Abstract
The goal of this study was to evaluate the contribution of ataxia telangiectasia mutated (ATM) gene promoter methylation to hepatocellular carcinoma (HCC) and the predictive value of radiotherapy outcome. ATM promoter methylation status was detected using methylation-specific PCR in 118 HCC, 50 adjacent liver, and 20 normal liver samples. PCR products were verified by bisulfite sequencing PCR. ATM expression was detected by quantitative PCR (qPCR) and immunohistochemistry (IHC) in 50 paired HCC and adjacent normal tissues and 68 locally advanced HCC biopsy tissues. Furthermore, radiotherapy outcomes in 68 locally advanced HCC patients were determined using European Association for the Study of Liver criteria and survival analysis. The results revealed that the methylation frequency of the ATM promoter was significantly higher in HCC tissues than in normal liver tissues (χ2 = 16.830, P < .001). Quantitative PCR (qPCR) and IHC results showed a significant association between ATM promoter methylation and ATM expression in HCC (χ2 = 10.510, P < .001), and methylated ATM was correlated with lower ATM expression compared with unmethylated ATM (r = 0.356, P < .001). Furthermore, methylation of the ATM promoter was significantly associated with superior outcomes in patients with locally advanced HCC who initially received radiotherapy. Together, these results indicate that ATM promoter methylation might increase the risk of HCC by regulating ATM expression, and thus may function as a potential biomarker for predicting radiotherapy outcomes in HCC patients.
Keywords: ATM, Biomarker, DNA methylation, hepatocellular carcinoma, radiotherapy
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide, and has a high degree of heterogeneity.[1] Although great progress has been made in the diagnosis and treatment of HCC in recent decades, the morbidity and mortality remain high and the survival rate, which is a key index of the effectiveness of therapeutic strategies, is still unsatisfactory, especially in Eastern Asia.[1,2] Radiotherapy plays an important role in the treatment of HCC, especially in patients with unresectable and advanced HCC, as well as in those who have shown a good response to immunotherapy.[3] However, irradiated patients have significantly different responses to ionizing radiation, which directly affect their treatment efficacy and survival. Previous studies have suggested that variability in radiotherapy efficacy results from genetic variants, for example, mutation, deletion, methylation, and polymorphism.[4] DNA damage induced by ionizing radiation generates complex cellular responses including DNA damage repair, cell cycle regulation, and apoptosis,.[5,6] Radiosensitivity and survival may be influenced by specific gene alterations including epigenetics changes.[7–11]
DNA methylation, a common epigenetic modification, is considered to be a hallmark of malignant tumors and is significantly correlated with multiple cancers.[12–20] Numerous studies have shown that human cancer cells lacking ataxia telangiectasia mutated (ATM) kinase are extremely radiosensitive,[9,10,21] because ATM kinase plays a central role in mediating the repair of DNA damage with transient cell cycle arrest to ensure the maintenance of genomic stability and cell viability.[6,22] Silencing of ATM expression by small interfering RNA contributes to glioma stem cell radiosensitivity in vitro and in vivo.[21] Furthermore, aberrant methylation of the ATM promoter in colorectal tumor cells correlates with increased radiosensitivity, which results at least partly from lower ATM expression induced by inappropriate methylation of the ATM promoter, as high radiosensitivity and lower ATM protein expression in colorectal tumor cells can be reversed by using the DNA demethylating agent 5-azacytidine.[7] A similar phenomenon has been observed in studies using glioma cells, in which methylation of the ATM promoter has been shown to influence ATM expression and alter its radiosensitivity.[9] The presence of methylated CpG islands in the promoter region of genes including ATM can suppress their expression, and methylation of the ATM promoter is a common event in many types of cancer including breast and colorectal cancers, and may correlate with superior radiosensitivity.[7,10,17,20,23] However, the association between ATM promoter methylation and HCC has not yet been investigated.
The goal of this study was to investigate the methylation status of the ATM promoter and the relationship between ATM promoter methylation and ATM protein level and clinical characteristics. Furthermore, the predictive value of radiotherapy outcome in HCC with differences in ATM methylation status was analyzed.
2. Materials and methods
2.1. Patients and specimen characteristics
From June 2017 to June 2018, a total of 50 pairs of HCC surgical specimens and corresponding adjacent liver tissues were collected from the Department of Pathology, the Affiliated Zhuzhou Hospital Xiangya Medical College Central South University (Zhuzhou, Hunan, China). The study included 38 male cases and 12 female cases with patient age ranging from 35 to 73 (53.2 ± 3.2) years old, including 25 cases of liver cirrhosis and 11 cases of portal vein tumor emboli. Tumor stage was confirmed according to the 7th IASLC/AJCC staging system. There were 22 stages I cases, 18 stages II cases, 10 stages III cases, and no stage IV cases; 20 cases of normal liver tissue were used as controls. From January 2015 to May 2016, 68 patients with locally advanced HCC, who underwent liver biopsy before treatment, were recruited to this study. The criteria for enrollment were as follows:
-
1)
patients suffering from pathologically confirmed HCC;
-
2)
patient age between 18 and 70 years old; and
-
3)
tumor node metastasis (TNM) stage of stage IIb, stage IIIa, or stage IIIb.
The exclusion criteria were as follows:
-
1)
biliary cell type or mixed type according to pathological examination;
-
2)
Karnofsky Performance Scale Index score below 70 before treatment;
-
3)
metastatic liver cancer; and
-
4)
lack of agreement from patients and/or their families to enter the trial.
None of the patients received radiotherapy or chemotherapy before initial treatment and all liver biopsy tissues were stored at −80°C immediately until testing. The pathological results of the tissues were confirmed by two pathologists in our hospital. Patients were followed up for 36 months and the median follow-up time was 18.5 months. During follow-up, 51 patients died and 11 patients were lost to follow-up. The study was approved by the Ethics Committee of the Affiliated Zhuzhou Hospital Xiangya Medical College CSU.
2.2. DNA extraction and bisulfite modification
The DNA was isolated using the TIANAMP FFPE DNA KIT(Tiangen Biotech, Beijing, China) according to the instruction manual, the optical density value (D260/D280) was determined by a microplate reader (Biotek, SYNERGY HTX, VT),and the extracted DNA quality was identified by 1% agarose gel electrophoresis (DYCP-32B, Liuyi, Beijing, China).Then, qualified DNA was subjected to bisulfite modification using DNA Bisulfite Conversion Kit (Tiangen Biotech, Beijing, China), according to the manufacturer's instruction.
2.3. Methylation-specific PCR and bisulfite sequencing PCR
Each methylation-specific PCR (MSP) reaction was performed using 2 μL DNA template in a 20 μL reaction volume containing 1 μL each of forward and reverse primers (concentration 10 μmol/L), which was designed using MethPrimers[24] (Table 1), 0.4 μL MSP DNA Polymerase, 1.7 μL 10× MSP PCR Buffer, and 1.4 μL dNTPs (Tiangen Biotech, Beijing, China). The PCR reaction conditions were as follows: denaturation at 95°C for 5 min → amplification at (94°C 20 s, 57°C 30 s, 72°C 20 s) for 35 cycles → extension at 72°C for 5 minutes. The reaction was set up with a blank control group (ddH2O group), a water blank, a methylated sample blank, an unmethylated sample blank as a negative control, a methylated control, and an unmethylated control (Tiangen Biotech). Then the PCR products were analyzed by 1% agarose gel electrophoresis (DYCP-32B, Liuyi, Beijing China) and photographed using the Tianneng UV system (Tanon-5200 Multi; Tanon Science and Technology Co. Ltd., Shanghai China). There was a clearly visible band in the gel of methylated samples, when using the methylated primers. MSP results were verified by the bisulfite sequencing PCR (BSP) assay, which was performed by a professional sequencing company (Genechem, Shanghai China).
Table 1.
Primers used for MSP and qPCR assay.
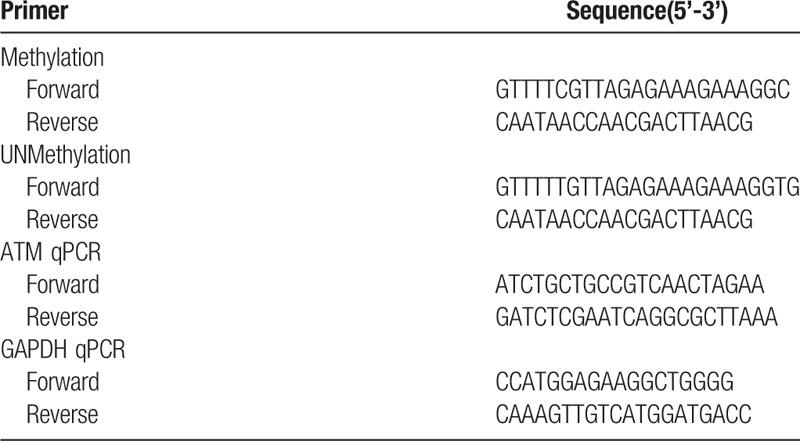
2.4. Total RNA extraction and quantitative PCR assay
Total RNA was extracted from 50 paired samples and 68 liver biopsy samples using a RnEASY mini kit according to the manufacturer's protocol (Qiagen, Beijing China). cDNA was reverse-transcribed by the High-Capacity cDNA Reverse Transcription Kit (Thermo, Shanghai China). Quantitative polymerase reaction (qPCR) was performed using the TaqMan Gene Expression Master Mix (Bio-Rad, Shanghai China) according to the manufacturer's protocol and TaqMan probes for human ATM and human GAPDH (Qingke, Changsha China),which is shown in Table 1.
2.5. Immunohistochemistry
The protein expression of ATM in 68 liver biopsy specimens was determined using the EnVision method. ATM rabbit monoclonal antibody (Bioworld Technology, St. Louis Park, MN) was used at the recommended ratio (dilution 1:200). Tissues were dewaxed in xylene and rehydrated in graded alcohol solutions. Then heat-induced epitope antigen retrieval was performed using 1 L citrate buffer solution (pH 6.0) boiled at full pressure for 3 minutes, 20 seconds. Then slides were incubated overnight with ATM monoclonal antibody (dilution 1:200), washed three times with phosphate-buffered saline (PBS), and incubated with secondary biotinylated anti-rabbit antibody (dilution 1:1000) at room temperature for 1 hour. Slides were washed three times with PBS, stained using DAB solution (Sigma Fast-DAB; Sigma, St. Louis, MO) for 1.5 min, again washed three times with PBS, and counterstained with hematoxylin (ZLI-9609; ZSGB Biotechnology, Beijing, China). Sections were finally dehydrated in serial alcohol solutions and coverslips were applied. Imaging was observed with a microscopy system (BX53 M; Olympus Corporation, Tokyo, Japan). The scoring system reference was mainly determined by the intensity of positive staining, and the proportion of positive cells as follows: 1) Staining intensity was defined as negative (0 point), positive (1 point), and strongly positive (2 points); 2) The percentage of positive cells was defined as 0% (0 point); 1%–25% (1 point); 26%–50% (2 points); 51%–75% (3 points); and 76%–100% (4 points). The positive expression of ATM protein was defined as a product score over 3.
2.6. Radiotherapy program and efficacy evaluation system
All 68 patients with locally advanced hepatocellular carcinoma had not anticancer treatment before radiotherapy. The radiotherapy plan was performed by two high-grade professors from our department, the details were as follows: The gross tumor volume (GTV) defined tumor visible lesion confirmed by imaging. The clinical tumor volume (CTV) defined extending 1.0 cm outside of the GTV. The planned target volume (PTV) defined extending 1.0 cm outside of the CTV, and the plan may be slightly modified because of clinical requirements. All patients accept conformal intensity-modulated radiotherapy (IMRT) (Elekta Compact, Beijing China) and radiotherapy prescription were as follows: a total dose of 60 Gy, 2 Gy per time, 1 time per day, 5 times per week. Blood routine, liver and kidney function were monitored weekly during radiotherapy, and liver protection and symptomatic supportive treatment were concurrently treated.
The evaluation of radiotherapy efficacy was performed by upper abdomen MRI or CT contrast, following the EASL evaluation criteria.[25]
2.7. Statistical analysis
All the statistical analysis were performed using SPSS V21.0 (SPSS Inc, Chicago, IL). A Chi-squared test or fisher's exact test was used to evaluate the difference in ATM promoter methylation frequency between different groups and the comparison of the ATM protein level was analyzed by student's t test. The association between ATM promoter methylation with ATM expression and radiotherapy efficacy were analyzed using Spearman test, which data were treated as binary variables. The overall survival curves were analyzed by Kaplan-Meier analysis and the prognostic value of ATM promoter methylation for locally advanced HCC patients were tested using Log-rank tests and multivariate Cox proportional hazard models.Two tailed P < .05 was considered statistically significant. Figures were drawn using the GraphPad Prism 7 software(GraphPad, San diego, CA).
3. Results
3.1. Methylation status of the ATM promoter in HCC and normal liver tissues
A representative agarose gel electrophoresis image and sequencing results are shown in Figure 1. Partial MSP products are verified by BSP and MSP results are consistent with BSP results. The methylation frequency of the ATM promoter was significantly greater in HCC tissues than in adjacent liver tissues and normal liver tissues (χ2 = 16.830, P < .001). The methylation rate of the ATM promoter in HCC tissues was 39.8% (47/118) compared to only 8.0% (4/50) and 0% (0/20) in adjacent liver tissues and normal liver tissues, respectively. Of those adjacent liver tissues with ATM promoter methylation, methylated ATM was also observed in the paired HCC tissues.
Figure 1.
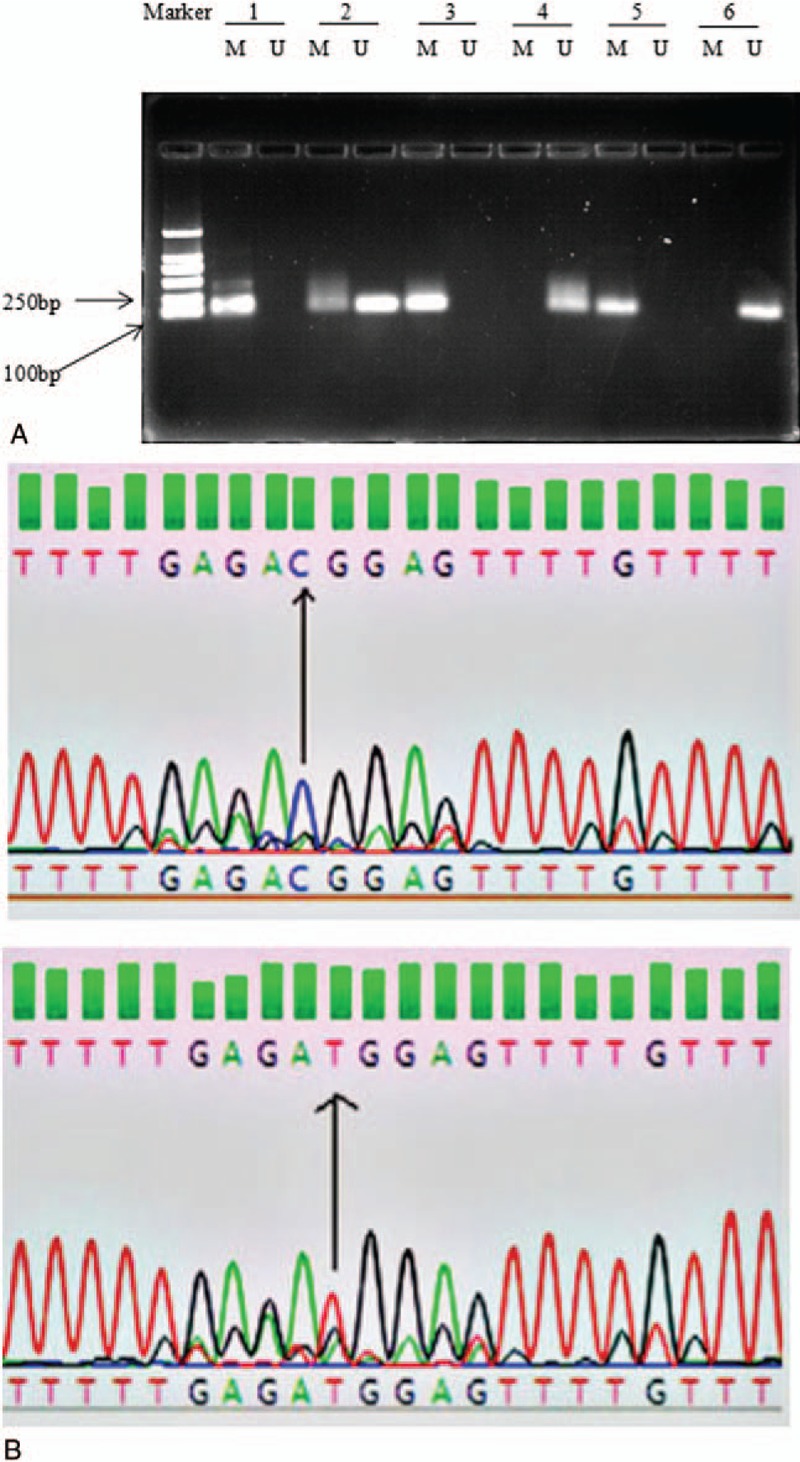
Methylation status of the ATM promoter in several samples. Marker: standard protein; 1–2: HCC surgical tissues; 3: HCC puncture specimens; 4: HCC adjacent liver tissue; 5: methylated control; 6: unmethylated control; M: methylated bands; U: unmethylated bands. BSP results of methylated and unmethylated PCR products. ATM: ataxia telangiectasia mutated gene.
3.2. Association between ATM promoter methylation and expression
The expression of ATM was evaluated using qPCR and IHC assays. Representative results are shown in Figures 2 and 3. The expression of ATM in HCC tissues was downregulated compared with adjacent liver tissues (χ2 = 10.510, P < .001). ATM promoter methylation was observed in 38% (19/50) of these patients, and ATM promoter methylation was correlated with lower ATM expression compared with those without ATM promoter methylation (r = 0.356, P < .001).
Figure 2.
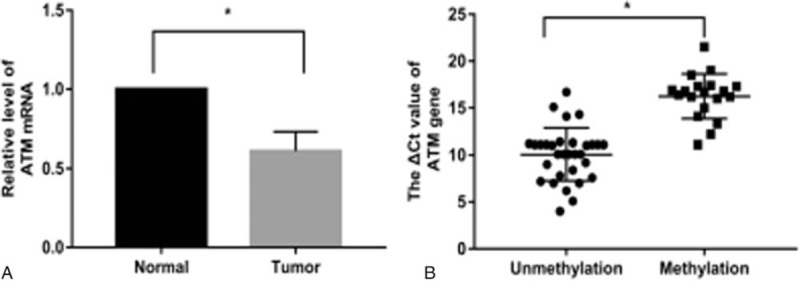
Expression of ATM determined by qPCR. A: Expression levels of ATM mRNA in HCC and adjacent normal liver tissues (n = 50). ATM expression level was significantly lower in HCC tissues than in adjacent liver tissues. B: Expression levels of ATM mRNA in the methylated group (n = 19) and unmethylated group (n = 31). The ATM expression level was significantly lower in the methylated group compared with the unmethylated group.
Figure 3.
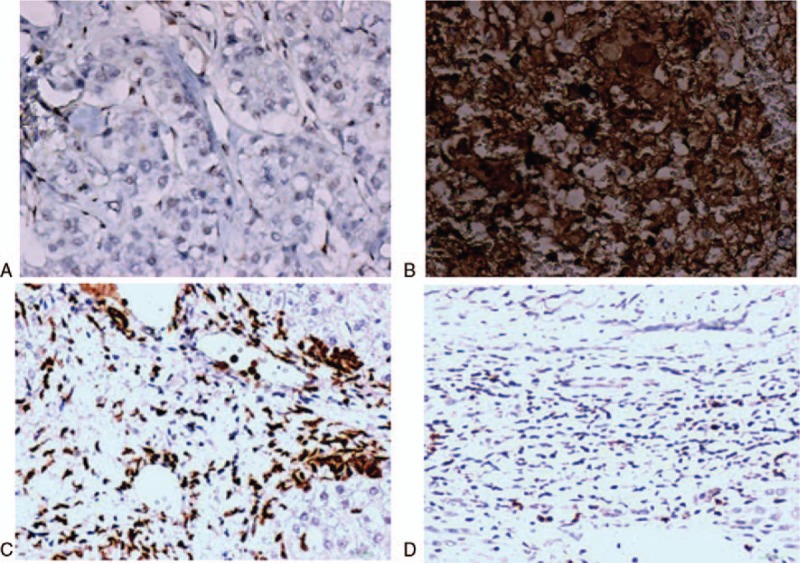
Expression of ATM protein as determined by the IHC assay. A: HCC tissue with ATM promoter methylation, ATM protein expression:(-) B: HCC tissue with ATM promoter unmethylation, ATM protein expression:(+++) C: Adjacent liver tissue, ATM protein expression:(+) D: Normal liver tissue, ATM protein expression:(−).
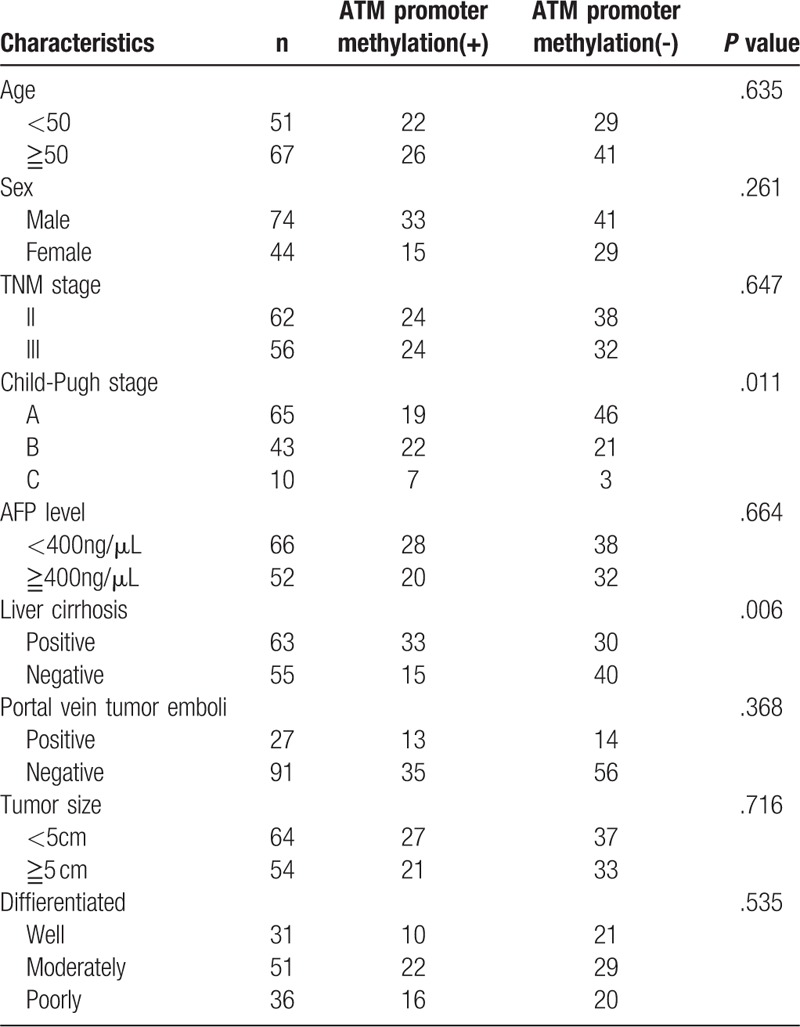
3.3. Association between ATM promoter methylation status and clinicopathological characteristics
The association between ATM promoter methylation status and clinicopathological characteristics of HCC patients including age, sex, TNM stage, Child-Pugh stage, alpha-fetoprotein (AFP) level, liver cirrhosis, portal vein tumor emboli, and tumor size and differentiation were analyzed (Table 2). ATM promoter methylation was correlated with liver cirrhosis and Child-Pugh stage, whereas this phenomenon was not observed between ATM promoter methylation and other parameters.
Table 2.
Association between ATM promoter methylation status and clinicopathological characteristics.
3.4. The correlation between ATM promoter methylation and radiotherapy efficacy in HCC patients
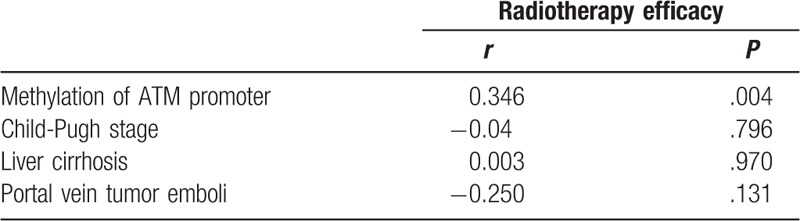
The correlation between ATM promoter methylation as well as Child-Pugh stage, liver cirrhosis, portal vein tumor emboli, and radiotherapy efficacy were analyzed by the Spearman's rank correlation test, which is shown in Table 3. The radiotherapy efficacy in patients with ATM promoter methylation was significantly better than that in those without ATM promoter methylation (χ2 = 8.150, P = .004). Spearman's rank correlation test showed that ATM promoter methylation was significantly correlated with radiotherapy efficacy in HCC patients (r = 0.346, P = .004), and there was no significant correlation between Child-Pugh stage, liver cirrhosis, portal vein tumor emboli, and radiotherapy efficacy (Table 4).
Table 3.
Radiotherapy efficacy statistics in patients with locally advanced HCC and differences in methylation status.
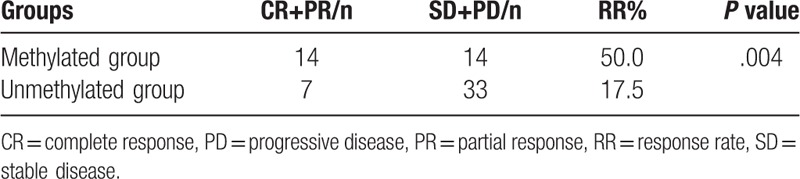
Table 4.
Correlation between ATM promoter methylation and radiotherapy efficacy in HCC patients.
3.5. The predictive value of ATM promoter methylation for radiotherapy outcomes in HCC patients
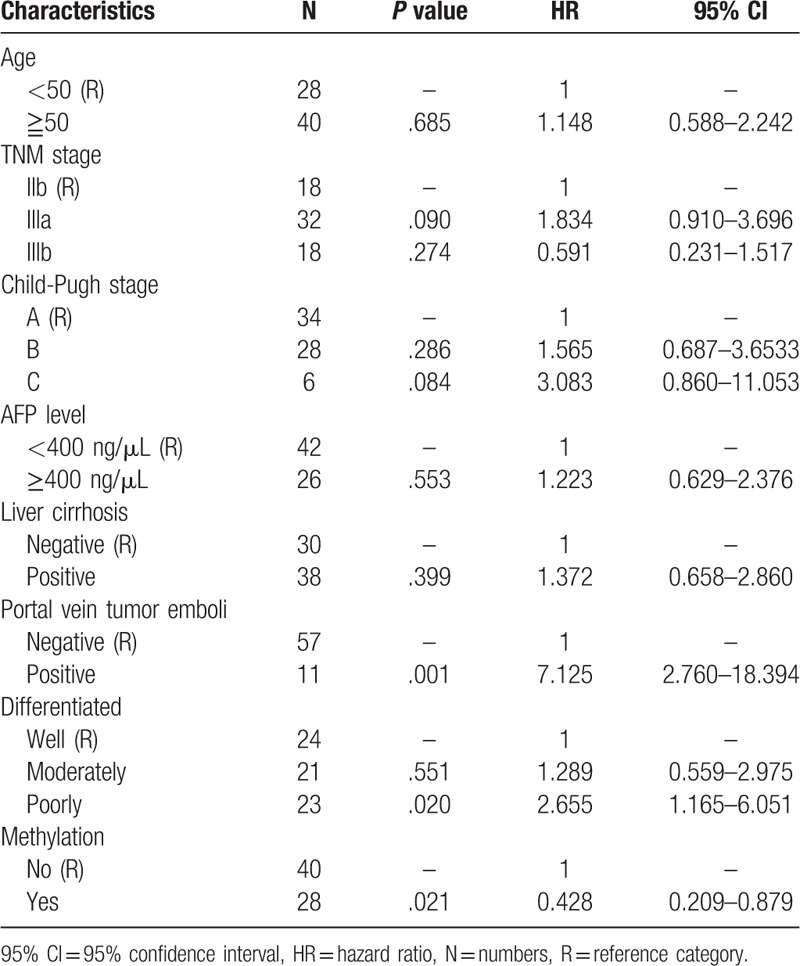
ATM promoter methylation was significantly correlated with radiotherapy outcomes in HCC patients (Fig. 4, Table 5). The survival curve revealed that methylation of the ATM promoter was significantly associated with superior outcomes in patients with locally advanced HCC who initially received radiotherapy (P = .032), and multivariate Cox proportional hazard analysis demonstrated that methylation of the ATM promoter was an independent favorable prognostic factor for locally advanced HCC patients (hazard ratio: 0.428, 95% confidence interval: 0.209–0.879, P = .021).
Figure 4.
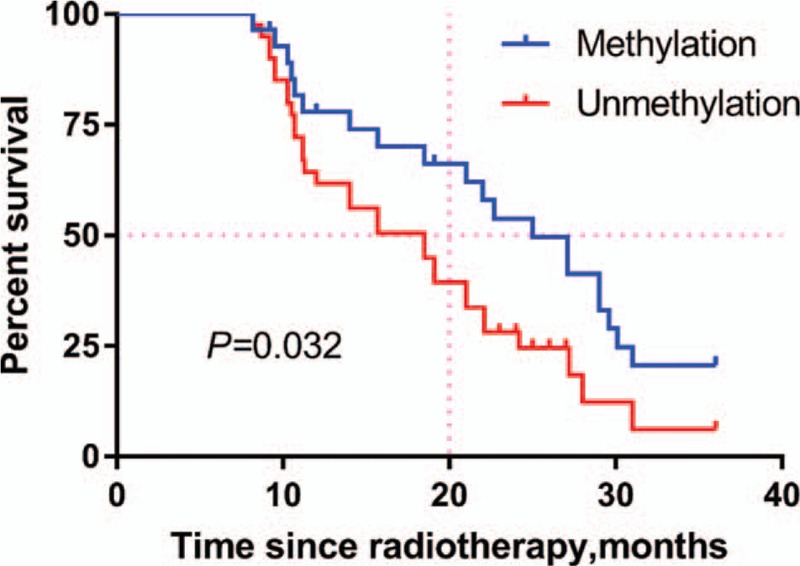
Survival curve of patient groups according to methylation status of ATM promoter. The ATM methylated group showed significantly superior survival rates compared to the ATM unmethylated group (P = .032).
Table 5.
Multivariate Cox proportional hazard analysis in 68 locally advanced HCC patients.
4. Discussion
Radiotherapy is one of the major treatments for hepatocellular carcinoma (HCC), especially advanced HCC, which can improve long-term survival when combined with other treatments, especially immunotherapy.[26] DNA methylation, one of the well-known epigenetic changes, has silencing effect on gene expression through multiple mechanisms. Methylation of tumor-suppressor genes can contribute to identifying biomakers that could serve as sensitive and specific markers for cancer diagnosis and prognosis.[27,28] ATM is a DNA damage repair gene that regulates the DNA damage response and cell survival, proliferation, metabolism, and differentiation.[6]
In previous studies, the aberrant methylation of ATM promoter has been found in many tumor types, serving as a promising biomarker for cancer detection and correlating with increased radiosensitivity.[7,9,27] To the best of our knowledge, this is the first study to detect the methylation status of ATM promoter in HCC tissue samples and normal liver tissue samples. Furthermore, ATM mRNA and protein expression, and the relationship between ATM promoter methylation and clinical characteristics and radiotherapy response in HCC patients were analyzed. It was found that the ATM promoter was partly methylated in HCC tissues but rarely methylated in normal liver tissues, consistent with previous reports.[15–17,29] Interestingly, ATM promoter methylation was more frequently observed in advanced Child-Pugh stages and in HCC patients with liver cirrhosis, indicating that the ATM promoter may be methylated in the early stage of HCC. Furthermore, ATM expression is significantly lower in HCC tissues than in adjacent liver tissues and methylation of the ATM promoter is associated with lower ATM expression in HCC compared with unmethylated ATM. These results suggest that ATM promoter methylation is targeted to special regions of the ATM genome and is regulated by methyl-CpG-binding proteins, which contributes to inhibiting transcriptional activity.[30] Thus, the downregulation of ATM may contribute to the methylation of promoter regions, which has been demonstrated in other types of cancer.[17,18,20,29]
ATM protein kinase is one of the central kinases involved in the cellular response to DNA double-strand breaks, which may arise intrinsically through the collapse of stalled replication forks or extrinsically through exposure to ionizing radiation.[6,22,31] The lower the ATM protein expression, the more sensitive to individuals are to radiotherapy. Several reports have shown that methylation of the ATM promoter correlates with higher radiosensitivity compared to unmethylated ATM, which may result from epigenetic silencing.[7,9,10] In this study, the phenomenon presented above was also observed, as radiotherapy efficacy in patients with ATM promoter methylation was significantly better than that in patients without ATM promoter methylation. ATM promoter methylation was significantly correlated with radiotherapy efficacy in patients with locally advanced HCC. Furthermore, it was strongly associated with superior 3-year overall survival (OS) in patients with locally advanced HCC who initially received radiotherapy, as the OS in these patients without methylated ATM was statistically lower than that in patients with methylated ATM, although other anti-cancer treatments such as transcatheter arterial chemoembolization, chemotherapy, and small molecule targeted therapy may be performed after radiotherapy. Considering the contribution of several factors (e.g., age, TNM stage, Child-Pugh stage, AFP level, liver cirrhosis, portal vein tumor emboli, histological differentiation, and ATM promoter methylation) to OS, multivariate Cox proportional hazard analysis was used to adjust for these factors, and the results confirmed that methylation of ATM promoter was an independent favorable prognostic factor for locally advanced HCC patients who initially received radiotherapy, which needs to be confirmed by multicenter clinical trials. We also found that diagnostic value of ATM promoter methylation for HCC, as determined by the receiver operating curve (data not shown), which has better diagnostic accuracy when combined with other tumor biomarkers. The results of our study should be verified in large Phase III clinical trials.
In conclusion, for the first time this study showed that the methylation status of ATM promoter in HCC has predictive value for radiotherapy outcome. We demonstrated that the ATM promoter is partly methylated in HCC, contributing to its transcriptional inactivation, and may be involved in the early stage of HCC. These results strongly indicate that methylation of the ATM promoter is a potential epigenetic biomarker for radiotherapy outcomes in HCC. Additional studies are needed to demonstrate the mechanisms underlying ATM promoter methylation in the pathogenesis of HCC.
Author contributions
Conceptualization: Xiaoping Yang, Gaofeng Li.
Data curation: Xiaoping Yang, Gaofeng Li.
Formal analysis: Xiaoping Yang, Gaofeng Li.
Funding acquisition: Xiaoping Yang, Gaofeng Li.
Investigation: Tianyu Wu, Mei Tang, Dongliang Chen, Meiyuan Huang.
Methodology: Tianyu Wu, Mei Tang, Dongliang Chen, Meiyuan Huang.
Project administration: Tianyu Wu, Sichun Zhou.
Resources: Xinjian Yan, Tianyu Wu, Mei Tang, Sichun Zhou, Huihui Zhang.
Software: Xinjian Yan, Tianyu Wu, Sichun Zhou, Huihui Zhang.
Supervision: Sichun Zhou, Huihui Zhang.
Validation: Sichun Zhou, Huihui Zhang.
Visualization: Sichun Zhou, Huihui Zhang.
Writing – original draft: Xinjian Yan.
Writing – review & editing: Xinjian Yan, Xiaoping Yang.
Footnotes
Abbreviations: ATM = ataxia telangiectasia mutated gene, BSP = bisulfite sequencing PCR, CR = complete response, DDR = DNA damage response, EASL criteria = European Association for the Study of Liver criteria, HCC = hepatocullular carcinoma, IHC = imunohistochemistry, MSP = methylation-specific polymerase chain reaction, PD = progressive disease, PR = partial response, RR = response rate, RT-qPCR = quantitative real-time PCR, SD = stable disease.
How to cite this article: Yan X, Wu T, Tang M, Chen D, Huang M, Zhou S, Zhang H, Yang X, Li G. Methylation of the Ataxia Telangiectasia Mutated Gene(ATM) Promoter as a Radiotherapy Outcome Biomarker in Patients with Hepatocellular Carcinoma. Medicine. 2020;99:4(e18823).
The work was supported by grants from the Hunan Provincial Natural Science Foundation of China (No. 2018JJ4097), the Start-up Funds of the Key Laboratory of Study and Discovery of Targeted Small Molecules of Hunan Province (2017TP020).
The authors declare that they have no conflicts of interest.
References
- [1].Yu WB, Rao A, Vu V, et al. Management of centrally located hepatocellular carcinoma: Update 2016. World J Hepatol 2017;9:627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chu KK, Cheung TT. Update in management of hepatocellular carcinoma in Eastern population. World J Hepatol 2015;7:1562–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hawkins MA, Dawson LA. Radiation therapy for hepatocellular carcinoma: from palliation to cure. Cancer 2006;106:1653–63. [DOI] [PubMed] [Google Scholar]
- [4].Borchiellini D, Etienne-Grimaldi MC, Thariat J, et al. The impact of pharmacogenetics on radiation therapy outcome in cancer patients. A focus on DNA damage response genes. Cancer Treat Rev 2012;38:737–59. [DOI] [PubMed] [Google Scholar]
- [5].Kuefner MA, Brand M, Engert C, et al. Radiation induced DNA double-strand breaks in radiology. Rofo 2015;187:872–8. [DOI] [PubMed] [Google Scholar]
- [6].Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther 2015;149:124–38. [DOI] [PubMed] [Google Scholar]
- [7].Kim WJ, Vo QN, Shrivastav M, et al. Aberrant methylation of the ATM promoter correlates with increased radiosensitivity in a human colorectal tumor cell line. Oncogene 2002;21:3864–71. [DOI] [PubMed] [Google Scholar]
- [8].Vo QN, Kim WJ, Cvitanovic L, et al. The ATM gene is a target for epigenetic silencing in locally advanced breast cancer. Oncogene 2004;23:9432–7. [DOI] [PubMed] [Google Scholar]
- [9].Roy K, Wang L, Makrigiorgos GM, et al. Methylation of the ATM promoter in glioma cells alters ionizing radiation sensitivity. Biochem Biophys Res Commun 2006;344:821–6. [DOI] [PubMed] [Google Scholar]
- [10].Ren J, Chu Y, Ma H, et al. Epigenetic interventions increase the radiation sensitivity of cancer cells. Curr Pharm Des 2014;20:1857–65. [DOI] [PubMed] [Google Scholar]
- [11].Ribeiro IP, Caramelo F, Esteves L, et al. Genomic and epigenetic signatures associated with survival rate in oral squamous cell carcinoma patients. J Cancer 2018;9:1885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ai L, Vo QN, Zuo C, et al. Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiol Biomarkers Prev 2004;13:150–6. [DOI] [PubMed] [Google Scholar]
- [13].Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006;31:89–97. [DOI] [PubMed] [Google Scholar]
- [14].Flanagan JM, Munoz-Alegre M, Henderson S, et al. Gene-body hypermethylation of ATM in peripheral blood DNA of bilateral breast cancer patients. Hum Mol Genet 2009;18:1332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brennan K, Garcia-Closas M, Orr N, et al. Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Res 2012;72:2304–13. [DOI] [PubMed] [Google Scholar]
- [16].Rengucci C, De Maio G, Casadei Gardini A, et al. Promoter methylation of tumor suppressor genes in pre-neoplastic lesions; potential marker of disease recurrence. J Exp Clin Cancer Res 2014;33:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Delmonico L, Moreira Ados S, Franco MF, et al. CDKN2A (p14(ARF)/p16(INK4a)) and ATM promoter methylation in patients with impalpable breast lesions. Hum Pathol 2015;46:1540–7. [DOI] [PubMed] [Google Scholar]
- [18].Begam N, Jamil K, Raju SG. Promoter Hypermethylation of the ATM Gene as a Novel Biomarker for Breast Cancer. Asian Pac J Cancer Prev 2017;18:3003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu J, Li H, Sun L, et al. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell Int 2017;17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hirakawa T, Nasu K, Aoyagi Y, et al. ATM expression is attenuated by promoter hypermethylation in human ovarian endometriotic stromal cells. Mol Hum Reprod 2019;25:295–304. [DOI] [PubMed] [Google Scholar]
- [21].Li Y, Li L, Wu Z, et al. Silencing of ATM expression by siRNA technique contributes to glioma stem cell radiosensitivity in vitro and in vivo. Oncol Rep 2017;38:325–35. [DOI] [PubMed] [Google Scholar]
- [22].Hennequin C, Quero L, Favaudon V. DNA repair and tumour radiosensitivity: focus on ATM gene. Bull Cancer 2011;98:239–46. [DOI] [PubMed] [Google Scholar]
- [23].van Eijk KR, de Jong S, Boks MP, et al. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics 2012;13:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fuso A, Ferraguti G, Scarpa S, et al. Disclosing bias in bisulfite assay: MethPrimers underestimate high DNA methylation. PLoS One 2015;10:e0118318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vincenzi B, Di Maio M, Silletta M, et al. Prognostic relevance of objective response according to EASL criteria and mRECIST criteria in hepatocellular carcinoma patients treated with loco-regional therapies: a literature-based meta-analysis. PLoS One 2015;10:e0133488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bruix J, Sherman M. American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dong Y, Zhao H, Li H, et al. DNA methylation as an early diagnostic marker of cancer (Review). Biomed Rep 2014;2:326–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schmitz M, Eichelkraut K, Schmidt D, et al. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer 2018;18:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mehdipour P, Karami F, Javan F, et al. Linking ATM promoter methylation to cell cycle protein expression in brain tumor patients: cellular molecular triangle correlation in ATM territory. Mol Neurobiol 2015;52:293–302. [DOI] [PubMed] [Google Scholar]
- [30].Schubeler D. Function and information content of DNA methylation. Nature 2015;517:321–6. [DOI] [PubMed] [Google Scholar]
- [31].Hammond EM, Muschel RJ. Radiation and ATM inhibition: the heart of the matter. J Clin Invest 2014;124:3289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]


