Abstract
Metabolic reprogramming, including the Warburg effect, is a hallmark of cancer. Indeed, the diversity of cancer metabolism leads to cancer heterogeneity, but accurate assessment of metabolic properties in tumors has not yet been undertaken. Here, we performed absolute quantification of the expression levels of 113 proteins related to carbohydrate metabolism and antioxidant pathways, in stage III colorectal cancer surgical specimens from 70 patients. The Warburg effect appeared in absolute protein levels between tumor and normal mucosa specimens demonstrated. Notably, the levels of proteins associated with the tricarboxylic citric acid cycle were remarkably reduced in the malignant tumors which had relapsed after surgery and treatment with 5‐fluorouracil‐based adjuvant therapy. In addition, the efficacy of 5‐fluorouracil also decreased in the cultured cancer cell lines with promotion of the Warburg effect. We further identified nine and eight important proteins, which are closely related to the Warburg effect, for relapse risk and 5‐fluorouracil benefit, respectively, using a biomarker exploration procedure. These results provide us a clue for bridging between metabolic protein expression profiles and benefit from 5‐fluorouracil adjuvant chemotherapy.
Keywords: absolute protein levels, colorectal cancer, relapse risk and chemotherapy benefit, tegafur‐uracil, Warburg effect
Using absolute quantification of protein levels, we identified that the behavior of Warburg effect‐related proteins was strongly associated with relapse prediction and the therapeutic indication of uracil‐tegafur (UFT)‐adjuvant therapy.
1. INTRODUCTION
In colorectal cancer (CRC), surgical resection alone is potentially curative, but local or distant relapse frequently develops in many patients. Indeed, patients with the highest risk of relapse are advised to receive 5‐fluorouracil (5‐FU)‐based systemic adjuvant chemotherapy, which has been determined to be beneficial in several cooperative group trials and analyses.1, 2, 3 Randomized trials have revealed that postoperative 5‐FU‐based adjuvant chemotherapy significantly improves relapse‐free survival and overall survival in patients with stage III CRC. However, this therapy remains ineffective in approximately 30% of patients.4 Although several potential predictive factors of relapse risk and chemotherapy benefit have been investigated,5, 6, 7, 8 they are not currently applied routinely in clinical practice.
Many studies have reported the relationship between cellular metabolism and therapeutic outcomes, including relapse and chemotherapy resistance.9, 10 Indeed, metabolic differences exist between malignant tumor cells and adjacent normal cells. In cancer cells, the glycolytic phenotype is caused by an impairment of mitochondrial oxidative phosphorylation (OXPHOS), a phenomenon called the Warburg effect,11 which has a critical role in cancer development. The activation of glycolysis imbues cancer cells with a powerful growth advantage that prevails over efficient ATP generation through OXPHOS where cell proliferation requires the synthesis of new molecules including nucleic acids, lipids and proteins. NADPH, a reducing equivalent, is increased with synthesis of nucleic acids via the oxidative branch of the pentose‐phosphate pathway (PPP).12 While the relationship between cellular metabolism related to the Warburg effect and therapeutic outcomes is implied, it is not still illustrated quantitatively at protein expression level.
Mass spectrometry has achieved the speed and sensitivity required to measure proteomes at a depth comparable to gene expression studies.13, 14 Moreover, selected/multiple reaction monitoring (SRM/MRM) in MS/MS devices is an essential analytical mode that allows the target peptides of target proteins to be distinguished in extremely complex samples and provides high selectivity and a high signal‐to‐noise ratio.15, 16 Combination of SRM/MRM and stable‐isotope‐labeled (internal standard, IS) peptides has led to the highly accurate absolute quantification of target proteins.17 Protein absolute quantitation detects small changes under various conditions to achieve a high‐throughput analysis of entire pathways. In addition to determining the global changes of absolute protein expression levels in tumor cells,17 proteomic analysis might help identify key molecules representing tumor malignancy in tumor tissues.
Here, we performed absolute quantification of expression levels of proteins in carbohydrate metabolism and antioxidant pathways, which were reported to be associated with cancer malignancy,18, 19, 20 in 70 CRC patients. Based on the protein quantification, we compared the expression levels of proteins in tumor and normal mucosa CRC patients, to investigate the relationship between the Warburg effect and malignancy. This relationship was also examined experimentally in cultured CRC cells and statistically by verifying the accuracy of markers for relapse risk and chemotherapy benefit. This is the first investigation of how the protein expression levels in carbohydrate metabolism are associated with the benefit from 5‐FU adjuvant chemotherapy in CRC.
2. MATERIALS AND METHODS
2.1. Human clinical samples
Patients with stage III colon cancer were randomly assigned and treated with uracil‐tegafur (UFT) in NSAS‐CC/RC. The study design and eligibility criteria have been reported previously.3 Enrolment of patients in the original trials occurred between October 1996 and April 2001. Patients were randomly assigned to receive either adjuvant chemotherapy with UFT or no chemotherapy treatment within 6 weeks after surgery. In the UFT group, UFT (tegafur 400 mg/m2/d; Taiho Pharmaceutical) was given orally, twice daily for 5 days/wk for 1 year. When this study was carried out, leucovorin tablets could not be used because they had not been approved in Japan; therefore, UFT alone was used. The stage was classified according to the General Rules for Clinical and Pathological Studies on Cancer of the Colon, Rectum and Anus.21 Cancers arising from the rectosigmoid colon were classified as rectal cancer. A diagnosis of recurrence was assessed by abdominal ultrasonography or computed tomography at 4‐month intervals during the first 2 years and at 6‐month intervals thereafter. In this study, we used all 70 freshly frozen tissues which were randomly preserved at the National Cancer Center Hospital. We developed a colon cancer survival classifier using 70 normal and 70 tumor region samples. Of the 70 patients, 31 (44%) underwent surgery alone, of which 11 (35%) relapsed, and 39 (56%) were treated with UFT, of which 10 (26%) relapsed (Table 1). The characteristics of patients did not differ significantly between surgery alone and UFT treatment groups.
Table 1.
Characteristics of 70 patients from the NSAS‐CC/RC study
| Surgery alone | UFT treatment | |
|---|---|---|
| Gender | ||
| Male | 15 | 24 |
| Female | 16 | 15 |
| Median age at diagnosis (range), y | 60.1 (35‐72) | 61.1 (41‐72) |
| Relapse | ||
| Yes | 11 (35.4%) | 10 (25.6%) |
| No | 20 (64.5%) | 29 (74.4%) |
Abbreviation: UFT, uracil‐tegafur.
This study was carried out in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of the National Cancer Center Hospital and the National Institute of Advanced Industrial Science and Technology.
2.2. Cell lines
HCT116 cells were purchased from the ATCC and grown and maintained in RPMI1640 media supplemented with 10% FBS. For non–adherent culture, dissociated single cells were seeded into PrimeSurface culture plates (Sumitomo Bakelite) and were cultured for 7 days. Quantification of cell viability was performed using the bioluminescence‐based CellTiter‐Glo assay (Promega), as described in the manufacturer’s manual.
2.3. Absolute quantification of expression levels of proteins in specimens
Absolute quantification of expression levels of proteins was carried out as described previously.16 Specimens were lysed with tissue lysis buffer and digested with Lysyl‐C and trypsin. Typically, known concentrations of stable‐isotope‐labeled internal standard peptides were added to digested lysate samples, and then measured by multiple reaction monitoring analysis using the QTRAP5500 instrument (SCIEX). The peptide sequences used for quantification of each protein level are listed in Table S1 (more details are provided in Appendix S1).
2.4. Protein‐based classifier
We built a protein‐based classifier using the following procedure. First, we performed logistic regression analysis for patients belonging to two groups: one group contained patients who had relapsed or had not relapsed after surgery alone; the second group contained patients who had relapsed or had not relapsed after treatment with UFT adjuvant therapy. In the regression analysis, variables included the combination of three proteins randomly selected from 66 proteins. In total, 3422 regressions of all combinations of three of 66 proteins were performed for the two groups. Second, we performed receiver operating characteristic (ROC) analysis and estimated the area under the curve (AUC) value in each regression. Third, we selected the best 100 sets of proteins according to the AUC and counted the frequency of proteins that appeared in these 100 sets (Table S2).
2.5. Evaluation of diagnosis models
We investigated the predictive accuracy of multi‐protein‐based classifier using ROC analysis. We used the AUC to measure predictive accuracy. The stability of the models was evaluated using a 10‐fold cross‐validation test. We used the “survivalROC” package of R software to perform the ROC curve analysis.
2.6. Statistical analysis
For hierarchical clustering, distances between the samples were defined as the Spearman correlation and were clustered using complete linkage with Mev software (Dana‐Farber Cancer Institute, Boston, MA, USA).
To determine the relapse‐free survival of patients in each clusters, we performed Kaplan‐Meier analysis with a log‐rank test using the “EZR” (Jichi Medical University, Saitama, Japan) package of R software.22
We used a logistic regression model to build classifiers to predict patients in the training set. We used the “glmnet” package (R Foundation for Statistical Computing, Vienna, Austria) to perform the logistic regression model analysis.
3. RESULTS
3.1. Absolute protein expression levels reflected benefit of uracil‐tegafur adjuvant therapy
We studied 70 patients randomly selected from the National Surgical Adjuvant Study of Colorectal Cancer (NSAS‐CC/RC) study (Table 1).3 All patients in this set had stage III colon cancer and had undergone surgical resection with histologically negative resection margins. Treatment with UFT, an oral prodrug of 5‐FU, showed efficacy in terms of relapse compared with patients treated with surgery alone.
Then, we measured the absolute expression levels of 113 proteins related to carbohydrate metabolism and antioxidant pathways in 70 pairs of freshly frozen tumor and adjacent normal mucosa tissues (Table S3). Among 140 specimens, the expression levels of 16 specimens could not be measured because of the poor quality of protein extraction, and 124 specimens (tumor, 66; normal mucosa, 58) remain to be studied.
Using hierarchical clustering based on the expression levels of 113 proteins, 124 specimens were clearly separated into two groups: tumor and normal mucosa groups. The tumor group samples were also separated into two subgroups: clusters 1 and 2 within the tumor group, and cluster 3 within the normal mucosa group (Figure 1A), with two exceptions of tumor specimens. We further characterized the members of the three clusters based on the existence of UFT treatment and relapse (Figure 1B). Overall, cluster 1 was composed of patients with poor prognosis irrespective of UFT treatment. The relapse rate of surgery alone was 41.7% (5/12) in cluster 1 and 31.3% (5/16) in cluster 2. Even in patients with UFT treatment, the relapse rate of cluster 1 (46.2%, 7/13) was larger than that of cluster 2 (13.0%, 3/23). These results suggest that the expression levels of some proteins among the 113 proteins investigated might reflect relapse risk and UFT adjuvant therapy benefit after surgery for CRC.
Figure 1.
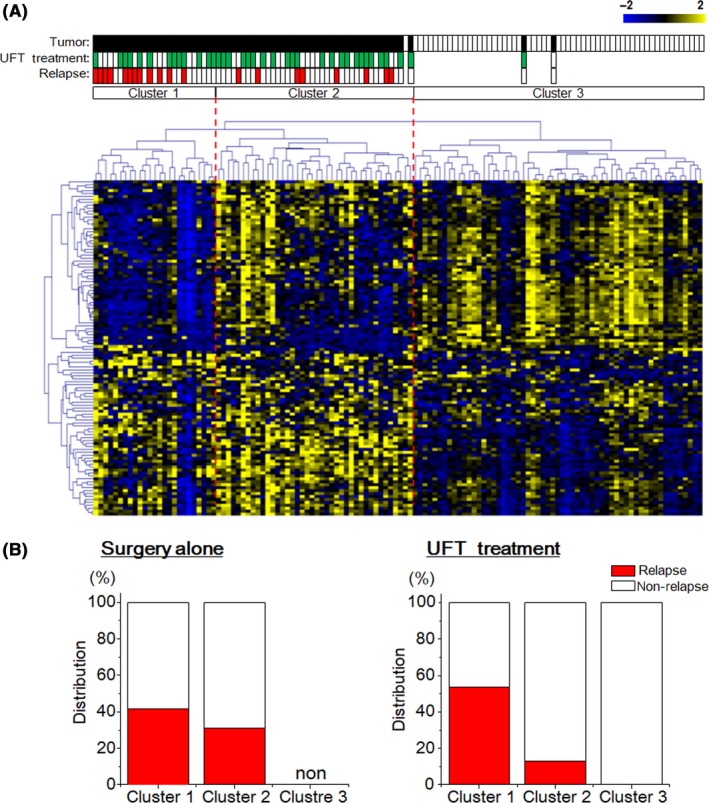
Classification of specimens based on the absolute expression levels of 113 proteins. A, Hierarchical clustering of 70 paired tumor tissues and adjacent normal mucosa with the 113 expressed proteins using Spearman’s correlation and complete linkage. Each dot represents an individual protein, and each column represents an individual specimen. Pseudo colors indicate expression levels from low‐to‐high on a z‐score from −2 to 2, indicating a low association strength. B, The relapse rate of tumor specimens treated with (left) or without (right) uracil‐tegafur (UFT) adjuvant therapy in three clusters
3.2. Protein levels of metabolic enzymes were associated with relapse and uracil‐tegafur treatment effects
Based on our classification (Figure 1A), we found similar numbers of upregulated and downregulated proteins in CRC; of 113 proteins, 55 proteins were upregulated and 58 proteins were downregulated. The changes in protein levels between tumor tissues and normal mucosa were projected onto two pathway maps (Figure 2A). Red and blue colors represent upregulated and downregulated proteins based on our classification, respectively. The distribution of proteins in the carbohydrate metabolism map was dependent on biological functions but appeared to be random in the antioxidant pathway. Indeed, in the carbohydrate metabolism map, proteins related to glycolysis, PPP and fatty acid synthesis were upregulated in tumor tissues, whereas proteins in the tricarboxylic citric acid (TCA) cycle and fatty acid metabolism were downregulated. Interestingly, these distribution patterns of protein expression levels were consistent with the Warburg effect. This is the first report to illustrate the Warburg effect at the protein level in surgical specimens, suggesting that the expression levels of proteins in carbohydrate metabolism might reveal the nature of CRC.
Figure 2.
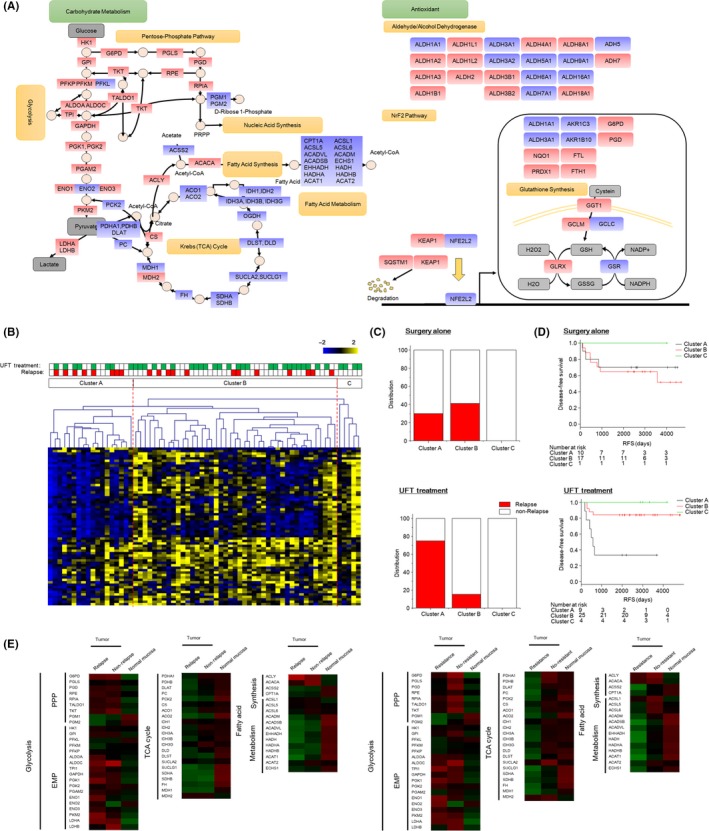
Association of the expression levels of carbohydrate metabolic enzymes with relapse risk and uracil‐tegafur (UFT) efficacy. A, Carbohydrate metabolism (left) and antioxidant (right) pathway maps. Red and blue colors represent upregulated and downregulated proteins, respectively. B, Hierarchical clustering of tumor specimens with carbohydrate metabolic enzymes using Spearman’s correlation and complete linkage. C, The relapse rate of tumor specimens treated without (upper) or with (lower) UFT adjuvant therapy in three clusters. D, Kaplan‐Meier survival curves of patients treated without (upper) or with (lower) UFT adjuvant therapy in three clusters. E, Heat map of the expression levels of metabolic proteins. Each protein was color coded by changes in relapse, non–relapse and normal mucosa (upper panel), and resistance, no‐resistant and normal mucosa (lower panel). Specimens that had relapsed were classified into the relapse group irrespective of UFT treatment and those that had not relapsed without UFT were classified into the non–relapse group. Specimens from those treated with UFT and with relapse were classified as the resistance group, and those treated with or without UFT and who had not relapsed were classified as the no‐resistant group. Pseudo colors indicate expression levels from low‐to‐high on a z‐score from −2 to 2
We investigated the relationship of the expression pattern of proteins in carbohydrate metabolism with CRC malignancy, relapse and UFT resistance. To do this, we performed hierarchical clustering analysis on the expression levels of carbohydrate metabolism proteins from tumor specimens. The specimens were separated into three clusters (Figure 2B). Patients with relapse without UFT treatment were distributed in clusters A and B, and patients with relapse following UFT treatment were concentrated in cluster A (Figure 2C). For cases of relapse after surgery without UFT treatment, the relapse rate was slightly increased in cluster B (7/17, 41.1%) compared with cluster A (3/10, 30.0%). For cases of relapse with UFT treatment, the rate was high in cluster 1 (6/8, 75.0%) compared with cluster B (4/28, 11.5%). Kaplan‐Meier analysis with a log‐rank test was used to determine relapse‐free survival (RFS) of patients treated with or without UFT adjuvant therapy (Figure 2D). In comparing the Kaplan‐Meier survival curve, patients divided into cluster A had a poor response from UFT adjuvant therapy, while the relapse rate after surgery alone did not change significantly in RFS. These results indicated that the protein expression patterns in carbohydrate metabolism were closely related to the malignancy of CRC, and in particular to UFT resistance.
We investigated the relationship between CRC malignancy and protein expression levels by comparing them between the three pathways of carbohydrate metabolism (Figure 2E). Specimens that had relapsed were classified into the relapse group irrespective of UFT treatment and those that had not relapsed without UFT were classified into the non–relapse group (left panel). Specimens from those treated with UFT and with relapse were classified as the resistance group, and those treated with or without UFT and who had not relapsed were classified as the no‐resistant group (right panel). The differences in protein expression between the relapse and non–relapse were ambiguous in the three pathways, although several proteins in each pathway showed marked differences. In contrast, differences between resistant (relapse with UFT treatment group) and no‐resistant (non–relapse with or without UFT treatment group) individuals were observed for proteins associated with the TCA cycle and fatty acid metabolism pathway but not for those associated with glycolysis. Taken together, expression levels of proteins in the three pathways were associated with efficacy of UFT treatment, whereas specific proteins were associated with relapse.
3.3. Relationship between colorectal cancer malignancy and Warburg effect‐related protein levels was reconstructed in colorectal cancer cell lines
The 3D culture of cells closely resembles the in vivo microenvironment of tissues. Cells in a 3D environment behave fundamentally differently from cells in monolayer culture.23 The spheroid formation causes resistance to anti–cancer drugs24 and metabolic heterogeneity25 in cultured cell lines of CRC. To investigate whether the relationship between the malignancy of CRC and the expression pattern of metabolic enzymes exists in cultured cells, we performed 3D sphere culture of HCT116 CRC cells. HCT116 cells formed spheroids by 7 days in culture (Figure 3A) and clearly acquired 5‐FU resistance compared with monolayer‐cultured cells (Figure 3B). Metabolic heterogeneity was also observed in spheroids (Figure S1). Next, we quantified the expression levels of metabolic enzymes in monolayer and 3D sphere‐cultured cells. The 3D sphere culture induced upregulation of five proteins and downregulation of 24. The upregulated proteins belonged to the Embden‐Meyerhof pathway (EMP) while those downregulated belonged in particular to the TCA cycle and mediators between the EMP and TCA cycle, resulting in representation of the Warburg effect (Figure 3C). These results indicated that the cells acquired 5‐FU resistance along with promotion of the Warburg effect, which was consistent with the results of specimens from CRC patients.
Figure 3.
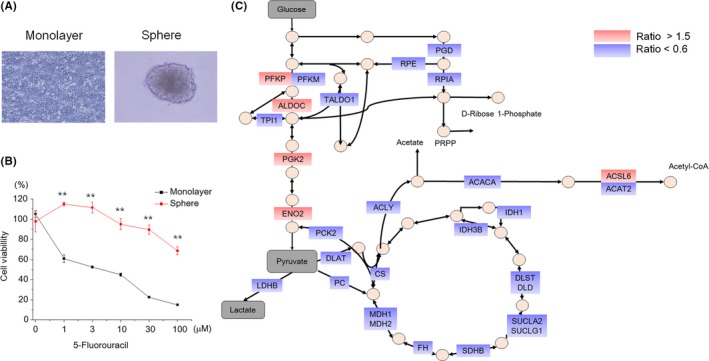
Association of efficacy of 5‐fluorouracil (5‐FU) and metabolic changes in cultured colorectal cancer cells. A, Representative image of monolayer and 3D sphere‐cultured HCT116 cells. B, Cytotoxic effect of 5‐FU in monolayer and 3D sphere‐cultured HCT116 cells. The cell viabilities were measured at 2 d after treatment with each concentration of 5‐FU. Data are represented as mean ± SEM. Statistical analysis was performed using Welch’s t test. **P < 0.01. C, Representation of the spheroid upregulated and downregulated proteins on the carbohydrate metabolism pathway. Red and blue colors represent upregulated and downregulated proteins, respectively
3.4. Warburg effect related proteins predicted the relapse risk and uracil‐tegafur adjuvant therapy benefit with high accuracy
We investigated which proteins might predict relapse risk and benefit of UFT adjuvant therapy among 66 carbohydrate metabolism proteins (see Section 2.4). We found nine proteins predictive of relapse risk and eight for UFT‐benefit prediction, which are illustrated in the pathway map (Figure 4A). The protein set for prediction of relapse risk (indicating in blue) and UFT benefit (indicating in red) were scattered over the three pathways. For relapse‐risk prediction, ALDOA, PKM2 and PDHB in glycolysis, ACO1, IDH1, SUCLA2 and FH in the TCA cycle, and HADH and ACAT1 in fatty acid metabolism were identified. For UFT‐benefit prediction, ENO2, TPI, G6PD and PDHB in glycolysis, SDHB and MDH2 in the TCA cycle, and HADH and EHHADH in fatty acid metabolism were identified. We then examined differences in the protein expression levels (Figure S2). Of note, the nine or eight protein‐based classifiers showed significantly higher prognostic accuracy than single protein classifiers. In the classifier for relapse prediction, expression of four of the nine proteins (fatty acid metabolism, HADH and ACAT1; TCA cycle, PDHB and ACO1) was decreased in tumor specimens of relapse after surgery, while the expression of five proteins (glycolysis, ALDOA and PKM2; TCA cycle, IDH1, SUCLA2 and FH) was increased. In the classifier for UFT efficacy, expression of six of the eight proteins (fatty acid metabolism, HADH and EHHADH; TCA cycle, SDHB, PDHB and MDH2; PPP, G6PD) was decreased in malignant tumor specimens, while the expression of two proteins (glycolysis, TPI and ENO2) was increased. Based on the protein frequency (Figure S2), we used these two sets of proteins to build the protein‐based classifiers for relapse risk and UFT benefit (Table S4). Then, we assessed the prognostic accuracy of the two protein‐based classifiers (Figure 4B,C). For comparing the accuracy of relapse predictions, we used a 41‐patient dataset consisting of 21 patients who relapsed with or without UFT treatment and 20 patients who did not relapse without UFT treatment. For comparing the accuracy of UFT efficacy prediction, we used a 57‐patient dataset consisting of 10 patients who relapsed and 47 patients who did not relapse with UFT treatment. We performed ROC analyses for regression of the data against relapse prediction (training test: AUC = 1.000, X‐fold cross‐validation test: AUC = 0.982) (Figure 4B) and UFT efficacy prediction (training test: AUC = 0.947, X‐fold cross‐validation test: AUC = 0.902) (Figure 4C). The ROC curves indicated that the two classifiers could predict the relapse and prognosis of UFT adjuvant therapy. These results suggest that the behavior of the Warburg effect‐related proteins was strongly associated with relapse prediction and the therapeutic indication of UFT adjuvant therapy.
Figure 4.
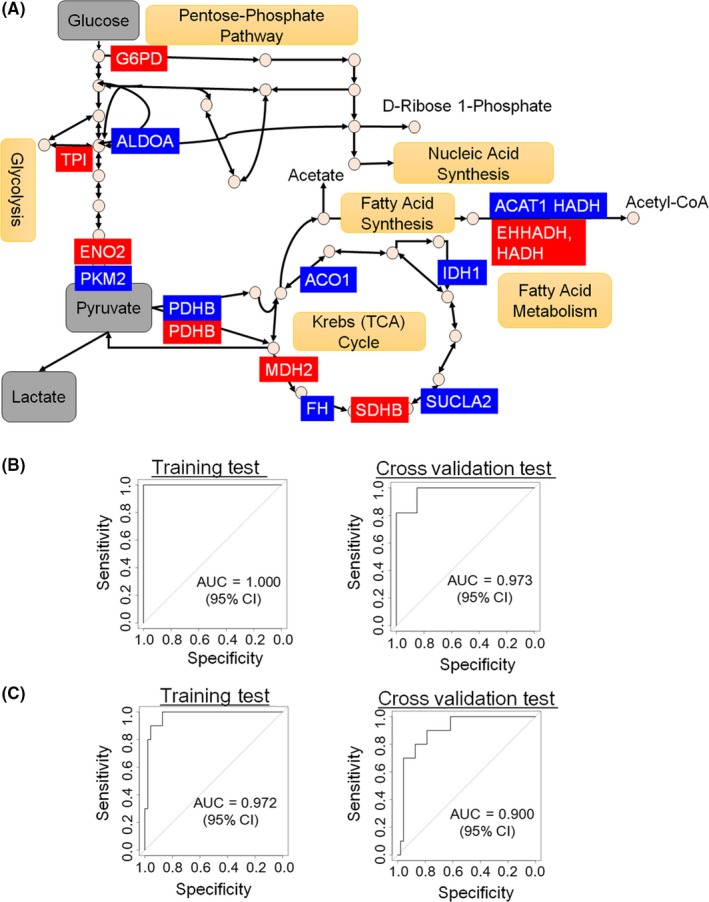
Prediction of relapse risk and uracil‐tegafur (UFT) adjuvant therapy efficacy based on selected protein levels. A, Representation of the selected nine (for relapse‐risk prediction) and eight (for UFT‐benefit prediction) proteins on the metabolic pathway map. The protein sets for relapse‐risk prediction are indicated in blue, and in red for UFT‐benefit prediction. B, Time‐dependent receiver operating characteristic (ROC) curves comparing the accuracy of the relapse‐risk prediction in 41 patients. C, Time‐dependent ROC curves comparing the accuracy of UFT‐benefit prediction in 57 patients
4. DISCUSSION
In this study, we measured the absolute expression levels of 113 proteins, which included those related to the Warburg effect, from 70 pairs of tumor and normal mucosa specimens. We demonstrated that the Warburg effect was present when comparing between the absolute expression levels of tumor and normal mucosa specimens, and especially within those tumor specimens when stratified by the progression of tumor tissue malignancy. The relationship between malignancy and the promotion of the Warburg effect was also observed in a cultured CRC cell line. Based on the demonstration of the Warburg effect at the absolute protein expression level, we built classifiers of nine and eight proteins to predict with high accuracy the relapse risk and UFT efficacy benefit, respectively, in CRC.
The expression patterns of proteins related to the Warburg effect showed a clear contrast dependent upon the progression of tumor malignancy in CRC. Conversely, recent studies have shown that a shift from glycolysis to re–established oxidative metabolism, which indicates that cells are transformed into normal‐like cells and that the Warburg effect is diminished in metabolism, wasS required for tumor malignancy progression.26, 27 An explanation for the inconsistency between our study and other recent studies is that we assumed that glycolytic tumor cells produced lactate and that oxidative tumor cells consumed lactate in tumor tissues. A cooperative system might be established between glycolytic and oxidative tumor cells for the growth of tumor tissues. Indeed, an intercellular lactate shuttle was shown to be involved in the progression of tumor malignancy.28, 29 In fact, spheroids from cultured cell lines consisted of glycolytic and oxidative metabolism‐dependent cells (Figure S1), and the expression pattern of metabolic enzymes represented progression of the Warburg effect (Figure 3C). Therefore, the Warburg effect might be advantageous for generating heterogeneity and tumor tissue growth in a multicellular environment.
All the proteins used to predict relapse risk and UFT efficacy in our classifiers were functional in cancer progression. ALDOA and ENO2 in the EMP are overexpressed in tumor cells compared with normal cells, leading to a significant induction of aerobic glycolytic metabolism.30, 31 G6PD, a rate‐limiting enzyme in the PPP responsible for the oxidation of glucose‐6‐phosphate to 6‐phosphoglucono‐δ‐lactone and the generation of NADPH, is a biomarker for prostate carcinoma.32 The protein expression of IDH1, SUCLA2, FH, SDHB and MDH2 in the TCA cycle change to regulate tumor growth.33, 34, 35 In particular, the loss of SDHB induces a Warburg‐like bioenergetics phenotype.36 ACAT1, HADH and EHHADH in the fatty acid β‐oxidation pathway are related to activation of the carbohydrate metabolism pathway of glycolysis and the TCA cycle. This is because NADH, FADH2 and acetyl CoA generated by β‐oxidation produce ATP through the electron transport chain,37 resulting in the induction of glycolysis and the suppression of OXPHOS and the TCA cycle. Furthermore, TPI, PKM2 and PDHB mediate the switch to aerobic glycolysis from OXPHOS.36, 38, 39 Based on these pathways, proteins in our classifiers were composed of representative factors in the activation of the energy metabolic pathway and key regulators in tumor growth via cancer metabolism.
Integrating multiple biomarkers into a single model substantially improved its prognostic value compared with a single biomarker. When developing a predictive classifier, we performed a new procedure that did not use the standard technique of variable selection in various regression models such as LASSO.40 Although automatic selections are easily handled, they have some limitations.41 Instead, we built classifiers of small numbers of proteins that had large AUC values, and then built an integrated classifier using proteins that emerged frequently in the limited set of classifiers. Our procedure required more steps than the single step for variable selection by regression models but is expected to be more robust for external data obtained in the future.
In conclusion, the two main contributions of this work were identifying the differential expression of carbohydrate metabolic enzymes between poor‐prognosis and good‐prognosis cancer by proteomic analysis, and the identification of a marker for resistance to 5‐FU‐based adjuvant therapy. Future larger studies will be required to test whether this signature can be combined with other analyses to construct multivariable classifiers to predict responses with greater accuracy. Our findings provide new insight into the contribution of the tumor microenvironment to CRC biology and may lead to the development of new therapies for CRC.
DISCLOSURE
The authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
We thank H. Kusano and K. Koike for their assistance in data analysis. We also thank Gillian Campbell from Edanz Group (https://www.edanzediting.com/ac) for editing a draft of this manuscript.
Kitazawa M, Hatta T, Sasaki Y, et al. Promotion of the Warburg effect is associated with poor benefit from adjuvant chemotherapy in colorectal cancer. Cancer Sci. 2020;111:658–666. 10.1111/cas.14275
REFERENCES
- 1. Macdonald JS. Adjuvant therapy for colon cancer. CA Cancer J Clin. 1997;47:243‐256. [DOI] [PubMed] [Google Scholar]
- 2. Moertel CG, Fleming TR, Macdonald JS, et al. Levamisole and fluorouracil for adjuvant therapy of resected colon carcinoma. N Engl J Med. 1990;322:352‐358. [DOI] [PubMed] [Google Scholar]
- 3. Hamaguchi T, Shirao K, Moriya Y, Yoshida S, Kodaira S, Ohashi Y. Final results of randomized trials by the National Surgical Adjuvant Study of Colorectal Cancer (NSAS‐CC). Cancer Chemother Pharmacol. 2011;67:587‐596. [DOI] [PubMed] [Google Scholar]
- 4. Malet‐Martino M, Martino R. Clinical studies of three oral prodrugs of 5‐fluorouracil (capecitabine, UFT, S‐1): a review. Oncologist. 2002;7:288‐323. [DOI] [PubMed] [Google Scholar]
- 5. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759‐767. [DOI] [PubMed] [Google Scholar]
- 6. Conlin A, Smith G, Carey FA, Wolf CR, Steele RJ. The prognostic significance of K‐ras, p53, and APC mutations in colorectal carcinoma. Gut. 2005;54:1283‐1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Connell MJ, Lavery I, Yothers G, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937‐3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17‐24. [DOI] [PubMed] [Google Scholar]
- 9. Lee C, Raffaghello L, Brandhorst S, et al. Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci Transl Med. 2012;4:124ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11‐20. [DOI] [PubMed] [Google Scholar]
- 11. Warburg O. The metabolism of carcinoma cells. J Cancer Res. 1925;9:148‐163. [Google Scholar]
- 12. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891‐899. [DOI] [PubMed] [Google Scholar]
- 13. Kim M‐S, Pinto SM, Getnet D, et al. A draft map of the human proteome. Nature. 2014;509:575‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilhelm M, Schlegl J, Hahne H, et al. Mass‐spectrometry‐based draft of the human proteome. Nature. 2014;509:582‐587. [DOI] [PubMed] [Google Scholar]
- 15. Picotti P, Aebersold R. Selected reaction monitoring‐based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555‐566. [DOI] [PubMed] [Google Scholar]
- 16. Kitazawa M, Hatta T, Ogawa K, Fukuda E, Goshima N, Natsume T. Determination of rate‐limiting factor for formation of beta‐catenin destruction complexes using absolute protein quantification. J Proteome Res. 2017;16:3576‐3584. [DOI] [PubMed] [Google Scholar]
- 17. Matsumoto M, Matsuzaki F, Oshikawa K, et al. A large‐scale targeted proteomics assay resource based on an in vitro human proteome. Nat Methods. 2016;14:251‐258. [DOI] [PubMed] [Google Scholar]
- 18. Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85‐95. [DOI] [PubMed] [Google Scholar]
- 19. Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carpentino JE, Hynes MJ, Appelman HD, et al. Aldehyde dehydrogenase‐expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69:8208‐8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Japanese Society for Cancer of the Colon and Rectum (ed) . General Rules for Clinical and Pathological Studies on Cancer of the Colon, Rectum, and Anus. Tokyo: Kanehara; 1997. [Google Scholar]
- 22. Kanda Y. Investigation of the freely available easy‐to‐use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601‐610. [DOI] [PubMed] [Google Scholar]
- 24. Hoffmann OI, Ilmberger C, Magosch S, Joka M, Jauch KW, Mayer B. Impact of the spheroid model complexity on drug response. J Biotechnol. 2015;205:14‐23. [DOI] [PubMed] [Google Scholar]
- 25. Russell S, Wojtkowiak J, Neilson A, Gillies RJ. Metabolic profiling of healthy and cancerous tissues in 2D and 3D. Sci Rep. 2017;7:15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation‐resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pavlides S, Whitaker‐Menezes D, Castello‐Cros R, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984‐4001. [DOI] [PubMed] [Google Scholar]
- 29. Sonveaux P, Végran F, Schroeder T, et al. Targeting lactate‐fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930‐3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou W, Capello M, Fredolini C, et al. Proteomic analysis reveals Warburg effect and anomalous metabolism of glutamine in pancreatic cancer cells. J Proteome Res. 2012;11:554‐563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ritterson LC, Tolan DR. Targeting of several glycolytic enzymes using RNA interference reveals aldolase affects cancer cell proliferation through a non–glycolytic mechanism. J Biol Chem. 2012;287:42554‐42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zampella EJ, Bradley EL Jr, Pretlow TG. Glucose‐6‐phosphate dehydrogenase: a possible clinical indicator for prostatic carcinoma. Cancer. 1982;49:384‐387. [DOI] [PubMed] [Google Scholar]
- 33. Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17:291‐293. [DOI] [PubMed] [Google Scholar]
- 34. King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675‐4682. [DOI] [PubMed] [Google Scholar]
- 35. Bayley JP, Devilee P. Warburg tumours and the mechanisms of mitochondrial tumour suppressor genes. Barking up the right tree? Curr Opin Genet Dev. 2010;20:324‐329. [DOI] [PubMed] [Google Scholar]
- 36. Cardaci S, Zheng L, MacKay G, et al. Pyruvate carboxylation enables growth of SDH‐deficient cells by supporting aspartate biosynthesis. Nat Cell Biol. 2015;17:1317‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grüning N‐M, Rinnerthaler M, Bluemlein K, et al. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 2011;14:415‐427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230‐233. [DOI] [PubMed] [Google Scholar]
- 40. Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression (with discussion). Ann Statist. 2004;32:407‐499. [Google Scholar]
- 41. Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Statist Soc B. 2005;67:301‐320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


