Abstract
High‐grade neuroendocrine lung cancer (HGNEC), which includes small cell lung cancer (SCLC) and large cell neuroendocrine carcinoma (LCNEC) of the lung is a rapidly proliferating, aggressive form of lung cancer. The initial standard chemotherapeutic regimens of platinum doublets are recommended for SCLC and have been frequently used for LCNEC. However, there are currently no molecularly targeted agents with proven clinical benefit for this disease. The deubiquitinating enzyme ubiquitin C‐terminal hydrolase‐L1 (UCHL1) is a neuroendocrine cell‐specific product that is known as a potential oncogene in several types of cancer, but little is known about the biological function of UCHL1 and its therapeutic potential in HGNEC. In this study, we found that preclinical efficacy evoked by targeting UCHL1 was relevant to prognosis in HGNEC. UCHL1 was found to be expressed in HGNEC, particularly in cell lines and patient samples of SCLC, and the combined use of platinum doublets with selective UCHL1 inhibitors improved its therapeutic response in vitro. Immunohistochemical expression of UCHL1 was significantly associated with postoperative survival in patients with HGNEC and contributed towards distinguishing SCLC from LCNEC. Circulating extracellular vesicles (EV), including exosomes isolated from lung cancer cell lines and serum from early‐stage HGNEC, were verified by electron microscopy and nanoparticle tracking analysis. Higher levels of UCHL1 mRNA in EV were found in the samples of patients with early‐stage HGNEC than those with early‐stage NSCLC and healthy donors’ EV. Taken together, UCHL1 may be a potential prognostic marker and a promising druggable target for HGNEC.
Keywords: exosomes, extracellular vesicles, high‐grade neuroendocrine carcinoma, large cell neuroendocrine carcinoma, small cell lung cancer, UCHL1
In this study, we found that preclinical efficacy evoked by targeting UCHL1 was relevant to prognosis in HGNEC. UCHL1 may be a potential prognostic marker and a promising druggable target for HGNEC.
1. INTRODUCTION
High‐grade neuroendocrine lung cancer (HGNEC), including small cell lung cancer (SCLC) and large cell neuroendocrine carcinoma (LCNEC) of the lung, is a rapidly proliferating, biologically aggressive type of lung cancer. SCLC is the most lethal form of lung cancer, comprising approximately 15% of lung cancers, and has a substantial initial response to platinum‐doublet combinations, which is followed by the rapid development of resistance.1 A large number of genomic aberrations in SCLC have been demonstrated, including inactivated p53 and RB1, overexpression of cyclin D1, c‐Myc amplification and other somatic mutations.2, 3, 4, 5, 6 However, the clinical benefit of targeting these genomic aberrations has not been confirmed to date. Large cell neuroendocrine carcinoma shares similar epidemiological features and genomic profiles with SCLC, despite the differences in cell size.7, 8, 9 The optimal treatment for LCNEC is now considered to be similar to that for SCLC, owing to their close biological relationship. Although extensive effort has been put into the therapeutic development of HGNEC, no targeted drugs are available. Hence, there is a need for new therapeutic approaches and a greater understanding of the disease.
Ubiquitin C‐terminal hydrolase‐L1 (UCHL1) is a neuroendocrine cell‐specific product that functions to remove ubiquitin from ubiquitinated proteins and is found to be normally expressed only in the neurons and testis.10, 11 UCHL1 is reported to be expressed at higher levels in smokers than nonsmokers, and to be potentially involved in smoking‐induced neoplastic transformation.12 Although UCHL1 is also upregulated in several tumor tissues and is regarded as an important regulator in the progression of many cancers, including lymphoma, colorectal cancer, breast cancer, multiple myeloma and non–small cell lung cancer (NSCLC), there is a lack of detailed information on UCHL1 expression in HGNEC, which is almost always smoking‐induced, particularly regarding its prognostic value and therapeutic potential.13, 14, 15, 16, 17, 18 In this study, we assessed the preclinical efficacy of targeting UCHL1 in vitro, the expression pattern and clinical significance of UCHL1 in patients with HGNEC, and the potential use of UCHL1 expression level in extracellular vesicles (EV) as a noninvasive tumor biomarker.
2. MATERIALS AND METHODS
2.1. Cell lines and cell culture
A549, H1299, PC9 and HCC827 NSCLC cell lines, H82, H69 and H526 SCLC cell lines, and a human bronchial epithelial cell (HBEC3‐KT) were purchased from ATCC, whereas human pulmonary fibroblasts (HPF‐c) were obtained from PromoCell. All cells except for HBEC3‐KT were cultured in RPMI‐1640 (Gibco, Thermo Fisher Scientific) with 10% exosome‐depleted FBS (SBI). HBEC3‐KT was cultured in Keratinocyte‐SFM (Gibco) with l‐Glutamine, EGF and BPE. All cell lines were confirmed to be mycoplasma free using the e‐Myco plus, Mycoplasma OCR Detection Kit (iNtRON Biotechnology).
2.2. Western blotting
Cells were lysed in RIPA lysis buffer (Thermo Fisher Scientific) containing a protease inhibitor cocktail (Roche). Western blot analyses were conducted with 20‐30 μg of total cell protein and 300 ng of EV protein. Protein concentration was determined by BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of total protein were loaded onto SDS‐PAGE on 4%‐20% gels and then transferred to PVDF membranes. The membranes were blocked with 5% milk and then incubated in Tris‐buffered saline with Tween 20 containing primary antibodies overnight, followed by incubation with HRP‐conjugated secondary antibody (1:5000; Sigma‐Aldrich). Membranes were imaged on the ChemiDoc Touch Imaging System (BIO‐RAD). The following were used as primary antibodies: UCHL1 (#13179; CST), p53 (#9282; CST), RB1 (MAB6495; R&D Systems), β‐actin (A5316; Sigma‐Aldrich), CD9 (EXOAB‐CD9A‐1; SBI) and CD63 (EXOAB‐CD63A‐1; SBI).
2.3. MTS assay
Relative cell growth was analyzed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay Kit (Promega) according to the manufacturer’s instructions. Cells were plated onto a 96‐well plate at a density of 1 × 104 cells per well with increasing concentrations of cisplatin (Fujifilm Wako Pure Chemical) plus etoposide (Sigma)/cisplatin plus irinotecan (Cayman Chemical) or in combination with the UCHL1 inhibitor LDN57444 (Sigma) or WP1130 (Santa Cruz), and incubated at 37°C for 72 hours in 5% CO2. Optical density at 490 nm was measured using a plate reader. Each experiment was performed independently at least three times using eight wells each.
2.4. Patients and clinical samples
We reviewed the cases of 72 consecutive patients with HGNEC who underwent complete surgical resection between January 2008 and December 2015 at Tokyo Medical University Hospital. All patients signed the Institutional Review Board‐approved informed consent form. TNM stage was determined in accordance with the 8th edition of the TNM Classification of Malignant Tumors.19 The tumors were histologically subtyped and graded according to the 4th edition of the World Health Organization guidelines.20 Clinical characteristics were retrieved from our clinical records.
2.5. Histopathology
After the specimens were fixed with formalin and embedded in paraffin, serial 4‐µm sections were stained with H&E. All slides were evaluated by a pulmonary pathology specialist (J.M).
2.6. Immunohistochemical analysis
Immunohistochemical (IHC) staining for UCHL1 (#13179; CST) was performed on whole‐section samples, as follows: 5‐μm‐thick formalin‐fixed, paraffin‐embedded tissue sections were deparaffinized and rehydrated. Antigen retrieval was performed using a pressure cooker containing 250 mL of citrate (pH 6.0). Slides were then incubated with the primary antibody overnight. The slides were incubated with HRP‐conjugated polymer secondary antibody (MAX‐PO; Nichirei Biosciences) developed with chromogenic substrates, and counterstained with hematoxylin. The IHC staining intensity and extensiveness of HGNEC were examined using a light microscope under a 40× magnification. Cytoplasmic expression of UCHL1 in lung tumor cells was quantified using a four‐value intensity score (0, no; 1, weak; 2, moderate; and 3, strong), we categorized immunoreactivity by the percentage of the immunopositive area (0%‐100%).
2.7. Quantitative RT‐PCR
The mRNA was isolated using a Qiagen kit (Qiagen) for cell lines and NucleoSpin total RNA FFPE XS (Macherey‐Nagel) for patient samples. The cDNA was generated with SuperScript VILO (Thermo Fisher Scientific). Gene‐specific TaqMan probes (Thermo Fisher Scientific) were utilized for quantitative analyses of mRNA transcript levels of UCHL1 and GAPDH gene as an internal reference. PCR reactions were run using StepOne (Thermo Fisher Scientific), and relative expression was calculated using the 2−ΔΔCT method.
2.8. Isolation of extracellular vesicles
Extracellular vesicles were recovered by a sequential centrifugation procedure using the Exosome Isolation Kit PS (MagCapture, Fujifilm Wako), as follows. Cells were grown in T75 culture flasks for 3‐4 days and the conditioned media was removed from the flasks. Cells were pelleted from the media by centrifugation at 300 g for 5 minutes followed by 1200 g for 20 minutes. To eliminate other cellular debris, the supernatant was spun at 10 000 g for 30 minutes. The sample was concentrated by filtration (Vivaspin 20; Sartorius). After sample preparation, EV were purified by MagCapture according to the manufacturer’s instructions. EV were verified by electron microscopy. EV size and particle numbers were analyzed using the LM10 Nanoparticle Characterization System (NanoSight, Malvern Instruments). The final EV pellet was eluted with elution buffer.
2.9. Electron microscopy
Isolated EV were prepared for examination by transmission electron microscopy. Briefly, 10 μL of EV suspension was placed on a piece of parafilm in a closed Petri dish and 200 mesh Formvar carbon grid (EM Resolutions) was placed on the sample drop for 1 minute. The sample was washed three times for 1 minute each in a 10 μL drop of water by placing the grid on top of the water and gently moving the grid in an up and down motion and then the grid was placed onto a 20‐μL drop of 2% uranyl acetate for 1 minute, followed by a water wash in a 10‐μL drop of water. The grids were dried for a few minutes and imaged using an H‐7500 electron microscope (Hitachi High‐Technologies).
2.10. Digital PCR
mRNA were isolated using a Total Exosome RNA and Protein Isolation Kit (Thermo Fisher Scientific), and the cDNA was generated using SuperScript (Thermo Fisher Scientific). PrimePCR ddPCR Gene Expression Probes (BIO‐RAD) were used for quantitative analyses of mRNA transcript levels of UCHL1, and β‐actin gene was used as an internal reference. PCR reactions were run using QX200 Droplet Generator (BIO‐RAD), and vector copy number was determined with the QX200 droplet digital PCR system as per the manufacturer’s instructions and obtained with the formula of UCHL1 concentration/β‐actin concentration × 2 copies.
2.11. Statistical analysis
Overall survival (OS) was measured from the day of surgery to the day of death from any cause or the day on which the patient was last known to be alive, whereas disease‐free survival (DFS) was measured from the day of surgery to the day until the first event (relapse or death from any cause) or the last follow‐up visit. OS and DFS curves were plotted using the Kaplan‐Meier method, and differences in variables were determined using the log‐rank test. Univariate analysis and multivariate logistic regression analysis with a backward stepwise selection method were performed to identify predictors of poor DFS. The Pearson χ2 test or Fisher’s exact test for categorical data and the Student t test for continuous data were performed to identify factors associated with high UCHL1 expression. Spearman rank correlation was used to measure the degree of association between two numerical variables. All tests were two‐sided, and P‐values of less than 0.05 were considered to indicate a statistically significant difference. The SPSS statistical software package (version 25.0; DDR3 RDIMM, SPSS) was used for statistical analysis.
3. RESULTS
3.1. UCHL1 is widely expressed in small cell lung cancer cell lines
In nearly all SCLC, biallelic inactivation of p53 and RB1 has been reported.2 In addition, several reports have suggested that UCHL1 is upregulated and is an important regulator in the progression of lung cancers.10, 18, 21 Hence, we performed western blot analysis with NSCLC, SCLC cell lines and HBEC3‐KT to confirm if p53, RB1, as well as UCHL1 expression could be detected (Figure 1A). In the three SCLC cell lines, no expression of p53 or RB1 was detected, whereas UCHL1 was widely expressed. As shown in the four NSCLC cell lines, UCHL1 was deactivated in HCC827 whereas p53 was overexpressed in HCC827 and PC9. The ratio of UCHL1 and β‐actin as an internal control showed heterogeneous expression patterns depending on the type of cell lines in NSCLC compared with SCLC (Figure 1B). We then assessed the quantification of the UCHL1 mRNA level. UCHL1 mRNA was also widely shown in the three SCLC cell lines and the four NSCLC cell lines except for HCC827 (Figure 1C).
Figure 1.
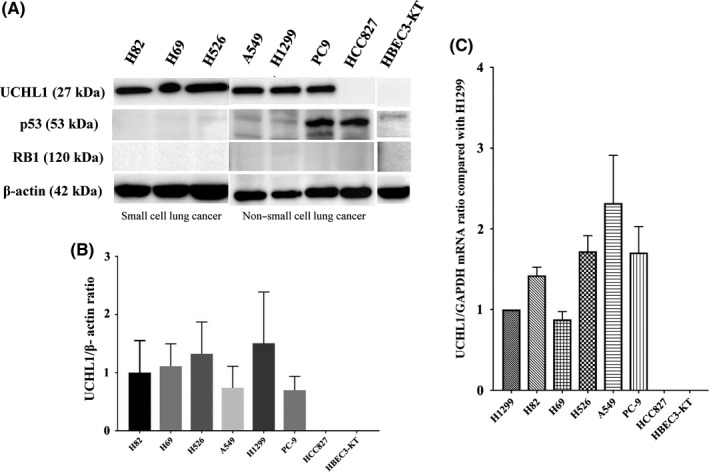
Protein expression of UCHL1, p53 and RB1, and mRNA expression of UCHL1 in various lung cancer cell lines and normal lung epithelial cells. A, Increased UCHL1 levels in all small cell lung cancers (SCLC) tested (H82, H69 and H526) and in three non–small cell lung cancer (NSCLC) cell lines (A549, H1299 and PC9) was confirmed by western blotting. B, Comparison of UCHL1/β‐actin protein expression ratio among the cell lines showed that SCLC consistently had high UCHL1 levels, whereas NSCLC showed different expression patterns depending on the cell line. C, Comparison of UCHL1 mRNA expression among the cell lines showed that all lung cancer cell lines except for HCC827 had high UCHL1 levels
3.2. UCHL1 inhibitors boost therapeutic response of platinum doublet against small cell lung cancer
To test whether UCHL1 could be used as a therapeutic target in SCLC, we analyzed the combined effects of initial treatment regimens for extensive‐disease SCLC, namely cisplatin plus etoposide (PE) or cisplatin plus irinotecan (PI), together with the UCHL1 inhibitors LDN57444 and WP1130, and the effect of these selective inhibitors alone using the MTS assay to compare the IC50 values (Figure 2A). PE or PI in combination with these UCHL1 inhibitors boosted its cytotoxic effects, whereas LDN57444 or WP1130 alone also had a therapeutic effect compared with a PE or PI regimen on H82 cells. PI plus the UCHL1 inhibitors demonstrated slightly increased therapeutic effects on H526 cells compared with PE or PI therapy. Interestingly, LDN57444 but not WP1130 showed a cytotoxic effect on these cells. In contrast, regardless of the degree of UCHL1 expression, combining the UCHL1 inhibitors with platinum doublets and the selective inhibitor monotherapies demonstrated no effects on NSCLC cells. Treatment responses that were summarized with the IC50 are shown in Figure 2B. Taken together, the use of PE or PI in combination with selective UCHL1 inhibitors improves its therapeutic response in SCLC lines.
Figure 2.
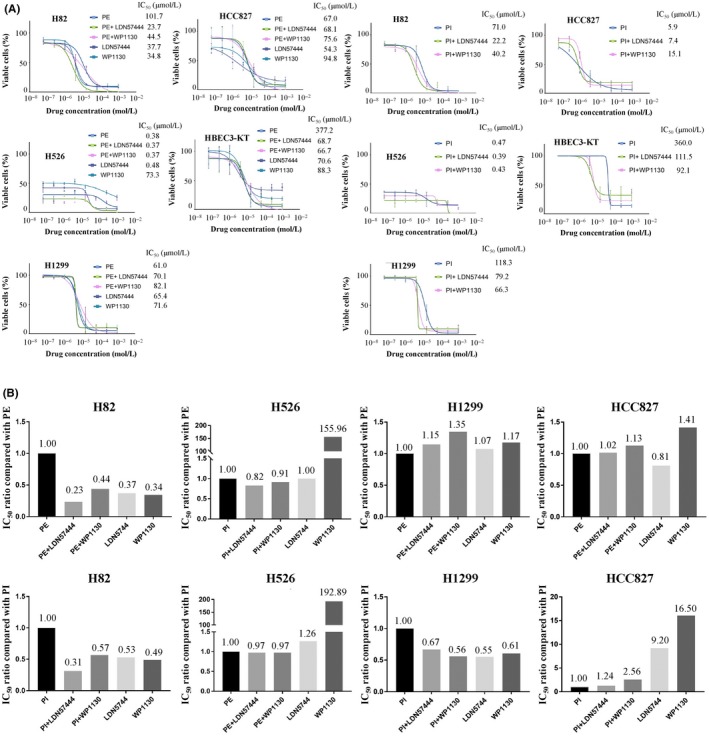
Combined cytotoxic effects of cisplatin plus etoposide (PE) or cisplatin plus irinotecan (PI) with selective UCHL1 inhibitors and the inhibitors alone on small cell lung cancer (SCLC) cells using the MTS assay. A, The combination of PE or PI and the selective UCHL1 inhibitors WP1130 and LDN57444 increased the therapeutic effects in H82 and H526 cells but not in non–small cell lung cancer (NSCLC) lines. LDN57444 or WP1130 alone had an effect compared with a PE or PI regimen on H82 cells. B, Treatment responses in SCLC and NSCLC were summarized with the IC50
3.3. UCHL1 is a prognostic marker in patients with high‐grade neuroendocrine lung cancer
We used IHC to evaluate the expression level of UCHL1 in primary tumors from 72 patients with HGNEC, including 38 LCNEC and 34 SCLC. Representative UCHL1 positive and negative tissues of SCLC and LCNEC that were evaluated using a four‐value intensity score multiplied by stained area (ranging from 0 to 300) resulted in an expression level of 270 for a positive SCLC case, 240 for a positive LCNEC case, 20 for a weak SCLC case, and 0 for a negative LCNEC case (Figure 3A). Comparison of the IHC and mRNA expression levels between SCLC and LCNEC showed a significantly higher IHC expression level (Figure 3B; P = 0.005) and mRNA level (Figure 3C; P = 0.026) of UCHL1, which allows UCHL1 to be a potential differential diagnostic marker for HGNEC. UCHL1 IHC expression level was positively correlated with UCHL1 mRNA expression (Figure 3D).
Figure 3.
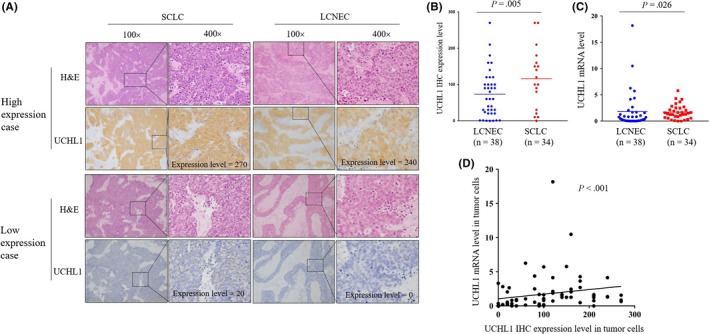
UCHL1 expression in resected high‐grade neuroendocrine lung cancer (HGNEC) patients analyzed by immunohistochemistry (IHC) and real‐time quantitative PCR. A, Consecutive paraffin slides from two representative small cell lung cancer (SCLC) tissues and two large cell neuroendocrine cancer (LCNEC) tissues were evaluated by H&E and UCHL1 staining. Representative H&E and IHC of UCHL1‐positive SCLC cases and LCNEC cases with high UCHL1 expression and those with low UCHL1 expression. Quantification of the staining intensity of sections as shown. B, SCLC tissues showed significantly higher IHC expression of UCHL1 than LCNEC (P = 0.005). C, SCLC tissues showed significantly higher mRNA levels of UCHL1 than LCNEC (P = 0.026). GAPDH was used as the corresponding control. D, UCHL1 IHC expression level was positively correlated with UCHL1 mRNA expression (P < 0.001)
Patient demographics and clinical characteristics are summarized in Table 1. The mean age of the patients was 69 ± 9 years, and 58 patients (81%) were men. The majority of patients underwent a lobectomy (n = 62, 86%). Lymph‐vascular invasions and lymph node metastasis were observed in 68 (94%) and 26 (36%) patients, respectively.
Table 1.
Patient characteristics
| Variable | Value (n = 72, %) |
|---|---|
| Age, y (mean ± SD) | 49‐84 (69 ± 9) |
| Sex | |
| Male | 58 (81) |
| Female | 14 (19) |
| Brinkman index (mean ± SD) | 0‐2940 (1038 ± 529) |
| Comorbidities | |
| No | 27 (38) |
| Yes | 45 (62) |
| FEV1.0%, (mean ± SD) | 38.4‐92.9 (67.0 ± 10.4) |
| Surgical procedure | |
| Pneumonectomy | 1 (1) |
| Lobectomy | 62 (86) |
| Limited resection | 9 (13) |
| Tumor size, cm (mean ± SD) | 1.0‐12.0 (3.4 ± 2.1) |
| Adjuvant chemotherapy | |
| Yes | 31 (43) |
| No | 41 (57) |
| Lymph‐vascular invasion | |
| Positive | 68 (94) |
| Negative | 4 (6) |
| Lymph node metastasis | |
| Positive | 26 (36) |
| Negative | 46 (64) |
| Histology | |
| SCLC | 34 (47) |
| LCNEC | 38 (53) |
| pTNM stage | |
| Stage IA | 20 (28) |
| Stage IB | 17 (24) |
| Stage II | 22 (31) |
| Stage III | 12 (17) |
| Stage IV | 1 (1) |
Abbreviations: FEV1.0, forced expiratory volume in 1 s; LCNEC, large cell neuroendocrine carcinoma; SCLC, small cell lung cancer; SD, standard deviation; TNM, pathological tumor, node and metastasis.
We then investigated the prognostic significance of UCHL1 positivity in HGNEC cancer cells and calculated the receiver operating characteristics curve for recurrence using the UCHL1 expression score. The area under the curve and the optimal cut‐off value relevant to recurrence were 0.699 (P < 0.001) and 70, respectively (data not shown). Univariate and multivariate analyses were performed to identify factors associated with DFS (Table 2). The UCHL1 expression level was significantly associated with DFS for both univariate (P = 0.006) and multivariate analysis (P = 0.006). The Kaplan‐Meier curves of UCHL1 expression of ≥70 and <70 for DFS (Figure 4A) and OS (Figure 4B) demonstrated a 5‐year DFS of 22.0% and 53.4% (P = 0.005), and a 5‐year OS of 40.1% and 55.6% (P = 0.081), respectively. Patients with LCNEC tended to show better prognosis in DFS (Figure 4C, P = 0.309) and OS (Figure 4D, P = 0.089) than those with SCLC but not significantly. We also evaluated the association between various clinicopathological factors and UCHL1 expression (Table 3). SCLC histology, decreased forced expiratory volume in 1 second, and the presence of chromogranin A expression were found to be associated with a higher expression level of UCHL1.
Table 2.
Univariate and multivariate analysis of disease‐free survival
| Variable |
Univariate analysis Hazard ratio (95% CI) |
P‐value |
|---|---|---|
| Age | 1.032 (0.999‐1.067) | 0.053 |
| Sex (men vs women) | 1.565 (0.698‐3.509) | 0.276 |
| Adjuvant chemotherapies (no vs yes) | 1.339 (0.740‐2.421) | 0.335 |
| Tumor size | 1.062 (0.905‐1.245) | 0.462 |
| Lymph‐vascular invasion (yes vs no) | 1.105 (0.342‐3.571) | 0.867 |
| Lymph node metastasis (yes vs no) | 1.282 (0.703‐2.338) | 0.417 |
| Histology (SCLC vs LCNEC) | 1.352 (0.754‐2.423) | 0.311 |
| pTNM stage (II‐IV vs I) | 1.011 (0.984‐1.038) | 0.425 |
| UCHL1 expression (≥70 vs <70) | 2.532 (1.301‐4.926) | 0.006 |
|
Multivariate analysis Hazard ratio (95% CI) |
P‐value | |
|---|---|---|
| Age | 1.033 (1.000‐1.067) | 0.053 |
| UCHL1 expression (≥70 vs <70) | 2.556 (1.312‐4.983) | 0.006 |
Abbreviations: CI, confidence interval; FEV1.0, forced expiratory volume in 1 s; LCNEC, large cell neuroendocrine carcinoma; SCLC, small cell lung cancer; TNM, pathological tumor, node, and metastasis.
The bold values are statistically significant with P‐value < 0.05.
Figure 4.
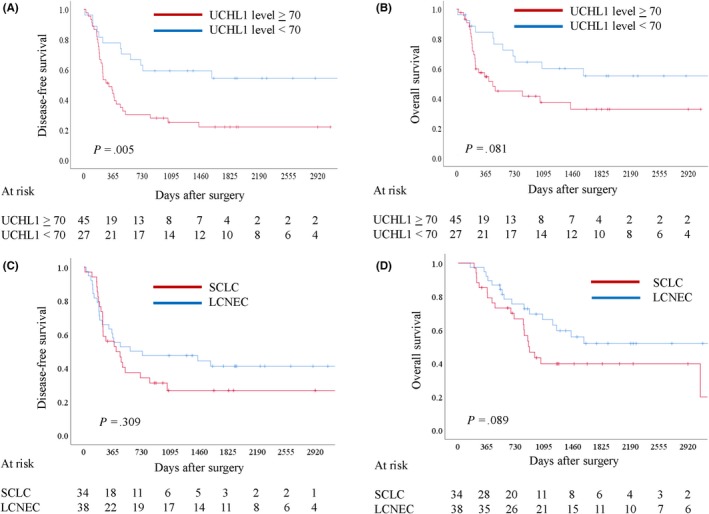
Survival analyses showing prognostic significance of UCHL1 expression in high‐grade neuroendocrine lung cancer (HGNEC) patients and the prognostic difference according to histology. The receiver operating characteristics curves for recurrence provided an immunohistochemical staining score of 70 as an adequate cut‐off value of UCHL1 levels. The prognostic value of a high UCHL1 level (≥70) in HGNEC patients was statistically significant for disease‐free survival (DFS, A; P = 0.005), and marginally significant for overall survival (OS, B; P = 0.081). There was no statistically significant difference between large cell neuroendocrine cancer and small cell lung cancer for DFS (C; P = 0.309) and there was a marginally significant difference for OS (P = 0.089)
Table 3.
Factors associated with high‐UCHL1 expression in patients with resected HGNEC
| Variable |
High UCHL1 level (≥70) |
Low UCHL1 level (<70) |
P‐value |
|---|---|---|---|
| Age, y (mean ± SD) | 70 ± 9 | 69 ± 9 | 0.777 |
| Sex | |||
| Men | 37 | 21 | |
| Women | 8 | 6 | 0.645 |
| Brinkman index (mean ± SD) | 1060 ± 561 | 1000 ± 477 | 0.641 |
| FEV1.0% (mean ± SD) | 65.1 ± 9.6 | 70.4 ± 11.0 | 0.040 |
| Tumor size, cm (mean ± SD) | 3.3 ± 2.2 | 3.7 ± 1.8 | 0.410 |
| Lymph‐vascular invasion | |||
| Positive | 43 | 25 | |
| Negative | 2 | 2 | 0.595 |
| Lymph node metastasis | |||
| Positive | 17 | 9 | |
| Negative | 28 | 18 | 0.704 |
| Histology | |||
| SCLC | 27 | 7 | |
| LCNEC | 18 | 20 | 0.005 |
| pTNM stage | |||
| I | 24 | 13 | |
| II‐IV | 21 | 14 | 0.670 |
| Synaptophysin | |||
| Positive | 35 | 17 | |
| Negative | 10 | 10 | 0.174 |
| CD56 | |||
| Positive | 45 | 27 | Not evaluable |
| Negative | 0 | 0 | |
| Chromogranin A | |||
| Positive | 35 | 13 | |
| Negative | 10 | 14 | 0.010 |
Abbreviations: FEV1.0, forced expiratory volume in 1 s; HGNEC, high‐grade neuroendocrine carcinoma; LCNEC, large cell neuroendocrine carcinoma; SCLC, small cell lung cancer; TNM, pathological tumor, node and metastasis.
The bold values are statistically significant with P‐value < 0.05.
3.4. Characterization of cancer‐derived extracellular vesicles
Cancer‐derived EV, including exosomes, are reported to play a crucial role in the metastatic process.22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33 As HGNEC have a high metastatic ability, UCHL1 enriched in patients’ cancer‐derived EV may serve as a promising blood‐based biomarker for the early detection of HGNEC. Figure 5A shows EV that were isolated from the serum of an SCLC patient and the two SCLC cell lines. EV were identified by transmission electron microscopy as small spherical vesicles and were characterized by the nanoparticle characterization system (Figure 5A). Microvesicle clusters from this patient as well as from H82 and H526 were observed as round vesicles measuring 133.7 nm, 123.9 nm and 140.0 nm in median diameter, respectively. The protein contents of EV were assayed by western blotting and were confirmed to express the common exosome markers CD9 and CD63 (Figure 5B). The quantity of EV released by the 2 SCLC cell lines, A549 and HPF‐c, demonstrated that SCLC cell lines produced significantly more EV than HPF‐c (Figure 5C).
Figure 5.
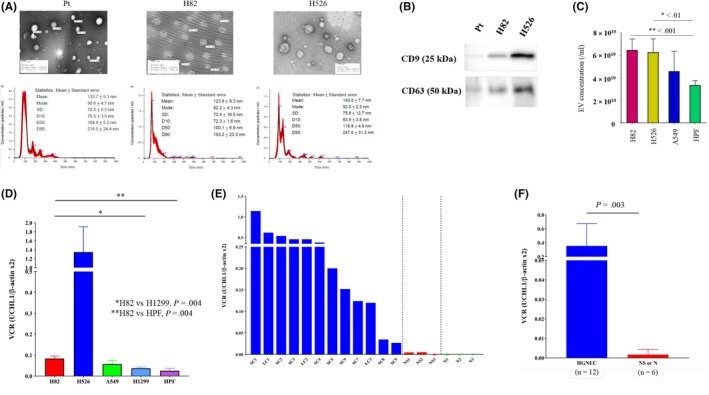
Characteristics of small cell lung cancer (SCLC)‐derived extracellular vesicles (EV) and quantification of UCHL1 levels in EV from SCLC patients and cell lines. A, Representative electron microscopic images and the results of nanoparticle characterization analysis providing the number of EV and the size distribution from the serum of an SCLC patient and two SCLC cell lines. B, Expression levels of CD9 and CD63 in EV from different cells were assayed by western blotting. C, Comparative analysis of EV production among the different cell lines. D, Comparative analysis of UCHL1 mRNA levels in cancer‐derived EV in vitro. SCLC cells, particularly H82 cells, showed higher UCHL1 levels in their EV compared with H1299 cells (P = 0.004) and HPF‐c cells (P = 0.004). E, UCHL1 mRNA levels in serum‐derived EV of p‐stage I‐II SCLC patients (n = 9), large cell neuroendocrine cancer (n = 3), non–small cell lung cancer (NSCLC) patients (n = 3) and healthy donors (n = 3). LC, large cell neuroendocrine carcinoma; NS, non–small cell lung cancer; SC, small cell lung cancer. F, UCHL1 mRNA levels in serum‐derived EV of p‐stage I‐II high‐grade neuroendocrine cancer patients (n = 12) was significantly higher than those of p‐stage I‐II NSCLC or healthy donors (n = 6, P = 0.393)
3.5. UCHL1 mRNA in extracellular vesicles as a potential biomarker for high‐grade neuroendocrine lung cancer
We next compared UCHL1 mRNA levels in cancer‐derived EV from different cell lines and among patient samples. Among the cell lines, SCLC cells, particularly H82 cells, showed higher UCHL1 levels in their EV compared with H1299 cells (P = 0.004) and HPF‐c cells (P = 0.004; Figure 5D). Likewise, UCHL1 mRNA levels in serum‐derived EV of p‐stage I‐II SCLC patients (n = 9; SC1‐9) and p‐stage I‐II LCNEC (n = 3; LC1‐3) were significantly higher than in p‐stage I‐II NSCLC patients (n = 3; NS1‐3) or healthy donors (n = 3; N1‐3; Figure 5E, and SC cases vs NS or N cases, P = 0.003; Figure 5F). Taken together, the increased EV‐derived UCHL1 mRNA levels in the EV of both SCLC cell lines and serum from patients with early‐stage SCLC suggest that UCHL1 is useful as a novel prognostic marker as well as for the early diagnosis of HGNEC.
4. DISCUSSION
No substantial advances have been achieved in the treatment/prevention of or in effective early detection methods for SCLC for the past 30 years.34 For resected early‐stage SCLC patients, clinical consensus on the application of adjuvant treatment based on the randomized trials has not been established. The initial standard chemotherapeutic regimens with platinum doublets which were recommended for SCLC and had been frequently used for LCNEC had not been changed for decades.35, 36, 37, 38 A recent epoch‐making study on the first‐line treatment of extensive‐stage SCLC demonstrated that the addition of a programmed death ligand 1 inhibitor, atezolizumab, to carboplatin and etoposide resulted in longer survival than chemotherapy alone. However, there are still no significant targeted therapies with proven clinical advantage for HGNEC.39 HGNEC is initially highly responsive to chemotherapy and radiotherapy but eventually results in a very quick relapse, with a 5‐year survival rate of less than 10% in SCLC and 16.8% in LCNEC.1, 40 Thus, a deeper understanding of the biological and molecular characteristics underlying the initiation, metastasis and acquisition of resistance of HGNEC is warranted.
Comprehensive genomic and proteomic approaches to discover novel biomarkers that are potentially helpful toward defining molecularly selected subpopulations of patients with HGNEC are key to identifying new molecular targets.2, 40 Genome sequencing profiles of SCLC demonstrated that approximately 25% of human SCLC tumors have inactivating mutations in NOTCH family genes.2 Activation of NOTCH signaling in a preclinical SCLC mouse model reduced the tumor burden and extended the survival of the mice, whereas Notch activity in SCLC cells abrogated neuroendocrine gene expression.2 The enhancer of zeste homolog (EZH2) is highly upregulated in SCLC compared with normal lung tissue and this increase is correlated with high methylation of the EZH2 promotor.41 EZH2 also promotes epigenetic silencing of SLFN11, which contributes to DNA damage repair deficiency, to drive the acquired resistance of chemotherapy.42 Proteomic analysis of SCLC tumors led to the discovery that Poly ADP ribose polymerase 1 (PARP1) inhibition had a therapeutic efficacy in a preclinical model and in a subset of SCLC patients, with proteomic markers of DNA repair and phosphatidylinositol 3‐kinase (PI3K) pathway activation being predictive for the response of PARP inhibition in SCLC.43 For LCNEC, sequencing‐based molecular profiling with resected HGNEC samples demonstrated that LCNEC and SCLC had similar genomic profiles.7, 8, 9 Genetic alterations in the PI3K/AKT/mTOR pathway were found in 15% of LCNEC, which, hence, may be a promising druggable target.7 These data demonstrated several potential therapeutic targets for HGNEC, and clinical trials to target these candidates are ongoing. However, the translational application of these preclinical findings has yet to succeed in offering real clinical benefit to patients with HGNEC.
UCHL1 acts as either an oncogene or a tumor‐suppressor gene depending on the cancer origin, partially because its nature of the deubiquitinating activity that inhibits proteasome‐mediated degradation is diverse, by the deubiquitination of proteins themselves that are affected in a variety of tumor tissues.13, 14, 15, 17, 44, 45 It has been documented that deubiquitinases (DUBs), including UCHL1, play a role in cancer development and progression, and several DUB inhibitors have been discovered to have profound effects in inhibiting tumorigenesis.46 In lung cancer cells, UCHL1 is markedly expressed and is a key regulator of cell migration, invasion and metastasis.10, 18, 21 UCHL1 is normally expressed only in neurons, neuroendocrine cells and testis, and is consistently upregulated in the large and small airway epithelium of smokers but not never‐smokers.10, 11, 12 Therefore, there was higher expression of UCHL1 shown in the airway epithelium of patients with chronic obstructive pulmonary disease.12 That may be a possible explanation for the result of our study that the high UCHL1 expression was correlated with low forced expiratory volume in 1 second. Given that UCHL1 is a marker of neuroendocrine cells and can be involved in the tobacco‐induced malignant transformation of normal cells, we hypothesized that UCHL1 plays a role in the carcinogenesis of HGNEC, which has a very high mutation load owing to its long‐term exposure to carcinogens in cigarette smoke. Therefore, the inhibition of UCHL1 is evolving as a potential anticancer treatment by synergistically preventing platinum resistance in cancer cells.2, 12
We showed that the use of PE/PI together with selective UCHL1 inhibitors improved its therapeutic response in SCLC lines but not in NSCLC lines, regardless of the degree of UCHL1 expression status. In a murine model, LDN57444 was reported to have an antimetastatic effect on UCHL1‐expressing cancer cells.14 WP1130‐mediated inhibition of tumor‐activated DUB was found to induce the downregulation of anti–apoptotic proteins and the upregulation of proapoptotic proteins, such as p53.47 However, little is known about how these selective UCHL1 inhibitors boosted the effect of platinum doublets exclusively for SCLC. Further studies to investigate the therapeutic mechanism of selective UCHL1 inhibitors in HGNEC are required.
We confirmed that UCHL1 expression levels were significantly correlated with poor prognosis and were statistically useful for the differential diagnosis of SCLC and LCNEC. Sasaki et al48 reported that UCHL1 expression was strongly associated with the pathological stage of NSCLC. The use of UCHL1 as a predictor of post–therapeutic survival or as a druggable target might be promising for overcoming chemo‐resistance in patients with HGNEC.
Extracellular vesicles, including exosomes, are known as intercellular messengers and can shuttle cargos, such as proteins, lipids, mRNA and microRNA between cells.23, 24, 25, 26 EV have been studied in the cancer diagnostic setting because cancer‐derived EV are found to be involved in every event of the metastatic cascade, such as invasion, migration and priming of metastatic niches.28, 31, 32, 49 As HGNEC metastasize rapidly to many sites, cancer‐derived EV are likely to serve as a promising noninvasive tool for the early detection of HGNEC. In the present study, EV from SCLC cell lines showed higher UCHL1 levels than those from others, whereas UCHL1 mRNA levels in serum‐derived EV of p‐stage I‐II HGNEC patients was demonstrated to be higher than in patients with identical stage NSCLC, and healthy donors. These studies demonstrated for the first time that UCHL1 mRNA levels in EV may be useful as a prognostic marker as well as for the early diagnosis of HGNEC. However, it remains unclear whether EV‐based liquid biopsy is a more suitable method for the early detection of HGNEC compared with other methods such as circulating‐tumor cells or cell‐free DNA. Moreover, the isolation and characterization methods of EV are still a matter of debate. Further studies are needed before this liquid biopsy approach for patients with HGNEC can be successfully applied in routine clinical practice.
In the present study, our findings demonstrated that UCHL1 is expressed widely among HGNEC, particularly in SCLC in vitro and in patient samples, and may be useful as a prognostic marker and a treatment target for HGNEC. Our blood‐based liquid biopsy study demonstrated that UCHL1 gene expression levels were higher in HGNEC samples than in NSCLC and healthy donor samples, even though the number of patients was small. The biological mechanism as to how UCHL1 is involved in the tumorigenesis, metastasis and therapeutic resistance of HGNEC is not yet known and merits further investigation.
DISCLOSURE
The authors have no conflicts of interest to disclose.
ACKNOWLEDGMENTS
The authors are indebted to the medical editors of the Department of International Medical Communications of Tokyo Medical University for their editorial review of the English manuscript. We thank Kumiko Nagase for her technical support. We also thank Mami Murakami for helping with statistical analyses of this work. This study was supported in part by a Grant‐in‐Aid for Cancer Research from the Ministry of Education, Culture, Sports, and Technology, Japan (#18K08799), and a grant from the Tokyo Medical University Cancer Research Foundation.
Shimada Y, Kudo Y, Maehara S, et al. Ubiquitin C-terminal hydrolase-L1 has prognostic relevance and is a therapeutic target for high‐grade neuroendocrine lung cancers. Cancer Sci. 2020;111:610–620. 10.1111/cas.14284
REFERENCES
- 1. Bunn PA Jr, Minna JD, Augustyn A, et al. Small cell lung cancer: can recent advances in biology and molecular biology be translated into improved outcomes? J Thorac Oncol. 2016;11:453‐474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peifer M, Fernandez‐Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small‐cell lung cancer. Nat Genet. 2012;44:1104‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small‐cell lung cancer. Nat Genet. 2012;44:1111‐1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nau MM, Brooks BJ, Battey J, et al. L‐myc, a new myc‐related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69‐73. [DOI] [PubMed] [Google Scholar]
- 6. Mollaoglu G, Guthrie MR, Bohm S, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. 2017;31:270‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miyoshi T, Umemura S, Matsumura Y, et al. Genomic profiling of large‐cell neuroendocrine carcinoma of the lung. Clin Cancer Res. 2017;23:757‐765. [DOI] [PubMed] [Google Scholar]
- 8. Ito M, Miyata Y, Hirano S, et al. Therapeutic strategies and genetic profile comparisons in small cell carcinoma and large cell neuroendocrine carcinoma of the lung using next‐generation sequencing. Oncotarget. 2017;8:108936‐108945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones MH, Virtanen C, Honjoh D, et al. Two prognostically significant subtypes of high‐grade lung neuroendocrine tumours independent of small‐cell and large‐cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363:775‐781. [DOI] [PubMed] [Google Scholar]
- 10. Wilkinson KD, Lee KM, Deshpande S, Duerksen‐Hughes P, Boss JM, Pohl J. The neuron‐specific protein PGP 9.5 is a ubiquitin carboxyl‐terminal hydrolase. Science. 1989;246:670‐673. [DOI] [PubMed] [Google Scholar]
- 11. Sekiguchi S, Yoshikawa Y, Tanaka S, et al. Immunohistochemical analysis of protein gene product 9.5, a ubiquitin carboxyl‐terminal hydrolase, during placental and embryonic development in the mouse. Exp Anim. 2003;52:365‐369. [DOI] [PubMed] [Google Scholar]
- 12. Carolan BJ, Heguy A, Harvey BG, Leopold PL, Ferris B, Crystal RG. Up‐regulation of expression of the ubiquitin carboxyl‐terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Res. 2006;66:10729‐10740. [DOI] [PubMed] [Google Scholar]
- 13. Hussain S, Bedekovics T, Chesi M, Bergsagel PL, Galardy PJ. UCHL1 is a biomarker of aggressive multiple myeloma required for disease progression. Oncotarget. 2015;6:40704‐40718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goto Y, Zeng L, Yeom CJ, et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF‐1alpha. Nat Commun. 2015;6:6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schroder C, Milde‐Langosch K, Gebauer F, et al. Prognostic relevance of ubiquitin C‐terminal hydrolase L1 (UCH‐L1) mRNA and protein expression in breast cancer patients. J Cancer Res Clin Oncol. 2013;139:1745‐1755. [DOI] [PubMed] [Google Scholar]
- 16. Zhong J, Zhao M, Ma Y, et al. UCHL1 acts as a colorectal cancer oncogene via activation of the beta‐catenin/TCF pathway through its deubiquitinating activity. Int J Mol Med. 2012;30:430‐436. [DOI] [PubMed] [Google Scholar]
- 17. Hussain S, Foreman O, Perkins SL, et al. The de‐ubiquitinase UCH‐L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia. 2010;24:1641‐1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim HJ, Kim YM, Lim S, et al. Ubiquitin C‐terminal hydrolase‐L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117‐127. [DOI] [PubMed] [Google Scholar]
- 19. Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39‐51. [DOI] [PubMed] [Google Scholar]
- 20. Travis W, Brambilla E, Burke AP, et al. WHO Classification of Tumors of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2015. [DOI] [PubMed] [Google Scholar]
- 21. Hibi K, Westra WH, Borges M, Goodman S, Sidransky D, Jen J. PGP9.5 as a candidate tumor marker for non–small‐cell lung cancer. Am J Pathol. 1999;155:711‐715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol. 2017;189:259‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vanni I, Alama A, Grossi F, Dal Bello MG, Coco S. Exosomes: a new horizon in lung cancer. Drug Discov Today. 2017;22:927‐936. [DOI] [PubMed] [Google Scholar]
- 24. Guo W, Gao Y, Li N, et al. Exosomes: new players in cancer (Review). Oncol Rep. 2017;38:665‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug‐resistance of cancer cells. Int J Cancer. 2017;141:220‐230. [DOI] [PubMed] [Google Scholar]
- 26. Wen SW, Sceneay J, Lima LG, et al. The biodistribution and immune suppressive effects of breast cancer‐derived exosomes. Cancer Res. 2016;76:6816‐6827. [DOI] [PubMed] [Google Scholar]
- 27. Paggetti J, Haderk F, Seiffert M, et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer‐associated fibroblasts. Blood. 2015;126:1106‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Costa‐Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816‐826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tickner JA, Urquhart AJ, Stephenson SA, Richard DJ, O’Byrne KJ. Functions and therapeutic roles of exosomes in cancer. Front Oncol. 2014;4:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91:431‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nat Med. 2012;18:883‐891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581‐593. [DOI] [PubMed] [Google Scholar]
- 34. Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer. 2015;121:664‐672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rudin CM, Ismaila N, Hann CL, et al. Treatment of small‐cell lung cancer: American Society of Clinical Oncology endorsement of the American College of Chest Physicians guideline. J Clin Oncol. 2015;33:4106‐4111. [DOI] [PubMed] [Google Scholar]
- 36. Fruh M, De Ruysscher D, Popat S, et al. Small‐cell lung cancer (SCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2013;24(suppl 6):vi99‐vi105. [DOI] [PubMed] [Google Scholar]
- 37. Niho S, Kenmotsu H, Sekine I, et al. Combination chemotherapy with irinotecan and cisplatin for large‐cell neuroendocrine carcinoma of the lung: a multicenter phase II study. J Thorac Oncol. 2013;8:980‐984. [DOI] [PubMed] [Google Scholar]
- 38. Iyoda A, Hiroshima K, Moriya Y, et al. Prospective study of adjuvant chemotherapy for pulmonary large cell neuroendocrine carcinoma. Ann Thorac Surg. 2006;82:1802‐1807. [DOI] [PubMed] [Google Scholar]
- 39. Horn L, Mansfield AS, Szczęsna A, et al. First‐line Atezolizumab plus chemotherapy in extensive‐stage small‐cell lung cancer. N Eng J Med. 2018;23:2220‐2229. [DOI] [PubMed] [Google Scholar]
- 40. Cao L, Li ZW, Wang M, Zhang TT, Bao B, Liu YP. Clinicopathological characteristics, treatment and survival of pulmonary large cell neuroendocrine carcinoma: a SEER population‐based study. PeerJ. 2019;7:e6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poirier JT, Gardner EE, Connis N, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015;34:5869‐5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2‐SLFN11 axis. Cancer Cell. 2017;31:286‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Byers LA, Wang J, Nilsson MB, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798‐811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ummanni R, Jost E, Braig M, et al. Ubiquitin carboxyl‐terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol Cancer. 2011;10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li L, Tao Q, Jin H, et al. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16:2949‐2958. [DOI] [PubMed] [Google Scholar]
- 46. Liu J, Shaik S, Dai X, et al. Targeting the ubiquitin pathway for cancer treatment. Biochim Biophys Acta. 2015;1855:50‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small‐molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265‐9276. [DOI] [PubMed] [Google Scholar]
- 48. Sasaki H, Yukiue H, Moriyama S, et al. Expression of the protein gene product 9.5, PGP9.5, is correlated with T‐status in non–small cell lung cancer. Jpn J Clin Oncol. 2001;31:532‐535. [DOI] [PubMed] [Google Scholar]
- 49. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569‐579. [DOI] [PubMed] [Google Scholar]


