Abstract
Regorafenib has improved the survival of patients with refractory metastatic colorectal cancer (mCRC), yet the mechanisms of inherited or acquired resistance are not well understood. A total of 50 patients with refractory mCRC were enrolled. Circulating tumor cell (CTC) enumeration was carried out at baseline, day 21 after initiation of regorafenib, and at the time of progression of disease (PD) using the CellSearch System (Veridex LLC, NJ, USA). Poly(A) mRNA was extracted from CTCs, and gene expression of epithelial and epithelial‐mesenchymal transition markers was analyzed by a multiplex‐PCR based DNA Chip. Patients with fewer than 3 CTCs at baseline and day 21 had a longer progression‐free survival than those with 3 or more CTCs (3.3 vs 2.0 months, P = .008 and 3.3 vs 2.0 months, P = .004, respectively). Patients with fewer than 3 CTCs at baseline and day 21 had a longer overall survival (OS) than those with 3 or more CTCs (10.0 vs 4.6 months, P < .001 and 8.7 vs 3.8 months, P = .003, respectively). In multivariable analysis, CTC counts remained significantly associated with OS at baseline and day 21 (P = .019 and P = .028). Circulating tumor cell EGFR gene expression was upregulated at day 21 and/or PD in 64% of patients. Patients had significantly increased EGFR expression at PD compared to baseline (P = .041) and at day 21 and/or PD compared to baseline (P = .004). Our findings suggest that CTC count and EGFR expression could be useful markers of regorafenib efficacy and outcomes. Upregulation of CTC EGFR expression might be a molecular escape mechanism under regorafenib therapy.
Keywords: circulating tumor cell, colorectal cancer, epidermal growth factor receptor, epithelial marker and EMT, regorafenib
We identified circulating tumor cell count as a prognostic marker in metastatic colorectal cancer patients treated with regorafenib. Circulating tumor cell EGFR expression was significantly increased with regorafenib at the time of disease progression, suggesting this to be a mode of resistance and lending further mechanistic evidence for the synergistic effects seen with regorafenib and anti‐EGFR mAbs.
![]()
1. INTRODUCTION
Regorafenib, an oral multikinase inhibitor, blocks the activity of several protein kinases, including v‐raf murine sarcoma viral oncogene homolog B1 (BRAF),1 and improves the progression‐free survival (PFS) and overall survival (OS) of chemorefractory metastatic colorectal cancer (mCRC) patients.1, 2 A retrospective exploratory analysis of the pivotal phase III CORRECT trial proposed BEAMing analysis of circulating DNA as a potential biomarker and viable approach to obtain real‐time tumor‐associated genotypic information in mCRC patients treated with regorafenib. Nonetheless, there are currently no validated predictive or prognostic biomarkers of regorafenib efficacy.
Circulating tumor cells (CTCs) are shed from the primary tumor, migrate to sites of metastases, and serve as a noninvasive means of monitoring the dynamic alterations driving treatment efficacy and disease progression. The most widely studied CTC detection methods are based on immunomagnetic enrichment with antiepithelial cell adhesion molecule (EpCAM) Abs and subsequent immunological identification with anticytokeratin (anti‐CK). The CellSearch system is one such method and is the only FDA‐approved assay for the enumeration of CTCs in peripheral blood.3, 4, 5 In a study by Cohen et al using CellSearch, the presence of 3 or more CTCs at baseline and follow‐up was an independent prognostic marker of inferior survival in mCRC patients.5, 6, 7 However, it is unclear whether CTC enumeration at baseline and over time is predictive or prognostic in mCRC patients specifically treated with regorafenib.
In addition to enumeration, protein expression and molecular profiling of CTCs might serve as more refined biomarkers and help inform a more personalized treatment approach. Under the pressure imposed by chemotherapy and mAbs, clonal selection and genomic instability emerge and provoke eventual treatment resistance. The ability to characterize such intratumoral heterogeneity could help to identify novel predictive and prognostic biomarkers and improve treatment decision‐making. To this end, our group has previously examined the prognostic role of epithelial‐mesenchymal transition (EMT) gene (PI3K, Akt‐2, and Twist1) expression within CTCs of patients with mCRC (Ning Y et al).8
In this study, we examined the ability of CTC enumeration and molecular characterization, using mRNA levels of epithelial (EGFR, EpCAM, and CEA) and EMT (PI3K, Akt‐2, and Twist1) markers, to predict outcomes and identify potential molecular escape mechanisms in mCRC patients undergoing regorafenib therapy.
2. MATERIALS AND METHODS
2.1. Study design and patient population
This was a retrospective analysis of a prospective single‐arm study examining the efficacy and toxicity of regorafenib in patients with refractory mCRC undertaken at the Cancer Institute Hospital of the Japanese Foundation for Cancer Research. Eligible patients were 18 years or older with histologically confirmed mCRC, who had progressed or were intolerant to standard therapy (including a fluoropyrimidine, irinotecan, oxaliplatin, bevacizumab, and an antiepidermal growth factor receptor [EGFR] agent if indicated), and were subsequently treated with regorafenib at a dose of 160 mg orally daily for 21 days of every 28‐day cycle. Treatment was continued until the development of intolerable toxicities or disease progression. Standard inclusion criteria applied, including ECOG performance status of 0 or 1, presence of measurable disease, life expectancy of at least 3 months, and adequate organ function. Between March 2013 and December 2014, a total of 62 patients were enrolled, of which 50 met inclusion/exclusion criteria and had sufficient blood samples for analysis. Study protocols were approved by the Institutional Review Board at the Cancer Institute Hospital of the Japanese Foundation for Cancer Research. All patients provided written informed consent for the evaluation of molecular correlates.
2.2. Isolation and enumeration of CTCs
Circulating tumor cell enumeration was carried out at baseline prior to initiation of regorafenib, day 21 after initiation of therapy, and at the time of treatment intolerance or progression of disease (PD) using the CellSearch system. A total of 7.5 mL whole blood was collected on a CellSave Preservative Tube, and the CellSearch Circulating Tumor Cell Kit (Veridex) was applied for CTC enrichment and enumeration. Immunomagnetic enrichment was undertaken using anti‐EpCAM ferrofluid.
Enriched cells were then fluorescently stained with the nucleic acid dye DAPI and labeled with mAbs specific for epithelial cells (anti‐CK8, 18, and 19) and leukocytes (anti‐CD45). A CTC was defined as an EpCAM, isolated, intact, round to oval cell, with a visible nucleus (DAPI‐positive), positive staining for CK, and negative staining for CD45. Results of CTC enumeration were then expressed as the number of cells per 7.5 mL blood.9
2.3. Gene expression analysis using quantitative RT‐PCR and multiplex PCR
The mRNA expression of epithelial (EGFR, EpCAM, and CEA) and EMT (PI3K‐α, Akt‐2 and, TWIST1) markers in the enriched CTC cartridges after counting the CTC number using the CellSearch system was measured using the HotStarTaq Master Mix (#203443; Qiagen), a thermocycler, and the Agilent 2100 Bioanalyzer on a DNA LabChip (Agilent Technologies).
Primers for epithelial (EGFR, EpCAM, and CEA) and EMT (PI3K‐α, Akt‐2, and TWIST1) markers were provided by AdnaGen. The analysis of tumor cell‐derived mRNA was carried out by quantitative RT‐PCR for the following transcripts: EGFR, EpCAM, CEA, PI3K‐α, Akt‐2, and TWIST1. AdnaTest ColonCancerDetect and AdnaTest EMT‐2/StemCell Detect kits (AdnaGen), containing oligo(dT)25‐coated beads, were used to isolate mRNA from tumor cells in the enriched CTC cartridges of CellSearch system. PrimerMix ColonDetect was first used to amplify 3 genes (EGFR, EpCAM, and CEA) and 1 control gene (Actin). PrimerMix EMT‐2/StemCell Detect was used to amplify 3 EMT‐related genes (PI3K‐α, Akt‐2, and TWIST1) and 1 control gene (Actin). Visualization of PCR fragments was carried out with the Agilent 2100 Bioanalyzer, with a limit of detection of 0.01 ng/μL or more.
2.4. Statistical analysis
The primary outcome measure was PFS, defined as the date of randomization to the date of first documented disease progression or death from any cause. If disease progression or death was not observed, PFS was censored on the day of the last computed tomography scan. Secondary end‐points were response rate and OS. Overall survival was defined as the period from randomization to the date of death or censored on the date of last contact if alive. Patients were dichotomized into responders (including complete or partial response) and nonresponders (including stable or progressive disease) as defined by RECIST 1.1 criteria.
Circulating tumor cell levels were dichotomized into low and high values based on the widely accepted cut‐off value of 3.5 The mRNA levels of CTC EGFR, EpCAM, CEA, PI3K‐α, Akt‐2, and TWIST1 were categorized into positive and negative groups. The cut‐off value of 0.05 for EGFR expression at baseline was chosen based on the maximum χ2 approach. Associations between CTC and gene expression levels, and PFS and OS were analyzed by Kaplan‐Meier curves and log‐rank test in univariable analysis, and the Cox regression model in a multivariable model, adjusting for baseline patient and tumor characteristics.
SAS 9.4 (SAS Institute) was used to perform all analyses. All tests were 2‐sided at a significance level of .05.
3. RESULTS
3.1. Patient and tumor characteristics
Clinicopathologic characteristics are presented in Table 1. The median follow‐up time was 180 days. The median PFS and OS were 69 days and 192 days, respectively. Six patients were deemed not evaluable; 2 patients had rapid disease progression, and 4 patients had adverse events. Associations between baseline characteristics and clinical outcomes were examined using the log‐rank test in univariate analysis. Using the Cox regression model in a multivariable model, the presence of liver metastases, metastases to other organs, and KRAS mutation status were significantly associated with PFS and OS.
Table 1.
Patients with metastatic colorectal cancer and tumor characteristics (N = 50)
| Characteristic | N | % |
|---|---|---|
| Age, y | ||
| Median (range) | 65 | 34‐78 |
| Sex | ||
| M | 22 | 44 |
| F | 28 | 56 |
| Performance status | ||
| 0 | 27 | 54 |
| 1 | 23 | 46 |
| Primary tumor site | ||
| Right | 14 | 28 |
| Left | 36 | 72 |
| Primary tumor resection | ||
| No | 42 | 84 |
| Yes | 8 | 16 |
| KRAS mutation status | ||
| Wild type | 34 | 69 |
| Mutant | 15 | 31 |
| Metastases | ||
| Liver | 32 | 64 |
| Lung | 26 | 52 |
| Lymph nodes | 28 | 56 |
| Peritoneal | 12 | 24 |
| Other organs | 13 | 26 |
| Line of chemotherapy | ||
| 2 | 1 | 2 |
| 3 | 11 | 22 |
| 4 | 35 | 70 |
| 5 | 3 | 6 |
| Response | ||
| Complete response | 0 | 0 |
| Partial response | 1 | 2 |
| Stable disease | 17 | 34 |
| Progressive disease | 26 | 52 |
| NE | 6 | 12 |
| Histological type | ||
| tub1 | 11 | 22 |
| tub2 | 33 | 66 |
| por | 5 | 10 |
| Unknown | 1 | 2 |
aA patient could have multiple sites of metastases.
NE, not evaluable; por, poorly differentiated adenocarcinoma; tub1, well‐differentiated adenocarcinoma; tub2, moderately differentiated adenocarcinoma;
3.2. Clinical outcomes by CTC count and gene expression in patients receiving regorafenib
The distribution of CTC count and CTC gene expression at baseline and day 21 is outlined in Table 2. At baseline, 64% of patients had detectable CTCs, all of whom had measurable CTC EGFR expression. Among patients without detectable CTCs, all had measurable mRNA expression of at least one of the tested genes at baseline.
Table 2.
Circulating tumor cell (CTC) count and gene expression distribution
| Marker | N | No detectable expression | Median | Range | Interquartile range | |
|---|---|---|---|---|---|---|
| Baseline | ||||||
| CTC count | 50 | 18 | 2.00 | 0‐105 | 0‐6 | |
| EGFR | 50 | 17 | 0.01 | 0.00‐0.83 | 0.00‐0.02 | |
| CEA | 50 | 24 | 0.01 | 0.00‐0.51 | 0.00‐0.01 | |
| EpCAM | 50 | 39 | 0.00 | 0.00‐2.11 | 0.00‐0.00 | |
| TWIST1 | 50 | 36 | 0.00 | 0.00‐0.06 | 0.00‐0.01 | |
| Akt2 | 49 | 38 | 0.00 | 0.00‐0.48 | 0.00‐0.00 | |
| PI3Kα | 50 | 42 | 0.00 | 0.00‐0.06 | 0.00‐0.00 | |
| Day 21 | ||||||
| CTC Count | 50 | 20 | 1.00 | 0‐346 | 0‐4 | |
| EGFR | 50 | 25 | 0.01 | 0.00‐1.48 | 0.00‐0.01 | |
| CEA | 50 | 37 | 0.00 | 0.00‐0.02 | 0.00‐0.01 | |
| EpCAM | 50 | 40 | 0.00 | 0.00‐0.02 | 0.00‐0.00 | |
| TWIST1 | 49 | 39 | 0.00 | 0.00‐0.21 | 0.00‐0.00 | |
| Akt2 | 50 | 39 | 0.00 | 0.00‐0.02 | 0.00‐0.00 | |
| PI3Kα | 50 | 43 | 0.00 | 0.00‐0.02 | 0.00‐0.00 | |
Associations between CTC count and outcomes are analyzed (Table 3). The CTC count at baseline and at day 21 was significantly associated with PFS and OS. Specifically, patients with fewer than 3 CTCs at baseline and day 21 had a longer median PFS of 3.3 and 3.3 months, respectively, compared to those with 3 or more CTCs, who had a median PFS of 2.0 and 2.0 months, respectively (hazard ratio [HR], 2.1 and 2.4; 95% confidence interval [CI], 1.1‐4.2 and 1.0‐5.6; P = .008 and P = .004, respectively) (Table 3, Figure 1A,1). Patients with less than 3 CTCs at baseline and day 21 had a longer median OS of 10.0 and 8.7 months, respectively, compared with those with 3 or more CTCs, who had a median OS of 4.6 and 3.8 months, respectively (HR, 3.0 and 2.5; 95% CI, 1.5‐5.8 and 1.3‐4.8; P < .001 and P = .003, respectively) (Table 3, Figure 1C,1). In multivariable analysis, CTC counts remained significantly associated with OS at baseline and day 21 (HR, 2.7 and 2.3, adjusted P = .019 and P = .028) (Table 3). Additionally, patients with less than 3 CTCs at baseline and day 21 had a significantly higher tumor response rate compared to those with 3 or more CTCs (Fisher’s exact test, P = .007 and P = .03, respectively) (Table 3).
Table 3.
Associations between circulating tumor cell (CTC) count and gene expression and outcomes in patients with metastatic colorectal cancer treated with regorafenib
| Marker | N | Tumor response | Progression‐free survivala | Overall survival | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PR + SD | PD | P b | Median (95%CI) | HR (95% CI) | P b | HR (95% CI)c | P*‡ | Median (95% CI) | HR (95% CI) | P b | HR (95% CI)c | P b,c | ||
| Baseline | ||||||||||||||
| CTC count | ||||||||||||||
| <3 | 27 | 14 (61%) | 9 (39%) | .007 | 3.3 (1.8,4.7) | 1 (ref.) | .008 | 1 (ref.) | .96 | 10.0 (6.5,13.9) | 1 (ref.) | <.001 | 1 (ref.) | .019 |
| ≥3 | 23 | 4 (19%) | 17 (81%) | 2.0 (1.7,2.5) | 2.12 (1.06,4.23) | 0.98 (0.41,2.50) | 4.6 (3.4,6.3) | 2.96 (1.51,5.83) | 2.68 (1.19,6.14) | |||||
| EGFR | ||||||||||||||
| Negative | 17 | 7 (50%) | 7 (50%) | .51 | 2.7 (1.8,3.7) | 1 (ref.) | .77 | 1 (ref.) | .44 | 6.3 (3.3,7.9) | 1 (ref.) | .23 | 1 (ref.) | .90 |
| Positive | 33 | 11 (37%) | 19 (63%) | 2.0 (1.8,3.3) | 1.10 (0.56,2.14) | 1.32 (0.67,2.73) | 8.0 (4.1,10.8) | 0.68 (0.36,1.29) | 0.95 (0.48,1.96) | |||||
| EGFR | ||||||||||||||
| <0.05 | 45 | 18 (46%) | 21 (54%) | .069 | 2.3 (1.8,3.3) | 1 (ref.) | .007 | 1 (ref.) | .007 | 7.8 (4.8,10.0) | 1 (ref.) | .25 | 1 (ref.) | .030 |
| ≥0.05 | 5 | 0 (0%) | 5 (100%) | 1.7 (0.6,2.5) | 3.20 (1.16,8.80) | 4.26 (1.41,11.24) | 5.2 (1.2,12.9) | 1.72 (0.67,4.46) | 3.14 (1.05,8.19) | |||||
| CEA | ||||||||||||||
| Negative | 24 | 4 (20%) | 16 (80%) | .015 | 2.0 (1.7,2.7) | 1 (ref.) | .16 | 1 (ref.) | .27 | 5.9 (4.1,8.0) | 1 (ref.) | .23 | 1 (ref.) | .56 |
| Positive | 26 | 14 (58%) | 10 (42%) | 2.7 (1.8,4.5) | 0.66 (0.34,1.28) | 0.68 (0.35,1.34) | 7.9 (4.0,11.8) | 0.69 (0.37,1.29) | 0.82 (0.43,1.58) | |||||
| EpCAM | ||||||||||||||
| Negative | 39 | 13 (37%) | 22 (63%) | .45 | 2.3 (1.8,3.2) | 1 (ref.) | .29 | 1 (ref.) | .66 | 6.7 (4.0,9.6) | 1 (ref.) | .71 | 1 (ref.) | .29 |
| Positive | 11 | 5 (56%) | 4 (44%) | 2.0 (1.7,7.6) | 0.69 (0.32,1.51) | 1.21 (0.49,2.70) | 7.6 (3.6,12.0) | 0.87 (0.41,1.83) | 1.53 (0.67,3.24) | |||||
| TWIST1 | ||||||||||||||
| Negative | 36 | 12 (35%) | 22 (65%) | .27 | 2.0 (1.8,2.7) | 1 (ref.) | .11 | 1 (ref.) | .61 | 6.7 (5.0,10.3) | 1 (ref.) | .60 | 1 (ref.) | .36 |
| Positive | 14 | 6 (60%) | 4 (40%) | 4.2 (1.1,7.6) | 0.56 (0.27,1.19) | 0.81 (0.35,1.73) | 5.8 (3.3,11.8) | 1.19 (0.61,2.34) | 1.40 (0.67,2.80) | |||||
| Akt2 | ||||||||||||||
| Negative | 38 | 15 (43%) | 20 (57%) | 1.00 | 2.5 (1.8,3.3) | 1 (ref.) | .62 | 1 (ref.) | .20 | 6.5 (4.6,9.6) | 1 (ref.) | .58 | 1 (ref.) | .30 |
| Positive | 11 | 3 (38%) | 5 (63%) | 2.2 (1.6,3.7) | 1.20 (0.54,2.68) | 1.76 (0.70,3.99) | 7.6 (3.3,11.8) | 1.24 (0.56,2.75) | 1.57 (0.63,3.51) | |||||
| PI3Kα | ||||||||||||||
| Negative | 42 | 13 (36%) | 23 (64%) | .24 | 2.2 (1.8,2.7) | 1 (ref.) | .51 | 1 (ref.) | .61 | 6.3 (4.6,9.6) | 1 (ref.) | .18 | 1 (ref.) | .15 |
| Positive | 8 | 5 (63%) | 3 (38%) | 3.3 (0.6,4.7) | 0.77 (0.34,1.75) | 0.80 (0.32,1.78) | 8.7 (1.2,24.9) | 0.54 (0.21,1.41) | 0.47 (0.15,1.19) | |||||
| Any EMT marker | ||||||||||||||
| Negative | 27 | 10 (38%) | 16 (62%) | .75 | 2.3 (1.8,3.2) | 1 (ref.) | .55 | 1 (ref.) | .82 | 6.3 (4.1,9.6) | 1 (ref.) | .74 | 1 (ref.) | .93 |
| Any positive | 22 | 8 (47%) | 9 (53%) | 2.3 (1.8,4.2) | 0.83 (0.44,1.57) | 1.08 (0.55,2.10) | 7.8 (3.6,11.8) | 0.90 (0.47,1.71) | 1.03 (0.52,2.00) | |||||
| Day 21 | ||||||||||||||
| CTC count | ||||||||||||||
| <3 | 32 | 15 (54%) | 13 (46%) | .030 | 3.3 (2.0,4.5) | 1 (ref.) | .004 | 1 (ref.) | .060 | 8.7 (6.5,10.8) | 1 (ref.) | .003 | 1 (ref.) | .028 |
| ≥3 | 18 | 3 (19%) | 13 (81%) | 2.0 (1.8,2.3) | 2.42 (1.04,5.63) | 2.39 (0.99,5.94) | 3.8 (2.8,5.1) | 2.49 (1.28,4.83) | 2.27 (1.10,4.66) | |||||
| EGFR | ||||||||||||||
| Negative | 25 | 8 (36%) | 14 (64%) | .76 | 2.5 (1.8,4.5) | 1 (ref.) | .37 | 1 (ref.) | .56 | 7.8 (4.6,12.0) | 1 (ref.) | .37 | 1 (ref.) | .63 |
| Positive | 25 | 10 (45%) | 12 (55%) | 2.3 (1.8,4.1) | 1.32 (0.65,2.65) | 1.25 (0.59,2.64) | 6.7 (4.0,10.0) | 1.32 (0.71,2.48) | 1.17 (0.62,2.25) | |||||
| EGFR | ||||||||||||||
| <0.05 | 45 | 16 (40%) | 24 (60%) | 1.00 | 2.3 (1.8,3.3) | 1 (ref.) | .69 | 1 (ref.) | .84 | 7.6 (5.0,9.6) | 1 (ref.) | .40 | 1 (ref.) | .25 |
| ≥0.05 | 5 | 2 (50%) | 2 (50%) | 2.7 (1.8,4.1) | 1.26 (0.38,4.18) | 1.14 (0.27,3.80) | 4.1 (1.6,12.9) | 1.55 (0.55,4.42) | 1.93 (0.57,5.44) | |||||
| CEA | ||||||||||||||
| Negative | 37 | 12 (38%) | 20 (63%) | .51 | 2.3 (1.8,3.7) | 1 (ref.) | .64 | 1 (ref.) | .91 | 6.7 (5.0,8.0) | 1 (ref.) | .42 | 1 (ref.) | .88 |
| Positive | 13 | 6 (50%) | 6 (50%) | 2.7 (1.8,4.2) | 0.85 (0.40,1.81) | 1.05 (0.45,2.29) | 8.7 (2.8,25.7) | 0.74 (0.35,1.58) | 0.94 (0.41,2.00) | |||||
| EpCAM | ||||||||||||||
| Negative | 40 | 13 (37%) | 22 (63%) | .45 | 2.3 (1.8,3.7) | 1 (ref.) | .98 | 1 (ref.) | .57 | 7.8 (5.0,10.8) | 1 (ref.) | .086 | 1 (ref.) | .25 |
| Positive | 10 | 5 (56%) | 4 (44%) | 3.0 (1.1,4.2) | 1.01 (0.43,2.38) | 0.78 (0.31,1.75) | 5.5 (1.7,8.0) | 1.86 (0.84,4.10) | 1.60 (0.70,3.41) | |||||
| TWIST1 | ||||||||||||||
| Negative | 39 | 13 (38%) | 21 (62%) | .46 | 2.0 (1.8,3.3) | 1 (ref.) | .50 | 1 (ref.) | .49 | 8.0 (4.6,10.3) | 1 (ref.) | .29 | 1 (ref.) | .11 |
| Positive | 10 | 5 (56%) | 4 (44%) | 3.7 (1.8,4.8) | 0.77 (0.35,1.72) | 0.73 (0.28,1.69) | 5.5 (2.8,7.9) | 1.46 (0.71,3.04) | 1.99 (0.83,4.51) | |||||
| Akt2 | ||||||||||||||
| Negative | 39 | 14 (40%) | 21 (60%) | 1.00 | 2.3 (1.8,3.2) | 1 (ref.) | .51 | 1 (ref.) | .39 | 6.7 (4.6,9.6) | 1 (ref.) | .93 | 1 (ref.) | .81 |
| Positive | 11 | 4 (44%) | 5 (56%) | 3.3 (1.7,6.6) | 0.77 (0.33,1.76) | 0.68 (0.27,1.55) | 8.7 (3.5,13.9) | 1.03 (0.51,2.09) | 0.91 (0.41,1.91) | |||||
| PI3Kα | ||||||||||||||
| Negative | 43 | 15 (41%) | 22 (59%) | 1.00 | 2.3 (2.0,3.3) | 1 (ref.) | .71 | 1 (ref.) | .67 | 7.8 (4.8,10.8) | 1 (ref.) | .078 | 1 (ref.) | .039 |
| Positive | 7 | 3 (43%) | 4 (57%) | 3.0 (1.7,4.7) | 1.17 (0.48,2.88) | 1.23 (0.44,3.01) | 5.9 (3.5,7.9) | 2.02 (0.85,4.78) | 2.61 (1.01,6.15) | |||||
| Change at day 21 | ||||||||||||||
| CTC count | ||||||||||||||
| Stable/decreased | 40 | 15 (44%) | 19 (56%) | .49 | 2.7 (1.8,4.1) | 1 (ref.) | .31 | 1 (ref.) | .25 | 6.7 (5.0,9.6) | 1 (ref.) | 1.00 | 1 (ref.) | .85 |
| Increased | 10 | 3 (30%) | 7 (70%) | 2.1 (1.8,2.7) | 1.47 (0.65,3.29) | 1.68 (0.67,3.92) | 8.3 (1.2,15.3) | 1.00 (0.45,2.20) | 1.08 (0.46,2.33) | |||||
| EGFR | ||||||||||||||
| Stable/decreased | 36 | 11 (33%) | 22 (67%) | .09 | 2.2 (1.8,3.3) | 1 (ref.) | .53 | 1 (ref.) | .42 | 6.5 (4.8,9.6) | 1 (ref.) | .74 | 1 (ref.) | .76 |
| Increased | 14 | 7 (64%) | 4 (36%) | 3.2 (1.8,4.5) | 0.76 (0.31,1.86) | 0.64 (0.20,1.74) | 6.7 (2.0,11.8) | 1.12 (0.57,2.21) | 0.89 (0.42,1.80) | |||||
| CEA | ||||||||||||||
| Stable/decreased | 47 | 17 (41%) | 24 (59%) | 1.00 | 2.5 (2.0,3.3) | 1 (ref.) | .62 | 1 (ref.) | .11 | 6.7 (4.8,8.7) | 1 (ref.) | .46 | 1 (ref.) | .61 |
| Increased | 3 | 1 (33%) | 2 (67%) | 1.8 (1.8,4.5) | 1.33 (0.40,4.42) | 3.17 (0.72,12.35) | 12.9 (3.5,12.9) | 0.59 (0.14,2.46) | 1.46 (0.28,5.34) | |||||
| EpCAM | ||||||||||||||
| Stable/decreased | 41 | 14 (39%) | 22 (61%) | .70 | 2.3 (1.8,3.7) | 1 (ref.) | .98 | 1 (ref.) | .68 | 7.8 (5.0,10.8) | 1 (ref.) | .018 | 1 (ref.) | .085 |
| Increased | 9 | 4 (50%) | 4 (50%) | 2.7 (1.1,4.2) | 1.01 (0.41,2.48) | 0.83 (0.31,1.91) | 4.8 (1.7,8.0) | 2.32 (1.05,5.16) | 2.02 (0.88,4.32) | |||||
| TWIST1 | ||||||||||||||
| Stable/decreased | 40 | 13 (38%) | 21 (62%) | .46 | 2.0 (1.8,3.3) | 1 (ref.) | .65 | 1 (ref.) | .53 | 7.6 (4.6,10.3) | 1 (ref.) | .44 | 1 (ref.) | .23 |
| Increased | 9 | 5 (56%) | 4 (44%) | 3.2 (1.8,4.8) | 0.84 (0.38,1.87) | 0.75 (0.29,1.77) | 6.3 (2.8,13.9) | 1.34 (0.63,2.84) | 1.72 (0.69,4.03) | |||||
| Akt2 | ||||||||||||||
| Stable/decreased | 40 | 14 (40%) | 21 (60%) | .70 | 2.3 (1.8,3.3) | 1 (ref.) | .57 | 1 (ref.) | .48 | 6.3 (4.6,7.9) | 1 (ref.) | .68 | 1 (ref.) | .51 |
| Increased | 9 | 4 (50%) | 4 (50%) | 4.2 (1.7,6.6) | 0.78 (0.32,1.90) | 0.72 (0.27,1.68) | 10.0 (3.1,15.3) | 0.86 (0.40,1.84) | 0.76 (0.32,1.65) | |||||
| PI3Kα | ||||||||||||||
| Stable/decreased | 44 | 16 (42%) | 22 (58%) | 1.00 | 2.5 (2.0,3.3) | 1 (ref.) | .38 | 1 (ref.) | .78 | 7.6 (4.8,10.3) | 1 (ref.) | .075 | 1 (ref.) | .092 |
| Increased | 6 | 2 (33%) | 4 (67%) | 1.8 (1.7,4.5) | 1.50 (0.57,3.97) | 1.16 (0.38,2.98) | 4.7 (3.5,8.7) | 2.13 (0.86,5.29) | 2.26 (0.83,5.44) | |||||
Epithelial‐mesenchymal transition (EMT) combines TWIST1, Akt2, and PI3Kα at baseline.
CI, confidence interval; HR, hazard ratio; ref., reference.
Seven patients with progression‐free survival (PFS) shorter than 21 d were excluded from analyses of associations between markers measured at day 21 and PFS.
P values based on Fisher’s exact test for tumor response, log‐rank test for PFS and overall survival (OS) in the univariable analysis, and the Wald test in the multivariable Cox regression model. P values were not adjusted for multiple testing.
Based on the multivariable Cox regression model adjusting for liver metastases, metastases of other organs, and KRAS mutation status for PFS and OS.
Figure 1.
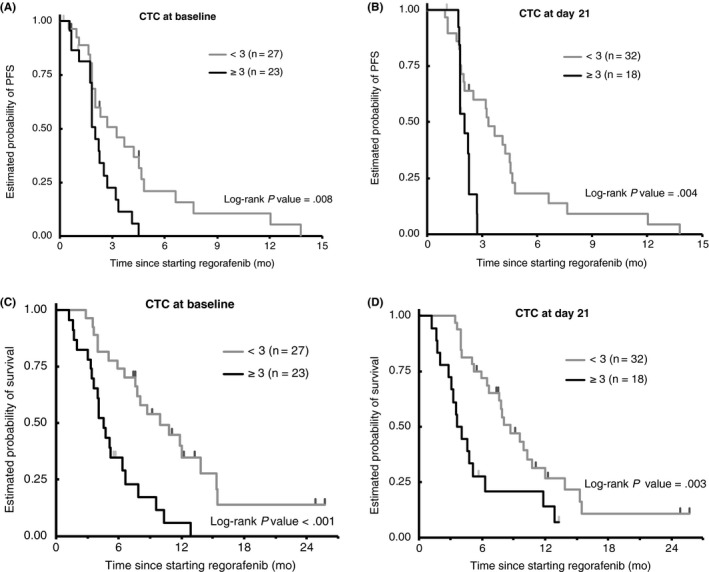
Comparison of clinical outcomes by circulating tumor cell (CTC) count at baseline and day 21 of treatment with regorafenib in patients with metastatic colorectal cancer. A,B, Progression‐free survival probability at (A) baseline and (B) day 21. C,D, Overall survival probability at (C) baseline and (D) day 21
We then examined associations between CTC gene expression levels and outcomes. We used the optimal cut‐off value method as previously described.10, 11, 12 The cut‐off value of 0.05 for EGFR expression at baseline was chosen based on the maximum χ2 approach. Patients with baseline CTC EGFR levels 0.05 or higher had a shorter PFS, compared to those with lower levels, both in univariable (HR 3.20, 95% CI, 1.16‐8.80, P = .007) and multivariable (HR 4.26, 95% CI, 1.41‐11.24, P = .007) analyses. All 5 patients with increased baseline CTC EGFR expression had disease progression as their best response. No significant associations were seen with the other markers that were analyzed (Table 3).
3.3. Changes in CTC count and CTC gene expression at day 21
Next, we assessed CTC count and CTC gene expression levels at day 21 and at the time of disease progression as compared to baseline. Circulating tumor cell EGFR expression was significantly increased at day 21 and/or PD (P = .004) in 64% of patients (Table 4). More specifically, CTC EGFR expression was significantly increased at PD compared to baseline (P = .041) in 48% of patients. No significant associations were seen with the other genes that were analyzed.
Table 4.
Circulating tumor cell EGFR upregulation compared to baseline value in patients with metastatic colorectal cancer treated with regorafenib (N = 50)
| Day 21 | PD | Day 21 and/or PD | |
|---|---|---|---|
| Increased ≥0.01 ng/µL, n (%) | 14 (28) | 24 (48) | 32 (64) |
| Stable/decreased, n (%) | 36 (72) | 17 (34) | 18 (36) |
| P a | .600 | .041 | .004 |
Abbreviation: PD, progressive disease.
Based on the sign test for paired data.
3.4. Associations between changes in CTC EGFR and EMT marker expression
We then hypothesized that increased CTC EGFR expression could result in an increase in the expression of CTC EMT markers.13 However, there were no significant associations between changes in CTC EGFR and EMT marker gene expression.
4. DISCUSSION
Our study is the first to show that CTC enumeration and mRNA gene expression levels could be useful markers of regorafenib efficacy in mCRC patients. Specifically, we found the presence of 3 or more CTCs at baseline and day 21 to be an independent prognostic factor for inferior survival. Moreover, upregulation of CTC EGFR expression induced by regorafenib was associated with disease progression.
Regorafenib blocks both WT and mutant BRAF (V600E) signaling. Preclinical studies have previously indicated that BRAF inhibition causes feedback activation of EGFR signaling and increased EGFR expression, allowing for ongoing tumor cell proliferation.14 In previous studies, tumor tissue EGFR expression has been shown to be a negative prognostic marker.15 Our data further support these findings, suggesting that regorafenib therapy could increase EGFR expression and signaling to an extent that leads to molecular escape and treatment resistance. Patients with baseline CTC EGFR levels 0.05 or higher had a shorter PFS and had disease progression at the time of their first radiographic evaluation. Although the findings of this study are hypothesis generating, if validated in future studies, CTC EGFR expression could serve as a useful marker to identify patients resistant to regorafenib prior to start of therapy.
Importantly, we investigated CTC gene expression levels in patients with and without detectable CTCs, as defined by the CellSearch assay. A subgroup of patients without detectable CTCs according to CellSearch criteria had expression of epithelial marker genes, suggesting the presence of circulating tumor load, CTCs or cell fragments like exosomes, which were not detected or recognized by the CellSearch system. Similarly, in a study by Mostert et al,16 CRC patients without detectable CTCs were found to have a circulating tumor mRNA profile, distinct from both patients with 3 or more detectable CTCs and healthy donors, further suggesting that the blood of patients with no CTCs by CellSearch can contain CTCs or fragments. In patients without detectable CTCs by CellSearch criteria, exosomes could still be present, because CTC isolation is preceded by a Ficoll‐based density gradient enrichment.16 Taken together, our findings highlight a limitation of relying on CTC count alone as a biomarker, and the importance of circulating mRNA expression as a potential prognostic marker in mCRC patients treated with regorafenib.
Furthermore, a considerable proportion of patients in our study did not have detectable CTCs at baseline or subsequent follow‐up. Previous reports suggest that CRC cells can lack CK8, 18, or 19 expression, the markers by which a CTC is defined in CellSearch, reflecting the EMT‐like phenotype of these cells.17, 18 A lack of CK8, 18, or 19 expression would not affect the isolation of CTCs by immunomagnetic enrichment, which is based on their EpCAM expression. However, the lack of cytokeratin expression could result in these cells not being identified as epithelial cells and thus not counted as CTCs in the subsequent enumeration step. Therefore, in addition to the presence of cell fragments (eg exosomes), another possible explanation for the lack of detectable CTCs according to CellSearch criteria could be the insufficient expression of epithelial markers (ie CK8, 18, and 19).
It has been reported that CTCs often exist in a state of EMT or could coexpress both epithelial and mesenchymal markers. During the EMT, epithelial cells downregulate epithelial‐related genes, acquire mesenchymal gene expression, and undergo major changes in their cytoskeleton that result in loss of cell‐cell contact and cell polarity, leading to increased motility and invasiveness.19 Here, we investigated the expression of CTC EMT markers, including TWIST1, Akt2, and PI3Kα, as well as associations between CTC EGFR and EMT gene expression, and found a nonsignificant trend towards an inverse association. The lack of a significant association could reflect challenges associated with CTC characterization using the CellSearch technique, including the presence of an excess of leukocytes, which express EMT marker genes, in CTC‐enriched blood fractions.
Certain limitations of our study should be acknowledged. Our data are limited by the small sample size and are only exploratory. The lack of a control group of patients not receiving regorafenib preclude any conclusions that can be drawn regarding the use of CTC count or associated EGFR expression as predictive markers.
In summary, we identified CTC count as a prognostic marker in mCRC patients treated with regorafenib. Circulating tumor cell EGFR expression was significantly increased with regorafenib at the time of disease progression, suggesting this to be a mode of resistance and lending further mechanistic evidence for the synergistic effects seen with regorafenib and anti‐EGFR mAbs.20 Our findings warrant further investigation and, if replicated in future investigations, could inform the selection of mCRC patients best suited for regorafenib in the chemorefractory setting.
DISCLOSURE
HJ Lenz has received commercial research grants from Bayer Yakuhin, Roshe, Bristol‐Myers Squibb, and Merck Serono. All remaining authors have declared no conflicts of interest.
ACKNOWLEDGEMENTS
The project described was supported in part by award number P30CA014089 from the NCI. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH. This project was also supported in part by the Wunderglo Project and Call to Cure Research Fund. S. Matsusaka is a recipient of the Takashi Tsuruo Memorial Fund. MD Berger received a grant from the Swiss Cancer League (BILKLS‐3334‐02‐2014).
Matsusaka S, Hanna DL, Ning Y, et al. Epidermal growth factor receptor mRNA expression: A potential molecular escape mechanism from regorafenib. Cancer Sci. 2020;111:441–450. 10.1111/cas.14273
Funding information
NCI (grant/award no. P30CA014089).
REFERENCES
- 1. Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metasttic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo‐controlled, phase 3 trial. Lancet. 2013;381:303‐312. [DOI] [PubMed] [Google Scholar]
- 2. Li J, Qin S, Xu R, et al. Regorafenib plus best supportive care versus placebo plus best supportive care in Asian patients with previously treated metastatic colorectal cancer (CONCUR): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet Oncol. 2015;16:619‐629. [DOI] [PubMed] [Google Scholar]
- 3. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781‐791. [DOI] [PubMed] [Google Scholar]
- 4. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res. 2008;14:6302‐6309. [DOI] [PubMed] [Google Scholar]
- 5. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression‐free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213‐3221. [DOI] [PubMed] [Google Scholar]
- 6. Tol J, Koopman M, Miller MC, et al. Circulating tumor cells early predict progression‐free and overall survival in advanced colorectal patients treated with chemotherapy and target agents. Ann Oncol. 2010;21:1006‐1012. [DOI] [PubMed] [Google Scholar]
- 7. Matsusaka S, Suenaga M, Mishima Y, et al. Circulating tumor cells as a surrogate marker for determining response to chemotherapy in Japanese patients with metastatic colorectal cancer. Cancer Sci. 2011;102:1188‐1192. [DOI] [PubMed] [Google Scholar]
- 8. Ning Y, Zhang W, Hanna DL, et al. Clinical relevance of EMT and stem-like gene expression in circulating tumor cells of metastatic colorectal cancer patients. Pharmacogenomics J. 2018;18(1):29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897‐6904. [DOI] [PubMed] [Google Scholar]
- 10. Vallbohmer D, Iqbal S, Yang DY, et al. Molecular determinants of irinotecan efficacy. Int J Cancer. 2006;119:2435‐2442. [DOI] [PubMed] [Google Scholar]
- 11. Vallbohmer D, Zhang W, Gordon M, et al. Molecular determinants of cetuximab efficacy. J Clin Oncol. 2005;23(15):3536‐3544. [DOI] [PubMed] [Google Scholar]
- 12. Wilson PM, Yang D, Azuma M, et al. Intratumoral expression profiling of genes involved in angiogenesis in colorectal cancer patients treated with chemotherapy plus the VRGFR inhibitor PTK787/ZK 222584 (vatalanib). Pharmacogenomics J. 2013;13:410‐416. [DOI] [PubMed] [Google Scholar]
- 13. Serrano MJ, Ortega FG, Alvarez‐Cubero MJ, et al. EMT and EGFR in CTCs cytokeratin negative non‐metastatic breast cancer. Oncotarget. 2014;5:7486‐7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100‐103. [DOI] [PubMed] [Google Scholar]
- 15. Ma CJ, Huang CW, Chang TK, et al. Oncologic outcome in metastatic colorectal cancer with regorafenib with FOLFIRI as a third‐ or fourth‐line setting. Transl Oncol. 2019;12(3):502‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mostert B, Sieuwerts AM, Bolt‐de Vries J, et al. mRNA expression profiles in circulating tumor cells of metastatic colorectal cancer patients. Mol Oncol. 2015;9(4):920‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joosse SA, Hannemann J, Spotter J, et al. Changes in keratin expression during metastatic progression of breast cancer: impact on the detection of circulating tumor cells. Clin Cancer Res. 2012;18:993‐1003. [DOI] [PubMed] [Google Scholar]
- 18. Moll R, Lowe A, Laufer J, Franke WW. Cytokeratin 20 in human carcinomas. A new histodiagnostic marker detected by monoclonal antibodies. Am J Pathol. 1992;40:427‐447. [PMC free article] [PubMed] [Google Scholar]
- 19. Moustakas A, Heldin CH. Signaling networks guiding epithelial‐mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98(10):1512‐1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Napolitano S, Martini G, Rinaldi B, et al. Primary and acquired resistance of colorectal cancer to anti‐EGFR monoclonal antibody can be overcome by combined treatment of Regorafenib with Cetuximab. Clin Cancer Res. 2015;21(13):2975‐2983. [DOI] [PubMed] [Google Scholar]


