Abstract
Artificial intelligence (AI) is now a trendy subject in clinical medicine and especially in gastrointestinal (GI) endoscopy. AI has the potential to improve the quality of GI endoscopy at all levels. It will compensate for humans’ errors and limited capabilities by bringing more accuracy, consistency, and higher speed, making endoscopic procedures more efficient and of higher quality. AI showed great results in diagnostic and therapeutic endoscopy in all parts of the GI tract. More studies are still needed before the introduction of this new technology in our daily practice and clinical guidelines. Furthermore, ethical clearance and new legislations might be needed. In conclusion, the introduction of AI will be a big breakthrough in the field of GI endoscopy in the upcoming years. It has the potential to bring major improvements to GI endoscopy at all levels.
Keywords: Artificial intelligence, Computer-assisted diagnosis, Deep learning, Gastrointestinal endoscopy
Introduction
With the incessant advances in information technology and its implications in all domains of our life, artificial intelligence (AI) algorithms started to emerge as a need for a better machine performance. Unlike machines, human brain's performance could be altered by fatigue, stress, or limited experience. AI technology would compensate for human's limited capability, prevent human errors, give machines some reliable autonomy, increase work productivity and efficiency. Therefore, when looking for a fast and reliable assistant to treat the continuously growing number of patients, AI can be the best option that we are looking for. The application of AI technology in gastrointestinal (GI) endoscopy could carry so many advantages. It can reduce inter-operator variability, enhance the accuracy of diagnosis, and help in taking on the spot rapid though accurate therapeutic decisions. Furthermore, AI would reduce the time, cost, and burden of endoscopic procedures.
AI-assisted endoscopy is based on computer algorithms that perform like human brains do. They react (output) to what they receive as information (input) and what they have learned when built. The fundamental principle of this technology is “machine learning” (ML) which is a general term for teaching computer algorithms to recognize patterns in the data. It provides them the ability to automatically learn and improve from experience without being explicitly programmed. The result is AI comparable or even superior to the performance of human brains. One of the fastest-growing machine-learning methods is deep learning (DL). This approach inspired by the biological neural network of the human brain uses a layered structure of algorithms called multi-layered artificial neural networks. In addition, just like our brains do, DL models can analyze data with logic, identify patterns, draw conclusions, and make decisions. This makes DL AI far more capable than that of standard ML.[1]
So basically, AI technology is based on a computer algorithm that is trained for a specific function such as to recognize or characterize defined lesions, colon polyps for example. This computer algorithm is trained using the previously described ML through exposure on numerous training elements such as a large number of predefined polyp-containing video frames for the previous example. These computer algorithms will extract and analyze specific features like micro-surface topological pattern, color differences, micro-vascular pattern, pit pattern, appearance under filtered light such as narrow band imaging (NBI), high-magnification, endocystoscopy appearance, and many other features from these video-frames allowing automated detection or diagnosis-prediction of lesions of interest. The result algorithm is validated thereafter using another test database and/or by prospective in vivo clinical trials.
Different types of AI computer systems exist to fulfill so many functions. The main two AI systems categories are computer-assisted detection (CADe) for lesion detection and computer-assisted diagnosis (CADx) for optical biopsy and lesion characterization. Other AI systems offer therapeutic assistance such as lesion delineation for complete endoscopic resection. Furthermore, other AI systems also exist to offer technical assistance for better GI endoscopy performance (such as scope insertion guidance), disease prediction based on patient data and more. The main AI systems under development are summarized in Table 1.
Table 1.
AI systems and related functions.
The main objective of this article is to introduce this new technology to every GI endoscopist to become familiar with as it will bring major changes to the way we perform endoscopy in the very near future. This will be as a general overview about AI in the domain of GI endoscopy: how does it work, what are the potential benefits and how it can help in improving both diagnostic and therapeutic endoscopy in each and every part of the GI tract, the esophagus, stomach, small bowel (SB), and colon. Some AI algorithms will be presented in a simplified way as examples to help readers understanding how it works and how it can be applied to endoscopy in real life.
We searched the internet using Google search engine, Google Scholar, and PubMed, for related abstracts and published articles using keywords: computer-assisted endoscopy, artificial intelligence and endoscopy, computer-aided diagnosis and endoscopy, computer-aided detection, CAD, automated systems in endoscopy, deep and convolutional neural networks in endoscopy. Structured and systematic global overview is made using most relevant and latest works on this topic. No exclusions were made on the study design. All articles included were in English language only.
Esophagus
The main advantage that AI can carry for esophageal endoscopy is to improve esophageal cancer screening: identification of dysplasia/cancer on Barrett esophagus (BE) and identification of squamous cell carcinoma.
Over the past four decades, the incidence of esophageal adenocarcinoma (EAC) has risen rapidly due to the increasingly prevalent excess body weight.[18] EAC is typically diagnosed at an advanced stage when it has a very poor prognosis.[19] Therefore, early detection of BE neoplastic changes is of great interest especially in the presence of highly curative endoscopic treatment such as endoscopic mucosal resection and radiofrequency ablation.[20,21] Currently, screening for EAC is based on direct endoscopic visualization coupled with guided or random-biopsies. Screening random-biopsies could be taken according to the well-known Seattle protocol which is currently considered as being relatively inefficient, time-consuming, and providing a low diagnostic rate, even if it remains the standard.[22]
To improve EAC screening, in terms of sensitivity and rapidity of the exam, AI-assisted endoscopy might have a great place here and that is in addition to the relief that it could offer to endoscopists who are facing the challenge and anxiety not to miss early cancer as well as.
Examples of CADx for EAC
Swager et al[23] conducted the first study in which a clinically inspired computer algorithm for BE neoplasia was developed based on volumetric laser endomicroscopy (VLE) images with direct histological correlates. VLE is an advanced imaging technique that provides a near-microscopic resolution scan of the esophageal wall layers up to 3-mm deep. The study objective was to investigate the feasibility of a computer algorithm to identify early BE neoplasia on ex vivo VLE images. It compared the performance of this computer algorithm in detecting VLE images containing BE neoplasia vs. VLE experts. In total, 60 different images were analyzed by the computer algorithm and VLE experts. The histological diagnosis, correlated with the images served as the reference standard. The algorithm showed good performance to detect BE neoplasia (area under the curve [AUC] 0.95 with best-performing features) and thus it might have the potential to assist endoscopists to detect early neoplasia on VLE. However, future studies on in vivo VLE scans are needed to further expand and validate this algorithm on in vivo data.
Van der Sommen et al[24] tested another computer algorithm for the detection of early neoplastic lesions on BE based on specific imaging details like carcinoma related mucosal color deviation and texture patterns. This automated computer algorithm, developed and tested using 100 endoscopic images from 44 patients with BE, was able to identify early neoplastic lesions with reasonable accuracy. In this study, AI-assisted endoscopy identified early neoplastic lesions with a sensitivity and specificity of 0.83 on a per-image analysis. At the patient level, the system achieved a sensitivity and specificity of 0.86 and 0.87, respectively. Results were validated by four international experts in the detection and treatment of early Barrett neoplasia who were blinded to the endoscopy findings and pathology.
In addition to the work of Swager et al[23] and van der Sommen et al,[24] several articles have been published on this subject and have demonstrated a great utility for the application of AI in detecting EAC. Horie et al[25] demonstrated the utility of AI using convolutional neural networks (CNNs) to make a diagnosis of esophageal cancer with a sensitivity of 95% and it was able to identify all small cancers of less than 10 mm. Shin et al[26] and Quang et al[27] tested their own algorithms to successfully diagnose esophageal squamous neoplasia with a sensitivity reaching 87% and 95%, respectively and a specificity of 97% and 91%.
In conclusion, AI-assistance shows great promising results in terms of improving the detection and diagnosis of EAC and thus reducing the mortality and morbidity related to this type of malignancy with a poor prognosis when diagnosed at an advanced stage.
Stomach
AI can offer invaluable assistance in the management of early gastric cancer (EGC) at different levels:
- (1)
-
(2)
CADx for cancer staging and estimation of invasion depth.[9]
-
(3)
AI systems for automated lesion delineation.[7]
-
(4)
And AI systems for Helicobacter pylori (H. pylori) infection prediction.[28–31]
Globally, gastric adenocarcinoma, with nearly one million incident cases annually, is the third leading cause of global cancer mortality.[32] Early detection and prompt treatment remain the best measure to improve patient survival. In the United States and Europe, EGC, defined as invasive gastric cancer that invades no more deeply than the submucosa, accounts for 10% to 20% of resected cancers.[33] EGC is sometimes hard to detect since it may not present any specific abnormalities and so it can be easily overlooked during endoscopy.
Currently, screening for EGC is based on direct visualization during gastroscopy with the help of some image-enhancing tools used to highlight mucosal abnormalities such as the traditional dye-based chromoendoscopy, NBI, and magnification.
Meticulous examination of the whole stomach using current techniques can be time-consuming. Furthermore, inter-operator variability could be significant. Since the fact that early cancer detection significantly improves the prognosis, the need for reliable automated detection-systems of EGC-related mucosal abnormalities becomes even more clear.
Several studies implicating AI in EGC endoscopy were published and others are ongoing to develop more efficient and reliable automated systems to help endoscopists not to miss any EGC. When diagnosed, dedicated AI systems could help to decide the best treatment strategy by predicting cancer-stage (depth of invasion) and in case it is resectable endoscopically, AI still can offer therapeutic assistance by suggesting reliable automated lesion-delineation for R0 resection. The result is less missed EGC diagnosis and a better therapeutic outcome.
Examples of CADx for EGC
Miyaki et al[8] developed an automated computer recognition system, called the support vector machine (SVM)-based analysis system, for use with blue laser imaging (BLI) endoscopy in differentiating between EGC, reddened lesions, and surrounding tissue, based on a software program that identifies and quantifies features of endoscopic images along with an analysis algorithm to detect and describe local features in images. The computer system was trained using 587 images of previously confirmed gastric cancer and 503 images of surrounding normal tissue. Validation images were gathered from 100 EGC in 95 patients who were examined by BLI magnification using LASEREO endoscopy system (Fujifilm Medical Co. Ltd., Tokyo, Japan) histologically confirmed after endoscopic sub-mucosal dissection or surgical resection. A set of images of flat or slightly depressed, reddened lesions from 40 gastric lesions that showed no evidence of malignancy on biopsies was also produced. In applying the above-mentioned software for quantitative analysis and identification of the various types of tissue, the average SVM output value was 0.846 ± 0.220 for cancerous lesions, 0.381 ± 0.349 for reddened lesions, and 0.219 ± 0.277 for surrounding tissue, with the SVM output value for cancerous lesions being significantly greater than that for reddened lesions or surrounding tissue. The average SVM output value for differentiated-type cancer was 0.840 ± 0.207 and for undifferentiated-type cancer was 0.865 ± 0.259. The authors concluded that their AI system used with BLI will identify gastric cancers quantitatively. Further development is needed.
Hirasawa et al[34] developed a computer-system for EGC automated diagnosis using DL. This system was trained using 13,584 endoscopic images of gastric cancer and tested on 2296 images collected from 69 patients with 77 gastric cancer lesions. The system took 47 s to analyze all the 2296 test images. This CADx correctly diagnosed 71 of 77 gastric cancer lesions with an overall sensitivity of 92.2%, and 161 non-cancerous lesions were detected as gastric cancer, resulting in a positive predictive value of 30.6%. Seventy of the 71 lesions (98.6%) with a diameter of 6 mm or more as well as all invasive cancers were correctly detected. All missed lesions were superficially depressed and differentiated-type intra-mucosal cancers that were difficult to distinguish from gastritis even for experienced endoscopists. Nearly half of the false-positive lesions were gastritis with changes in color tone or an irregular mucosal surface. The authors concluded that their system could process numerous stored endoscopic images in a very short time with a clinically relevant diagnostic ability.
Another CADx was developed by Kubota et al[35] for automated diagnosis of depth of wall-invasion of gastric cancer. The primary database included 902 endoscopic images of 344 patients who underwent gastrectomy or endoscopic resection for gastric cancer. The images were divided into ten groups with almost equal number of patients and T staging. Computer learning was performed using about 800 images from all but one group which was used for accuracy testing (90 images). The overall diagnostic accuracy rate of the CADx was 64.7%. The authors concluded that computer-aided diagnosis is useful for diagnosing depth of wall invasion of gastric cancer on endoscopic images.
Several other studies were published on the benefit of AI in the detection, diagnosis, and treatment of EGC. These studies showed that automated systems can offer reliable assistance to endoscopists in managing EGC. In 2018, Kanesaka et al[7] published their work about a CADx system for EGC. This automated system was built using magnifying NBI images. It showed accuracy of 96.3%, sensitivity of 96.7%, specificity of 95%, and positive predictive value (PPV) of 98.3%. Zhu et al[36] tested an automated AI system for the diagnosis of depth of invasion of EGC: confined to the mucosa, upper-third of the mucosa (SM1), or deeper than SM1 using white light (WL) endoscopy. The accuracy of this system was 89.2% along with a sensitivity of 74.5% and specificity of 95.6%. One should note that almost all of these studies were retrospective. Therefore, prospective head to head trials between the available standard and the different AI algorithms still needed before the application of these automated systems in our daily practice for the management of EGC for which we are in need for reliable assistance in terms of detection/diagnosis and treatment, for patients’ better outcome.
On the other hand, another use for AI in upper GI endoscopy is for the diagnosis of H. Pylori. Starting in 2004 by Huang et al,[28] several teams have tested their automated diagnostic systems in prospective clinical studies. These systems showed sensitivity and specificity data between 85% and 90%.[29,31] With more improvement in the accuracy of these systems, we can avoid a huge number of unnecessary biopsies just for the diagnosis of H. Pylori gastritis. This can reduce the cost and burden of endoscopic procedures for this disease along with a real-time fast diagnosis given to the patient just at the end of the procedure.
AI in wireless video capsule endoscopy (VCE)
It has been recommended by several guidelines that VCE is the first-line diagnostic exam for SB exploration and especially before any complementary-diagnostic or therapeutic enteroscopy.[37,38] VCE is an excellent well-tolerated exam. That is why ongoing researches are in place now to develop pan-enteric VCE to explore the whole GI tract. The major drawback of this technology is that a large amount of data collected by the VCE must be analysed, which can be time-consuming and burdensome. Here again, AI can be a savior and a reliable assistant for clinicians. Several conditions were attempted for the automated diagnosis including bleeding angioectasia, celiac disease, or intestinal hookworms.[39–41]
AI in VCE has already been shown to be effective in the detection of SB bleeding. The first several developed CADe systems for bleeding detection used color-based feature extraction, using ratios of the intensity values of the images in the red, green, and blue or hue, saturation, and intensity domain, to help distinguish bleeding-containing frames from those without bleeding.[42,43] Other CADe systems are based on textural content of bleeding and non-bleeding images to perform the classification.[44] Combination use of both color and texture descriptors demonstrated more accurate bleeding detection.[45]
Two examples of CAD in VCE
Hassan et al[5] aimed to draw inferences (bleeding or non-bleeding) using CAD system based on the spatial domain of an image by extracting features in the frequency domain using complex DL to achieve sensitivities and specificities as high as 99% for detection of GI bleeding.
Xiao et al[6] developed another CAD system to reach >99% F1 score or performance score for GI bleeding detection in wireless capsule endoscopy (WCE). F1 score is calculated using precision and recall scores. They used DL to build this CAD using a dataset consisted of 10,000 WCE images, including 2850 GI bleeding frames and 7150 normal frames.
AI systems in VCE were among the first to be used in GI endoscopy and this was mainly for bleeding detection. Although these systems can be helpful in pointing to possible bleeding areas which could help in reducing the reading time, these systems still need improvement to reduce non-specific results by pointing to bleeding-like red artifacts. AI systems are not yet a standard of care and we still need to read and look at all the VCE video frames to give the final result.
Colon
Here, AI can offer the greatest help on multiple levels. So many computer assistant systems are under development and many of them have already been tested and shown attractive results. We can cite some of these AI systems for which studies have already been published:
- (1)
-
(2)
CADx for polyp characterization and classification (also called optical biopsy or histology prediction).[11,12]
-
(3)
CADx for mucosal inflammatory activity prediction in inflammatory bowel disease (IBD) patients.[13]
-
(4)
AI systems helping to make decisions as to whether additional surgery is needed after endoscopic resection of T1 colorectal cancers (CRCs) or not, by predicting lymph node metastasis (LNM).[16]
-
(5)
And last but not least, AI systems for colonoscope guidance and loop prevention during insertion, especially helpful for difficult colonoscopy or trainees.[2]
CRC is the third most diagnosed cancer and it has the second cancer-related death rate in 2018.[46] Therefore, CRC remains a critical health concern and a significant financial burden worldwide. Colonoscopy is considered to be an effective method for CRC screening and prevention. Benefit is based on the detection and R0 resection of any neoplastic polyps. For instance, a 1% increase in the adenoma detection rate was associated with a 3% decrease in interval CRC incidence.[47] Unfortunately, adenoma miss rate remains high (6%–27%), despite novel technologies and devices.[48] Moreover, a large prospective trial of optical biopsy of small colon polyps using NBI showed that the accuracy of trained physicians was only 80% in diagnosing detected polyps as adenomas.[49]
Therefore, as a solution to address human imperfection, automated colon polyp detection has been one of the primary areas of interest for the application of AI in GI endoscopy. The most promising of these efforts have been in computer-aided detection and diagnosis of colorectal polyps, with recent systems demonstrating high sensitivity and accuracy even when used by novel endoscopists.
Screening colonoscopy with complete adenoma resection can reduce the incidence of CRC by as much as 80% and the associated mortality by 50%.[50] However, endoscopic resection of hyperplastic polyps exacerbates medical costs, including those for resection and unnecessary pathologic evaluation, because malignant transformations are rare. Also, resection of every hyperplastic polyp could be time-consuming for little or no benefits at all. Therefore, AI-assisted polyps’ characterization (ie, histology prediction) could offer reassuring support to endoscopists not to remove hyperplastic polyps without the fear of leaving probable neoplastic ones. This technology will lead to reduction of the costs related to unnecessary polypectomy and for sure it will shorten the time of the procedure.
Examples of AI support systems for colonoscopy
For colon polyps’ detection: Misawa et al[4] developed an algorithm designed on a large number of routine colonoscopy videos. Training and test video frames samples were extracted from retrospectively collected colonoscopy videos performed between April and October 2015. Two expert endoscopists retrospectively annotated the presence of polyps in each frame of each video, and this annotation was treated as the gold standard for the presence of polyps. The tested CADe system presented the probability of the presence of polyps as a percentage of 0%–100% in each frame. This probability value simulated the confidence level of human endoscopists on a given image frame. In this study, the CADe detected 94% of the test polyps (47 of 50), and the false-positive detection was 60% (51 of 85). The study showed that AI has the potential to provide automated detection of colorectal polyps. Such CADe systems are expected to fill the gap between endoscopists with different levels of experience. An example of a real-time output of this CADe is shown in Figure 1.
Figure 1.
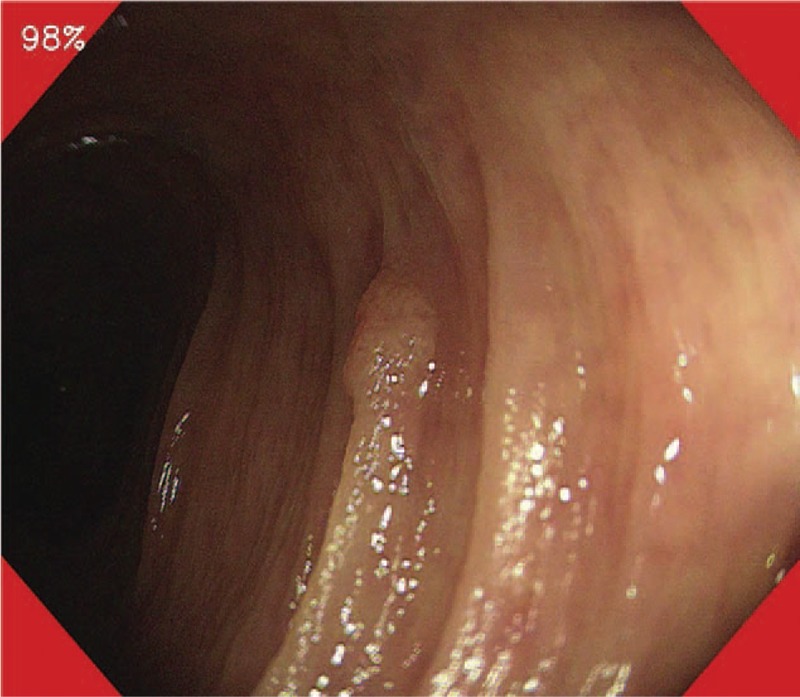
The system presented the probability of the presence of polyps as a percentage in the upper left corner of the endoscopic image. When the probability exceeded the cut-off set at 75%, the CAD system warned of the possibility of the presence of polyps by changing the color in the four corners of the endoscopic image to red. This figure is originally published by Misawa et al.[4]
Other than the previously detailed example of CADe of Misawa et al,[4] many other CADe were developed and tested for their efficacy and accuracy for colon polyp detection. Fernandez-Esparrach et al[51] developed a CADe using WL endoscopy. They tested it in a retrospective study published in 2018. This CADe showed a sensitivity of 70.4% and specificity of 72.4%. In the same year, Urban et al[52] published the result of their work on a CADe. This CADe developed using WL and NBI showed an accuracy of 96.4% and an area under the receiver operating characteristic curve of 0.991. Another CADe developed by Wang et al[53] using WL endoscopy, and tested in a retrospective study on multiple image and video datasets, showed a sensitivity between 88.2% and 100% and specificities around 95% for polyps’ detection. Finally and most recently Klare et al,[54] published in 2019 their results for another CADe developed using WL endoscopy and tested in a small prospective study including 55 patients, this CADe showed a polyp detection rate of 50.9% and an adenoma detection rate of 29.1%.
As a summary of the available published data, AI is a very promising tool to increase colon polyps’ detection. In other terms it would decrease the rate of missed polyps which is directly associated with interval CRC development. For the moment the results are not consistent between the studies and it varies according to the type of the AI algorithms and the type of endoscopic material (WL, NBI, or combination) used for CADe learning. For sure more work is needed to decide about the best and the most practical CADe that can be used in screening colonoscopy.
Another use of AI technology during colonoscopy is to offer diagnostic assistance: CADx. The ability to diagnose small polyps in-situ via “optical-biopsy” may reduce unnecessary polypectomies of non-neoplastic polyps. In other terms, this may help to apply the application of “diagnose and disregard” strategy for hyperplastic polyps.
Misawa et al[12] developed a CADx system based on endocytoscopy (EC). This system automatically provides highly accurate diagnosis in real-time as endoscopists concurrently take EC images. Vital stain such as methylene blue is required to visualize glandular structural and cellular atypia. In contrast, the endocytoscopic vascular pattern can effectively evaluate micro-vessel findings using EC with NBI (EC-NBI) without using any dye. EC-NBI has shown a highly accurate diagnostic ability, similar to other modalities allowing endoscopists to use it more easily without the cumbersome dye staining during colonoscopy.[55]
The custom software is called EndoBRAIN (Cybernet Systems Co., Ltd., Tokyo, Japan). To develop this CAD system, an image-database was created using 1079 EC-NBI images (431 non-neoplasms and 648 neoplasms) from 85 lesions colorectal lesions that had been detected during colonoscopy using EC and subsequently resected between December 2014 and April 2015. From this image-database, 979 images (381 non-neoplasms, 598 neoplasms) were used for ML and the remaining randomly selected 100 images were used for ML. The CAD system provided diagnosis of 100% (100/100) of the validation samples with a diagnosis time of 0.3 s per image.
The major limitation of this system was that it cannot diagnose cancers and sessile serrated adenomas/polyps (SSA/Ps), because there were few EC-NBI images of invasive cancers and SSA/Ps for training. If there were more suitable images, better system that provides 4-class diagnosis (non-neoplastic, adenoma, invasive cancer, and SSA/P) could be developed.[12] An example of a corresponding video-output is shown in Figure 2.
Figure 2.
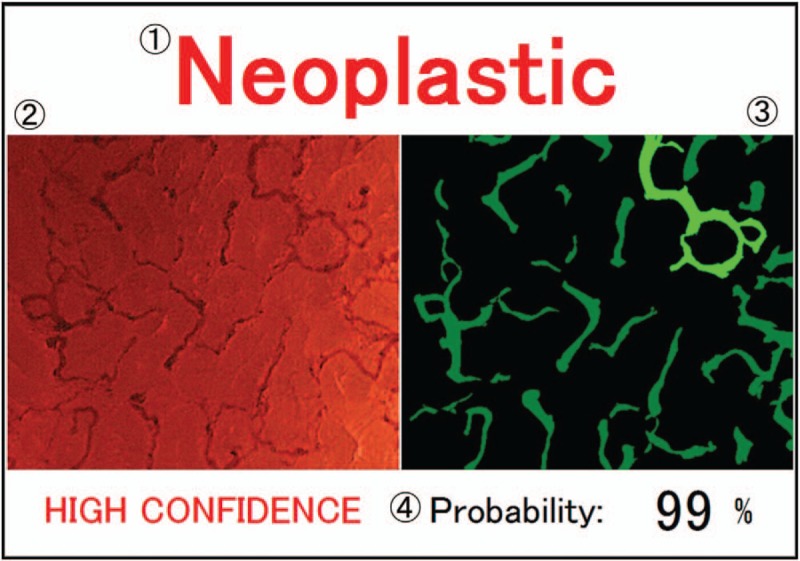
Output image. (1) Computer diagnosis. (2) Input endocytoscopy with narrow band imaging. (3) Extracted vessel image, in which the green area denotes the extracted vessels. The light-green vessel has the maximum diameter. (4) Probability of computer diagnosis is calculated by the support vector machine classifier. This figure is originally published by Misawa et al.[12]
In this category of AI systems, or CADx for real-time optical biopsy, several other studies were published by different teams, showed very promising results for different types of CADx. In 2011, Gross et al[56] conducted a prospective study to test their CADx for the differentiation of colon polyps less than 10 mm in size. The AI system was developed using magnifying NBI images. This CADx was accurate in 93.1% with a sensitivity and specificity of 95% and 90.3%, respectively. Another CADx developed by Kominami et al[57] using also magnifying NBI images and tested in a prospective study published in 2016, showed an accuracy of 93.2% a sensitivity of 93.0%, and a specificity of 93.3% for the classification of histology of colorectal polyps. Similarly, Chen et al[58] published in 2018 the results of their CADx that were developed using magnifying NBI images. This CADx was tested in a retrospective study for the identification of neoplastic or hyperplastic polyps less than 5 mm in size. The sensitivity was 96.3%, the specificity was 78.1% with a PPV of 89.6% and negative predictive value (NPV) of 91.5%. More CADx were developed and studied to diagnose CRC such as the previously detailed CADx developed by Misawa et al[12] or the CADx developed by Takeda et al[59] to diagnose invasive CRC that showed an accuracy of 94.1%, a sensitivity of 89.4%, a specificity of 98.9%, a PPV 98.8%, and a NPV of 90.1%. Finally, diagnostic AI-assistance can be used in IBD patients like the CADx developed by Maeda et al[13] for the prediction of persistent histologic inflammation in ulcerative colitis patients. It showed an accuracy of 91%, a sensitivity of 74%, and a specificity of 97%. In conclusion, AI can be of great utility again, it can give the endoscopist quick histologic diagnosis prediction that can be of high importance while performing endoscopic procedure. The available data showed already very good accuracy for the tested CADx, though more prospective in vivo studies are needed to confirm optical biopsy analysis through every CADx is reliable enough and non-inferior to the gold standard tissue biopsy before its introduction to clinical use to replace the time-consuming traditional biopsy.
On the therapeutic level, AI can offer helpful assistance too: en bloc endoscopic resection of T1 CRC without evident signs of deep invasion seems justified to prevent surgery.[60] Pre-operative prediction of LNM could be sometimes challenging as LNM is present in approximately 10% of these patients who; therefore, subsequently require surgical resection with lymph node dissection.[61,62] To find a solution for this challenge, Ichimasa et al[16] investigated whether AI can predict LNM presence, thus minimizing the need for additional surgery. They developed an AI system based on data collected from 690 patients with T1 CRCs divided into two groups. A group of 590 patients was used for ML for the AI model, and the second group of the remaining 100 patients was used for model validation. The AI model analyzed 45 clinico-pathological factors (height, weight, age, sex, comorbidities, endoscopic appearance, histology details, etc) and then predicted positivity or negativity for LNM. Operative specimens were used as the gold standard for the presence of LNM. The AI system accuracy in predicting LNM presence was calculated in comparison with the American, European, and Japanese guidelines that are applicable for making decisions about further surgery based on the results of histopathological examination of the endoscopically resected specimen. The rates of unnecessary additional surgery attributable to misdiagnosing LNM-negative patients as having LNM were: 77% for the AI model, and 85%, 91%, and 91% for the American, European, and Japanese guidelines, respectively. This study showed that AI significantly reduced unnecessary additional surgery after endoscopic resection of T1 CRC without missing LNM positivity. Such AI systems can offer valuable assistance to interventional endoscopists in taking rapid and appropriate therapeutic decisions.
Future perspectives
We think that AI will be available in the very near future as many AI systems proved highly accurate for the tasks they were built and tested for. Some functions need aggressive testing before they can be applied in clinical use, especially those that can have major impact on patients’ outcomes in case of AI failures such as CADx that deals with cancer diagnosis and treatment. We think that CADe and CADx for colon polyps’ detection and characterization are among the most promising systems that can be introduced soon in our practice. A new endoscopic system developed by Olympus Corp. called endocytoscope (a prototype endoscope CFH290ECI; Olympus Corp., Tokyo, Japan) has already been tested in one of the first large-scale prospective trials of AI-assisted endoscopy in a clinical setting for optical assessment of diminutive colorectal polyps. Overall, 466 polyps from 325 patients were assessed by CAD, with a pathologic prediction rate of 98.1% (457 of 466).[3] We think that for any AI system to be easily accepted and introduced for clinical use, besides its safety and efficacy, it should be easy to use, does not require special training, does not require more complicated and time-consuming technical maneuvers and not extremely expensive. A transition time frame is needed for endoscopists to feel comfort and confidence to rely on AI in their practice.
There is a huge area of creativity in this field, any team can develop and for each category (CADx, CADe, etc) its unique AI system use different computer algorithms, different ML methods, different endoscopic materials (video-frames, video segments), and different endoscopic techniques (WL, NBI, magnification, endoscystoscopy, etc). This is of advantage that we can build and select the best AI system for any function we can imagine that we need in GI endoscopy, starting from disease prediction, to the scope introduction, and to endoscopic diagnosis and treatment. On the other hand, this will put big responsibility and pressure on the clinical and endoscopic societies to standardize and develop the proper guidelines for the use of this new technology in clinical practice.
We think that the future of GI endoscopy will face for sure a dramatic change in the upcoming years with the introduction of AI into this domain. Huge investments are raised by manufacturers for this promising technology and results started to come out. Currently, an endoscopist performs, detects, analyzes, decides, and treats all alone by himself. In addition, he must have all the experience and the enormous knowledge to be able to offer the best care for his patients. He has to maintain a good memory and practical skills over the time and to keep updated for every new information, recommendation, and guideline. Something that can be really complicated nowadays due to the lack of free time and rapid change in medical knowledge. Now, with the help of computer algorithms, machines have got AI and human-brain like functions. AI proved considerable efficiency and reliability in GI endoscopy though it still needs more development for perfection. Currently, AI will be given the role of assistance so the last decision will remain for endoscopists. However, who knows if AI one day will replace human intelligence to judge and decide while endoscopists become the assistants that assure the correct performance of machines? That is why many questions could come out here: who will be responsible for machine's medical errors? What if this error could be fatal? What about machine crash or malfunctions in critical situations? All of this should not prevent us to continue the development of AI, because the optimal goal in our daily practice will always be the same: the best for the patients in the first place and the physicians in the second place. More research still needed to face our concerns about AI, keeping in mind the need for regulations that ensure security and ethical standards are upheld.
Conclusions
The application of AI in GI endoscopy is attracting a growing amount of attention because it has the potential to improve the quality of endoscopy at all levels. It will make a major breakthrough and a big revolution in the development of GI endoscopy. AI has the advantage to limit inter-operator variability. It can compensate the limited-experience of novice endoscopists and the errors of even the most experienced endoscopists. AI can also compensate for humans’ unsteady performance that can be caused by lack of attention or forgetfulness due to fatigue, anxiety, or any other physical or emotional stress. Since AI is basically computer algorithms, the door is open to develop as many different AI systems as we might need to improve our clinical practice for the best of both patients and physicians. Further researches and regulations are still needed before its introduction in GI endoscopy that can be very soon.
Conflicts of interest
None.
Footnotes
How to cite this article: El Hajjar A, Rey JF. Artificial intelligence in gastrointestinal endoscopy: general overview. Chin Med J 2020;133:326–334. doi: 10.1097/CM9.0000000000000623
References
- 1.Arel I, Rose D, Karnowski T. Deep machine learning - a new frontier in artificial intelligence research. IEEE Comp Int Mag 2010; 5:13–18. doi: 10.1109/MCI.2010.938364. [Google Scholar]
- 2.Eickhoff A, van Dam J, Jakobs R, Kudis V, Hartmann D, Damian U, et al. Computer-assisted colonoscopy (the NeoGuide Endoscopy System): results of the first human clinical trial (“PACE study”). Am J Gastroenterol 2007; 102:261–266. doi: 10.1111/j.1572-0241.2006.01002.x. [DOI] [PubMed] [Google Scholar]
- 3.Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, et al. Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy: a prospective study. Ann Intern Med 2018; 169:357–366. doi: 10.7326/M18-0249. [DOI] [PubMed] [Google Scholar]
- 4.Misawa M, Kudo S, Mori Y, Cho T, Kataoka S, Yamauchi A, et al. Artificial intelligence-assisted polyp detection for colonoscopy: initial experience gastroenterology 2018; 154:2027–2029.e3. doi: 10.1053/j.gastro.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Hassan AR, Haque MA. Computer-aided gastrointestinal hemorrhage detection in wireless capsule endoscopy videos. Comput Methods Programs Biomed 2015; 122:341–353. doi: 10.1016/j.cmpb.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Xiao Jia, Meng MQ. A deep convolutional neural network for bleeding detection in wireless capsule endoscopy images. Conf Proc Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Annu Conf 2016; 2016:639–642. doi: 10.1109/embc.2016.7590783. [DOI] [PubMed] [Google Scholar]
- 7.Kanesaka T, Lee TC, Uedo N, Lin KP, Chen HZ, Lee JY, et al. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc 2018; 87:1339–1344. doi: 10.1016/j.gie.2017.11.029. [DOI] [PubMed] [Google Scholar]
- 8.Miyaki R, Yoshida S, Tanaka S, Kominami Y, Sanomura Y, Matsuo T, et al. A computer system to be used with laser-based endoscopy for quantitative diagnosis of early gastric cancer. J Clin Gastroenterol 2015; 49:108–115. doi: 10.1097/MCG.0000000000000104. [DOI] [PubMed] [Google Scholar]
- 9.Yoon HJ, Kim S, Kim JH, Keum JS, Jo J, Cha JH, et al. Sa1235 application of artificial intelligence for prediction of invasion depth in early gastric cancer: preliminary study. Gastrointest Endosc 2018; 87:AB176.doi: 10.1016/j.gie.2018.04.273. [Google Scholar]
- 10.Lee B-I, Matsuda T. Estimation of invasion depth: the first key to successful colorectal ESD. Clin Endosc 2019; 52:100–106. doi: 10.5946/ce.2019.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori Y, Kudo SE, Chiu PWY, Singh R, Misawa M, Wakamura K, et al. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy 2016; 48:1110–1118. doi: 10.1055/s-0042-113609. [DOI] [PubMed] [Google Scholar]
- 12.Misawa M, Kudo S, Mori Y, Nakamura H, Kataoka S, Maeda Y, et al. Characterization of colorectal lesions using a computer-aided diagnostic system for narrow-band imaging endocytoscopy. Gastroenterology 2016; 150:1531–1532.e3. doi: 10.1053/j.gastro.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Maeda Y, Kudo S, Mori Y, Misawa M, Ogata N, Sasanuma S, et al. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc 2019; 89:408–415. doi: 10.1016/j.gie.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 14.Ruffle JK, Farmer AD, Aziz Q. Artificial intelligence-assisted gastroenterology- promises and pitfalls. Am J Gastroenterol 2019; 114:422–428. doi: 10.1038/s41395-018-0268-4. [DOI] [PubMed] [Google Scholar]
- 15.de Groof J, van der Sommen F, van der Putten J, Struyvenberg MR, Zinger S, Curvers WL, et al. The Argos project: the development of a computer-aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United Eur Gastroenterol J 2019; 7:538–547. doi: 10.1177/2050640619837443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichimasa K, Kudo S, Mori Y, Misawa M, Matsudaira S, Kouyama Y, et al. Artificial intelligence is a powerful tool to determine the need for addtional surgery after endoscopic resection of t1 colorectal cancer − analysis based on a big data for machine learning. Gastrointest Endosc 2018; 87:AB497.doi: 10.1055/s-0043-122385. [Google Scholar]
- 17.Das A, Ben-Menachem T, Cooper GS, Chak A, Sivak MV, Gonet JA, et al. Prediction of outcome in acute lower-gastrointestinal haemorrhage based on an artificial neural network: internal and external validation of a predictive model. Lancet 2003; 18:1261–1266. doi: 10.1016/S0140-6736(03)14568-0. [DOI] [PubMed] [Google Scholar]
- 18.Lagergren J, Lagergren P. Oesophageal cancer. BMJ 2010; 341:c6280.doi: 10.1136/bmj.c6280. [DOI] [PubMed] [Google Scholar]
- 19.Sharma P, Hawes RH, Bansal A, Gupta N, Curvers W, Rastogi A, et al. Standard endoscopy with random biopsies versus narrow band imaging targeted biopsies in Barrett's oesophagus: a prospective, international, randomised controlled trial. Gut 2013; 62:15–21. doi: 10.1136/gutjnl-2011-300962. [DOI] [PubMed] [Google Scholar]
- 20.Dent J. Barrett's esophagus: a historical perspective, an update on core practicalities and predictions on future evolutions of management. J Gastroenterol Hepatol 2011; 26 Suppl 1:11–30. doi: 10.1111/j.1440-1746.2010.06535.x. [DOI] [PubMed] [Google Scholar]
- 21.Phoa KN, Pouw RE, Bisschops R, Pech O, Ragunath K, Weusten BLAM, et al. Multimodality endoscopic eradication for neoplastic Barrett oesophagus: results of an European multicentre study (EURO-II). Gut 2016; 65:555–562. doi: 10.1136/gutjnl-2015-309298. [DOI] [PubMed] [Google Scholar]
- 22.Abrams JA, Kapel RC, Lindberg GM, Saboorian MH, Genta RM, Neugut AI, et al. Adherence to biopsy guidelines for Barrett's esophagus surveillance in the community setting in the United States. Clin Gastroenterol Hepatol 2009; 7:736–710. doi: 10.1016/j.cgh.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swager A-F, van der Sommen F, Klomp SR, Zinger S, Meijer SL, Schoon EJ, et al. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc 2017; 86:839–846. doi: 10.1016/j.gie.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 24.van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten Blam, et al. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy 2016; 48:617–624. doi: 10.1055/s-0042-105284. [DOI] [PubMed] [Google Scholar]
- 25.Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc 2019; 89:25–32. doi: 10.1016/j.gie.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 26.Shin D, Protano M-A, Polydorides AD, Dawsey SM, Pierce MC, Kim MK, et al. Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol 2015; 13:272–279.e2. doi: 10.1016/j.cgh.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quang T, Schwarz RA, Dawsey SM, Tan MC, Patel K, Yu X, et al. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc 2016; 84:834–841. doi: 10.1016/j.gie.2016.03.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang CR, Sheu BS, Chung PC, Yang HB. Computerized diagnosis of Helicobacter pylori infection and associated gastric inflammation from endoscopic images by refined feature selection using a neural network. Endoscopy 2004; 36:601–608. doi: 10.1055/s-2004-814519. [DOI] [PubMed] [Google Scholar]
- 29.Itoh T, Kawahira H, Nakashima H, Yata N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open 2018; 6:E139–E144. doi: 10.1055/s-0043-120830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, et al. Application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine 2017; 25:106–111. doi: 10.1016/j.ebiom.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging-bright and linked color imaging: a single-center prospective study. Ann Gastroenterol 2018; 31:462–468. doi: 10.20524/aog.2018.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 33.Everett SM, Axon AT. Early gastric cancer in Europe. Gut 1997; 41:142–150. doi: 10.1136/gut.41.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018; 21:653–660. doi: 10.1007/s10120-018-0793-2. [DOI] [PubMed] [Google Scholar]
- 35.Kubota K, Kuroda J, Yoshida M, Ohta K, Kitajima M. Medical image analysis: computer-aided diagnosis of gastric cancer invasion on endoscopic images. Surg Endosc 2012; 26:1485–1389. doi: 10.1007/s00464-011-2036-z. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, Wang QC, Xu M-D, Zhang Z, Cheng J, Zhong YS, et al. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc 2019; 89:806–815.e1. doi: 10.1016/j.gie.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Gerson LB, Fidler JL, Cave DR, Leighton JA. ACG clinical guideline: diagnosis and management of small bowel bleeding. Am J Gastroenterol 2015; 110:1265–1287. doi: 10.1038/ajg.2015.246. [DOI] [PubMed] [Google Scholar]
- 38.Rondonotti E, Spada C, Adler S, May A, Despott EJ, Koulaouzidis A, et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Technical Review. Endoscopy 2018; 50:423–446. doi: 10.1055/a-0576-0566. [DOI] [PubMed] [Google Scholar]
- 39.Leenhardt R, Vasseur P, Li C, Saurin JC, Rahmi G, Cholet F, et al. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc 2019; 89:189–194. doi: 10.1016/j.gie.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 40.Zhou T, Han G, Li BN, Lin Z, Ciaccio EJ, Green PH, et al. Quantitative analysis of patients with celiac disease by video capsule endoscopy: a deep learning method. Comput Biol Med 2017; 85:1–6. doi: 10.1016/j.compbiomed.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 41.He JY, Wu X, Jiang YG, Peng Q, Jain R. Hookworm detection in wireless capsule endoscopy images with deep learning. IEEE Trans Image Process Publ IEEE Signal Process Soc 2018; 27:2379–2392. doi: 10.1109/TIP.2018.2801119. [DOI] [PubMed] [Google Scholar]
- 42.Pan G, Yan G, Qiu X, Cui J. Bleeding detection in wireless capsule endoscopy based on probabilistic neural network. J Med Syst 2011; 35:1477–1484. doi: 10.1007/s10916-009-9424-0. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Yuan X. Obscure bleeding detection in endoscopy images using support vector machines. Optim Eng 2009; 10:289–299. doi: 10.1007/s11081-008-9066-y. [Google Scholar]
- 44.Coimbra MT, Cunha JPS. MPEG-7 visual descriptors—contributions for automated feature extraction in capsule endoscopy. IEEE Trans Circuits Syst Video Technol 2006; 16:628–637. doi: 10.1109/TCSVT.2006.873158. [Google Scholar]
- 45.Li B, Meng MQH. Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Trans Inf Technol Biomed Publ IEEE Eng Med Biol Soc 2012; 16:323–329. doi: 10.1109/TITB.2012.2185807. [DOI] [PubMed] [Google Scholar]
- 46.Estimated Number of New Cancer Cases and Deaths in 2018, Worldwide, all Cancers, Both Sexes, All Ages. Lyon, France: International Agency for Research on Cancer, World Health Organization, 2018. Available from: http://gco.iarc.fr/today/home [Accessed November 22, 2019]. [Google Scholar]
- 47.Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014; 370:1298–1306. doi: 10.1056/NEJMoa1309086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn SB, Han DS, Bae JH, Byun TJ, Kim JP, Eun CS. The miss rate for colorectal adenoma determined by quality-adjusted, back-to-back colonoscopy. Gut Liver 2012; 6:64–70. doi: 10.5009/gnl.2012.6.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rees CJ, Rajasekhar PT, Wilson A, Close H, Rutter MD, Saunders BP, et al. Narrow band imaging optical diagnosis of small colorectal polyps in routine clinical practice: the detect inspect characterise resect and discard 2 (DISCARD 2) study. Gut 2017; 66:887–895. doi: 10.1136/gutjnl-2015-310584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiis-Evensen E, Hoff GS, Sauar J, Langmark F, Majak BM, Vatn MH. Population-based surveillance by colonoscopy: effect on the incidence of colorectal cancer. Telemark Polyp Study I Scand J Gastroenterol 1999; 34:414–420. doi: 10.1080/003655299750026443. [DOI] [PubMed] [Google Scholar]
- 51.Fernández-Esparrach G, Bernal J, López-Cerón M, Córdova H, Sánchez-Montes C, Rodríguez de Miguel C, et al. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy 2016; 48:837–842. doi: 10.1055/s-0042-108434. [DOI] [PubMed] [Google Scholar]
- 52.Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, et al. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 2018; 155:1069–1078.e8. doi: 10.1053/j.gastro.2018.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P, Xiao X, Glissen Brown JR, Berzin TM, Tu M, Xiong F, et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng 2018; 2:741–748. doi: 10.1038/s41551-018-0301-3. [DOI] [PubMed] [Google Scholar]
- 54.Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, et al. Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc 2019; 89:576–582.e1. doi: 10.1016/j.gie.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 55.Dekker E, Kara M, Offerhaus J, Fockens P. Endocytoscopy in the colon: early experience with a new real-time contact microscopy system. Gastrointest Endosc 2005; 61:AB224.doi: 10.1016/S0016-5107(05)01230-7. [Google Scholar]
- 56.Gross S, Trautwein C, Behrens A, Winograd R, Palm S, Lutz HH, et al. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc 2011; 74:1354–1359. doi: 10.1016/j.gie.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, et al. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc 2016; 83:643–649. doi: 10.1016/j.gie.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HHS, Tseng VS. Accurate classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology 2018; 154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 59.Takeda K, Kudo SE, Mori Y, Misawa M, Kudo T, Wakamura K, et al. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy 2017; 49:798–802. doi: 10.1055/s-0043-105486. [DOI] [PubMed] [Google Scholar]
- 60.Overwater A, Kessels K, Elias SG, Backes Y, Spanier BWM, Seerden TCJ, et al. Endoscopic resection of high-risk T1 colorectal carcinoma prior to surgical resection has no adverse effect on long-term outcomes. Gut 2018; 67:284–290. doi: 10.1136/gutjnl-2015-310961. [DOI] [PubMed] [Google Scholar]
- 61.Bosch SL, Teerenstra S, de Wilt JHW, Cunningham C, Nagtegaal ID. Predicting lymph node metastasis in pT1 colorectal cancer: a systematic review of risk factors providing rationale for therapy decisions. Endoscopy 2013; 45:827–834. doi: 10.1055/s-0033-1344238. [DOI] [PubMed] [Google Scholar]
- 62.Miyachi H, Kudo SE, Ichimasa K, Hisayuki T, Oikawa H, Matsudaira S, et al. Management of T1 colorectal cancers after endoscopic treatment based on the risk stratification of lymph node metastasis. J Gastroenterol Hepatol 2016; 31:1126–1132. doi: 10.1111/jgh.13257. [DOI] [PubMed] [Google Scholar]


