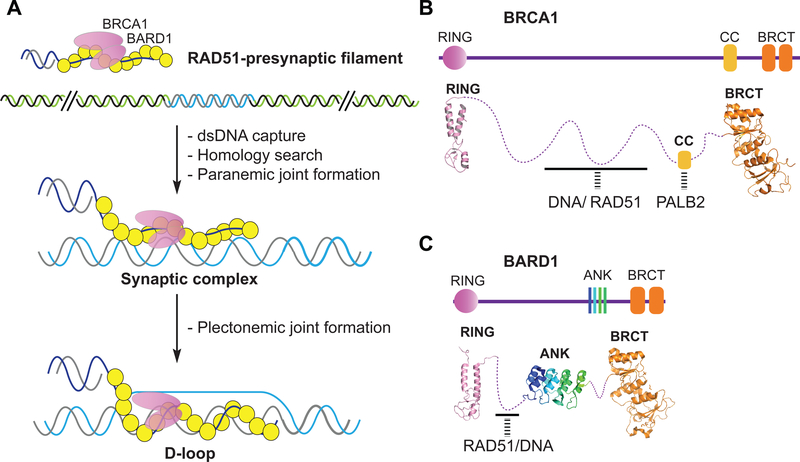
The presynaptic filament possesses a site, termed the secondary site, that engages duplex DNA to enable the search for sequence homology in the latter (83). Work done with E. coli RecA has provided evidence that DNA homology search occurs by way of random collisions between the presynaptic filament and the duplex DNA (83). Upon the location of homology in the duplex partner, the presynaptic filament forms a triple-stranded structure that entails limited base pairing between the ssDNA and dsDNA molecules. This nascent DNA joint is called the “paranemic joint”, and the three-stranded nucleoprotein ensemble is referred to as the “synaptic complex” ((69, 83, 84); Figure 3A). Even though the paranemic joint is transient in nature, its formation allows for iterative three-dimensional search through sequence space to facilitate downstream events ((85); see Section 4.3b). Biochemical and biophysical methods have been developed to study paranemic joint formation (36, 86, 87). As revealed in single-molecule imaging of homologous DNA pairing by RecA/Rad51 family recombinases, a contiguous homology tract of eight nucleotides is sufficient for paranemic joint formation (85). HR factors that enhance the assembly of the synaptic complex have been identified (36, 69, 86, 88). Below (see Section 5.5), we discuss recent findings that identify the BRCA1-BARD1 complex as such a stimulatory factor of RAD51 (89).
Figure 3.
BRCA1-BARD1 structure and function.
(A) Cartoon illustrating the role of BRCA1-BARD1 in the promotion of paranemic and plectonemic joint formation (89). BRCA1-BARD1 aids in the capture of the recombining duplex molecule to facilitate paranemic joint formation and the assembly of the triple-stranded synaptic complex. Subsequent plectonemic joint formation yields the D-loop intermediate. (B) Functional motifs and domains in BRCA1 (B) and BARD1 (C) including the RING (Really Interesting New Gene) domain (PDB: 1JM7), CC (Coiled Coil) domain, ANK (Ankyrin) motifs (PDB: 3C5R), and BRCT (BRCA1 Carboxy-Terminal) repeats (PDB: 1T2U for BRCTs of BRCA1; PDB: 2NTE for BRCTs of BARD1). The known atomic structures are linked by the dash lines depicting protein regions of which no structural information is available.
4.3b. Formation of the post-synaptic complex:
In DSB repair, upon synaptic complex formation (see Section 4.3a), strand exchange between the 3´-tailed ssDNA and the homologous dsDNA partner initiates from the free end of the former. This gives rise to the “plectonemic joint” in which the two DNA strands are topologically intertwined (Figure 3A). The nucleoprotein ensemble harboring the plectonemic joint has been termed the “post-synaptic” complex, with the corresponding DNA structure that harbors a displaced ssDNA region being referred to as the displacement loop, or D-loop (69, 83, 84). In congruence with the organization of ssDNA as base triplets in the presynaptic filament, DNA strand exchange occurs in 3-nucleotide steps (85, 90). Analysis by cryo-EM has revealed the hierarchy of protein-DNA interactions that help stabilize the plectonemic joint within the context of base pair triplets ((74); Figure 2D–F).
4.4. Repair DNA Synthesis and Resolution of HR Intermediates
Following D-loop formation, DNA synthesis commences from the terminus of the invading strand to copy sequence information from the donor DNA molecule (Figure 1). In both yeast and metazoans, DNA polymerase (Pol) δ catalyzes the repair synthesis reaction (91). The extended D-loop structure can be resolved by one of several mechanistically distinct pathways, namely, canonical DSB repair (DSBR), synthesis-dependent DNA strand annealing (SDSA), and double Holliday Junction (dHJ) dissolution. Of these pathways, only DSBR is capable of generating recombinants that harbor an exchange of chromosome arms, or crossover (92, 93). As was first revealed in genetic studies done in S. cerevisiae, during mitotic DSB repair, there is a strong tendency for cells to employ SDSA or dHJ dissolution to form non-crossover recombinants. Shunting the D-loop away from the DSBR pathway helps prevent chromosome arm translocations. The mechanistic details and regulation of HR pathways are highly complex, and we refer interested readers to recent reviews on this topic (8, 32, 93).
5. MULTIFACETED ROLE OF BRCA1-BARD1 IN GENOME MAINTENANCE
5.1. BRCA1: Discovery and Protein Domain Structure
The BRCA1 (Breast Cancer 1) gene was identified based on genetic linkage to familial breast cancer susceptibility (94, 95). Importantly, BRCA1 mutations cause familial ovarian cancer and sporadic cancer in different organs as well (96, 97). Deletion of Brca1 in mice engenders embryonic lethality (38–40). The human protein comprises 1,863 amino acid residues and harbors a RING (Really Interesting New Gene) domain at its N-terminus and two copies of the BRCT (BRCA1 Carboxy-Terminal) repeats at its C-terminus (Figure 3B). Through its RING domain, BRCA1 combines with BARD1 to form a heterodimer that possesses E3 ubiquitin ligase activity (98, 99). The tandem BRCT repeats confer the ability to associate with DNA damage response factors such as CtIP and FANCJ (100). Immediately preceding the BRCT repeats is a coiled coil domain that mediates complex formation with PALB2 (33, 101, 102). The central region of BRCA1 that is encoded by exon 11 accounts for >60% of the protein and is disordered based on secondary structure prediction (100). This large central region harbors sequences that confer capabilities for binding DNA (89, 103, 104) and interaction with various DNA damage response and repair factors, including RAD51 (89, 100, 105).
5.2. BARD1: Discovery, Protein Domain Structure, and Tumor Suppression Activity
BARD1 was identified as a BRCA1 interactor in a yeast two-hybrid screen (98). BARD1 comprises 777 amino acid residues and, like BRCA1, possesses an N-terminal RING domain and tandem BRCT repeats at its C-terminus (Figure 3C). The RING domain mediates heterodimer formation with BRCA1 (98, 106), and the BRCT repeats interact with proteins distinct from those recognized by the equivalent domain in BRCA1 (107). There are four ankyrin motifs that are involved in protein-protein interactions (107). Like BRCA1, BARD1 binds DNA and physically interacts with RAD51 (89). BARD1 mutations have been identified in breast, ovarian, and uterine cancers (107, 108). Importantly, BARD1 variants are also associated with neuroblastoma and other cancer types (107). In mouse studies, the ablation of Bard1 engenders embryonic lethality (42), while its conditional deletion in mammary epithelial cells induces breast cancer (107, 109).
5.3. Involvement of BRCA1 and BARD1 in Chromosome Damage Repair by HR
Following the discovery of BRCA1, intense efforts by different research groups provided concrete evidence for its involvement in chromosome damage repair. In one study, BRCA1 was found to colocalize with RAD51 in S phase cells and in human meiotic spermatocytes, and to also coimmunoprecipitate with RAD51 from cell extracts (105). Other endeavors showed that BRCA1-deficient cells are hypersensitive to DNA damaging agents and display chromosomal instability (110–112). Importantly, with the use of a HR reporter, an impairment of HR-mediated DSB repair in Brca1 deficient mouse embryonic stem cells was revealed (112, 113). Likewise, genetic ablation of BARD1 engenders a defect in chromosome damage repair, genome instability, and an inability to execute HR (42, 89, 114).
5.4. Role of BRCA1-BARD1 in DNA End Resection and RAD51 Recruitment
DNA damage-induced RAD51 focus formation becomes impaired when BRCA1 or BARD1 is absent, thus implicating the BRCA1-BARD1 complex in DNA end resection and RAD51 recruitment (89, 115, 116). As mentioned earlier, cells in the G1 cell cycle phase are poorly equipped to resect DSBs. The G1 restriction on end resection occurs at several levels, namely, (i) general shielding of the DNA lesion by an ensemble of proteins being anchored by 53BP1; (ii) occlusion of DNA ends by Ku and associated factors; and (iii) absence of cyclin-dependent kinase (CDK) activity needed for the recruitment or activation of key resection factors, such as CtIP (53, 61, 63–67, 117). In particular, BRCA1-BARD1 is believed to help counter the inhibitory effect of the 53BP1-associated ensemble when cells enter S phase. In BRCA1 mutant cells, ablation of 53BP1, RIF1, or any of the subunits of Shieldin or the CST complex (Table 1) partially suppresses the DNA end resection deficiency of these cells and their hypersensitivity to DNA damaging agents (61–67, 118, 119). Importantly, acting as E3 ligase, BRCA1-BARD1 conjugates ubiquitin to three lysine residues (K125/127/129) in histone H2A, a modification that leads to the recruitment of the chromatin remodeler SMARCAD1 to mobilize chromatin-bound 53BP1 and to facilitate DNA end resection (106, 116, 120).
5.5. Function of BRCA1-BARD1 in Synaptic Complex Assembly
An insect cell-based platform has been developed to assemble and purify wild type and mutant variants of the BRCA1-BARD1 complex (89). Biochemical testing confirmed previous findings that BRCA1 binds DNA (103, 104) and physically interacts with RAD51 (105). Interestingly, BARD1 was found to possess DNA binding and RAD51 interaction attributes as well (89). In DNA binding, BRCA1-BARD1 has the highest affinity for the D-loop structure, followed by the replication fork structure, dsDNA, and then ssDNA (89). Importantly, BRCA1-BARD1 was shown to enhance the ability of RAD51 to form D-loops, although not to promote RPA-RAD51 exchange on ssDNA (89). Biochemical and single-molecule imaging analyses provided evidence that stimulation of D-loop formation stems from an ability of BRCA1-BARD1 to promote synaptic complex assembly ((89); Figure 3A). Control experiments revealed that BRCA1-BARD1 does not associate with or affect the activity of yeast Rad51 or E. coli RecA. Several BARD1 mutations that weaken or ablate RAD51 interaction were constructed and shown to compromise the ability of BRCA1-BARD1 to promote D-loop and synaptic complex formation, in a manner that mirrors the degree of RAD51 interaction deficiency imparted by the mutations (89). Cell-based analysis of one of the BARD1 mutants provided evidence that the BARD1-RAD51 interaction is indispensable for chromosome damage repair and the execution of HR (89).
5.6. Knowledge Gaps Regarding BRCA1-BARD1 in Genome Repair
Much remains to be learned regarding how BRCA1-BARD1 helps mediate crucial steps in chromosome damage repair (see Section 5.4). Below, we outline some of the prevailing issues key to achieving holistic understanding of the multifaceted role of BRCA1-BARD1 in genome maintenance.
5.6a. Role in DNA end resection.
How BRCA1-BARD1 helps relieve the restriction imposed by the 53BP1-RIF1-Shieldin-CST ensemble on DNA end resection in S phase cells remains largely undefined. While published work has implicated BRCA1-BARD1 in the recruitment of the chromatin remodeler SMARCAD1 to facilitate resection, it will be important to determine whether BRCA1-BARD1 also recruits other factors, e.g. additional chromatin remodelers, or conjugates ubiquitin to components of the 53BP1-associated ensemble to further enhance access of the resection machinery to DNA damage. Even though BRCA1-BARD1 forms a complex with the DNA end resection factor CtIP (121), cells expressing a CtIP mutant defective in this interaction show only a mild resection deficiency (122). It is possible that BRCA1-BARD1 directly influences the activity of the nuclease entities, namely, the MRN complex, EXO1, BLM-DNA2 or WRN-DNA2, that catalyze the resection process. This premise awaits future investigations.
5.6b. Function in presynaptic filament assembly.
Existing evidence suggests that BRCA1-BARD1 aids in the assembly of the presynaptic filament (33, 63, 66, 101, 102). Since purified BRCA1-BARD1 is devoid of HR mediator activity (89), it likely functions within the context of the pentameric BRCA Mediator Complex that harbors PALB2 and BRCA2-DSS1 ((4); Figure 1). In this regard, one would expect the DNA binding and RAD51 interaction attributes of BRCA1-BARD1, BRCA2, and PALB2 (36, 103–105, 123) to provide a reservoir of RAD51 protomers and to increase the dwell time of the BRCA Mediator Complex on ssDNA to maximize the likelihood and efficiency of RPA replacement (Figure 1). This model awaits experimental validation. A recent study suggests that Shieldin interferes with the loading of RAD51 onto resected DNA ends (63). The basis for this effect of Shieldin on RAD51 presynaptic filament assembly and how it may be overcome by BRCA1-BARD1 remain to be delineated.
5.6c. Relationship with other RAD51 co-factors.
Like BRCA1-BARD1 (89), PALB2, RAD54, and the RAD51AP1-UAF1 complex each can enhance Rad51-mediated D-loop formation (8, 36, 37, 88). While RAD54 uses the free energy from ATP hydrolysis to generate negative supercoiling in the duplex target to facilitate DNA strand invasion, PALB2 and RAD51AP1-UAF1 enhance synaptic complex formation (8, 36, 37, 88). Given the association of PALB2 with BRCA1-BARD1 (Figure 1), it will be important to test whether they work together in synaptic complex assembly, and if BRCA2-DSS1 influences the synaptic activity of BRCA1-BARD1-PALB2. Moreover, it will be of considerable interest to examine whether BRCA1-BARD1-PALB2 functions in parallel or as a single entity with RAD54 or RAD51AP1-UAF1 in DNA strand invasion.
5.6d. Role in R-loop resolution.
R-loops are RNA-DNA hybrids that harbor a displaced ssDNA region. This structure arises mostly during transcription and serves important biological roles, e.g. in gene promoter regulation and transcription termination (124). However, R-loops that form upon perturbation of transcription or transcription-coupled mRNA splicing can interfere with DNA replication and lead to replication fork collapse. Cells possess at least three distinct mechanisms to avoid such transcription-replication conflicts, namely (i) avoidance of R-loop formation via topoisomerase I-mediated removal of negative DNA supercoiling; (ii) digestion of the RNA moiety in R-loops by RNase H; and (iii) dissociation of R-loops by putative RNA-DNA helicases, such as Senataxin (SETX) (124, 125). BRCA1 interacts with SETX and has been postulated to regulate the R-loop dissociative activity of the latter (124, 125). It will be important to test whether BRCA1 partners with BARD1 in the recognition of R-loops and in the enhancement of SETX to resolve these pathological structures.
5.6e. Function of the ubiquitin E3 ligase activity.
The E3 ubiquitin ligase activity of BRCA1-BARD1 is required for the protection of cells from the harmful effects of a variety of genotoxic agents but dispensable for the response to DNA replication stress or to treatment with cisplatin that induces ICLs (116). Thus far, the BRCA1-BARD1-mediated ubiquitination of histone H2A has been linked to the DNA end resection step of HR ((106, 116); see Section 5.4). It remains to be determined whether H2A ubiquitination is also germane for subsequent HR stages and whether there are other targets of the BRCA1-BARD1 E3 ligase activity relevant for the execution of HR and chromosome damage repair.
6. ROLE OF BRCA2 IN PRESYNAPTIC FILAMENT ASSEMBLY
6.1. Discovery of BRCA2 and Domain Structure
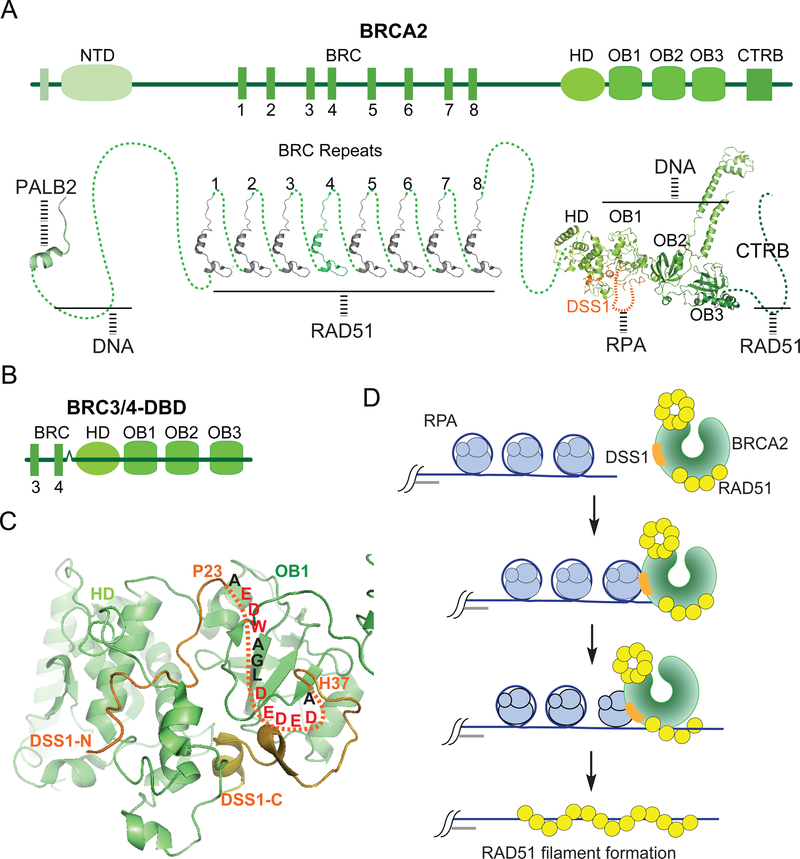
BRCA2 was identified based on genetic linkage analysis of non-BRCA1 families afflicted with breast cancer at a young age (126, 127). Subsequently, BRCA2 mutations were shown to engender a high risk to familial ovarian cancer and other cancer types (97). BRCA2 protein is exceptionally large, comprising 3,418 amino acid residues. The N-terminus of BRCA2 is involved in complex formation with PALB2 ((33); Figure 1). There are eight BRC (Breast Cancer) repeats (with each harboring ~30 residues) that interact with RAD51 individually (128, 129), and an additional, unrelated C-terminal RAD51-binding (CTRB) domain (41). BRCA2 possesses two DNA binding domains, one of which comprises three oligonucleotide/oligosaccharide binding (OB-1, 2, and 3) folds and is called the DBD (123), and the second, N-terminally proximal DNA binding module has been named the NTD (130, 131). A portion of OB1 and a neighboring helical domain constitute the interface for association with the small acidic protein DSS1 ((123, 132); Figure 4A; see Section 6.5).
Figure 4.
BRCA2 structure and function.
(A) Functional motifs and domains in BRCA2. The BRC (Breast Cancer) repeats, HD (helical domain), NTD (N-terminal DNA Binding Domain), DBD (DNA Binding Domain) comprising three OB (oligonucleotide/oligosaccharide binding) folds, and CTRB (C-terminal RAD51-binding) domain are shown. The HD and a portion of OB1 constitute the interface for association with DSS1 that physically interacts with RPA via a solvent-exposed acidic loop (77). The known atomic structures (PDB: 3EU7,1N0W and 1MIU) are linked by the dash lines depicting protein regions of which no structural information is available. (B) Schematic of the BRCA2-derived polypeptide composed of BRC3, BRC4 and the DBD. (C) Structure of the BRCA2-DSS1 complex ((123); PDB: 1IYJ)) with the BRCA2 parts in green. Residues in the solvent-exposed loop of DSS1 targeted for mutagenesis in the construction of a mutant defective in RPA interaction are in red (77). (D) Cartoon illustrating the HR mediator function of BRCA2-DSS1 in which BRCA2 provides the DNA binding and RAD51 interaction attributes, while DSS1 facilitates presynaptic filament assembly through its ability to interact with RPA and to act as a DNA mimetic to attenuate the affinity of RPA for ssDNA (77).
6.2. BRCA2 in Chromosome Damage Repair and Replication Fork Maintenance
Evidence that BRCA2 plays a role in chromosome damage repair and is essential for life came first from mouse studies (41). Specifically, mouse embryos deleted for exon 27 of the Brca2 gene were found to be hypersensitive to ionizing radiation and to arrest in early development. Moreover, yeast two-hybrid analysis revealed an interaction between the CTRB of Brca2 with the N-terminal region of Rad51 (41). Subsequently, the BRCA2 deficient CAPAN-1 cell line, derived from a pancreatic cancer patient, and a mouse cell line that harbors alleles of Brca2 deleted for the CTRB were shown to be deficient in HR-mediated gene targeting and also in the repair of a site-specific DSB by HR (133). More recently, BRCA2 was implicated in preventing the accumulation of pathogenic R-loops and also in the protection of stressed DNA replication forks against nucleolytic attack (15, 134). The replication fork protection activity of BRCA2 is thought to stem from its ability to nucleate the formation and stabilization of RAD51 filaments on the “chicken foot” structure derived from the enzymatic regression of a stressed fork (14, 135). Within this context, RAD51 likely shields regressed forks from various nucleases (14, 15, 135).
6.3. HR Mediator Activity of BRCA2 and its Ustilago maydis Ortholog Brh2
BRCA2 mutant cells are proficient in DNA end resection but fail to assemble RAD51 foci upon the occurrence of DNA damage (136), indicating that BRCA2 functions in the assembly of the RAD51 presynaptic filament. In this regard, biochemical testing of Brh2, the BRCA2 ortholog in U. maydis (a fungus that causes corn smut) that possesses a single BRC repeat, in complex with Dss1 furnished the first evidence for a HR mediator activity able to promote Rad51 presynaptic filament assembly on RPA-bound ssDNA (137). Notably, Brh2-Dss1 is devoid of HR mediator activity with E. coli RecA, being reflective of the fact that the BRC repeat in Brh2 has no affinity for RecA (137). In concordance with the above findings, fusing the BRC3 and BRC4 repeats and the DBD of human BRCA2 generates a polypeptide, BRC1/2-DBD (Figure 4B), that can physically interact with human RAD51 but not with yeast Rad51 or RecA. Importantly, the BRCA2 polypeptide could deliver human RAD51, but neither of the heterologous recombinases, to ssDNA pre-coated with RPA to form the presynaptic filament (138). Interestingly, BRC3/4-DBD is just as adept at mediating RAD51 presynaptic filament assembly on ssDNA decorated with the E. coli ssDNA binding protein SSB (138). Unlike RAD51, which binds ssDNA and dsDNA equally well (139), BRC3/4-DBD has higher affinity for ssDNA (138). Accordingly, BRC3/4-DBD specifically targets RAD51 to ssDNA when an excess of dsDNA is present (138). Thus, studies on Brh2-Dss1 and the BRCA2 polypeptide BRC3/4-DBD revealed two key attributes expected of a HR mediator, namely, preferential RAD51 targeting to ssDNA and seeding the assembly of the presynaptic filament on ssDNA pre-occupied by a ssDNA binding protein (137, 138).
The eventual purification of full-length human BRCA2 (77, 140–143) afforded the opportunity to test its HR mediator activity biochemically (77, 140–142) and by electron microscopy (143). Results from these endeavors have shown that the full-length protein is more adept than the BRC3/4-DBD polypeptide in RAD51 presynaptic filament assembly (77, 140). Like BRC3/4-DBD, BRCA2 is specific for its cognate recombinase and is equally efficacious with ssDNA bound by E. coli SSB (77, 140). In congruence with the latter finding, no physical interaction of BRCA2 with RPA has been detected (77, 140). As we will discuss in Section 6.5, DSS1 confers to the BRCA2-DSS1 complex specificity for RPA. As was first shown for BRC3/4-DBD, full length BRCA2 helps target RAD51 to ssDNA when dsDNA is present (140, 142). Analysis by electron microscopy of negatively stained BRCA2 has yielded an 18.5 Å reconstruction revealing a homodimeric structure (143).
6.4. Distinct Functions of the BRC Repeats
A systematic biochemical analysis has placed the BRC repeats in human BRCA2 into two distinct categories (139, 144). The first repeat class, comprising BRC1, 2, 3, and 4, is characterized by its high affinity for RAD51 that is not DNA bound. In contrast, BRC5, 6, 7, and 8, members of the second class, possess little affinity for free RAD51 but associate with the presynaptic filament avidly. Both types of BRC repeats stabilize the presynaptic filament, but they do so via a different mechanism. Specifically, while members of the first class work by attenuating ATP hydrolysis by RAD51, repeats from the second class do not affect this RAD51 attribute. Moreover, repeats of the first class suppress binding of RAD51 to dsDNA and enhance the ability of the presynaptic filament to catalyze DNA strand exchange. Thus, multiple BRC repeats with distinctive properties synergize to ensure the timely assembly and maintenance of the presynaptic filament, and also to enhance the catalytic potential of the filament. However, it should be noted that conjugation of selected BRC repeats to the DBD and/or CTRB yields polypeptides that are capable of complementing BRCA2 deficiency in cells (130, 145, 146), indicating that the eight BRC repeats function in an additive fashion in chromosome damage repair.
6.5. RPA-targeting and DNA Mimicry by DSS1
DSS1 comprises 70 amino acid residues and is highly acidic. Aside from its role in proteasome biogenesis and RNA metabolism (147), DSS1 also forms a stoichiometric complex with BRCA2 that is indispensable for chromosome damage repair (148, 149). Specifically, depletion of DSS1 engenders phenotypic changes reminiscent of BRCA2 deficiency, including hypersensitivity to agents that damage DNA or interfere with DNA replication and an impairment of RAD51 focus formation upon the occurrence of DNA damage (148, 149). Even though BRCA2 becomes destabilized when DSS1 is absent (77, 149), overexpression of BRCA2 fails to complement DSS1 deficiency (77), indicating that the latter fulfills at least one other role in HR distinct from BRCA2 stabilization. Importantly, the cancer-associated BRCA2-D2723H mutation, which affects HR and chromosome damage repair adversely (150), has been found to attenuate the interaction of BRCA2 with DSS1 and to cause mis-localization of BRCA2 to the cytoplasm due to the exposure of a nuclear export signal in the latter (151). As summarized below, a recent study documents how DSS1 influences BRCA2-dependent presynaptic filament assembly (77).
The BRCA2-DSS1 complex has been assembled in insect cells and purified for biochemical testing (77). In a side-by-side comparison, BRCA2-DSS1 was shown to be significantly more active than BRCA2 in the mediation of presynaptic filament assembly on ssDNA coated with human RPA. Importantly, while BRCA2 alone is equally adept at nucleating presynaptic filament assembly on ssDNA coated with E. coli SSB, the enhanced efficacy of BRCA2-DSS1 is specific for RPA-bound ssDNA. By affinity pulldown, DSS1 was found to physically interact with the N domain and all three DNA binding OB folds (OB-A, B, and C) of RPA70, the largest of the three RPA subunits. NMR analysis revealed that DSS1 makes contacts with DNA binding residues within the basic cleft of the OB folds, thus providing evidence for a DNA mimetic function in DSS1. Indeed, DSS1 was found to inhibit ssDNA binding by RPA but have no such effect on E. coli SSB. The X-ray crystal structure of the Brca2-Dss1 complex solved earlier (123) not only identifies the interfaces involved in protein complex formation, but also a solvent-exposed, disordered loop in Dss1 that is lined with acidic residues ((123); Figure 4C). To test the relevance of this solvent-exposed acidic loop in RPA interaction, a compound DSS1 point mutant that changes eight of the loop residues to alanine was constructed. The DSS18A mutant, while being proficient in BRCA2 interaction, is greatly compromised for the ability to associate with RPA or to attenuate ssDNA binding by RPA. Importantly, the BRCA2-DSS18A mutant complex is no more efficacious than BRCA2 in the promotion of presynaptic filament assembly. Accordingly, cells expressing the DSS18A mutant are hypersensitive to DNA damaging agents, defective in DNA damage-induced RAD51 focus formation, and impaired for HR. Thus, DSS1 targets BRCA2-RAD51 to RPA-bound ssDNA and also functions as a DNA mimetic to facilitate RPA-RAD51 exchange ((77); Figure 4D).
6.6. Questions Regarding the Genome Maintenance Role of BRCA2-DSS1
As mentioned earlier (see Section 5.6b and Figure 1), it remains to be determined how BRCA2-DSS1 synergizes with BRCA1-BARD1 and PALB2 within the context of the BRCA Mediator Complex in the assembly and maintenance of the presynaptic filament. Likewise, given that BRCA1 and BRCA2 function together to protect stressed replication forks from nucleolytic attack (15, 16), it will be important to verify that the other subunits of the BRCA Mediator Complex work in the same pathway of replication fork protection, and that DSS1 acts in the same manner as in presynaptic filament assembly.
BRCA2 possesses two DNA binding domains, the DBD and NTD (123, 130, 131). In one study, the NTD was found to have a higher affinity for DNA than the DBD (131), but the opposite conclusion was drawn in a separate investigation (130). Future work will resolve the discrepancy regarding DNA binding affinity of the NTD and DBD (130, 131), and will determine whether these domains fulfill different roles or function additively in presynaptic filament assembly and in replication fork protection.
Finally, like BRCA1 (125), BRCA2 suppresses the accumulation of pathogenic R-loops (134). It will be important to ascertain the role of DSS1 in this process and whether BRCA2 acts in conjunction with BRCA1-BARD1 and PALB2 in the recruitment of SETX and other R-loop processing enzymes, or if separate pathways of R-loop resolution exist that are dependent on subsets of these tumor suppressor proteins.
7. EPILOGUE
Deleterious mutations in HR genes have been documented for many cancer types and this common trait has been termed “BRCAness” (5). Notably, a recent pan-cancer analysis of the Cancer Genome Atlas data set revealed that over 5% of all cancers have bi-allelic pathogenic alterations in genes that mediate or regulate HR, and that the HR pathway is genetically compromised in 25% and 10% of all ovarian and breast cancers, respectively (152). Those cancer patients with an abnormality in HR may respond favorably to HR-directed therapies that entail the use of inhibitors of PARP, of cell cycle regulators, or of DNA damage checkpoints (5, 153, 154). With the availability of purified BRCA complexes (77, 89, 140–142), the field is now poised to delineate the mechanistic underpinnings of BRCA-mediated chromosome damage repair and replication forks protection. Such endeavors can be expected to provide the knowledge base to help drive the development of targeted cancer therapeutics, predict patient response to therapy, and explain acquired drug resistance.
ACKNOWLEDGEMENTS
The studies in our laboratories have been supported by grants RO1 CA220123, RO1 ES007061, PO1 CA092584, RO1 ES021454, RO1 CA205224, RO1 GM109645 and P30 CA054174 from the U.S. National Institutes of Health and by a Basser Innovation Award from the Basser Center for BRCA at Penn Medicine’s Abramson Cancer Center. We apologize to colleagues whose work is not cited because of article length restriction.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holding that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Lindahl T, Barnes DE. 2000. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 65: 127–33 [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. 2009. DNA damage, aging, and cancer. N Engl J Med 361: 1475–85 [DOI] [PubMed] [Google Scholar]
- 3.Garaycoechea JI, Crossan GP, Langevin F, Mulderrig L, Louzada S, et al. 2018. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 553: 171–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash R, Zhang Y, Feng W, Jasin M. 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 7: a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashworth A, Lord CJ. 2018. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol 15: 564–76 [DOI] [PubMed] [Google Scholar]
- 6.Her J, Bunting SF. 2018. How cells ensure correct repair of DNA double-strand breaks. J Biol Chem 293: 10502–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pannunzio NR, Watanabe G, Lieber MR. 2018. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 293: 10512–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright WD, Shah SS, Heyer WD. 2018. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 293: 10524–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zickler D, Kleckner N. 2015. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb Perspect Biol 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sallmyr A, Tomkinson AE. 2018. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J Biol Chem 293: 10536–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasero P, Vindigni A. 2017. Nucleases Acting at Stalled Forks: How to Reboot the Replication Program with a Few Shortcuts. Annu Rev Genet 51: 477–99 [DOI] [PubMed] [Google Scholar]
- 12.Vaisman A, Woodgate R. 2017. Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol 52: 274–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. 2003. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev 17: 3017–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22: 106–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, et al. 2015. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell 59: 478–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clauson C, Scharer OD, Niedernhofer L. 2013. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol 5: a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Walter JC. 2014. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 19: 135–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michl J, Zimmer J, Tarsounas M. 2016. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J 35: 909–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto S, Anai H, Hanada K. 2016. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ 38: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood RD. 2010. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ Mol Mutagen 51: 520–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amunugama R, Willcox S, Wu RA, Abdullah UB, El-Sagheer AH, et al. 2018. Replication Fork Reversal during DNA Interstrand Crosslink Repair Requires CMG Unloading. Cell Rep 23: 3419–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutreja K, Krietsch J, Hess J, Ursich S, Berti M, et al. 2018. ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Rep 24: 2629–42e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nalepa G, Clapp DW. 2018. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer 18: 168–85 [DOI] [PubMed] [Google Scholar]
- 26.Game JC, Mortimer RK. 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat Res 24: 281–92 [DOI] [PubMed] [Google Scholar]
- 27.Game JC. 1993. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol 4: 73–83 [PubMed] [Google Scholar]
- 28.Bai Y, Symington LS. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev 10: 2025–37 [DOI] [PubMed] [Google Scholar]
- 29.Klein HL. 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shor E, Weinstein J, Rothstein R. 2005. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics 169: 1275–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Symington LS. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66: 630–70, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Symington LS, Rothstein R, Lisby M. 2014. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198: 795–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, et al. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–29 [DOI] [PubMed] [Google Scholar]
- 34.Tischkowitz M, Xia B. 2010. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res 70: 7353–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nepomuceno TC, De Gregoriis G, de Oliveira FMB, Suarez-Kurtz G, Monteiro AN, Carvalho MA. 2017. The Role of PALB2 in the DNA Damage Response and Cancer Predisposition. Int J Mol Sci 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dray E, Etchin J, Wiese C, Saro D, Williams GJ, et al. 2010. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol 17: 1255–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, et al. 2010. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol 17: 1247–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. 1996. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet 12: 191–4 [DOI] [PubMed] [Google Scholar]
- 39.Hakem R, de la Pompa JL, Sirard C, Mo R, Woo M, et al. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85: 1009–23 [DOI] [PubMed] [Google Scholar]
- 40.Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH. 1996. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev 10: 1835–43 [DOI] [PubMed] [Google Scholar]
- 41.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, et al. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–10 [DOI] [PubMed] [Google Scholar]
- 42.McCarthy EE, Celebi JT, Baer R, Ludwig T. 2003. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol Cell Biol 23: 5056–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rantakari P, Nikkila J, Jokela H, Ola R, Pylkas K, et al. 2010. Inactivation of Palb2 gene leads to mesoderm differentiation defect and early embryonic lethality in mice. Hum Mol Genet 19: 3021–9 [DOI] [PubMed] [Google Scholar]
- 44.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–9 [DOI] [PubMed] [Google Scholar]
- 45.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, et al. 2007. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 39: 162–4 [DOI] [PubMed] [Google Scholar]
- 46.Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, et al. 2015. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov 5: 135–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golmard L, Castera L, Krieger S, Moncoutier V, Abidallah K, et al. 2017. Contribution of germline deleterious variants in the RAD51 paralogs to breast and ovarian cancers. Eur J Hum Genet 25: 1345–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brosh RM Jr., Cantor SB. 2014. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front Genet 5: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martino J, Bernstein KA. 2016. The Shu complex is a conserved regulator of homologous recombination. FEMS Yeast Res 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor MRG, Spirek M, Chaurasiya KR, Ward JD, Carzaniga R, et al. 2015. Rad51 Paralogs Remodel Pre-synaptic Rad51 Filaments to Stimulate Homologous Recombination. Cell 162: 271–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaines WA, Godin SK, Kabbinavar FF, Rao T, VanDemark AP, et al. 2015. Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52. Nat Commun 6: 7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen RB, Ozes A, Kim T, Estep A, Kowalczykowski SC. 2013. BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Repair (Amst) 12: 306–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Symington LS. 2016. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 51: 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, et al. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cejka P 2015. DNA End Resection: Nucleases Team Up with the Right Partners to Initiate Homologous Recombination. J Biol Chem 290: 22931–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cannavo E, Cejka P, Kowalczykowski SC. 2013. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci U S A 110: E1661–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, et al. 2014. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J Biol Chem 289: 27314–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinto C, Kasaciunaite K, Seidel R, Cejka P. 2016. Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Y, Lee SH, Williamson EA, Reinert BL, Cho JH, et al. 2015. EEPD1 Rescues Stressed Replication Forks and Maintains Genome Stability by Promoting End Resection and Homologous Recombination Repair. PLoS Genet 11: e1005675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, et al. 2015. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol 208: 545–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panier S, Boulton SJ. 2014. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15: 7–18 [DOI] [PubMed] [Google Scholar]
- 62.Bunting SF, Callen E, Wong N, Chen HT, Polato F, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dev H, Chiang TW, Lescale C, de Krijger I, Martin AG, et al. 2018. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta R, Somyajit K, Narita T, Maskey E, Stanlie A, et al. 2018. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 173: 972–88e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mirman Z, Lottersberger F, Takai H, Kibe T, Gong Y, et al. 2018. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noordermeer SM, Adam S, Setiaputra D, Barazas M, Pettitt SJ, et al. 2018. The shieldin complex mediates 53BP1-dependent DNA repair. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barazas M, Annunziato S, Pettitt SJ, de Krijger I, Ghezraoui H, et al. 2018. The CST Complex Mediates End Protection at Double-Strand Breaks and Promotes PARP Inhibitor Sensitivity in BRCA1-Deficient Cells. Cell Rep 23: 2107–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sung P 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–3 [DOI] [PubMed] [Google Scholar]
- 69.San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–57 [DOI] [PubMed] [Google Scholar]
- 70.Kowalczykowski SC. 2015. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb Perspect Biol 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. 2001. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci U S A 98: 8419–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.VanLoock MS, Yu X, Yang S, Lai AL, Low C, et al. 2003. ATP-mediated conformational changes in the RecA filament. Structure 11: 187–96 [DOI] [PubMed] [Google Scholar]
- 73.Short JM, Liu Y, Chen S, Soni N, Madhusudhan MS, et al. 2016. High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. Nucleic Acids Res 44: 9017–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu J, Zhao L, Xu Y, Zhao W, Sung P, Wang HW. 2017. Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat Struct Mol Biol 24: 40–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z, Yang H, Pavletich NP. 2008. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453: 489–4 [DOI] [PubMed] [Google Scholar]
- 76.Zelensky A, Kanaar R, Wyman C. 2014. Mediators of homologous DNA pairing. Cold Spring Harb Perspect Biol 6: a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao W, Vaithiyalingam S, San Filippo J, Maranon DG, Jimenez-Sainz J, et al. 2015. Promotion of BRCA2-Dependent Homologous Recombination by DSS1 via RPA Targeting and DNA Mimicry. Mol Cell 59: 176–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sung P 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–21 [DOI] [PubMed] [Google Scholar]
- 79.Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. 2001. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 15: 3308–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, et al. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–9 [DOI] [PubMed] [Google Scholar]
- 81.Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. 2011. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479: 245–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, et al. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol 21: 2858–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Renkawitz J, Lademann CA, Jentsch S. 2014. Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol 15: 369–83 [DOI] [PubMed] [Google Scholar]
- 84.Bianchi M, DasGupta C, Radding CM. 1983. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell 34: 931–9 [DOI] [PubMed] [Google Scholar]
- 85.Qi Z, Redding S, Lee JY, Gibb B, Kwon Y, et al. 2015. DNA sequence alignment by microhomology sampling during homologous recombination. Cell 160: 856–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao W, Sung P. 2015. Significance of ligand interactions involving Hop2-Mnd1 and the RAD51 and DMC1 recombinases in homologous DNA repair and XX ovarian dysgenesis. Nucleic Acids Res 43: 4055–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ito K, Murayama Y, Takahashi M, Iwasaki H. 2018. Two three-strand intermediates are processed during Rad51-driven DNA strand exchange. Nat Struct Mol Biol 25: 29–36 [DOI] [PubMed] [Google Scholar]
- 88.Liang F, Longerich S, Miller AS, Tang C, Buzovetsky O, et al. 2016. Promotion of RAD51-Mediated Homologous DNA Pairing by the RAD51AP1-UAF1 Complex. Cell Rep 15: 2118–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao W, Steinfeld JB, Liang F, Chen X, Maranon DG, et al. 2017. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 550: 360–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee JY, Terakawa T, Qi Z, Steinfeld JB, Redding S, et al. 2015. DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases. Science 349: 977–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prindle MJ, Loeb LA. 2012. DNA polymerase delta in DNA replication and genome maintenance. Environ Mol Mutagen 53: 666–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. 1983. The double-strand-break repair model for recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- 93.West SC, Blanco MG, Chan YW, Matos J, Sarbajna S, Wyatt HD. 2015. Resolution of Recombination Intermediates: Mechanisms and Regulation. Cold Spring Harb Symp Quant Biol 80: 103–9 [DOI] [PubMed] [Google Scholar]
- 94.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, et al. 1990. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250: 1684–9 [DOI] [PubMed] [Google Scholar]
- 95.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71 [DOI] [PubMed] [Google Scholar]
- 96.Friedman LS, Ostermeyer EA, Szabo CI, Dowd P, Lynch ED, et al. 1994. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet 8: 399–404 [DOI] [PubMed] [Google Scholar]
- 97.Petrucelli N, Daly MB, Pal T. 1993–2018. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer In GeneReviews((R)), ed. Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, et al. Seattle (WA) 1998 Sep 4 [updated 2016 Dec 15] [Google Scholar]
- 98.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, et al. 1996. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet 14: 430–40 [DOI] [PubMed] [Google Scholar]
- 99.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, et al. 2001. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276: 14537–40 [DOI] [PubMed] [Google Scholar]
- 100.Christou CM, Kyriacou K. 2013. BRCA1 and Its Network of Interacting Partners. Biology (Basel) 2: 40–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang F, Ma J, Wu J, Ye L, Cai H, et al. 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19: 524–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang F, Fan Q, Ren K, Andreassen PR. 2009. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res 7: 1110–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. 2001. Direct DNA binding by Brca1. Proc Natl Acad Sci U S A 98: 6086–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Simons AM, Horwitz AA, Starita LM, Griffin K, Williams RS, et al. 2006. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res 66: 2012–8 [DOI] [PubMed] [Google Scholar]
- 105.Scully R, Chen J, Plug A, Xiao Y, Weaver D, et al. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265–75 [DOI] [PubMed] [Google Scholar]
- 106.Densham RM, Morris JR. 2017. The BRCA1 Ubiquitin ligase function sets a new trend for remodelling in DNA repair. Nucleus 8: 116–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Irminger-Finger I, Ratajska M, Pilyugin M. 2016. New concepts on BARD1: Regulator of BRCA pathways and beyond. Int J Biochem Cell Biol 72: 1–17 [DOI] [PubMed] [Google Scholar]
- 108.Thai TH, Du F, Tsan JT, Jin Y, Phung A, et al. 1998. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum Mol Genet 7: 195–202 [DOI] [PubMed] [Google Scholar]
- 109.Shakya R, Szabolcs M, McCarthy E, Ospina E, Basso K, et al. 2008. The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc Natl Acad Sci U S A 105: 7040–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shen SX, Weaver Z, Xu X, Li C, Weinstein M, et al. 1998. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene 17: 3115–24 [DOI] [PubMed] [Google Scholar]
- 111.Cortez D, Wang Y, Qin J, Elledge SJ. 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286: 1162–6 [DOI] [PubMed] [Google Scholar]
- 112.Moynahan ME, Cui TY, Jasin M. 2001. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res 61: 4842–50 [PubMed] [Google Scholar]
- 113.Moynahan ME, Chiu JW, Koller BH, Jasin M. 1999. Brca1 controls homology-directed DNA repair. Mol Cell 4: 511–8 [DOI] [PubMed] [Google Scholar]
- 114.Laufer M, Nandula SV, Modi AP, Wang S, Jasin M, et al. 2007. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J Biol Chem 282: 34325–33 [DOI] [PubMed] [Google Scholar]
- 115.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem 275: 23899–903 [DOI] [PubMed] [Google Scholar]
- 116.Densham RM, Garvin AJ, Stone HR, Strachan J, Baldock RA, et al. 2016. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat Struct Mol Biol 23: 647–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferretti LP, Lafranchi L, Sartori AA. 2013. Controlling DNA-end resection: a new task for CDKs. Front Genet 4: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, et al. 2015. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521: 541–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, et al. 2015. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5’ end resection. Nature 521: 537–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kalb R, Mallery DL, Larkin C, Huang JT, Hiom K. 2014. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep 8: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem 273: 25388–92 [DOI] [PubMed] [Google Scholar]
- 122.Cruz-Garcia A, Lopez-Saavedra A, Huertas P. 2014. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep 9: 451–9 [DOI] [PubMed] [Google Scholar]
- 123.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, et al. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–48 [DOI] [PubMed] [Google Scholar]
- 124.Santos-Pereira JM, Aguilera A. 2015. R loops: new modulators of genome dynamics and function. Nat Rev Genet 16: 583–97 [DOI] [PubMed] [Google Scholar]
- 125.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, et al. 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 57: 636–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, et al. 1994. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12–13. Science 265: 2088–90 [DOI] [PubMed] [Google Scholar]
- 127.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, et al. 1995. Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–92 [DOI] [PubMed] [Google Scholar]
- 128.Bork P, Blomberg N, Nilges M. 1996. Internal repeats in the BRCA2 protein sequence. Nat Genet 13: 22–3 [DOI] [PubMed] [Google Scholar]
- 129.Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. 1998. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A 95: 5287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chatterjee G, Jimenez-Sainz J, Presti T, Nguyen T, Jensen RB. 2016. Distinct binding of BRCA2 BRC repeats to RAD51 generates differential DNA damage sensitivity. Nucleic Acids Res 44: 5256–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.von Nicolai C, Ehlen A, Martin C, Zhang X, Carreira A. 2016. A second DNA binding site in human BRCA2 promotes homologous recombination. Nat Commun 7: 12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. 1999. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol 19: 4633–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moynahan ME, Pierce AJ, Jasin M. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 7: 263–72 [DOI] [PubMed] [Google Scholar]
- 134.Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511: 362–5 [DOI] [PubMed] [Google Scholar]
- 135.Kolinjivadi AM, Sannino V, de Antoni A, Techer H, Baldi G, Costanzo V. 2017. Moonlighting at replication forks - a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett 591: 1083–100 [DOI] [PubMed] [Google Scholar]
- 136.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res 59: 3547–51 [PubMed] [Google Scholar]
- 137.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. 2005. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433: 653–7 [DOI] [PubMed] [Google Scholar]
- 138.San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. 2006. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem 281: 11649–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, et al. 2009. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136: 1032–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jensen RB, Carreira A, Kowalczykowski SC. 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu J, Doty T, Gibson B, Heyer WD. 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, et al. 2010. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17: 1263–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shahid T, Soroka J, Kong E, Malivert L, McIlwraith MJ, et al. 2014. Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nat Struct Mol Biol 21: 962–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carreira A, Kowalczykowski SC. 2011. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci U S A 108: 10448–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Saeki H, Siaud N, Christ N, Wiegant WW, van Buul PP, et al. 2006. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci U S A 103: 8768–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Siaud N, Barbera MA, Egashira A, Lam I, Christ N, et al. 2011. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet 7: e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kragelund BB, Schenstrom SM, Rebula CA, Panse VG, Hartmann-Petersen R. 2016. DSS1/Sem1, a Multifunctional and Intrinsically Disordered Protein. Trends Biochem Sci 41: 446–59 [DOI] [PubMed] [Google Scholar]
- 148.Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A. 2004. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep 5: 989–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q. 2006. DSS1 is required for the stability of BRCA2. Oncogene 25: 1186–94 [DOI] [PubMed] [Google Scholar]
- 150.Wu K, Hinson SR, Ohashi A, Farrugia D, Wendt P, et al. 2005. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res 65: 417–26 [PubMed] [Google Scholar]
- 151.Jeyasekharan AD, Liu Y, Hattori H, Pisupati V, Jonsdottir AB, et al. 2013. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat Struct Mol Biol 20: 1191–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Riaz N, Blecua P, Lim RS, Shen R, Higginson DS, et al. 2017. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun 8: 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–21 [DOI] [PubMed] [Google Scholar]
- 154.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, et al. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–7 [DOI] [PubMed] [Google Scholar]