Abstract
Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer with a high mortality disease which has been positioned the first and second cancer morbidity of men and women in China, separately. Our study was to assess the prognostic meaningful of ubiquitin conjugating enzyme E2 T (UBE2T) expression in LUAD dependent on data acquired from The Cancer Genome Atlas (TCGA) and so as to increase further knowledge into the biological pathways involved in LUAD pathogenesis related to UBE2T.
Information on gene expression and comparing clinical data were recognized and downloaded from TCGA. Gene set enrichment analysis (GSEA) created an arranged list of all genes s indicated by their connection with UBE2T expression.
Our study cohort included 265 (54.5%) female and 221 (36.0%) male patients. The scatter plot and paired plot showed the difference of UBE2T expression between normal and tumor samples (P < .01). Overall survival (OS) analysis demonstrated that LUAD with UBE2T-high had a more terrible prognosis than that with UBE2T-low (P < .01). Multivariate analysis with the cox proportional hazards model indicated that the expression of UBE2T (hazard ratio [HR]: 1.28; 95% Confidence Interval (CI): 1.06–1.56; P = .011) and stage (HR: 2.02; 95% CI: 1.27–3.21; P = .003) were independent prognostic factors for patients with LUAD. The GSEA results showed that cell cycle, DNA replication, RNA degradation, oxidative phosphorylation, pathogenic Escherichia coli infection, citrate cycle tricarboxylic acid cycle, Alzheimer's disease, P53 signaling pathway, and purine metabolism are differentially enriched in UBE2T high expression phenotype.
Our study found that the expression of UBE2T was significantly increased in LUAD patients and associated with several clinical features. UBE2T may be a potentially useful prognostic molecular biomarker of bad survival in LUAD, while further experimental ought to be performed to demonstrate the biologic effect of UBE2T.
Keywords: lung adenocarcinoma, prognosis, TCGA, ubiquitin conjugating enzyme E2 T
1. Introduction
Lung adenocarcinoma (LUAD), the leading cause of cancer-related mortality worldwide shows the highest and second highest mortality amongst the male and female Chinese population, respectively.[1,2] The causal factors associated with it, viz., tobacco, aging, and atmosphere pollution, has resulted with a significant incidence of lung cancer patients, records show an estimated 2.1 million new cases and 1.8 million deaths annually.[3–5] LUAD, the most common subtype of lung cancer has a 5-year overall survival rate is 15%, due to its late diagnoses.[6] Though a study reported that 60% of LUAD patients harboring targetable gene alterations, have improved survival rate,[7] nevertheless, due to the high rate of metastasis and drug resistance as well as the lack of specific and sensitive early biomarkers, of LUAD, contributes to the high mortality of LUAD.[8] Thus, it is critical to discover sensitive and specific novel target molecules for developing effective diagnosis and treatment strategies of LUAD.
It is known that posttranscriptional modifications are vital for the initiation and the progression of tumors. The ubiquitin conjugating enzyme E2T (UBE2T; otherwise called FANCT; PIG50; HSPC150), is a member of the ubiquitin-proteasome family and catalyzes the covalent attachment of ubiquitin to different protein substrates. It was initially reported in Fanconi anemia and is critical for the DNA damage repair pathway.[9] UBE2T regulates cellular processes such as signal transduction, cell cycle control, and tumorigenesis by triggering the degradation of relevant substrates.[10] Studies have shown that UBE2T is amplified in a wide variety of tumors. Ueki et al[11] showed that UBE2T regulates the progression of breast cancer by interacting with the BRCA1 DNA repair associated/BRCA1 associated RING domain 1 complex. In addition, Hu et al[12] demonstrated that UBE2T promotes the development and progression of the nasopharyngeal carcinoma by activating the AKT/GSK3β/β-catenin pathway. Similarly, Gong et al[13] reported that the knockdown of UBE2T significantly diminished the bladder cancer cell proliferation and colony formation. The study of Liu et al[14] showed that the UBE2T complex, miR-543/UBE2T/p53 might represent a new significant potential therapeutic target in hepatocellular carcinoma. Moreover, Luo et al[15] reported that UBE2T facilitates the invasion and metastasis of gastric cancer by triggering the epithelial mesenchymal transition process. Another study by Lian et al[16] found that UBE2T regulated the PI3K/Akt signaling pathway to promote the proliferation of the renal cell carcinoma cells. However, the mechanism of the role of UBE2T in LUAD prognosis still needs to be elucidated. In the current study, we assessed the expression of UBE2T in LUAD from the The Cancer Genome Atlas (TCGA) database to understand the role of UBE2T in LUAD pathogenesis. To gain a more in-depth understanding of the biological mechanisms involved, the GSEA analysis was carried. Further, we analyzed the relationship between the expression of the UBE2T with the clinical features and with the immune infiltration level in LUAD based on an online tumor infiltrating immune cells tool.
2. Materials and methods
2.1. Gene information and bioinformatics analysis
Information on the gene expression and comparing clinical data (486 cases, Data Format: BCR XML) was downloaded from the level 3 gene-expression information (FPKM normalized) of the TCGA LUAD cohort. Boxplots were utilized to analyze the expression differences of the discrete variables. The clinicopathological data collected included sex, age, stage, grade, T-classification, M-classification, N-classification, survival status, and survival duration in days.
2.2. GSEA enrichment
The gene set enrichment analysis (GSEA) created a list of all genes connected with the expression of the UBE2T. Then the samples were categorized as the high- and low-UBE2T training groups to elucidate the potential function and the significant survival difference utilizing GSEA of the UBE2T. The annotated gene sets c2.cp.kegg.v6.0.symbols.gmt was selected as the reference gene sets, which includes the terms with false discovery rate (FDR < 0.05). Gene set permutations were executed multiple times for every examination. The expression degree of UBE2T was applied as the phenotype label. The normalized enrichment score (NES) and the nominal P value was used to find the pathways enriched in every phenotype.
2.3. Statistical analysis
The listwise deletion technique was utilized to deal with any missing data, which excluded the entire sample from the investigation if any single value was absent. The connection between the clinical factors and UBE2T was examined with the logistic regression, the Wilcoxon signed-rank test and the Kruskal test. Clinical factors were related to the overall survival using the Cox regression and the Kaplan–Meier approach. Multivariate Cox analysis was utilized to evaluate the impact of the expression of theUBE2T on survival alongside other clinical attributes (such as age, sex, stage, distant metastasis). Benjamini–Hochberg method was used to transform the P values to FDRs. Data were examined with the R (version 3.5.3) and R Bioconductor packages. Pearl language was used for the data matrix and the data processing according to P <.5.
3. Results
3.1. Patients’ characteristics
The 486 patients from the TCGA database and thier clinicopathological attributes are shown in Table 1. The middle age at diagnosis in the TCGA database was 60.5 years (range 33–88 years) and the median follow-up time for the subjects was 9.3 years (range 0–18.7 years). The follow-up data showed that 162 (33.3%) patients were alive while 324 (66.7%) were death patients. Our study cohort included 265 (54.5%) female and 221 (36.0%) male patients. Stage I disease was located in 262 patients (53.9%), stage II in 112 (23.1%), stage III in 79 (23.0%), and stage IV in 25 (5.1%). Tumor stage was found T1 in 260 patients (53.5%), T2 in 95 (19.5%), T3 in 41 (8.4%), and T4 in 19 (3.9%). Node stage contained N0 in 321 (64.2%), N1 in 90 (18.5%), N2 in 70 (14.4%), N3 in 2 (0.4%). From the study total of 486 adenomas or adenocarcinomas patients, 24 (4.9%) cases had distant metastases.
Table 1.
TCGA lung adenocarcinoma patient characteristics.
3.2. Association between UBE2T expression and the clinicopathologic factors
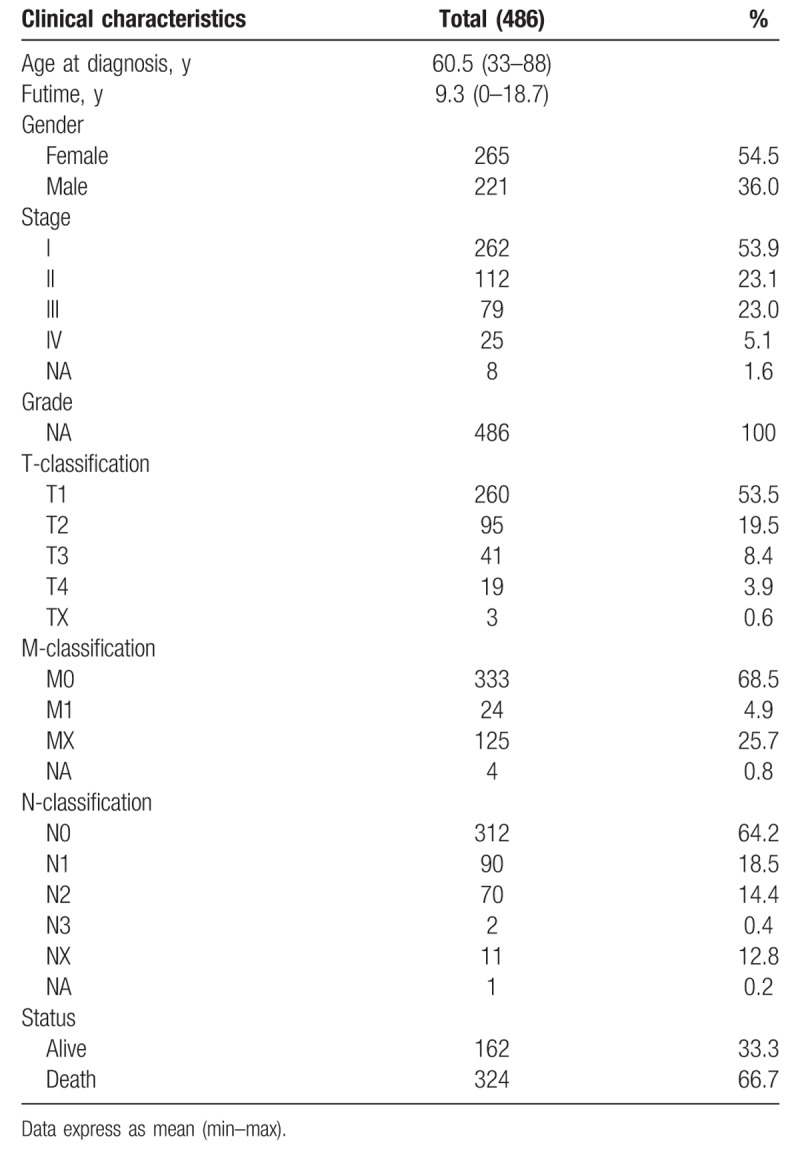
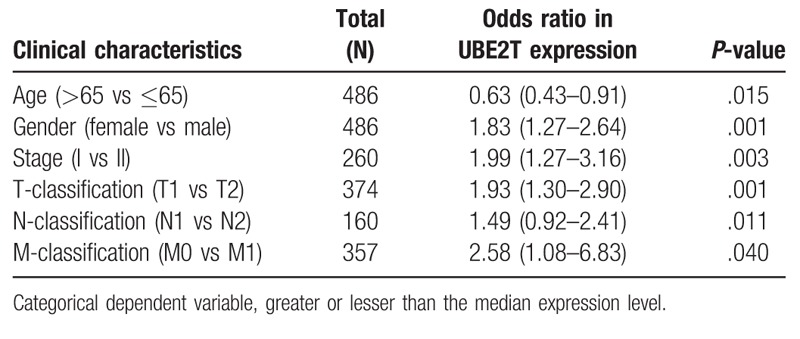
The scatter plot shows the UBE2T expression difference between the normal and the tumor samples (P < .01), Fig. 1A. We observed a significant difference in the expression of UBE2T, suggesting it might play a regulatory role in the development of cancer. There was significant correlation between the expression of UBE2T with the patient age, sex, clinical stage, T-classification, and M-classification (P < .05), Fig. 2B–F. Univariate and logistic regression analysis showed that expression of UBE2T had a negative correlation to the prognostic clinicopathologic factors (Table 2). The UBE2T expression in LAUD according to the different clinicopathological factors, such as, age (OR = 0.63; 95% Confidence Interval (CI) = 0.43–0.91, >65 vs ≤65), sex (OR = 1.83; 95% CI = 1.27–2.64, female vs male), stage (OR = 1.99; 95% CI = 1.27–3.16, I vs II), T-classification (OR = 1.93; 95% CI = 1.30–2.90, T1 vs T2), M-classification (OR = 2.58; 95% CI = 1.08–6.83, M1 vs T1), and N-classification (OR = 1.49; 95% CI = 0.92–2.41, N1 vs N2) clearly indicate that patients with a higher UBE2T mRNA expression are more prone to progress to a more advanced stage than those with a lower UBE2T mRNA expression.
Figure 1.
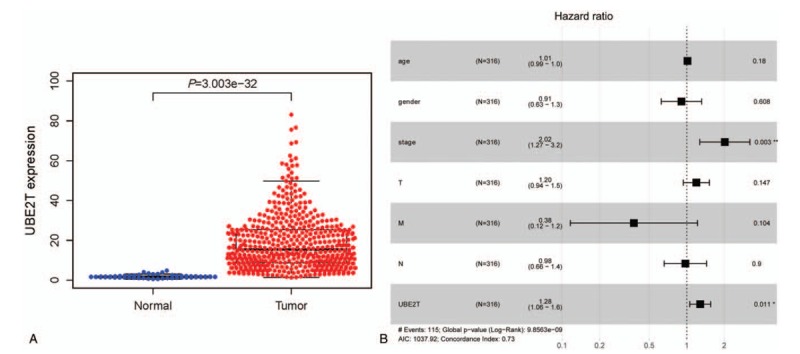
(A) The scatter plot showed the difference of UBE2T expression between normal and tumor samples (P < .01). (B) The forest plot about the multivariate analysis results. UBE2T = ubiquitin conjugating enzyme E2 T.
Figure 2.
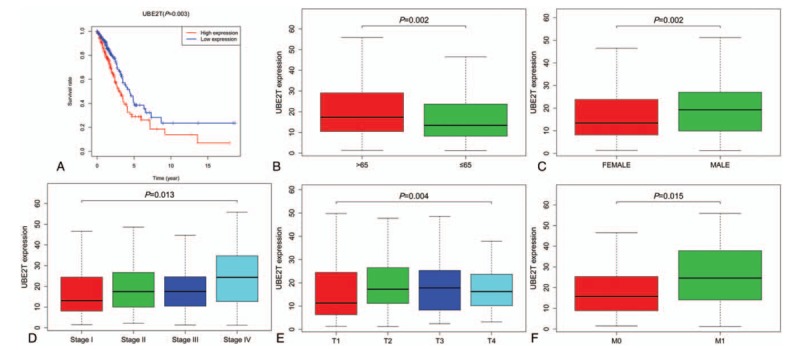
Survival analysis and the association with UBE2T expression and clinicopathologic factors. (A) Overall survival (OS) analysis demonstrated that LUAD with UBE2T-high had a more terrible prognosis than that with UBE2T-low (P < .01). (B) Age association with UBE2T expression. (C) Sex association with UBE2T expression. (D) Stage association with UBE2T expression. (E) T-stage association with UBE2T expression. (F) M-stage association with UBE2T expression. LUAD = lung adenocarcinoma, UBE2T = ubiquitin conjugating enzyme E2 T.
Table 2.
UBE2T expression associated with clinical pathological characteristics (logistic regression).
3.3. Survival results and multivariate examination
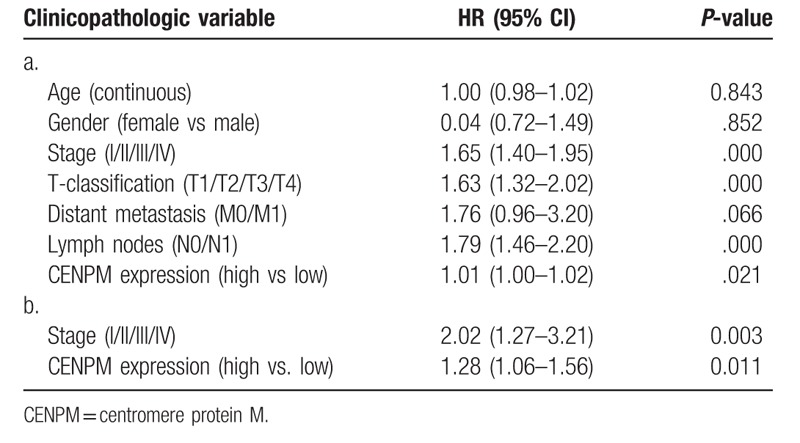
The overall survival (OS) analysis demonstrate that LUAD with higher levels of UBE2T expression had a more negative prognosis than that with a lower level of UBE2T expression (P < .01), Fig. 2A. The univariate analysis showed is that UBE2T associated essentially with stage (HR: 1.65; 95% CI: 1.40–1.95; P < .01), T-classification (HR: 1.63; 95% CI: 1.32–2.02; P < .01), and N-classification (HR: 1.79; 95% CI: 1.46–2.20; P < .01). Multivariate analysis with the cox proportional hazards model indicated that the expression of UBE2T (HR: 1.28; 95% CI: 1.06–1.56; P = .011) and stage (HR: 2.02; 95% CI: 1.27–3.21; P = .003) were the 2 independent prognostic factors for the patients with LUAD, Table 3. The forest plot on the multivariate analysis results are shown in Fig. 1B.
Table 3.
a. Associations with overall survival and clinicopathologic characteristics in TCGA patients using Cox regression. b. Multivariate survival model after variable selection.
3.4. GSEA recognizes the UBE2T related signaling pathway
In order to recognize signaling pathways, which might be differentially initiated in LUAD, we did the GSEA analysis in the high and the low UBE2T expression data sets (FDR P < .05, NOM P < .05) and choose the most significantly enriched signaling pathways dependent on the normalized enrichment score (NES), Table 4. We observed the following signaling pathways to be differentially enriched in UBE2T high expression phenotype, viz., cell cycle, DNA replication, RNA degradation, oxidative phosphorylation, pathogenic Escherichia coli infection, citrate cycle Tricarboxylic Acid Cycle (TCA) cycle, Alzheimer's disease, P53 signaling pathway, and purine metabolism (Fig. 3).
Table 4.
Gene sets enriched in phenotype high.
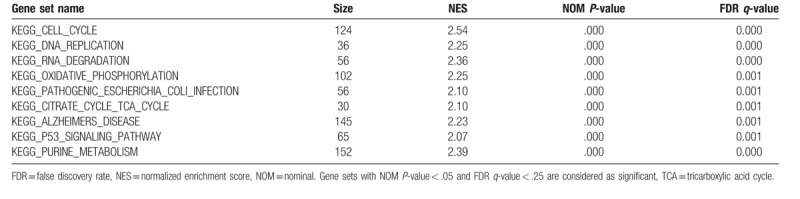
Figure 3.
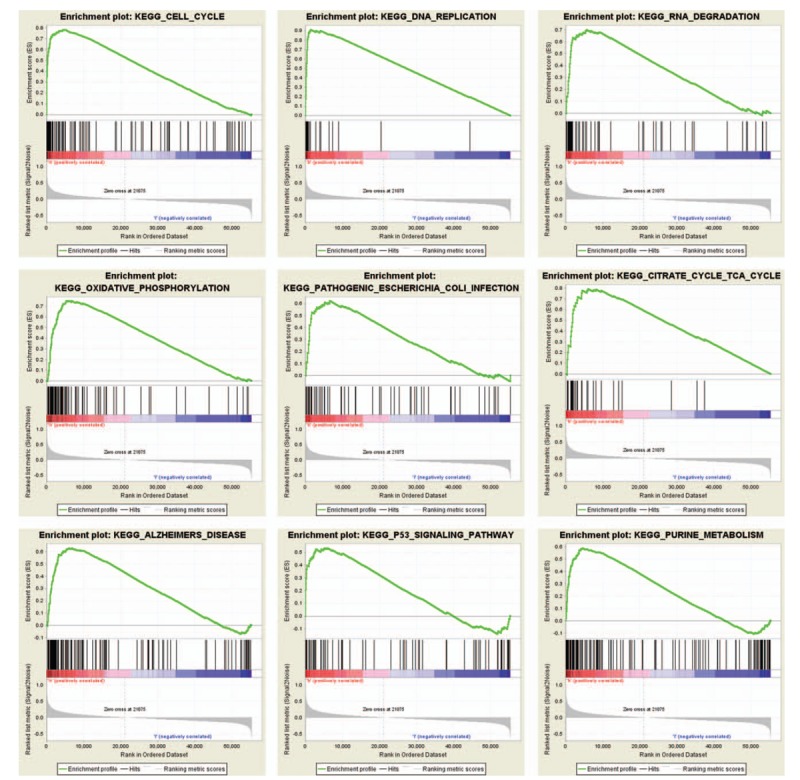
Enrichment plots from gene set enrichment analysis (GSEA).
4. Discussion
In the current study, we conducted a comprehensive and detailed assessment of the expression of UBE2T in LUAD from the TCGA database to explore its association with clinicopathologic characteristics, survival, function, and expression difference. This will help us get a better understand of the mechanistic details of whether the highly-expressed biomarkers in LUAD c have any correlation with the observed clinical survival patterns. We observed that UBE2T had a significant expression in the tumor compared with the normal samples. This indicates that UBE2T may play an important regulatory role in the progression of cancer. Thus, UBE2T can be used as a novel potential target molecule to develop an effective diagnosis and treatment strategy for LUAD.
UBE2T is a member of E2 the family of the ubiquitin-proteasome pathway, a complex protein degradation system that participates in many biological processes, including signal transduction, tumorigenesis, cell proliferation, differentiation, and cell cycle control.[9] Numerous studies have identified that the overexpression of UBE2T results in a variety of tumorigenesis such as osteosarcoma, diffuse large B-cell lymphoma, and malignant pleural mesothelioma.[17–19] However, till now, the expression of UBE2T and its potential prognostic effect on LUAD has not yet been investigated. Our study observed that the expression of UBE2T in LUAD was associated with advanced clinical pathologic factors (age, sex, clinical stage, T-classification, and M-classification). Survival analysis also showed a similar trend, where higher expression of UBE2T was associated with a poor prognosis as compared with the lower expressed UBE2T samples (P < .01). Univariate and logistic regression analysis showed the association between the expression of UBE2T to poor prognostic clinicopathologic factors, which are known to play critical roles in the advancement of the disease
The GSEA analysis data showed the cell cycle, DNA replication, RNA degradation, oxidative phosphorylation, pathogenic E coli infection, citrate cycle TCA cycle, Alzheimer's disease, p53 signaling pathway, and purine metabolism pathways to be differentially enriched in the UBE2T high expression phenotype. UBE2T may affect cell cycle, DNA replication, RNA degradation then regulates the occurrence and development of cancer cells. Hao et al[20] who was first to investigate the expression of UBE2T mRNA in normal human tissues and 8 lung cancer cell lines, found that UBE2T was significantly upregulated in the lung cancer tissue and cell lines, suggesting involvement of UBE2T in the malignant cell phenotype. The ubiquitin-proteasome system exerts a crucial role in extensive biological processes, and UBE2T, its crucial member, may affect tumorigenesis and cell cycle. Studies on the function of UBE2T in cancer will undoubtedly provide new insights regarding the role of UBE2T in both cell cycle regulation and tumorigenesis. Veziant et al[21] reported that infection with the pathogenic E coli strain causes inflammation and reactive oxygen species production in tumors much before the growth of the tumor indicating that the disorder of the intestinal microflora may affect the UBE2T expression in LUAD. Through enhancing the ubiquitination-mediated degradation of the p53 protein, the UBE2T can as an independent prognostic factor in hepatocellular carcinoma.[14] The ubiquitin proteasome pathway is critical in restraining the activities of the p53 tumor suppressor. When alterations of these proteins result in increased p53 activity, cells arrest in the cell cycle, senesce, or apoptosis and when alterations that result in decreased p53 levels yield tumor-prone phenotypes.[22] So UBE2T may play a role via the p53 signaling pathway in the LAUD associated pathology. Thus, more research is needed to learn about its role in the mechanism of LAUD tumorigenesis. An impaired ubiquitin-proteasome system could lead to negative consequences for protein regulation, including loss of function and incidence of different diseases such as Alzheimer disease and Parkinson disease.[23,24] Our study found that the expression of UBE2T was significantly increased in the LUAD patients and is associated with several clinical features. UBE2T is a potentially useful prognostic molecular biomarker of poor prognosis in LUAD; however more research needs to be done to demonstrate the biologic effect of UBE2T.
Acknowledgments
This work is not supported by grants. As the work is a bioinformatics article, ethical approval was not necessary.
Author contributions
Data curation: Hai-Ying Sun.
Investigation: Zeng-Hong Wu.
Methodology: You-Jing Zhang.
Project administration: Hai-Ying Sun.
Resources: Zeng-Hong Wu.
Software: Zeng-Hong Wu.
Supervision: Zeng-Hong Wu
Validation: Zeng-Hong Wu.
Visualization: Zeng-Hong Wu.
Writing – original draft: Zeng-Hong Wu, You-Jing Zhang.
Writing – review & editing: Hai-Ying Sun.
Footnotes
Abbreviations: GSEA = gene set enrichment analysis, LUAD = lung adenocarcinoma, NES = normalized enrichment score, UBE2T = ubiquitin conjugating enzyme E2 T.
How to cite this article: Wu ZH, Zhang Yj, Sun HY. High ubiquitin conjugating enzyme E2 T mRNA expression and its prognostic significance in lung adenocarcinoma: A study based on the TCGA database. Medicine. 2020;99:4(e18543).
W-ZH and Z YJ was the co-author.
Competing financial interests: No financial (no Funding, Employment and Personal financial interests) and Non-financial competing interests.
The authors have no conflicts of interest to disclose.
References
- [1].Youlden DR, Cramb SM, Baade PD. The International Epidemiology of Lung Cancer: geographical distribution and secular trends. J Thorac Oncol 2008;3:819–31. [DOI] [PubMed] [Google Scholar]
- [2].Tan WL, Jain A, Takano A, et al. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol 2016;17:e347–62. [DOI] [PubMed] [Google Scholar]
- [3].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [4].She J, Yang P, Hong Q, et al. Lung cancer in China: challenges and interventions. Chest 2013;143:1117–26. [DOI] [PubMed] [Google Scholar]
- [5].Zheng M. Classification and pathology of lung cancer. Surg Oncol Clin N Am 2016;25:447–68. [DOI] [PubMed] [Google Scholar]
- [6].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [7].Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Aris VM, Cody MJ, Cheng J, et al. Noise filtering and nonparametric analysis of microarray data underscores discriminating markers of oral, prostate, lung, ovarian and breast cancer. BMC Bioinformatics 2004;5:185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alpi A, Langevin F, Mosedale G, et al. UBE2T, the fanconi anemia core complex, and Fancd2 are recruited independently to chromatin: a basis for the regulation of Fancd2 monoubiquitination. Mol Cell Biol 2007;27:8421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lim KH, Song MH, Baek KH. Decision for cell fate: deubiquitinating enzymes in cell cycle checkpoint. Cell Mol Life Sci 2016;73:1439–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ueki T, Park JH, Nishidate T, et al. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res 2009;69:8752–60. [DOI] [PubMed] [Google Scholar]
- [12].Hu W, Xiao L, Cao C, et al. UBE2T promotes nasopharyngeal carcinoma cell proliferation, invasion, and metastasis by activating the AKT/GSK3beta/beta-catenin pathway. Oncotarget 2016;7:15161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gong Y, Peng D, Ning X, et al. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol Lett 2016;12:4485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu LP, Yang M, Peng QZ, et al. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun 2017;493:20–7. [DOI] [PubMed] [Google Scholar]
- [15].Luo C, Yao Y, Yu Z, et al. UBE2T knockdown inhibits gastric cancer progression. Oncotarget 2017;8:32639–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lian JH, Wang WH, Wang JQ, et al. MicroRNA-122 promotes proliferation, invasion and migration of renal cell carcinoma cells through the PI3K/Akt signaling pathway. Asian Pac J Cancer Prev 2013;14:5017–21. [DOI] [PubMed] [Google Scholar]
- [17].Gordon GJ, Appasani K, Parcells JP, et al. Inhibitor of apoptosis protein-1 promotes tumor cell survival in mesothelioma. Carcinogenesis 2002;23:1017–24. [DOI] [PubMed] [Google Scholar]
- [18].Kosari F, Parker AS, Kube DM, et al. Clear cell renal cell carcinoma: gene expression analyses identify a potential signature for tumor aggressiveness. Clin Cancer Res 2005;11:5128–39. [DOI] [PubMed] [Google Scholar]
- [19].Wang Y, Leng H, Chen H, et al. Knockdown of uBe2T inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing the Pi3K/akt signaling pathway. Oncol Res 2016;24:361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hao J, Xu A, Xie X, et al. Elevated expression of UBE2T in lung cancer tumors and cell lines. Tumor Biol 2008;29:195–203. [DOI] [PubMed] [Google Scholar]
- [21].Veziant J, Gagnière J, Jouberton E, et al. Association of colorectal cancer with pathogenic, Escherichia coli: Focus on mechanisms using optical imaging. World J Clin Oncol 2016;7:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pant V, Lozano G. Limiting the power of p53 through the ubiquitin proteasome pathway. Gen Dev 2014;28:1739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bing G, Miroslav R, Figueiredo-Pereira ME, et al. The Ubiquitin-Proteasome system: potential therapeutic targets for Alzheimer's disease and spinal cord injury. Front Mol Neurosci 2016;9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. Bioessays 2008;30:1172–84. [DOI] [PubMed] [Google Scholar]


