Abstract
The detection of Epstein‐Barr virus (EBV) DNA load in nasopharyngeal (NP) brushing samples for diagnosis of nasopharyngeal carcinoma (NPC) has attracted great attention. Further improvements that eliminate the need for clinical settings will greatly extend its application. A total of 250 participants were recruited to obtain NP brushing samples. Brush sampling both with and without the guide of endoscopy was conducted in 38 NPC patients. EBV DNA load, EBV RNA transcript and EBV DNA C promoter methylation status were, respectively, evaluated. Typical latency II transcripts were observed in brushing samples from NPC patients but not controls. Unlike in tissues, multiple lytic gene transcripts were observed not only in NPC patients but also in controls. Apart from EBV RNA transcript, samples from NPC patients also showed higher levels of EBV DNA load and C promoter methylation degree than their controls. Qualitative analysis further showed that EBV DNA C promoter was methylated in all NPC patients but in only 18.4% of the control group. Combined analysis of EBV DNA methylated degree and EBV DNA load increased the sensitivity to 100% in the detection of NPC. Using qualitative methylated type as the criteria, up to 89.5% of samples collected via blind brushing showed consistent results with samples collected via endoscopy‐guided brushing from NPC patients. Detection of the methylation status of EBV DNA C promoter in NP brushing samples shows great potential in diagnosing NPC and may provide an appealing alternative for the non–invasive detection and screening of NPC without the need for clinical settings.
Keywords: diagnosis, Epstein‐Barr virus DNA load, Epstein‐Barr virus DNA methylation, nasopharyngeal brushing, nasopharyngeal carcinoma
A significant difference in EBV DNA methylation was observed between NPC patients and the control group in high‐risk areas. Therefore, detection of methylation status of EBV DNA C promoter in NP brushing samples shows great potential in diagnosing NPC and may provide an appealing alternative for non–invasive detection and screening of NPC without the need for clinical settings.
1. INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a highly invasive and metastatic cancer that is widely prevalent in southern China. Although the overall survival rate is approximately 90% in patients diagnosed with early clinical stage disease, unfortunately, most patients are diagnosed with advanced stage diseased at their first visit to the hospital, and the survival rate decreases to <50%.1 Previous studies have demonstrated that genetic susceptibility, endemic environmental factors and Epstein‐Barr virus (EBV) infection constitute the three etiological contributors to NPC.2 Although EBV is ubiquitous, it is so closely linked to NPC that nearly 100% of tumor lesions possess EBV genomes in undifferentiated type NPC (WHO type III), the predominant type of NPC in endemic areas. Squamous cell (WHO type I) and non–keratinizing (WHO type II) NPC are also frequently associated with EBV in endemic areas.3 The infection of epithelial cells by EBV is a typical characteristic of NPC in high‐risk areas.4, 5
To date, substantial efforts have been made to develop a simple and reliable method to facilitate diagnosis and screening of NPC by testing EBV‐related biomarkers. Antibody titers against EBV serum antigens, including viral capsid antigen (VCA), EBV nuclear antigen 1 (EBNA1) and early antigen (EA), have been frequently used in high‐risk areas. However, the results of these serological tests alone have proven to be insufficient to accurately diagnose NPC, because IgA antibodies are also often present in those who are not NPC patients.6
Since the late 1990s, detection of EBV DNA load in plasma or serum has gradually been established as a powerful biomarker of NPC.7, 8, 9, 10 EBV DNA in plasma from NPC patients is from tumor cells although it is considered as fragmented DNA released into blood.11 Recently, a large‐scale study involving 20 174 asymptomatic male subjects in an endemic region directly confirmed the role of plasma EBV DNA for screening of NPC,12 although some studies show that EBV DNA appears to be of limited value in diagnosing NPC patients with early clinical stage and local recurrence.13
Besides the detection of EBV DNA load in plasma, nasopharyngeal (NP) brushing/swab samples are also used for qualitative and quantitative detection of EBV DNA, because NP brush sampling could be used to accurately and less invasively obtain samples from the nasopharynx. High sensitivity (87.3%‐96.4%) and specificity (90%‐98.4%) in NPC diagnosis are observed in people from multiple high‐risk areas, including those in the present study.14, 15, 16, 17, 18, 19, 20, 21 NP brush/swab sampling combined with EBV DNA load detection brings hope for diagnosing NPC patients with early stage disease and local recurrence, due to its original lesion at the site of the nasopharynx. Additional tumor makers such as mRNA,18 microRNA22 and tumor suppressor gene methylation14, 23, 24, 25, 26 can be assessed by NP brush sampling.
Recently, the high levels of EBV DNA loads in NP brushing samples from NPC patients have proven to directly reflect tumor origin.27 However, EBV DNA load is also present in some NP brushing samples from normal NP, although with low levels,15, 18, 19, 20 which challenges the hypothesis that normal NP epithelial cells are negative for EBV infection.28 Our study showed that EBV DNA load was detectable in 87.8% of NP brushing samples (n = 82) from the control group in the high‐risk area.20 Another study revealed that 89% of NP brushing samples (n = 905) showed positive EBV DNA load and this increased with increasing serum VCA‐IgA titers.15 Currently, the EBV DNA in NP brushing samples from the control group is not clearly defined.
Nasopharyngeal brush sampling usually requires the insertion of a brush by clinical experts into the nasopharynx under the guide of nasal endoscopy to accurately obtain samples from suspected areas. However, brush sampling without the need for clinical settings will greatly extend its application in NPC diagnosis and screening. In this study, the methylation status, including quantitative and qualitative analysis of EBV DNA C promoter (Cp), was found to both have a significant difference between NPC patients and the control group from a high‐risk area. This difference can be used to further improve the detection of NPC. More importantly, our study revealed that detecting the methylated type of EBV DNA C promoter in NP samples via blind brushing showed great potential in diagnosing NPC and may provide an appealing alternative for the non–invasive detection and screening of NPC without the need for clinical settings.
2. MATERIALS AND METHODS
2.1. Patients and controls
A total of 250 participants undergoing NP brush sampling were recruited in this study: 130 NP brushing samples were collected from participants when they underwent NP biopsy at Sun Yat‐Sen University Cancer Center (SYSUCC); 104 samples were obtained from patients who exhibited biopsy‐proven NPC, and 26 samples were obtained from patients who were finally diagnosed with chronic nasopharyngitis (n = 23) or other non–NPC tumors (n = 3). Meanwhile, 50 NP brushing samples were collected from healthy individuals at Sihui City of Guangdong Province. All of these 76 people were defined as the control group in a high–risk area. A total of 70 NP brushing samples were collected from people in a low‐risk area at Yangquan City of Shanxi Province. This study was approved by the Human Ethics Committee of the Sun Yat‐Sen University Cancer Center, and all participants provided informed consent.
2.2. Sampling procedures
Nasopharyngeal brush sampling has been described in our previous studies.20, 22, 29 Briefly, it was conducted under the guidance of endoscopy by experienced specialists. Endoscopy was used to evaluate the entire nasopharynx to find the sites of suspicious tumors. Before the biopsy, an NP brush (Copan Diagnostics) was inserted via the nose until the NP cavity was reached. Subsequently, the brush was rotated several times over the NP epithelium at the site of the suspected lesion and quickly removed. Two brushing samples were collected for each participant. One was used for DNA extraction and the other was for RNA extraction. In addition, brushing without the guidance of endoscopy (defined as blind brushing) was concurrently conducted in 38 NPC patients. Immediately after sampling, the brush tip (1.5 cm) was cut and placed in 1 mL of RNAlater (Invitrogen) and stored at −80°C until use.
2.3. DNA and RNA extraction
As described in our previous study,20 total DNA from NP brushing samples was extracted using an automated workstation (Hamilton Robotic) following the protocol recommended by the manufacturer. A final elution volume of 150 μL was obtained after the whole extraction procedure. As described in our previous study,22 total RNA from biopsy tissue and NP brush samples was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. DNA and RNA concentration was quantified using a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies).
2.4. Quantitative PCR of Epstein‐Barr virus DNA load
The EBV DNA loads in NP brushing samples from all participants were quantified by quantitative PCR (Q‐PCR). Our real‐time Q‐PCR was based on a previously mature fluorogenic PCR reaction system.7 In this system, amplification primers targeting the BamHI‐W region of the EBV DNA genome and a dual‐labeled hybridization probe were included. Two primers and a probe targeting the β‐globin gene were used as a reference for quality control. All the sequences of primers and probes used are listed in Table S1. The standard sample ladders (103, 104, 105, 106 and 107 copies/μL) were used to obtain the standard curve. Each PCR reaction was set up in a volume of 8 μL, including 4 μL PCR master mix, 1 μL primer, 0.2 μL probe, 0.8 μL water and 2 μL DNA template. Thermal cycling was initiated with a denaturation step of 5 minutes at 95°C, then 45 cycles of 95°C for 15 seconds, 60°C for 30 seconds and 72°C for 15 seconds, and then 72°C for 5 minutes. The EBV DNA and β‐globin levels in brushing samples were expressed as copy/ng DNA.
A comparative EBV DNA Q‐PCR analysis was also performed, as described in previous studies,27 to distinguish the intact DNA or the fragmented DNA. In brief, dye‐based Q‐PCR, respectively, targeting a 99 and 213 bp region of Epstein‐Barr virus nuclear antigen‐1 (EBNA1) was simultaneously performed in one sample. The sequences of the two paired primers targeting EBNA1 are listed in Table S1. During PCR amplification, EBV DNA from EBV positive cell lines C666 was used as the positive control. The standard sample ladders (101, 102, 103 and 104 copies/μL) were used to obtain the standard curve. Each PCR reaction was set up in a volume of 10 μL including 5 μL PCR master mix, 1 μL primer, 3 μL water and 1 μL DNA template. Thermal cycling was initiated with a denaturation step of 5 minutes at 95°C, then 40 cycles of 95°C for 10 seconds, 55°C for 15 seconds and 72°C for 30 seconds, and then 72°C for 7 minutes. The EBV DNA in brushing samples was expressed as copy/μL. When the ratio (99 bp EBNA1 expression divided by 213 bp EBNA1 expression) was over 1.5, the sample was thought to have fragmented EBV DNA.
2.5. cDNA synthesis and quantitative PCR of Epstein‐Barr virus RNA transcript
Due to insufficient RNA quality, 10 brushing samples were excluded from the RNA analysis. Finally, 99 out of 104 brushing samples from NPC patients and 71 out of 76 brushing samples from the control group were tested for EBV RNA transcripts. Biopsy tissues from 36 NPC patients and 13 patients with chronic nasopharyngitis obtained from the biobank of our cancer center were also used. Relative quantification of six latent genes (EBER1, BART, EBNA1, EBNA2, LMP1 and LMP2A) and five lytic genes (two immediately early genes, Zebra and Rta, two early genes, PK and TK, and one late gene, VCA‐p18) were measured. The level of U1A housekeeping gene in each sample was used for EBV RNA level normalization. The sequences of the EBV RNA primers and U1A housekeeping gene are listed in Table S1, as described in a previous study.27 A total of 500 ng RNA was first used for cDNA synthesis. Each reaction contained 2 μL reverse transcription buffer and 8 μL template water. After the reaction, 40 μL of water was added so that the concentration became 10 ng/μL. Each Q‐PCR reaction was set up in a volume of 10 μL, including 5 μL PCR master mix, 1 μL primer, 3 μL water and 1 μL cDNA template. Thermal cycling was initiated with a denaturation step of 5 minutes at 95°C, then 45 cycles of 95°C for 10 seconds, 58°C for 15 seconds and 72°C for 30 seconds, and then 72°C for 7 minutes were carried out.
2.6. Methylation‐specific PCR of Epstein‐Barr virus DNA C promoter region
The methylation status of the C promoter of EBV DNA was detected by methylation‐specific PCR as described previously.27 In brief, genomic DNA from NP brushing samples was first modified by bisulfate treatment and purified using the methylation kit (ZYMO RESEARCH) according to the manufacturer’s instructions. The modified DNA was used as a template for PCR amplification using primers specific for either methylated or modified unmethylated DNA. The primer sequences are showed in Table S1. Each PCR reaction was set up in a reaction volume of 15 μL including 7.5 μL PCR master mix, 1 μL primer, 5.5 μL water and 1 μL DNA template. PCR condition was set up as follows: 10 minutes at 95°C, and than 40 cycles of 95°C for 30 seconds, 60°C for 35 seconds and 72°C for 30 seconds, and, finally, 72°C for 7 minutes. The brightness of the methylated (M) and unmethylated (U) bands of one sample in the electrophoresis was quantitatively calculated with Quantity One Software (Bio‐Rad Laboratories). The quantitative value of band brightness was used to reveal the methylated or unmethylated degree of EBV DNA C promoter. The epigenetic type of 1 sample, as a qualitative indicator, was defined by comparing the brightness of methylated and unmethylated bands. In brief, 1 sample with only M band or bands showing M > U (fold change ≥1.2) were defined as the methylated type. In contrast, 1 sample with only U band or bands showing U > M (fold change ≥ 1.2) were defined as the unmethylated type.
2.7. Statistical analyses
The Mann‐Whitney test (two‐tailed) was used to compare the differences in quantitative EBV DNA loads, EBV RNA transcripts and methylated degree between NPC and control groups. Correlations between EBV DNA load and frequency of gene transcripts were assessed by applying Spearman correlation coefficients and were subjected to two‐tailed significance tests. Differences in the qualitative epigenetic type of EBV DNA C promoter between NPC and controls were evaluated using a χ2 test. Receiver operating characteristic (ROC) curves were used to select cut‐off values (COV) of methylated degree with maximum sensitivity and specificity. P < 0.05 was considered significant. The statistical analyses were performed using the SPSS 16 software.
3. RESULTS
3.1. Epstein‐Barr virus DNA loads in nasopharyngeal brushing samples
All NP brushing samples (100%) from NPC patients were positive for EBV DNA detection, with high DNA loads (mean = 21 168.1 copies/ng DNA, range from 78 to 1 054 852). The EBV DNA loads and positive proportion in control groups from high‐risk and low‐risk areas both had a significant decrease (Figure 1). The EBV DNA loads (mean: 60.3 vs 7.5 copies) and positive proportion (86.8% vs 44.3%) in control groups also had a significant difference between the high‐risk and low‐risk areas. In the following study, NP brushing samples from the control group in the low‐risk area were excluded due to the low EBV DNA loads. Detailed characteristics of the study population are presented in Table S2.
Figure 1.
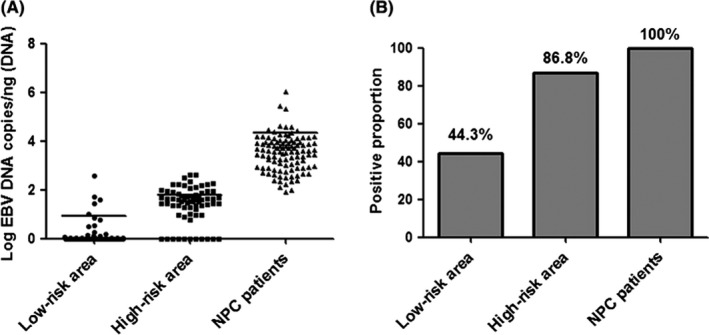
Epstein‐Barr virus (EBV) DNA loads in nasopharyngeal (NP) brushing samples were different in three groups. A, EBV DNA loads in brushing samples from NP carcinoma (NPC) patients had higher EBV DNA loads compared to the load in the control group. There was also a significant difference in EBV DNA loads in control groups between the high‐risk and low‐risk areas. B, The positive proportion of EBV DNA loads (86.8% vs 44.3%) in control groups also had a significant difference between the high‐risk and low‐risk areas
To determine whether the EBV DNA was intact or fragmented, we also performed a comparative EBV DNA Q‐PCR analysis. The results revealed that most of the EBV DNA in NP brushing samples from NPC patients was intact while higher levels of fragmented DNA indicated that an apoptotic origin was observed in 12 samples (Figure S1). Comparative PCR analysis was not done in the control group due to the low EBV DNA loads and positive rate.
3.2. Epstein‐Barr virus RNA transcripts in nasopharyngeal brushing samples
To further clarify the source of the EBV DNA in NP brushing samples, the EBV RNA profile including six latent gene (EBER1, BART, EBNA1, EBNA2, LMP1 and LMP2A) and five lytic gene (Zebra, Rta, TK, PK and VCA‐p18) transcripts was measured. EBV RNA transcripts in some biopsy tissues were also included. Quantitatively, all the gene transcripts in the control group were much less than their transcripts in NPC patients (Table S3). Qualitatively, the latency II transcription profile with EBER1, BART, LMP1, LMP2A, EBNA1 but not EBNA2 expression was observed in both NP brushing samples and tissues from NPC patients (Figure 2 and Figure S2). EBER1 transcript was found in both biopsy and brushing samples in the control group. Different from their expression in biopsy, a significant difference was found in the lytic gene transcripts. The results revealed higher frequency of lytic gene transcripts in NP brushing samples in NPC patients and the control group.
Figure 2.
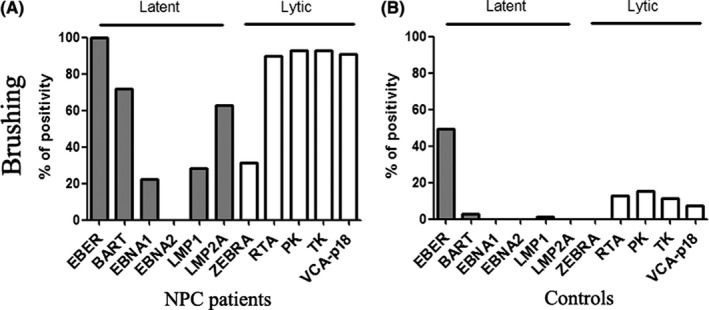
There was a significant difference in the RNA transcript of Epstein‐Barr virus (EBV) DNA between nasopharyngeal carcinoma (NPC) patients and the control group. Positivity of EBV latent and lytic gene detection in brushing samples from NPC patients (A) and control (B)
Furthermore, a comparison of EBV latent and lytic transcripts with the amount of EBV DNA in all NP brushing samples was conducted. First, the results revealed that samples with high EBV DNA load tended to reveal broader viral gene expression compared to samples with low EBV DNA load (Figure S3A,D). Second, a significant positive correlation was observed between the number of latent gene transcript and EBV DNA load in NP brushing samples from both NPC patients and controls (Figure S3B,E). Third, significant correlation was also found for the lytic gene transcript and EBV DNA load (Figure S3C,F).
3.3. Epstein‐Barr virus DNA C promoter methylation in nasopharyngeal brushing samples
The methylation status of EBV C promoter was further examined in NP brushing samples from both NPC patients and controls. A significant difference was observed between NPC patients and the control group in the high‐risk area (Figure 3 and Table 1). In NPC patients, the predominant status of EBV C promoter was methylated. There were 39 samples with only methylated bands (defined as M) and 65 samples with both methylated and unmethylated bands, but the methylated degree were significantly greater than the unmethylated degree (defined as M > U), according to the band brightness. In contrast, the predominant type of EBV DNA C promoter was unmethylated in the control group. There were 14 samples with only unmethylated bands (defined as U), 9 samples with the band brightness U > M, 6 samples with only methylated bands (M) and eight samples with the band brightness M > U. In addition, there were 39 samples with no PCR bands, probably due to the low EBV DNA loads.
Figure 3.
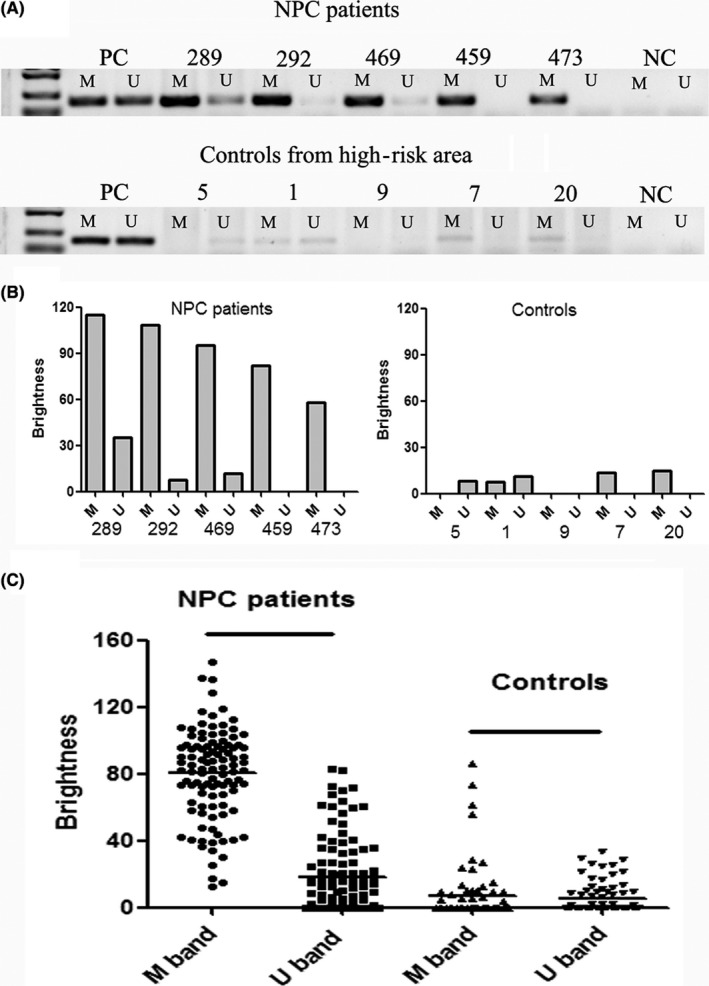
There was a significant difference in methylation status of EBV DNA C promoter between nasopharyngeal carcinoma (NPC) patients and the control group. A, Representative results of methylation‐specific PCR in NP brushing samples between NPC patients and the control group. In NPC patients, all the samples showed only methylated bands (ID 459 and 473) or showed both methylated and unmethylated bands (ID 289, 292 and 469), but the brightness of methylated bands was more obvious. In the control group, there were samples with only unmethylated band (ID 5), samples with the band brightness U > M (ID 1), samples with only the methylated bands (ID 7 and 20) and samples with no PCR bands (ID 9). NC, negative control; PC, positive control. B, Quantitative analysis of the methylated and unmethylated band brightness in representative samples. C, Quantitative analysis of the methylated and unmethylated band brightness in all the NP brushing samples
Table 1.
Methylation status of EBV DNA C promoter in the nasopharyngeal brushing samples from NPC patients and controls from the high‐risk area
| Status of EBV DNA methylation | NPC patients (n = 104) | Controls (n = 76) |
|---|---|---|
| M | 39 | 6 |
| M > U | 65 | 8 |
| U | 0 | 14 |
| U > M | 0 | 9 |
| ND | 0 | 39 |
| P | P < 0.001 | |
Abbreviations: ND, not detected; NPC, nasopharyngeal carcinoma
3.4. Combination of Epstein‐Barr virus DNA load and methylation detection can improve the diagnosis of nasopharyngeal carcinoma
In our previous study, the cut‐off value (COV) for EBV DNA loads in NP brushing was defined as 225 copy/ng DNA.20 This was used to calculate the sensitivity and specificity, as shown in Table 2: 95.2% sensitivity and 96.1% specificity were obtained by EBV DNA load detection. Under this condition, there were also 5 NPC cases with EBV DNA load below the COV and 3 controls with EBV DNA load above the COV. The EBV DNA loads in these samples were from 78 to 418 copies, located near the COV (Table S4).
Table 2.
Diagnosis by EBV DNA load, EBV DNA methylated degree, EBV DNA methylated type and their combination
| Controls (N = 76) | NPC patients (N = 104) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|
| Neg. | Pos. | Neg. | Pos. | |||||
| EBV DNA load | 73 | 3 | 5 | 99 | 95.2 | 96.1 | 97.1 | 93.6 |
| EBV DNA methylated degree | 72 | 4 | 5 | 99 | 95.2 | 94.7 | 96.1 | 93.5 |
| EBV DNA methylated type | 62 | 14 | 0 | 104 | 100 | 81.6 | 88.1 | 100 |
| Combinationa | 72 | 4 | 2 | 102 | 98.1 | 94.7 | 96.2 | 97.3 |
| Combinationb | 71 | 5 | 0 | 104 | 100 | 93.4 | 95.4 | 100 |
In our previous study, the cut‐off value for EBV DNA load was defined as 225 copy/ng DNA. The cut‐off value for EBV DNA methylated degree was defined as 33.49 (the brightness of EBV DNA methylation band), determined using receiver operating characteristic curves. EBV DNA methylated type was defined when the sample with only a methylated band or bands showing methylation > unmethylation (fold change ≥ 1.2). Neg, negative; NPV, negative predictive value; Pos, positive; PPV, positive predictive value.
Combination, using either EBV DNA load or EBV methylated degree as a panel.
Combination, using either EBV DNA load or EBV methylated type as a panel.
The cut‐off value for EBV DNA C promoter methylated degree was defined as 33.49 (the brightness of EBV DNA methylated band), determined by receiver operating characteristic (ROC) curves. Similarly, 95.2% sensitivity and 94.7% specificity were obtained. There was a significant correlation between quantitative EBV DNA load and EBV DNA methylated degree (P < 0.001). Using qualitative methylated type as the criteria, 100% sensitivity and 81.6% specificity was obtained (Table 2 ).
A combined analysis was conducted to explore whether the accuracy could be increased. Samples with EBV DNA loads located from 78 to 418 copies were further selected for the methylated analysis of EBV DNA C promoter. There were 13 samples from NPC patients and 17 samples from controls (Table S4). Using either EBV DNA load or EBV DNA methylated degree as the criteria, there were still 2 patients with false negativity and 4 controls with false positivity. Therefore, 98.1% sensitivity and 94.7% specificity was obtained. Using either EBV DNA load or EBV DNA methylated type as the criteria, the sensitivity increased from 95.2% to 100% and the specificity was 93.4%.
3.5. Detection of Epstein‐Barr virus C promoter methylation showed great potential in the diagnosis of nasopharyngeal carcinoma via blind brushing
Brush sampling both with and without the guide of endoscopy (named blind brushing) was conducted in 38 NPC patients. With the guide of endoscopy, 94.7% (36/38) of brushing samples had EBV DNA load (from 94 to 40 095 copies) above the defined COV (225 copies), and the median load was 5144 copies. In contrast, only 55.3% (21/38) of NP samples via blind brushing had EBV DNA load (from 1 to 8744 copies) above the COV, and the median load decreased to 275 copies. Similarly, 100% of brushing samples with the guide of endoscopy had EBV DNA methylated degree (from 36.79 to 146.57) above the defined COV (33.48), and the median brightness of M band was 80.99. However, only 57.9% (22/38) of samples via blind brushing had EBV DNA methylated degree above the COV (from 0 to 90.36), and the median brightness decreased to 42.61 (Table 3 and Table S5).
Table 3.
Comparisons of EBV DNA load, EBV DNA methylated degree and methylated type in the detection of NPC in brushing samples obtained with or without the guide of endoscopy
| Under the endoscopy | In blind brushing | P‐value | |||
|---|---|---|---|---|---|
| Number of patients | % | Number of patients | % | ||
| EBV DNA load | |||||
| Median (range) | 5144 (94‐40095) | 275 (1‐8744) | <.001[Link] | ||
| Negative | 2 | 5.3 | 17 | 44.7 | |
| Positive | 36 | 94.7 | 21 | 55.3 | |
| EBV DNA methylated degree | |||||
| Median (range) | 80.99 (36.79‐146.57) | 42.61 (0‐90.36) | <.001[Link] | ||
| Negative | 0 | 0.0 | 16 | 42.1 | |
| Positive | 38 | 100.0 | 22 | 57.9 | |
| EBV DNA methylated type | |||||
| M | 10 | 26.3 | 30 | 79.0 | |
| M > U | 28 | 73.7 | 4 | 10.5 | |
| ND | 0 | 0.0 | 4 | 10.5 | |
Abbreviations: M, methylation; ND, not detected; U, unmethylation.
P‐value from paired Wilcoxon signed—rank test
Different from the quantitative EBV DNA load and EBV DNA C promoter methylated degree, up to 89.5% (34/38) of samples collected via blind brushing showed consistent results of qualitative methylated type with samples collected via endoscopy‐guided brushing from NPC patients. All the samples were of methylated type with M or M > U (Table 3 and Table S5).
4. DISCUSSION
The application of EBV DNA load detection in NP brushing/swab samples for NPC auxiliary diagnosis and screening has been extensively studied and validated in different areas, including Hong Kong,14 Taiwan,17, 21 Canada,16 Indonesia,18, 19 Guangxi15 and Guangdong20. Improvement of its reliability and convenience is of great importance, especially when the use is not restricted to clinical settings, which may greatly extend its use in NPC diagnosis and screening.
In this study, a significant increase of EBV DNA load in NP brushing samples from NPC patients was observed compared to its load from the control group (Figure 1). This result has been validated in previous studies, including ours.20 Furthermore, it is interesting that the load (Figure 1A) and the positive proportion (Figure 1B) of EBV DNA also had a more significant increase in the control group from the high‐risk area than from the low‐risk area. This increase is consistent with the observation of a high incidence of NPC in the high‐risk area.
In the present study, the characteristics of the EBV DNA in NP brushing samples were focused on. First, whether the EBV DNA was intact or fragmented was evaluated by comparative Q‐PCR. This showed there were only a few samples (12 out of 104) with fragmented DNA, consistent with the results of a previous study (Figure S1). In that study, 6 out of 33 brushing samples were found to have fragmented EBV DNA.27 Second, the source of the EBV DNA was further investigated by detecting the EBV RNA transcripts in the NP brushing samples (Figure 2, Table S3 and Figure S3). As expected, typical latency II transcripts, including EBER1, BART, EBNA1, EBNA2 AND LMP2A, were found in brushing samples from NPC patients but not controls. It was interesting that greater frequency of lytic gene transcripts was found in brushing samples from NPC patients as well as controls (Figure 2) compared to the RNA transcript in biopsy tissues (Figure S2). Significant correlation between EBV DNA load and gene transcript (including latent gene and lytic gene) was also found. These results suggest that EBV genomic DNA from cells and free EBV virion both contribute to the EBV DNA load in the NP brushing samples.
It is known the C promoter of EBV DNA is unmethylated within the virion, while it becomes methylated when it is latent in NPC cells.30 Subsequently, the methylation status of EBV DNA C promoter in our study indicated that EBV DNA in NP brushing samples from NPC patients was mainly from EBV latent tumor cells, which is consistent with results of previous studies (Figure 3 and Table 1).27 In contrast, unmethylation was predominant in most samples from the control group. It seemed that most of the EBV DNA in the samples was probably from free EBV virion. A previous study reported that EBV DNA load in nasopharynx increased with the increasing serum VCA/IgA titers in healthy individuals with a large sample size (n = 905).15 The increase of serologic antibodies has been demonstrated to be closely related with the risk of NPC in prospective studies.31, 32 Therefore, the nature of EBV DNA in NP brushing samples from healthy individuals needs to be further studied in the future.
Nasopharyngeal brush sampling is a subjective method. Its success depends on fully contacting with the NP tumor and obtaining enough tumor cells. Then a high level of EBV DNA load can be observed. However, previous studies have shown that NP brush sampling fails to capture enough tumor cells in some patients.19, 20, 22 Further improvement in identifying these false negative patients is important in clinical practice. In this study, the quantitative methylated degree and qualitative methylated type were both analyzed. Similar sensitivity and specificity were observed, compared to the EBV DNA load. To further increase the sensitivity, a parallel combined analysis was conducted. As expected, it increased the sensitivity from 95.2% to 100% using either EBV DNA load or qualitative methylated type as the criteria (Table 2). Therefore, detection of methylation status is strongly recommended in samples with EBV DNA load located near the COV (78‐418 copies).
It is assumed that blind brushing without the guide of nasopharyngoscope will decrease the quantity of tumor cells, leading to low levels of EBV DNA load as well as low EBV DNA methylated degree. A pilot study was conducted as part of the present study. As expected, the median EBV DNA load decreased from 5144 copies to 275 copies (Table 3). At COV = 225 copies, the positive proportion of blind brushing was only 55.3% (21/38), far from the positive proportion of brushing under the guide of endoscopy. A similar result was observed considering the EBV DNA methylated degree, because there was a significant correlation between the two indicators.
Unlike EBV DNA load and EBV DNA methylated degree, detection of EBV DNA C promoter methylated type is a qualitative method. In contrast, up to 89.5% (34/38) of samples collected through blind brushing showed consistent results for qualitative methylated type with samples collected via endoscopy‐guided brushing from NPC patients (Table 3). This suggested that this test could be effective without prior knowledge of tumor location. Encouragingly, this qualitative test might be much easier to apply in a non–clinical setting due to its low cost and independence of Q‐PCR equipment.
However, positive rates in controls have not yet been evaluated in this study. On one hand, negative results of methylated detection were already observed in 51.3% (39/76) of brushing samples under the guide of endoscopy (Table 1). Most of these samples had very low levels of EBV DNA loads. In contrast, blind brushing greatly decreased the EBV DNA load, as shown in NP brushing samples from NPC patients (Table 3). It is concluded most of samples from healthy individuals would not have PCR amplification of EBV DNA C promoter. Therefore, the specificity was not further evaluated.
In conclusion, the methylation status of EBV DNA C‐promoter in nasopharyngeal brushing samples was found to have a significant difference between NPC patients and the control group. This difference could be used to make a better diagnosis of NPC along with the EBV DNA load. More importantly, detection of EBV DNA methylation shows great potential in the diagnosis of NPC through blind brushing, a brush sampling technique that does not require clinical settings. Therefore, it has great potential to be applied for the screening of NPC in the future.
DISCLOSURE
The authors have no potential conflicts of interest to disclose.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant number: 81802708), the Key Area Research and Development Program of Guangdong Province, China (grant number: 2019B110233004), the Science and Technology Planning Project of Guangdong Province, China (grant number 2019B030316031), the Science and Technology Planning Project of Guangzhou City, China (grant number: 201804020094), the Sino‐Sweden Joint Research Program (grant number: 81861138006) and the Special Support Program for flexible high‐level talents introduction of Xinjiang Uygur Autonomous Region, China.
Zheng X‐H, Wang R‐Z, Li X‐Z, et al. Detection of methylation status of Epstein‐Barr virus DNA C promoter in the diagnosis of nasopharyngeal carcinoma. Cancer Sci. 2020;111:592–600. 10.1111/cas.14281
Contributor Information
Ruo‐Zheng Wang, Email: wrz8526@vip.163.com.
Wei‐Hua Jia, Email: jiawh@sysucc.org.cn.
REFERENCES
- 1. Razak AR, Siu LL, Liu FF, Ito E, O’Sullivan B, Chan K. Nasopharyngeal carcinoma: the next challenges. Eur J Cancer. 2010;46:1967‐1978. [DOI] [PubMed] [Google Scholar]
- 2. Young LS, Rickinson AB. Epstein‐Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757‐768. [DOI] [PubMed] [Google Scholar]
- 3. Pathmanathan R, Prasad U, Chandrika G, Sadler R, Flynn K, Raab‐Traub N. Undifferentiated, nonkeratinizing, and squamous cell carcinoma of the nasopharynx. Variants of Epstein‐Barr virus‐infected neoplasia. Am J Pathol. 1995;146:1355‐1367. [PMC free article] [PubMed] [Google Scholar]
- 4. Niedobitek G, Young LS. Epstein‐Barr virus persistence and virus‐associated tumours. Lancet. 1994;343:333‐335. [DOI] [PubMed] [Google Scholar]
- 5. Pathmanathan R, Prasad U, Sadler R, Flynn K, Raab‐Traub N. Clonal proliferations of cells infected with Epstein‐Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693‐698. [DOI] [PubMed] [Google Scholar]
- 6. Pickard A, Chen C‐J, Diehl SR, et al. Epstein‐Barr virus seroreactivity among unaffected individuals within high‐risk nasopharyngeal carcinoma families in Taiwan. Int J Cancer. 2004;111:117‐123. [DOI] [PubMed] [Google Scholar]
- 7. Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell‐free Epstein‐Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188‐1191. [PubMed] [Google Scholar]
- 8. Mutirangura A, Pornthanakasem W, Theamboonlers A, et al. Epstein‐Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998;4:665‐669. [PubMed] [Google Scholar]
- 9. Hsiao JR, Jin YT, Tsai ST. Detection of cell free Epstein‐Barr virus DNA in sera from patients with nasopharyngeal carcinoma. Cancer. 2002;94:723‐729. [DOI] [PubMed] [Google Scholar]
- 10. Chan KC, Lo YM. Circulating EBV DNA as a tumor marker for nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:489‐496. [DOI] [PubMed] [Google Scholar]
- 11. Chan KC, Zhang J, Chan AT, et al. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res. 2003;63:2028‐2032. [PubMed] [Google Scholar]
- 12. Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein‐Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377:513‐522. [DOI] [PubMed] [Google Scholar]
- 13. Yip TTC, Ngan RKC, Fong AHW, Law SCK. Application of circulating plasma/serum EBV DNA in the clinical management of nasopharyngeal carcinoma. Oral Oncol. 2014;50:527‐538. [DOI] [PubMed] [Google Scholar]
- 14. Tong JHM, Tsang RKY, Lo KW, et al. Quantitative Epstein‐Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin Cancer Res. 2002;8:2612‐2619. [PubMed] [Google Scholar]
- 15. Chen Y, Zhao W, Lin L, et al. Nasopharyngeal Epstein‐Barr virus load: an efficient supplementary method for population‐based nasopharyngeal carcinoma screening. PLoS ONE. 2015;10:e0132669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tune CE, Liavaag P‐G, Freeman JL, et al. Nasopharyngeal brush biopsies and detection of nasopharyngeal cancer in a high‐risk population. J Natl Cancer Inst. 1999;91:796‐800. [DOI] [PubMed] [Google Scholar]
- 17. Hao SP, Tsang NM, Chang KP. Screening nasopharyngeal carcinoma by detection of the latent membrane protein 1 (LMP‐1) gene with nasopharyngeal swabs. Cancer. 2003;97:1909‐1913. [DOI] [PubMed] [Google Scholar]
- 18. Stevens SJC, Verkuijlen SAWM, Hariwiyanto B, et al. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein‐Barr virus DNA load and carcinoma‐specific viral BARF1 mRNA. Int J Cancer. 2006;119:608‐614. [DOI] [PubMed] [Google Scholar]
- 19. Adham M, Greijer AE, Verkuijlen S, et al. Epstein‐Barr virus DNA load in nasopharyngeal brushings and whole blood in nasopharyngeal carcinoma patients before and after treatment. Clin Cancer Res. 2013;19:2175‐2186. [DOI] [PubMed] [Google Scholar]
- 20. Zheng XH, Lu LX, Li XZ, Jia WH. Quantification of Epstein‐Barr virus DNA load in nasopharyngeal brushing samples in the diagnosis of nasopharyngeal carcinoma in southern China. Cancer Sci. 2015;106:1196‐1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin S‐Y, Tsang N‐M, Kao S‐C, et al. Presence of Epstein‐Barr virus latent membrane protein 1 gene in the nasopharyngeal swabs from patients with nasopharyngeal carcinoma. Head Neck‐J Sci Spec. 2001;23:194‐200. [DOI] [PubMed] [Google Scholar]
- 22. Zheng XH, Lu LX, Cui C, Chen MY, Li XZ, Jia WH. Epstein‐Barr virus mir‐bart1‐5p detection via nasopharyngeal brush sampling is effective for diagnosing nasopharyngeal carcinoma. Oncotarget. 2016;7:4972‐4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hutajulu SH, Indrasari SR, Indrawati LPL, et al. Epigenetic markers for early detection of nasopharyngeal carcinoma in a high risk population. Mol Cancer. 2011;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nawaz I, Moumad K, Martorelli D, et al. Detection of nasopharyngeal carcinoma in Morocco (North Africa) using a multiplex methylation‐specific PCR biomarker assay. Clin Epigenetics. 2015;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Z, Sun DI, Hutajulu SH, et al. Development of a non–invasive method, multiplex methylation specific PCR (MMSP), for early diagnosis of nasopharyngeal carcinoma. PLoS ONE. 2012;7:e45908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang HW, Chan A, Kwong DLW, Wei WI, Sham JST, Yuen APW. Evaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patient. Int J Cancer. 2003;105:851‐855. [DOI] [PubMed] [Google Scholar]
- 27. Ramayanti O, Juwana H, Verkuijlen SAMW, et al. Epstein‐Barr virus mRNA profiles and viral DNA methylation status in nasopharyngeal brushings from nasopharyngeal carcinoma patients reflect tumor origin. Int J Cancer. 2017;140:149‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liavaag PG, Cheung RK, Kerrebijn JD, Freeman JL, Irish JC, Dosch HM. The physiologic reservoir of Epstein‐Barr virus does not map to upper aerodigestive tissues. Laryngoscope. 1998;108:42‐46. [DOI] [PubMed] [Google Scholar]
- 29. Zhang P‐F, Zheng X‐H, Li X‐Z, et al. Nasopharyngeal brushing: a convenient and feasible sampling method for nucleic acid‐based nasopharyngeal carcinoma research. Cancer Commun. 2018;38:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Woellmer A, Hammerschmidt W. Epstein‐Barr virus and host cell methylation: regulation of latency, replication and virus reactivation. Curr Opin Virol. 2013;3:260‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chien Y‐C, Chen J‐Y, Liu M‐Y, et al. Serologic markers of Epstein‐Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N Engl J Med. 2001;345:1877‐1882. [DOI] [PubMed] [Google Scholar]
- 32. Cao S‐M, Liu Z, Jia W‐H, et al. Fluctuations of Epstein‐Barr Virus serological antibodies and risk for nasopharyngeal carcinoma: a prospective screening study with a 20‐year follow‐up. PLoS ONE. 2011;6:e19100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


