Abstract
Kinetochores are the multiprotein complexes that link chromosomal centromeres to mitotic-spindle microtubules. Budding yeast centromeres comprise three sequential centromere-determining elements, CDEI, II, and III. CDEI (8 bp) and CDEIII (~25 bp) are conserved between Kluyveromyces lactis and Saccharomyces cerevisiae, but CDEII in the former is twice as long (160 bp) as CDEII in the latter (80 bp). The CBF3 complex recognizes CDEIII and is required for assembly of a centromeric nucleosome, which in turn recruits other kinetochore components. To understand differences in centromeric nucleosome assembly between K. lactis and S. cerevisiae, we determined the structure of a K. lactis CBF3 complex by electron cryomicroscopy at ~4 Å resolution and compared it with published structures of S. cerevisiae CBF3. We show differences in the pose of Ndc10 and discuss potential models of the K. lactis centromeric nucleosome that account for the extended CDEII length.
Keywords: point centromere, kinetochore, centromeric nucleosome assembly, budding yeast, electron cryomicroscopy
Introduction
Kinetochores are multiprotein complexes that assemble on centromeres to ensure the faithful segregation of sister chromatids during mitosis. In addition to coupling microtubule dynamics to sister chromatid movement, kinetochores also regulate entry into anaphase by monitoring and correcting erroneous chromatid attachments. Budding yeast have a ~120- to 200-bp point centromere” on each chromatid that recruits a single centromere-specific nucleosome containing a specialized histone H3 variant (Cse4 in budding yeast, CENP-A in metazoans) [1]. The Cse4 nucleosome directs assembly of the kinetochore superstructure and subsequent attachment of a single spindle microtubule. Most other eukaryotes, including fission yeast, have longer, regional centromeres” with multiple centromere-specific nucleosomes [2–5]. The corresponding kinetochores, although they contain homologs of most of the yeast kinetochore components, are more extended along each chromatid than their budding yeast counterparts, with substantially more copies of each of the more than 50 structural proteins.
In contrast to the DNA sequence-independent propagation of centromeric nucleosomes in organisms with regional centromeres, the budding yeast centromere has a defined sequence, which enables formation of a Cse4 nucleosome after each round of DNA replication [6–9]. Indeed, yeast cellular extracts are competent to construct Cse4 particles on naked centromeric DNA, demonstrating the sufficiency of this sequence motif for conferring centromere identity [10]. Early studies defined three centromere-determining elements” (CDEs) in budding yeast centromeres: CDEI, CDEII, and CDEIII [11–13]. The flanking elements, CDEI and CDEIII, are ~8 and ~25 bp long, respectively, and their nucleotide sequences are conserved across budding yeast species [14]. The central element, CDEII, wraps around the centromeric core octamer [15], It is 80 bp long in Saccharomyces cerevisiae but twice that length in Kluyveromyces lactis [16]. The only similarity between the two CDEII sequences is their high adenine–thymidine nucleotide content (80%–90%). Although changes in CDEII length can be tolerated in either species to a limited extent, the centromeres cannot be interchanged between them [16–19]. DNase footprinting experiments show protection of 123–135 bp for S. cerevisiae centromeres, but ~270 bp for K. lactis, suggesting structural differences in their respective centromeric nucleosomes [20–22].
The CBF3 complex accounts for the sequence-specific Cse4 nucleosome assembly pathway. It is a biochemically defined entity containing two copies each of Ndc10 and Cep3 and one copy each of Ctf13 and Skp1. The heterohexamer binds CDEIII, in a register determined by a conserved CCG, recruits Scm3, the chaperone for a Cse4:H4 heterodimer, and sets the position of the centromeric nucleosome [22,23], DNA–protein crosslinking experiments show that Cep3, Ctf13, and Ndc10 have defined contacts with CDEIII [24], In particular, a Zn2Cys6 domain of Cep3 makes sequence-specific contacts with the conserved CCG motif in CDEIII. A recent structure of the S. cerevisiae CBF3 bound to a 147-bp DNA duplex of CEN3 (centromere from chromosome 3) is fully consistent with those results [25].
We report here the structure of K. lactis CBF3, as part of an effort to understand differences in organization or assembly of centromeric nucleosomes between yeast species with different CDEII lengths. We compare our structure with that of S. cerevisiae CBF3 and show differences in the pose of Ndc10 [25,26]. Finally, we discuss models of centromere organization in K. lactis that account for the differences in CDEII length.
Results and Discussion
We co-expressed the K. lactis CBF3 core (Cep3)2–Ctf13-Skp1 and Ndc10 and purified the heterohexameric CBF3 complex by size exclusion chromatography (Fig. S1a and b). Preliminary electron cryomicroscopy (cryo-EM) images showed heterogeneous particles, unsuitable for high-resolution structural studies (data not shown). We therefore expressed a monomeric fragment of Ndc10 (residues 1–403; Ndc10 D1D2), combined it with recombinant CBF3 core (Fig. S1c and d), stabilized the complex by crosslinking with a bifunctional PEG-NHS ester, and purified it by size exclusion chromatography (Fig. 1a). We confirmed the molecular mass of the crosslinked complex using multi-angle light scattering (MALS); the measured molecular mass (262 ± 8 kDa) corresponded to that of a single copy of Ndc10 D1D2 bound to CBF3 core (Fig. 1b).
Fig 1.
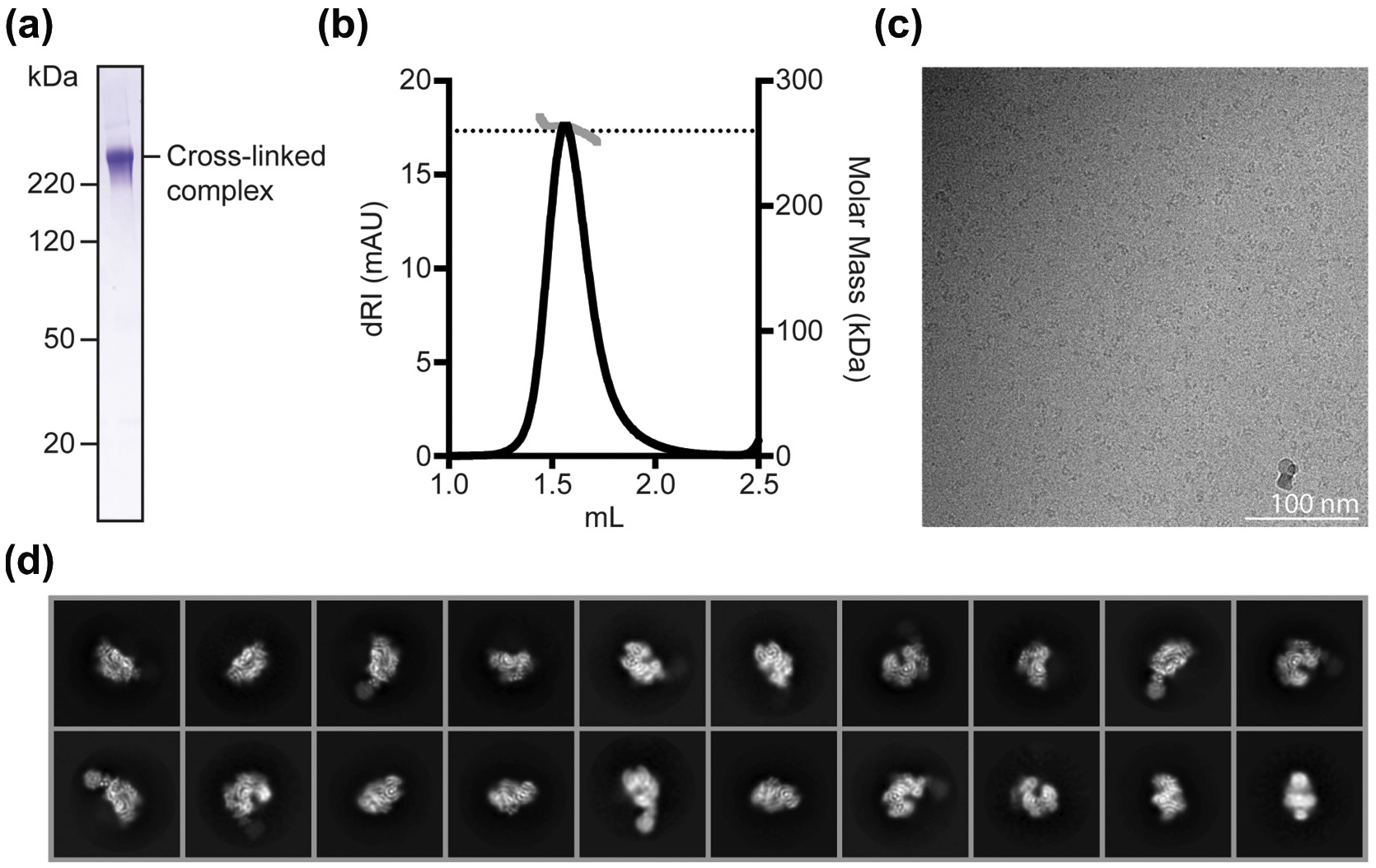
Purification, cryo-EM data collection and 2D averages for K. lactis CBF3–Ndc10 D1D2 complex, (a) SDS-PAGE analysis of cross-linked CBF3 complex, (b) Mass determination by MALS of cross-linked CBF3 complex, (c) Representative micrograph from grids prepared with Spotiton, showing cross-linked CBF3 particles embedded in vitreous ice. (d) Representative results of 2D class averages obtained with RELION-3 from ~ 1.6 million particles picked from motion-corrected and summed movies. Box size for particles is 288 × 288 (pixels).
Because reproducible ice thickness was critical to high-resolution imaging of the particle, we prepared self-wicking cryo-grids using the Spotiton instrument (New York Structural Biology Center) [27], collected 6946 movies, which we motion-corrected and summed, and picked approximately 1.6 million particles by automated particle picking in cisTEM (Fig. 1c) [27–29], Two-dimensional (2D) classification showed secondary-structure features for most of the complex (Fig. 1d). Following standard classification procedures described in Materials and Methods and illustrated in Fig. S2, we obtained three reconstructions from the motion-corrected and summed movies: (1) the CBF3 core at a resolution of 4.0 Å, (2) CBF3 core bound to Ndc10 domain I (CBF3–Ndc10 D1) at 4.1 Å resolution, and (3) CBF3 core bound to Ndc10 domains I and II (CBF3–Ndc10 D1D2) at 4.3 Å resolution (Fig. 2a and Fig. S3a, e, and i). Both the CBF3 core and CBF3–Ndc10 D1 maps had density corresponding to secondary-structure elements and amino-acid side chains, enabling us to build and refine atomic models (Fig. S3d and h). The CBF3–Ndc10 D1D2 map showed secondary-structure elements in all regions except those corresponding to Ndc10 D2, for which the density was weak and fragmented. For all maps, we also observed segments of broken density corresponding primarily to surface loops, most of which we did not attempt to model (See Table 1).
Fig. 2.
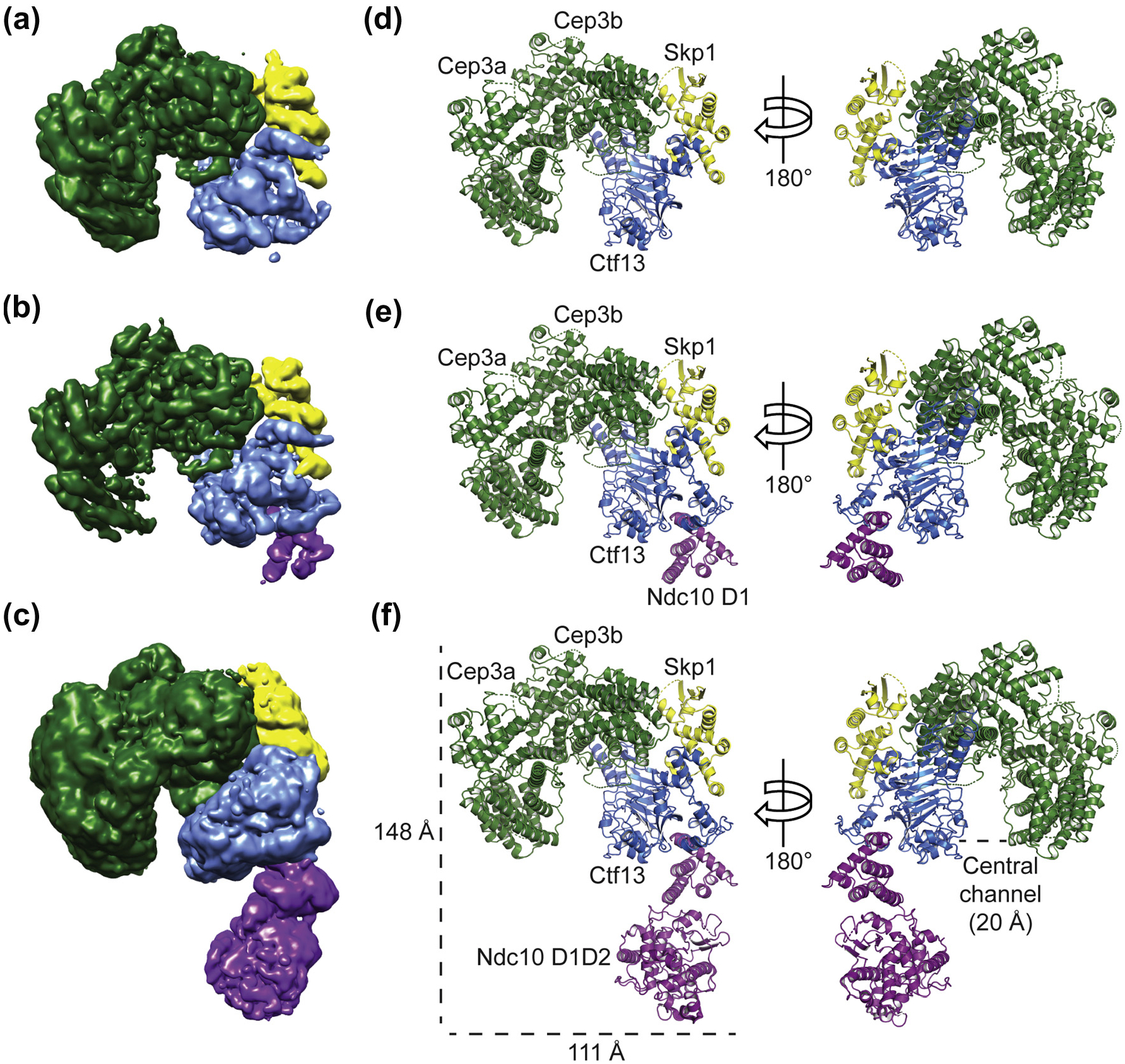
Structures of K. lactis CBF3 core, CBF3–Ndc10 D1, and CBF3–Ndc10 D1D2. Reconstructions of (a) CBF3 core, (b) CBF3–Ndc10 D1, and (c) CBF3–Ndc10 D1D2. Models of (d) CBF3 core, (e) CBF3–Ndc10 D1, and (f) CBF3–Ndc10 D1D2. For reconstructions and models, Cep3 is in green, Skp1 in yellow, Ctf13 in blue, and Ndc10 in purple.
Table 1.
Cryo-EM data collection, reconstruction, and modeling statistics
| Data collection | Data Collection 1 | Data Collection 2 | |
|---|---|---|---|
| Microscope | Titan Krios | Titan Krios | |
| Voltage (kV) | 300 | 300 | |
| Magnification | 130,000X | 130,000X | |
| Camera | K2 Summit | K2 Summit | |
| Energy filter slit | 20 eV | 20 eV | |
| Defocus range | 1.2–2.2 (underfocus) | 1.5–2.5 (underfocus) | |
| Pixel size | 1.0605 | 1.0605 | |
| Images (number) | 3260 | 3711 | |
| Reconstruction | CBF3 core | CBF3 Ndc10 D1 | CBF3 Ndc10 D1D2 |
| Particles | 130,143 | 63,177 | 63,177 |
| Box size | 288 | 288 | 288 |
| Accuracy of rotations | 2.56 | 3.32 | 2.27 |
| Accuracy of translations | 0.91 | 1.23 | 1.41 |
| Map resolution—FSC threshold 0.143 | 3.97 | 4.07 | 4.30 |
| Map sharpening B factor | 100 | 80 | 10 |
| Model refinement and statistics | |||
| Refinement resolution | 4.0 | 4.1 | |
| CC (mask) | 0.7985 | 0.78 | |
| R.M.S. deviations | |||
| Bond lengths | 0.011 | 0.007 | |
| Bond angles | 1.199 | 0.942 | |
| Validation | |||
| MolProbity scores | 1.91 | 1.96 | |
| Clashscore | 6.57 | 6.86 | |
| Favored rotamers | 95.34% | 95.81% | |
| Poor rotamers | 0.08% | 0.07% | |
| Cbeta deviations 0.25Å | 0 | 0 | |
| Ramachandran plot | |||
| Favored (%) | 89.99% | 88.63% | |
| Allowed (%) | 9.79% | 11.10% | |
| Disallowed (%) | 0.22% | 0.27% | |
| Peptide omega | |||
| Cis prolines (%) | 0.00% | 0.00% | |
| Twisted peptides | 0.15% | 0.20% |
FSC, Fourier shell correlation
To model the K. lactis CBF3 core, we generated homology models of Cep3, Ctf 13, and Skp1 based on published S. cerevisiae structures (PDB 6GSA) and docked them into our CBF3 core map [26,30]. Using these homology models as templates, we built and refined an atomic model of K. lactis CBF3 core (Fig. 2d). We then used the K. lactis CBF3 core model, together with the atomic coordinates describing the crystal structure of K. lactis Ndc10 D1 (PDB 3SQI) [31], to interpret the density of our CBF3–Ndc10 D1 map (Figs. 2e and Fig. S3d). The CBF3–Ndc10 D1 map contains density that could not be assigned to either the homology models of CBF3 core components or to Ndc10 D1. Guided by side-chain density, we assigned this region to Ctf 13 residues 28–85. Real-space refinement yielded an atomic model of K. lactis CBF3–Ndc10 D1 (Fig. 2e). We docked our CBF3–Ndc10 D1 model and the published crystal structure of K. lactis Ndc10 D2 (PDB 3SQI) [31] into our CBF3–Ndc10 D1D2 map to generate a model of the full complex (Fig. 2f). Ndc10 D2 fit well into the density, but weak and fragmented regions of density for this subdomain prevented us from rebuilding or refining the model (Fig. S3k). With the exception of density for Ndc10 D2, the maps of CBF3–Ndc10 D1 and CBF3–Ndc10 D1D2 were essentially identical (Table 1).
K. lactis CBF3 core has a horseshoe shape resulting from the orientation with which a Cep3 homodimer binds the heterodimer of Ctf13 and Skp1 (Fig. 2f). Only one Cep3 subunit (designated Cep3b in our coordinate set) interacts with Ctf 13 and Skp1 (blue box, Fig. S4a). Cep3a, Cep3b, and Ctf13 surround a central channel, approximately 20 Å in diameter. Conformational flexibility in the sample, which lacks DNA, may account for absence of density for either of the Zn2Cys6 domains of Cep3, one of which recognizes the CDEIII CCG (Fig. S4a). Ctf 13 has an N-terminal F-box, which binds Skp1 and Cep3b, and a leucine-rich repeat (LRR) with eight, parallel -strands (Fig. S4b and 3a–c). Between the F-box and the LRR is a small, intervening domain. Long loops between strands 6 and 7 and strands 7 and 8 of the LRR together form a subdomain that caps the extended sheet (Fig. S4b). This subdomain forms polar and non-polar interactions with Cep3b (Fig. 3d).
Fig. 3.
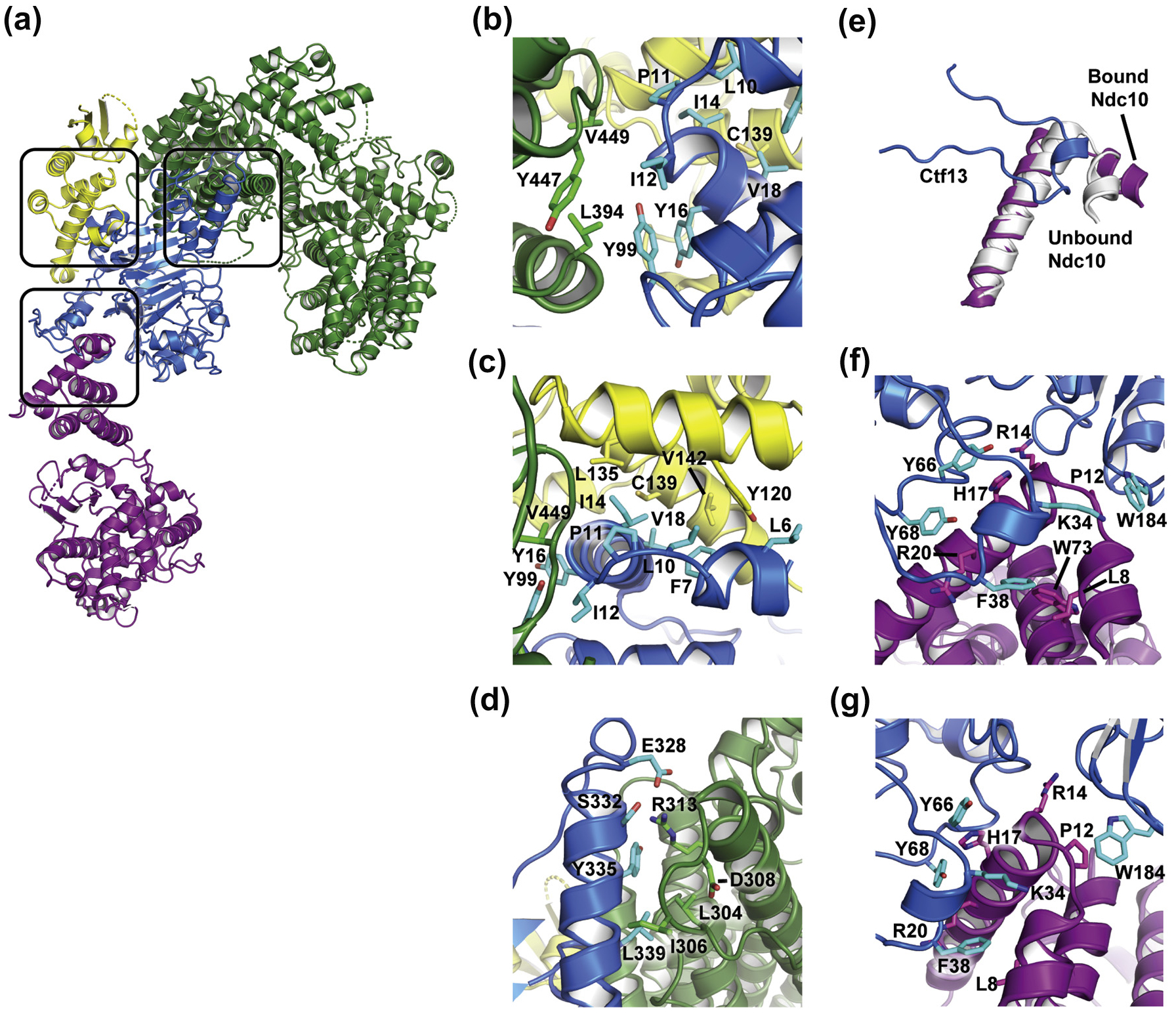
Interactions between subunits of the K. lactis CBF3–Ndc10 D1D2 complex, (a) Overall structure with Ctf 13–Skp1–Cep3b (F-box) interface, separate Ctf13–Cep3b interface, and Ctf13–Ndc10 D1D2 interface highlighted in black boxes, (b–c) Two views of residues at the F-box interface, (d) The Ctf13–Cep3b interface, (e) Isolated view of Ctf 13 residues 28–48 (blue) with the first two helices of Ndc10 D1D2 from the complex (purple) and from the published crystal structure (PDB 3SQI, gray), showing a reorientation of the first, shorter helix to accommodate interactions with Ctf13. (f–g) Two views of residues at the interface between Ctf13 and Ndc10 D1D2.
A canonical F-box, as found in E3 ubiquitin ligases, is a bundle of three α-helices of about 10 residues each (Fig. S5a). Skp1 clamps around this bundle in Ctf 13 with three helices of its own. The Ctf13 F-box is anomalous in several respects. Instead of the second helix, it has a long, 60-residue loop, which contributes to the binding surface for domain 1 of Ndc10, while a ~ 7-residue helix augments the bundle at the N-terminal end. These variations do not allow the final helix of K. lactis Skp1 to create the distal jaw of its usual clamp, and the residues at its C-terminus do not appear to be well ordered. The metazoan Skp2 is also an F-box-LRR protein. Visual comparison of the Skp1–Skp2 structure with that of Skp1–Ctf 13 shows little resemblance between the two, either of the structure of the LRR domains themselves or of their orientation with respect to the Skp1–F-box module (Fig. S5b–c).
The monomeric Ndc10 D1D2 extends from the CBF3 core complex (Fig. 3a). It interacts with Ctf 13 through its N-terminal domain (domain 1: residues 1–100), in accord with published biochemical data [31]. This interaction does not involve extensive contacts and buries a surface area of only −826 Å2. When it binds Ctf 13, the N-terminal helix of Ndc10 undergoes a small conformational change relative to the crystal structure (Fig. 3e), creating a set of polar and non-polar interactions with Ctf 13 (Fig. 3f–g). The interaction between Ctf 13 and Ndc10 D1 positions the DNA binding surface of Ndc10 D2 about 50 Å from the entrance to the CBF3 core central channel, which is proximal to the Cep3a Zn2Cys6 domain that recognizes the CDEIII CCG motif. The potential dimer interface seen in crystals of Ndc10 D1D2 with DNA is incompatible with the interaction we now see for Ndc10 bound to CBF3 core, because the second Ndc10 molecule would collide with Ctf13 [31].
The CBF3 cores of S. cerevisiae and K. lactis superpose well on each other (Fig. 4a). The principal differences between the two structures are the orientations of the extended Ctf13 F-box loop (residues 28–85) and of Ndc10 D1D2 (Fig. 4a). In K. lactis the DNA binding interface of Ndc10 is closer to CBF3 core than in S. cerevisiae. This difference in Ndc10 D1D2 orientation is due to interactions with Ctf13. In particular, the K. lactis Ctfl 3 F-box loop extends downward by an additional 15 Å relative to its S. cerevisiae counterpart in order to interact with the Ndc10 N-terminal helices. This contact is further stabilized by an interaction between Ndc10 and a loop that extends from the third strand of the Ctf 13 LRR. We docked the crystal structure of Ndc10 D1D2 bound to a 30-bp fragment of DNA and found that the K. lactis Ndc10 D1D2 positions DNA in plane with the central CBF3 core channel (Fig. 4b). The orientation of the K. lactis Ndc10 DNA binding domain would probably create a more substantial bend in the DNA than in the structure of S. cerevisiae CBF3 bound to CDEIII (Fig. 4b–c). Because the domains of Ndc10 responsible for dimerization (residues 410–540) and for Scm3 binding (residues 541–736) were absent in our construct [31], we cannot determine how Ndc10 positions Scm3–Cse4–H4 near CDEII to induce the assembly of a centromeric nucleosome. The DNA bend that K. lactis Ndc10 may generate at CDEIII could affect how Cse4–H4 is deposited onto CDEII.
Fig. 4.
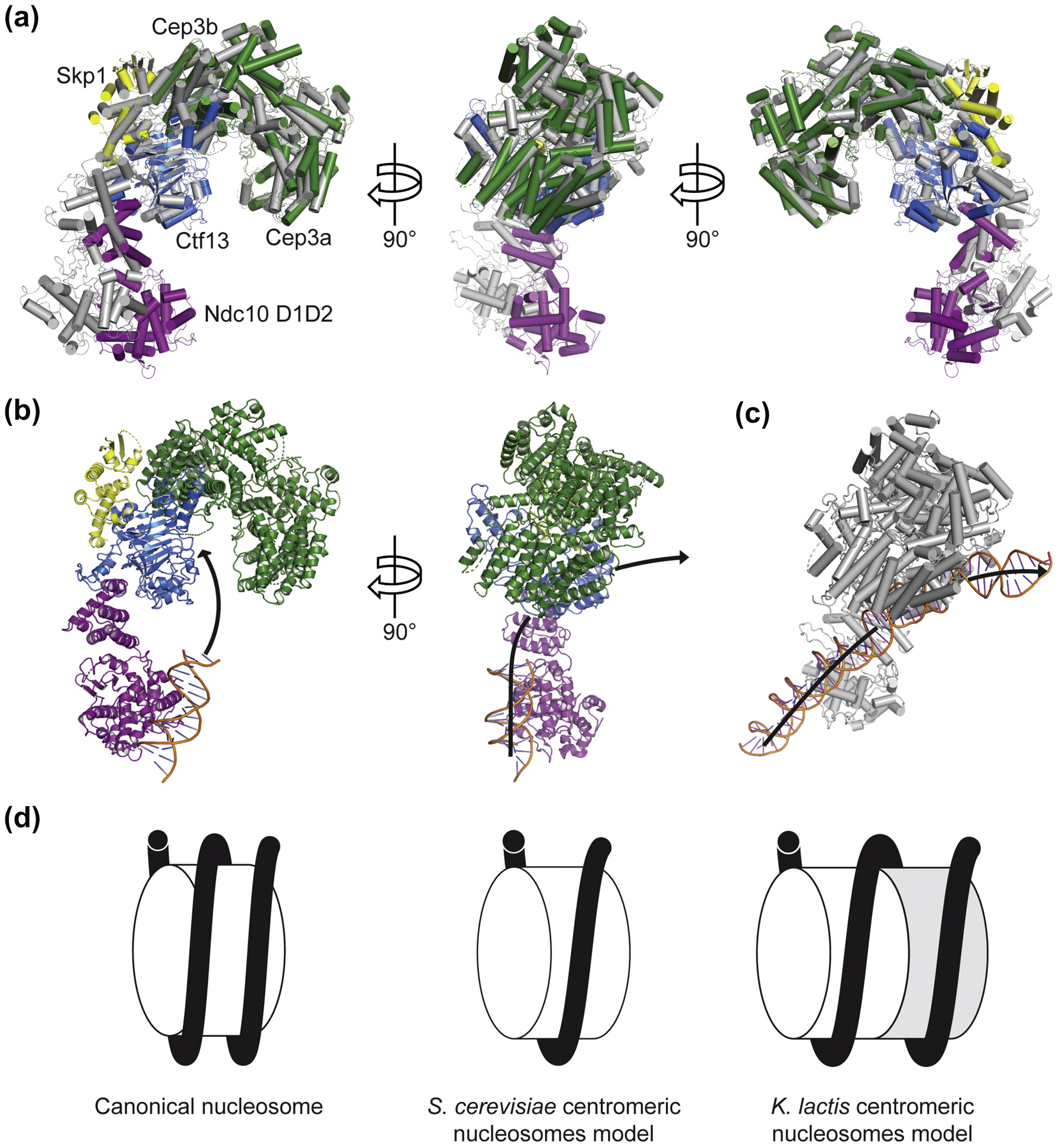
Comparison of the K. lactis CBF3–Ndc10 D1D2 structure with that of S. cerevisiae [25] (a) Overlay of K. lactis (green, yellow, blue, and purple) and S. cerevisiae CBF3–Ndc10 D1D2 (gray) structures aligned on Cep3b, viewed at 0°, 90°, and 180° as shown and illustrating the different orientations of Ndc10 D1D2 with respect to CBF3 core. All helices are represented as cylinders, (b) K. lactis CBF3–Ndc10 D1D2 structure with proposed path of DNA drawn, showing how the relative orientation of CBF3 core and Ndc10 D1D2 impacts positioning of DNA between Ndc10 binding site and CBF3 core channel, (c) Structure of S. cerevisiae CBF3 showing the path of DNA. (d) Comparison of models of a canonical nucleosome, S. cerevisiae centromeric nucleosome, and K. lactis centromeric nucleosome showing face-to-face packing of two centromeric octamers.
K. lactis and S. cerevisiae have point centromeres of different lengths, yet their kinetochore components are similar. In particular, we find no striking differences between the CBF3 structures from K. lactis and S. cerevisiae, nor are there noteworthy differences between the Scm3 chaperones from the two species. These similarities suggest that the mechanisms of centromeric nucleosome assembly are conserved among budding yeast. The centromeres cannot be interchanged between the two species, however, presumably because of the differences in CDEII lengths [16]. Therefore, how does the K. lactis kinetochore encompass 80 bp more DNA than its S. cerevisiae counterpart?
In considering possible models, we note that almost all known sequences of yeast point centromeres have either one length or the other, and that those with longer CDEIIs (~ 160-bp) are phylogenetically related (Fig. S6). Within any one of these related species, all chromosomes have centromeres of nearly identical length (Fig. S6) [14,32,33], We infer that a well-defined, structural feature accounts for this pattern. Moreover, additional evidence for a compact organization of some kind comes from micrococcal nuclease digestion experiments showing full protection of the entire centromere in both K. lactis and S. cerevisiae [20,22], For the latter, protection of 123–135 bp (depending on the centromere) shows precise localization from 1 bp upstream of CDEI to a few base pairs downstream of CDEIII [22], Reconstituted Cse4 nucleosomes also show partly unwrapped DNA ends, even on the nucleosome-favored 601 sequence [34], consistent with inferences from the structure of the Ctf19 complex (Ctf19c) that probably no more than one DNA wrapping (about 80 bp; i.e., CDEII) is in direct contact with the histone core [35]. Protection of 123–135 bp thus likely includes contributions from both Cbf1 and CBF3 and potentially also from Ctf19c.
We considered a model in which the additional DNA would wrap around a single histone octamer, but it would either position the DNA entry and exit sites on opposite ends of the nucleosomes or result in a mostly unsupported extra turn on one face of the octamer. The former scenario is unlikely given the conservation of kinetochore components such as Mif2 and Ctf19c, and the latter is equally unlikely because it would require an additional kinetochore component not present in yeasts with shorter CDEII sequences. Furthermore, DNAse I footprinting of S. cerevisiae chromatin with CDEII mutations that double its length results in a ~ 300-bp protected fragment, suggesting that expanded Cse4 nucleosomes can form without additional kinetochore factors and even with S. cerevisiae CBF3 [17]. Therefore, the most plausible explanation for both conservation and full protection is one full additional turn of DNA wrapped like the first around a second centromeric core octamer (Fig. 4d). The model is essentially a continuous, two-turn coil of DNA on two histone octamers stacked face-to-face, in a configuration that minimizes any residual electrostatic repulsion [36]. Except for the asymmetry conferred on one side by CDEI and Cbf1 and on the other side by CDEIII and CBF3, the abutting particles would be essentially 2-fold-related copies of each other. In support of this idea, numerous stacked canonical nucleosome structures have been determined [37].
CBF3 deposits Cse4–H4 heterodimers most likely by recruiting Scm3–Cse4–H4 complexes through binding of Scm3 to Ndc10. The Ndc10 dimer in CBF3 could thus nucleate a full octameric complement, by recruiting two copies of Scm3–Cse4–H4. Deposition of a second octamer would require either exchange of Cse4–H4 from an upstream chaperone onto Ndc10-bound Scm3 or exchange of empty” Scm3 for a copy loaded with its Cse4–H4 client. The number of octamers loaded would be limited by CDEI-bound Cbf1, which would block deposition of a third octamer, in K. lactis, or of a second, in S. cerevisiae. To test this model, it will be necessary to isolate or reconstitute the K. lactis centromeric nucleosome and to measure the number of Cse4 at centromeres in dividing K. lactis cells.
Materials and Methods
Cloning expression and purification of CBF3 core Ndc10 and Ndc10 D1D2
K. lactis CBF3 core components were cloned as pairs (Cep3–Sgt1 and Ctf13–Skp1), each with an N-terminal His tag, into pFastBacDuel plasmids modified for ligation-independent cloning. CBF3 core was expressed by co-infection of Hi5 cells with both baculoviruses and harvested after 72 h. Zinc acetate was added to the medium at a final concentration of 1 μM at the time of co-infection. Cells were pelleted and resuspended in binding buffer (500 mM NaCI, 30 mM Tris–HCI, 1 mM TCEP, pH 8), supplemented with protease inhibitors (1 mM PMSF and 1 μg/mL each of aprotinin, leupeptin, pepstatin). Cells were disrupted by sonication and debris removed by two rounds of centrifugation at 19,500g for 30 min. Imidazole was added to the supernatant to a final concentration of 10 mM, and the supernatant was applied to Cobalt Talon Resin (Takara Bio USA). The column was washed with binding buffer containing 10 mM imidazole and eluted with elution buffer (150 mM NaCI, 30 mM Tris–HCI, 600 mM imidazole, 1 mM TCEP, pH 8). Protein was loaded onto a 5-mL HiTrap Q HP column (GE) equilibrated with loading buffer (150 mM NaCI, 30 mM Tris–HCI, pH 8). Sgt1 dissociated from CBF3 core during the loading step. CBF3 core was eluted from the column by increasing the salt concentration to 1M. Fractions containing the target proteins were pooled and tobacco etch virus (TEV) protease added to the elution fraction in approximately 1:100 (w/w) ratio. After overnight incubation with rotation at 4 °C, the protease-treated sample was concentrated and applied to a Superdex 200 size exclusion column (GE) equilibrated with gel filtration buffer (150 mM NaCI, 30 mM Hepes, 1 mM TCEP, pH 8). Peak fractions were pooled, concentrated, flash frozen in liquid nitrogen, and stored at 80 °C.
The full-length K. lactis Ndc10 gene was codon optimized for bacterial expression (GenScript) and sub-cloned into a pET vector modified for ligation-independent cloning and encoding an N-terminal maltose binding protein tag and TEV protease cleavage site. The construct was transformed into Rosetta 2(DE3)pLys-competent cells (Novagen). Cells were grown at 37 °C until OD600 = 0.6, and expression was induced by the addition of IPTG to a final concentration of 1 mM (Gold Biotechnology). Cells were then grown at 18 °C overnight and centrifuged, and the pellet was harvested and resuspended in binding buffer. Protein was purified using cobalt talon affinity. The maltose binding protein tag was removed by incubation with TEV protease in approximately 1:100 (w/w) ratio at 4 °C overnight, and the protein was purified using ion-exchange and size-exclusion chromatography. Peak fractions were pooled, concentrated and stored at 80 °C for further use. Protein was in a final buffer of 150 mM NaCI, 30 mM Hepes, and 1 mM TCEP (pH 8).
K. lactis Ndc10 D1D2 (residues 1–410) with an N-terminal His tag was expressed and purified as previously described [31]. In brief, Ndc10 D1D2 was expressed in Rosetta 2(DE3)pLysS-competent cells (Novagen) and expression was induced by the addition of IPTG to a final concentration of 1 mM. Cells were grown at 18 °C overnight and centrifuged, and the pellet was harvested and resuspended in binding buffer. Protein was purified using cobalt talon affinity and ion-exchange chromatography. The His tag was removed by incubation with TEV protease in approximately 1:100 (w/w) ratio at 4 °C overnight. Protein was purified using size-exclusion chromatography in a final buffer of 150 mM NaCI, 30 mM Hepes, and 1 mM TCEP (pH 8).
MALS
Crosslinked CBF3 complex was analyzed by size-exclusion chromatography coupled to MALS using a Wyatt MALS system. Sample was injected onto a Superdex 200 5/150 gel-filtration column (GE) equilibrated in 150 mM NaCI, 30 mM Hepes, 1 mM TCEP, and 0.02% NaN3 (pH 8). Data were processed using standard pipelines in the Astra software package (Wyatt).
Assembly of the CBF3 complex and Spotiton vitrification
For initial in vitro CBF3 assembly reactions, CBF3 core and either Ndc10 or Ndc10 D1D2 were added in a 1:5 molar ratio and incubated at room temperature for 60 min. Sample was run over a size-exclusion chromatography column in 150 mM NaCI, 30 mM Hepes, and 1 mM TCEP (pH 8), and peak fractions were analyzed by SDS-PAGE.
In order to assemble the CBF3 complex for structural studies, CBF3 core and Ndc10 D1D2 were added in a 1:1.5 molar ratio and incubated at room temperature for 30 min. Bis-PEG2-NHS ester solubilized in DMSO (BroadPharm) was added to a final concentration of 1.5 mM and incubated for an additional 30 min. The crosslinker was neutralized by the addition of Tris–HCI (pH 8) to a final concentration of 100 mM. The sample was dialyzed overnight at 4 °C into 80 mM potassium acetate, 30 mM Tris–HCI, and 1 mM TCEP (pH 8) and applied to a size-exclusion chromatography column (GE) equilibrated with the same buffer. Selected fractions were collected and concentrated to 1.5–2.0 mg/mL.
Samples were vitrified in liquid ethane using the Spotiton robot at the New York Structural Biology Center [27,28]. Samples were applied to either copper or gold lacey carbon nanowire grids [38]. The spot-to-freeze time was set between 170 ms and 1.0 s. Grids were screened for particle density and vitreous ice conditions on a Technai T12 (FEI) or Technai F20 (FEI) electron microscope prior to data collection.
Cryo-EM data collection
Two cryo-EM data sets were collected on a Titan Krios electron microscope (Thermo Fisher Scientific) operating at 300 keV equipped with a post-column, Bio-Quantum energy filter and K2 Summit detector (Gatan). Movies were recorded using a nominal defocus of 1.2 to 2.5 μm and the energy filter slit set to a width of 20 eV. The total dose was ~ 65 e per A2 for each movie. A total of 6926 micrographs were collected semi-automatically using Leginon [39,40].
Image processing
Movies were aligned and summed using Motion-Cor2 within RELION-3 [41,42], CTF parameters were estimated using CTFFIND4 [43]. Corrected micrographs were exported to cisTEM, and particles selected by automated picking [29], Particle coordinates were imported into RELION-3, confirmed by eye, extracted from the images (1,653,364 particles), and subjected to five rounds of 2D classification. Particles in the best 2D classes were selected for three-dimensional (3D) classification (753,528 particles). After one round of 3D classification with three classes, the 367,572 particles in the best class were subjected to 3D auto-refinement in RELION-3, Bayesian polishing, and a subsequent round of 3D auto-refinement [44]. We noticed weak density for domain 2 (D2) of Ndc10, subtracted out the corresponding signal in real space, and carried out a further round of 3D auto-refinement and masked classification with three classes. The 227,684 particles in the best class were subjected to 3D auto-refinement, CTF refinement, and an additional round of 3D auto-refinement in RELION-3. These 227,684 particles were processed using two separate strategies. In strategy 1, we performed focused classification on CBF3 core without alignment and selected the 130,143 particles belonging to the best class for 3D auto-refinement. This procedure resulted in a reconstruction for CBF3 core at 4.0 Å resolution. In strategy 2, we performed focused classification without alignment on Cep3b–Skp1–Ctf13–Ndc10 D1 and selected for 3D auto-refinement the 63,177 particles showing clear density for Ndc10 D1. These steps resulted in a reconstruction of CBF3–Ndc10 D1 at 4.1 Å resolution. We restored the signal for D2 (previously subtracted from the particle images), performed a round of 3D auto-refinement, and obtained a map of the CBF3 complex at 4.3 Å resolution, although the density corresponding to D2 was at a lower resolution.
Model building
We created homology models using SWISS-MODEL, based on the published S. cerevisiae CBF3 core structure (PDB: 6GSA) [26,30,45–48]. Regions were rebuilt de novo using Coot to fit the experimental density of either the CBF3 core or CBF3–Ndc10 D1 maps, and both structures were refined with PHENIX real-space refinement, applying standard secondary constraints and using Cep3b coordinates as a reference for Cep3a [49,50]. We fit the CBF3–Ndc10 D1 structure (as a single, rigid body) and the crystal structure of Ndc10 D2 into the CBF3–Ndc10 D1D2 map in Coot [31], The D2 density did not allow additional real-space refinement of this structure.
Structure analysis
Buried surface area between Ctf 13 and Ndc10 was calculated by measuring the solvent-accessible surface area of Ctf 13 and Ndc10 separately and together in PyMol, then subtracting the surface area of both subunits together from the sum of the individual surface areas and dividing by two. Atomic distances were measured in PyMol using the measurement tool.
Supplementary Material
Acknowledgments
We thank Thomas Walz, Simon Jenni, and Stephen Hinshaw for helpful discussions and advice during data collection, processing, model building, and manuscript preparation; Uhn-soo Cho for providing plasmids used in this work; Zongli Li and Melissa Chambers for cryo-EM support at Harvard Medical School; Edward Eng and Laura Yen for cryo-EM support at the New York Structural Biology Center; and Clarence Cheng for helpful feedback on this manuscript prior to publication. Grid preparation and data collection were conducted at the National Resource for Automated Molecular Microscopy at the New York Structural Biology Center, supported by grants from the National Institutes of Health, National Institute of General Medical Sciences (GM103310) and the Simons Foundation (SF349247). We acknowledge support from the National Science Foundation (P.D.L.) and HHMI (S.C.H.).
Abbreviations used:
- CDE
centromere-determining element
- cryo-EM
electron cryomicroscopy
- K. lactis
Kluyveromyces lactis
- LRR
leucine-rich repeat
- S. cerevisiae
Saccharomyces cerevisiae
- TEV
tobacco etch virus
Footnotes
Accession numbers
The final maps (and half maps) for K. lactis CBF3 core, CBF3–Ndc10 D1, and CBF3–Ndc10 D1D2 have been deposited with accession codes EMD-20270, EMD-20271, and EMD-20272. The models have been deposited with accession codes 6P7V, 6P7W, and 6P7X.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2019.08.003.
References
- [1].Furuyama S, Biggins S, Centromere identity is specified by a single centromeric nucleosome in budding yeast, Proc. Natl. Acad. Sei. U. S. A 104 (2007) 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Clarke L, Baum MP, Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences, Mol. Cell. Biol 10 (1990) 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Murphy TD, Karpen GH, Localization of centromere function in a Drosophila minichromosome, Cell 82 (1995) 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pardue ML, Gall JG, Chromosomal localization of mouse satellite DNA, Science 168 (1970) 1356–1358. [DOI] [PubMed] [Google Scholar]
- [5].Malik HS, Henikoff S, Major evolutionary transitions in centromere complexity, Cell 138 (2009) 1067–1082. [DOI] [PubMed] [Google Scholar]
- [6].Musacchio A, Desai A, A molecular view of kinetochore assembly and function, Biology (Basel) 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Karpen GH, Allshire RC, The case for epigenetic effects on centromere identity and function, Trends Genet 13 (1997) 489–496. [DOI] [PubMed] [Google Scholar]
- [8].Wiens GR, Sorger PK, Centromeric chromatin and epigenetic effects in kinetochore assembly, Cell. 93 (1998) 313–316. [DOI] [PubMed] [Google Scholar]
- [9].Wisniewski J, Hajj B, Chen J, Mizuguchi G, Xiao H, Wei D, et al. , Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres, eLife 3 (2014), e02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lang J, Barber A, Biggins S, An assay for de novo kinetochore assembly reveals a key role for the CENP-T pathway in budding yeast, eLife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Clarke L, Carbon J, Isolation of a yeast centromere and construction of functional small circular chromosomes, Nature. 287 (1980) 504–509. [DOI] [PubMed] [Google Scholar]
- [12].Fitzgerald-Hayes M, Clarke L, Carbon J, Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs, Cell. 29 (1982) 235–244. [DOI] [PubMed] [Google Scholar]
- [13].Hieter P, Pridmore D, Hegemann JH, Thomas M, Davis RW, Philippsen P, Functional selection and analysis of yeast centromeric DNA, Cell. 42 (1985) 913–921. [DOI] [PubMed] [Google Scholar]
- [14].Gordon JL, Byrne KP, Wolfe KH, Mechanisms of chromosome number evolution in yeast, PLoS Genet 7 (2011), e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiao H, Wang F, Wisniewski J, Shaytan AK, Ghirlando R, FitzGerald PC, et al. , Molecular basis of CENP-C association with the CENP-A nucleosome at yeast centromeres, Genes Dev. 31 (2017) 1958–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Heus JJ, Zonneveld BJ, de Steensma HY, van den Berg JA, The consensus sequence of Kluyveromyces lactis centromeres shows homology to functional centromeric DNA from Saccharomyces cerevisiae, Mol Gen Genet 236 (1993) 355–362. [DOI] [PubMed] [Google Scholar]
- [17].Saunders M, Fitzgerald-Hayes M, Bloom K, Chromatin structure of altered yeast centromeres, Proc. Natl. Acad. Sei. U. S. A 85 (1988) 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Heus JJ, Zonneveld BJ, Steensma HY, Van den Berg JA, Centromeric DNA of Kluyveromyces lactis, Curr. Genet 18 (1990)517–522. [DOI] [PubMed] [Google Scholar]
- [19].Sreekrishna K, Webster TD, Dickson RC, Transformation of Kluyveromyces lactis with the kanamycin (G418) resistance gene of Tn903, Gene. 28 (1984) 73–81. [DOI] [PubMed] [Google Scholar]
- [20].Heus JJ, Bloom KS, Zonneveld BJ, Steensma HY, Van den Berg JA, Chromatin structures of Kluyveromyces lactis centromeres in K lactis and Saccharomyces cerevisiae, Chromosoma. 102 (1993) 660–667. [DOI] [PubMed] [Google Scholar]
- [21].Funk M, Hegemann JH, Philippsen P, Chromatin digestion with restriction endonucleases reveals 150–160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae, Mol Gen Genet 219 (1989) 153–160. [DOI] [PubMed] [Google Scholar]
- [22].Cole HA, Howard BH, Clark DJ, The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere, Proc. Natl. Acad. Sei. U. S. A 108 (2011) 12687–12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lechner J, Carbon J, A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere, Cell. 64 (1991) 717–725. [DOI] [PubMed] [Google Scholar]
- [24].Espelin CW, Kaplan KB, Sorger PK, Probing the architecture of a simple kinetochore using DNA–protein crosslinking, J. Cell Biol 139 (1997) 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yan K, Zhang Z, Yang J, McLaughlin SH, Barford D, Architecture of the CBF3–centromere complex of the budding yeast kinetochore, Nat. Struct. Mol. Biol 25 (2018) 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang W, Lukoyanova N, Miah S, Lucas J, Vaughan CK, Insights into centromere DNA bending revealed by the cryo-EM structure of the core centromere binding factor 3 with Ndc10, Cell Rep. 24 (2018) 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dandey VP, Wei H, Zhang Z, Tan YZ, Acharya P, Eng ET, et al. , Spotiton: new features and applications, J. Struct. Biol 202 (2018) 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jain T, Sheehan P, Crum J, Carragher B, Potter CS, Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM, J. Struct. Biol 179 (2012) 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Grant T, Rohou A, Grigorieff N, cisTEM, user-friendly software for single-particle image processing, eLife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. , SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res. 46 (2018) W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cho US, Harrison SC, Ndc10 is a platform for inner kinetochore assembly in budding yeast, Nat. Struct. Mol. Biol 19 (2012) 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kobayashi N, Suzuki Y, Schoenfeld LW, Muller CA, Nieduszynski C, Wolfe KH, et al. , Discovery of an unconventional centromere in budding yeast redefines evolution of point centromeres, Curr. Biol 25 (2015) 2026–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hedtke SM, Townsend TM, Hillis DM, Resolution of phylogenetic conflict in large data sets by increased taxon sampling, Syst. Biol 55 (2006) 522–529. [DOI] [PubMed] [Google Scholar]
- [34].Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K, Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes, Nat. Commun 2 (2011) 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hinshaw SM, Harrison SC, The structure of the Ctf 19c/CCAN from budding yeast, eLife. 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gebala M, Johnson SL, Narlikar GJ, Herschlag D, Ion counting demonstrates a high electrostatic field generated by the nucleosome, eLife. 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Korolev N, Lyubartsev AP, Nordenskiold L, A systematic analysis of nucleosome core particle and nucleosome-nucleosome stacking structure, Sei. Rep 8 (2018) 1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wei H, Dandey VP, Zhang Z, Raczkowski A, Rice WJ, Carragher B, et al. , Optimizing self-wicking” nanowire grids, J. Struct. Biol 202 (2018) 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, et al. , Automated molecular microscopy: the new Leginon system, J. Struct. Biol 151 (2005) 41–60. [DOI] [PubMed] [Google Scholar]
- [40].Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, et al. , Leginon: an automated system for acquisition of images from vitreous ice specimens, J. Struct. Biol 132 (2000) 33–45. [DOI] [PubMed] [Google Scholar]
- [41].Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA, MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy, Nat. Methods 14 (2017) 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3, eLife. 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs, J. Struct. Biol 192 (2015)216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zivanov J, Nakane T, Scheres SHW, A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis, lUCrJ. 6 (2019) 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Benkert P, Biasini M, Schwede T, Toward the estimation of the absolute quality of individual protein structure models, Bioinformatics. 27 (2011) 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T, Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology, Sei. Rep 7 (2017) 10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, et al. The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res 2017;45: D313–D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Guex N, Peitsch MC, Schwede T, Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective, Electrophoresis. 30 (Suppl. 1) (2009) S162–S173. [DOI] [PubMed] [Google Scholar]
- [49].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot, Acta Crystallogr D Biol Crystallogr. 66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, et al. , Real-space refinement in PHENIX for cryo-EM and crystallography, Acta Crystallogr D Struct Biol. 74 (2018) 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


