Abstract
The transcription factor MafB regulates macrophage differentiation. However, studies on the phenotype of Mafb-deficient macrophages are still limited. Recently, it was shown that the specific expression of MafB permits macrophages to be distinguished from dendritic cells. In addition, MafB has been reported to be involved in various diseases related to macrophages. Studies using macrophage-specific Mafb-deficient mice show that MafB is linked to atherosclerosis, autoimmunity, obesity, and ischemic stroke, all of which exhibit macrophage abnormality. Therefore, MafB is hypothesized to be indispensable for the regulation of macrophages to maintain systemic homeostasis and may serve as an innovative target for treating macrophage-related diseases.
Keywords: homeostasis, large Maf family, macrophage, MafB, transcription factor
Introduction
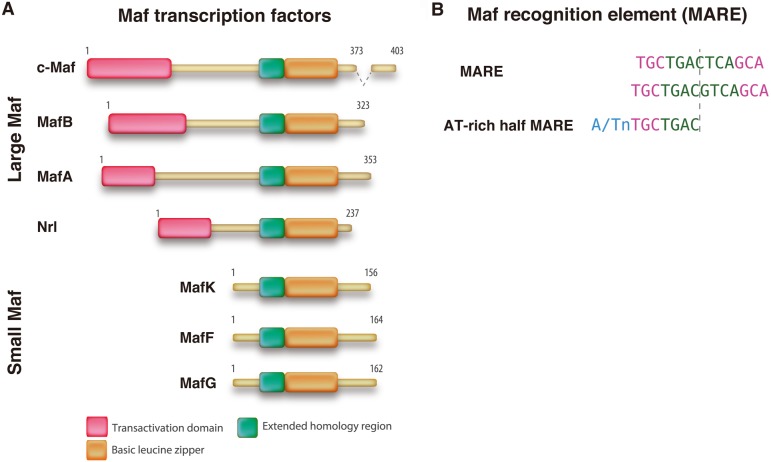
The Maf transcription factor family is a homolog of the v-Maf oncogene, which was identified from the AS42 virus and causes musculoaponeurotic fibrosarcoma in chicks [58]. The Maf transcription factor is separated into two subfamilies. Large Maf family proteins have a basic domain and a leucine zipper domain (b-Zip) that mediate dimerization and DNA binding and an acidic domain to regulate target gene transcription [32]. In contrast, the small Maf family only contains a b-Zip domain and lacks an acidic domain. Homodimers of small Mafs inhibit target gene transcription, whereas heterodimers regulate target gene transcription depending on the partner protein (Fig. 1) [28]. In mammals, four large Maf proteins (MafA, MafB, c-Maf, and NRL) and three small Maf proteins (MafK, MafG, and MafF) have been identified. All Maf proteins bind to the Maf recognition element (MARE), which is a palindromic sequence, or a MARE half-site in the promoter region of their target gene (Fig. 1b) [32, 80].
Fig. 1.
Structural features of Maf proteins. A. Schematic diagrams of four large Maf and three small Maf protein structures. Their amino acid lengths are indicated on the right side. The small Maf proteins are essentially composed of a carboxy-terminal basic leucine zipper (b-Zip) domain and a DNA-binding domain, whereas the large Maf proteins contain an additional transactivation domain. B. Maf proteins bind as homodimers to two types of target sequences, the classical Maf recognition element (MARE) site and the 5’-AT-rich MARE half-site.
The expression of large Maf transcription factors has been observed in various tissues. To date, studies using animal models have explained the function of large Mafs in tissue development and cellular differentiation. Expression of MafA and NRL in specific cells, pancreatic β cells and retinal cells, respectively, has been observed, while c-Maf and MafB are expressed in diverse cell types [49, 82]. c-Maf regulates the development of lens fiber cells, mechanoreceptors, T cells, natural killer (NK) T cells, and macrophages [34, 76]. MafB is also expressed in pancreatic α cells, renal podocytes, epidermal keratinocytes, hair follicles, and hematopoietic stem cells and functions in embryonic urethral formation [33, 51,52,53, 65, 71]. Importantly, large Mafs have different target genes depending on the cell type they are expressed in. For instance, c-Maf regulates the interleukin (IL) 4 (IL4) gene in helper T cells, the IL10 gene in macrophages, and crystallin genes in lens fiber cells [7, 34, 38]. MafB also regulates glucagon gene expression in pancreatic α cells, the PTH gene in the parathyroid gland, and C1q genes in macrophages [30, 33, 74]. Thus, this indicates that large Mafs regulate critical genes that define their cell types. Although large Mafs are indispensable for tissue development and cell differentiation, the ectopic expression of large Mafs has an oncogenic function that induces several types of cancer, such as multiple myeloma [5]. In addition, human diseases caused by a point missense mutation in large Mafs have been reported. Mutations in c-MAF cause cataracts [60] and abnormalities in Pacinian corpuscles [76], mutations in MAFA cause insulinomatosis or diabetes mellitus [27], and mutations in NRL induce enhanced S-cone syndrome [45] and retinitis pigmentosa [49]. Mutations in MAFB also cause multicentric carpotarsal osteolysis (MCTO) and Duane retraction syndrome (DRS) with focal segmental glomerulosclerosis (FSGS) [66, 81]. In this way, an abnormal function in a large Maf is responsible for disease onset in various tissues or cell types. Currently, there are limited treatments for most diseases caused by large Mafs, since the precise functions of large Mafs remain unknown. In particular, the functions of c-Maf and MafB in adulthood are unclear due to the embryonic lethality observed in deficient mice. Therefore, further analysis by using conditional knockout mice is required.
Macrophages exist in all tissues and have multiple functions, such as the clearance of cell debris and waste products, the maintenance of homeostasis, tissue repair, and innate and adaptive immunity. In various diseases, macrophages are treated as the therapeutic target because of their contribution to disease progression or repair [78]. Although an in vitro analysis was able to categorize macrophages into two subtypes, classical activated macrophages (M1 type) and alternative activated macrophages (M2 type), it seems that this arrangement is not truly applicable in vivo [46, 56]. Considering the multidimensional regulators of macrophages, including ontogeny of the yolk sac and definitive hematopoiesis, disease-derived cytokine signals, and fatty acid and cholesterol signals, it is difficult to understand the whole picture of macrophage functions [18].
In 2000, Kelly et al. reported that the overexpression of MafB induced macrophage differentiation from chick myeloblasts [35]. This finding led us to hypothesize in early 2000 that Mafb deficiency reduces the number of macrophages; thus, we generated Mafb-deficient mice to confirm the relationship between macrophage differentiation and MafB. Although our group and others analyzed the functions of MafB in macrophages using Mafb-deficient mice in 2006, the number of macrophages was not impaired in MafB-null mice, making its phenotype unclear [4, 53]. However, a recent study we performed showed that the functions of MafB are indispensable in disease-related macrophages [22, 69, 74]. In addition, recent evidence indicates that the expression of MafB is also observed in anti-inflammatory-type macrophages. In this review, we summarize the expression and homeostatic functions of MafB in macrophages related to disease development in humans and mice. Moreover, we report that recent evidence indicates that the functions of MafB in macrophages may allow it to serve as a new drug target for diseases related to macrophage accumulation or infiltration; therefore, a complete understanding of MafB in macrophages may help complete the puzzle of macrophage functions.
MafB is Specifically Expressed in Macrophages
In 1994, the Mafb gene was first identified as the causative gene of behavioral defects from impaired inner ear development in Kreisler mice [10]. Kataoka et al. also identified Mafb as a homolog of the oncogene v-Maf in chickens [31]. Sieweke et al. used cell lines and and peritoneal macrophages to show that, out of all the cells in the hematopoietic system, MafB is specifically expressed in macrophages [70]. In the 2000s, transgenic mice with the Mafb promoter labeled with green fluorescent protein (GFP) and mice with GFP knocked-in in the Mafb locus were generated by our group. GFP fluorescence was observed in both CD11b-positive and F4/80 (a macrophage marker)-positive cells within the fetal liver, bone marrow, and peripheral blood [21, 53].
Recent evidence has also validated the expression of Mafb in macrophages. A transcriptome analysis using subtypes of tissue macrophages and dendritic cells (DCs) showed that DCs and alveolar macrophages had lower Mafb expression, while Mafb was strongly expressed in splenic macrophages [15]. Originally, it was hypothesized that DCs phagocytose in a similar manner to that of macrophages and that they share common marker genes, making it difficult to distinguish DCs from macrophages [25]. Under normal conditions, MafB is one of the macrophage markers that is reduced in DCs, being a suitable signal to distinguish DCs and macrophages [19, 67]. In addition, macrophages, but not DCs, within tumors also express MafB [14]. MafB lineage-tracing mice with mCherry-Cre knocked-in in the Mafb locus were crossed with Rosa26-stop-YFP mice to identify the cell type that specifically expresses MafB in vivo. YFP expression clearly distinguished macrophage and other cell lineages, including DCs [77]. In human tissue, MAFB is expressed in macrophages in the skin and colon and in melanoma tumors [11]. MAFB expression has also been observed in CD16-positive human monocyte-derived macrophages [26]. Given these results, MafB is a specific macrophage marker in the myeloid lineage and can be used as a substantial marker that distinguishes DCs and macrophages. Genome-wide assays, such as RNA-seq and chromatin immunoprecipitation-sequencing (ChIP-seq) for transposase-accessible chromatin (ATAC-seq) using macrophages, monocytes and neutrophils, have shown that MafB targets the maf-binding motifs in promoter or enhancer regions specific to macrophage genes [43]. This data suggests that large Mafs play a role in macrophages after differentiating from monocytes. In summary, MafB expression is distinctive in macrophages and could be used as an ideal marker for high-throughput analysis.
Expression of MafB in Disease-related Macrophages
Macrophages exist as tissue-resident macrophages in various tissues. However, under abnormal conditions, such as wounds or infection, tissue macrophages induce inflammation, or monocytes infiltrate to the inflammatory region from peripheral blood. Previous reports have shown that MafB is expressed in specific disease-related macrophages or infiltrating macrophages. MafB expression has been observed in infiltrating macrophages at the site of bacterial infections from peripheral blood [2]. In myocardial infarction models, MafB expression was also observed in monocyte-derived MHC-II-positive macrophages [13]. Moreover, MafB is expressed in tumor-associated macrophages in breast cancer [14] and in atherosclerotic regions but not in DCs [8, 9, 22]. In the hind limb ischemia model, MafB was detected in the macrophages that promote angiogenesis, and in renal artery stenosis models, MafB was observed in kidney-resident macrophages that inhibit stenotic kidney injury [42, 61]. In lung fibrosis induced by bleomycin, profibrotic macrophages express MafB [1].
In the brain, MafB is also expressed in tissue-resident microglia in the central nervous system, where a recent report showed that MafB is expressed in activated microglia after peripheral nerve injury of the spinal cord [41, 47, 64]. In human patients with peritoneal dialysis, CD14-positive peritoneal macrophages express MafB [44]. In an in vitro analysis, we showed that MafB is specifically expressed in M-CSF-, IL-10-, and IL-4-derived M2-type macrophages [12]. These reports on in vitro and in vivo evidence of Mafb expression suggest that MafB may play a role in tissue repair or anti-inflammation in various macrophage-related diseases.
The Phenotype of Mafb-deficient Macrophages and MafB Target Genes
As described above, accumulating evidence indicates that MafB is specifically expressed in macrophages in many conditions. However, there have been only a few studies using Mafb-deficient mice. In 2006, our group and others published about Mafb-deficient macrophages simultaneously. In these studies, the development and number of macrophages were similar to those of wild-type mice under in vivo conditions [4, 53]. In contrast, our in vitro analysis showed that F4/80 expression was strongly reduced in Mafb-deficient macrophages using a nonadherent culture dish [53]. In addition, enhanced actin organization was observed in Mafb-deficient macrophages in vitro [4].
In 2009, it was identified that macrophages with double knockout (DKO) of MafB and c-Maf have self-renewal ability, even in their terminal differentiation because of the increase in c-Myc and Klf4 in the presence of M-CSF [3]. This report was indeed surprising, yet a phagocytotic analysis using bacteria showed that Mafb and c-Maf DKO macrophages were not different from the control, raising the question of whether other functions of Mafb and c-Maf DKO macrophages are normal or not. In fact, Mafb-deficient macrophages under normal conditions do not have a clear phenotype.
An analysis of various diseases using Mafb-deficient mice specifically in hematopoietic cells was recently reported. In atherosclerotic lesions, macrophages uptake oxidized low-density lipoprotein and differentiate into foam cells. To investigate the functions of MafB under these conditions, we transplanted Mafb−/− or control fetal liver cells of embryonic day 14.5 mice into X-ray-irradiated LDL receptor KO mice (Ldlr−/−) and then fed them with high-cholesterol diet to induce atherosclerosis. Mafb-deficient transplanted mice exhibited a reduced atherosclerotic lesion area compared with that of the control. Detailed analysis showed that the expression of apoptosis inhibitor of macrophage (AIM) was strongly reduced in foam cells within the atherosclerotic lesions of Mafb−/− mice. This finding explains why Mafb−/− foam cells induce apoptosis and reduce the area of atherosclerotic lesions. Thus, MafB promotes atherosclerosis by regulating foam cell apoptosis [22]. Moreover, we investigated whether macrophage-specific Mafb-deficient mice were obese after feeding them a high-fat diet. Under these conditions, the size and weight of white adipose tissue increased, and AIM inhibited fat synthesis after uptake by adipocytes. As a reduction in AIM expression was observed in the adipose tissue macrophages of Mafb-deficient mice, AIM reduction may be the cause of obesity in Mafb-deficient mice [73]. Another recent report demonstrated that after inducing ischemic stroke, macrophage-specific Mafb-deficient mice (Mafbf/f::LysM-Cre) showed excessive inflammation. Because MafB regulates mouse scavenger receptor 1 (MSR1) and macrophage receptor with collagenous structure (MARCO), which clears damage-associated molecular patterns (DAMPs), Mafb-deficient macrophages could not inhibit inflammation in the infarct area [69]. Our group also investigated whether MafB is important for efferocytosis (the clearance of apoptotic cells). One million apoptotic cells per second are produced in the human body; however, macrophages immediately engulf apoptotic cells to inhibit autoimmune disease. In our study, Mafb-deficient macrophages could not engulf apoptotic cells due to the reduction in the C1qa, C1qb, and C1qc genes that compose the complement component C1q complex. Luciferase assay and ChIP assay results indicated that MafB directly regulates all C1q genes, since the promoter regions of C1qa, C1qb, and C1qc contain a MARE site. C1q is the first protein involved in the complement component classical pathway, and its deficiency causes autoimmune disease in both humans and mice [6, 68]. Consistent with this evidence, Mafb-deficient mice, under conditions in which apoptotic cells accumulate as a result of X-ray irradiation, exhibited increased autoantibodies and glomerulonephritis. In addition, C1q genes were specifically expressed in macrophages, while other complement components were expressed in hepatocytes. Western blot analysis and a hemolysis assay using serum from macrophage-specific Mafb-deficient (Mafbf/f::LysM-Cre) mice showed that the amount and activity of C1q were drastically reduced [74]. These data indicate that MafB is indispensable for the regulation of complement component classical pathways. Another group also reported that single-cell RNA sequencing of microglia from Mafb conditional KO mice crossed with Csf1r-Cre transgenic mice showed an increase in the genes related to inflammation and viral infection [47].
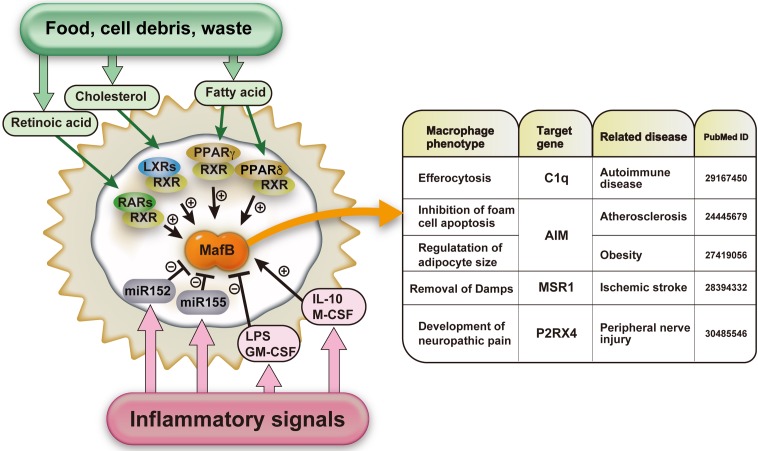
Given these findings, we identified the direct target genes of MafB, including F4/80, AIM, C1qa, C1qb, C1qc, and MSR1, by using Mafbf/f::LysM-Cre mice (Fig. 2) [22, 53, 69, 74]. Most of these genes have distinct roles in macrophage function. In addition to macrophages, Maf transcription factors also directly regulate genes that define the identity of other cell types, such as insulin in β cells, glucagon in alpha cells, crystallin in lens fiber cells, PTH in the parathyroid gland, Il-4 in T helper cells, and rhodopsin in rod cells [30, 33, 34, 38, 49, 82]. Consequently, if the target genes of MafB in macrophages are found, the unknown functions of macrophages can be clarified. Further analyses using various disease-related macrophages of Mafb-deficient mice would open the next door of macrophage research.
Fig. 2.
Schematic model of the major regulatory factors of MafB induced by an external stimulus. In macrophages, MafB is regulated by food, cell debris, and waste via the regulation of nuclear receptor transcription factors, and it is also controlled by inflammatory signals such as microRNAs or cytokines (left panel). The right table shows macrophage phenotypes and target genes in macrophages involved in several diseases.
Because MafB targets a specific gene, we identified a common point between the target genes, the large Mafs, and evolution. The sea lamprey, which belongs to the jawless vertebrates, has one large Maf. All fishes other than the sea lamprey have four large Mafs, which seem to increase through genome modifications such as gene duplication [37]. In addition to large Mafs, the sea lamprey has immature marco, msr1 (sr-a), AIM, and C1q-like genes in its genome [48, 64, 79]. Fishes that evolved after the lamprey have acquired their gene functions as their innate immunity has evolved. Consistent with these results, it was reported that the phagocytes of the sea lamprey have less protection against microbes, as protection against microbes was acquired after fishes that evolved after the lamprey, and heterogeneity to distinguish the proinflammatory type and maintain the homeostatic type [23]. Thus, acquiring the polarity of macrophages may be related to the timing of the gene duplication of large Mafs. In addition, MARE sequences in the promoter or enhancer region of target genes must be important. The comparison of not only target genes but also promoter or enhancer regions may be important for understanding the mechanism of macrophage evolution.
Regulators of MafB Expression
MafB functions in various tissues and stages of development. However, it is critical for it to be expressed at the right time and location depending on the physiological condition. Retinoic acid is the ligand of the nuclear receptor-type transcription factor retinoic acid receptor (RAR). An analysis of the rhombomere revealed the importance of the concentration of retinoic acid in regulation of MafB, which receives signals from RAR [20]. Moreover, recent studies have illustrated the importance of several nuclear receptor-type transcription factors, especially in macrophages. Macrophages at atherosclerotic lesions are reported to take in oxLDL, which leads to the production of oxysterol the ligand of liver X receptor (LXR). In turn, a heterodimer of LXR and retinoid X receptor (RXR) activated with oxysterol regulates the expression of MafB. In fact, LXR-deficient macrophages also exhibit reduced MafB expression [22]. In addition, the MafB target gene C1q is activated by nuclear receptors such as PPARδ, PPARγ and RXR [54, 62]. A study we performed further proved that PPARδ, LXR, RAR, and GR regulate C1q by increasing the expression of MafB [74]. Furthermore, RXR homodimers are involved in osteoclast differentiation through regulation of the expression of MafB [50]. MafB is also upregulated by certain cytokine signals, including M-CSF, IL-10, IL-4, and IL-13, and downregulated by GM-CSF and LPS [11, 12, 17]. In humans, vitamin D3/Hox-A10 regulates MafB expression during the differentiation of CD34+ cells to monocytes [16]. In the sense of monocyte and macrophage differentiation, transcription factor for immunoglobulin heavy-chain enhancer 3 (TFE3) binds to the human MAFB promoter region, and retinoic acid and TNF-alpha regulate the expression of MafB in the human monocyte cell line, THP-1 [83]. MafB is a target of miR-155, and miR-155 induces inflammatory macrophages [29, 36]. In contrast, miR-152 is reported to target MafB in microglia [72]. MafB is similarly regulated by various signals, including lipid metabolism, immunological regulators, and microRNAs (Fig. 2). Overall, these reports indicate the importance of MafB in various functions that maintain the homeostasis of macrophages.
Pathologies
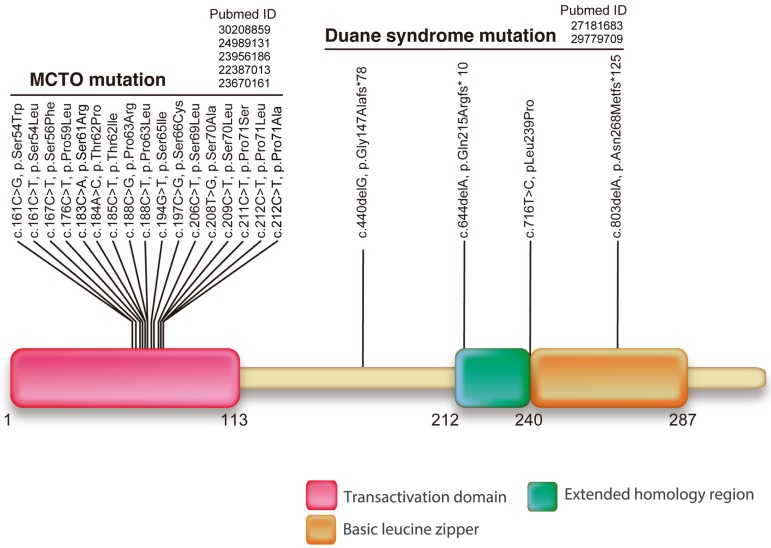
There have been a few studies regarding pathologies that are caused by a missense mutation at the transactivation domain of the Mafb gene. MCTO is a rare skeletal and nephropathic disorder, with its classical symptoms appearing as specific osteolysis at the carpal and tarsal bones [55, 81]. Since all carpal bones are present in MCTO patients during childhood and osteolysis progressively occurs as patients age, MCTO is hypothesized to be caused by excessive bone resorption [40, 84]. The expression of MafB reported by Kim et al. in 2007 also supports this hypothesis, revealing the need to analyze the link between MCTO and the functions of MafB in osteoclasts [39]. Moreover, the missense mutation in the transactivation domain of MafB is thought to interact with the functions of MafB as a transcription factor. However, studies on several point mutations in this domain of MAF proteins have shown that these mutations are clustered at the phosphorylation sites targeted by GSK3, which in turn affect the stability of MafB itself (Fig. 3) [24, 57]. Our group generated homozygous mice harboring the MCTO mutation that exhibited nephropathy similar to that of human MCTO patients [75]. Future work will focus on the bone and macrophage phenotypes of these mice to elucidate the potential risks and ideal therapeutic approaches against MCTO.
Fig. 3.
Domain structure and the location of mutations in the human MAFB protein. Each position indicates the amino acid substitution of the observed mutations in patients where mutations were previously reported for multicentric carpotarsal osteolysis (MCTO) and Duane syndrome. The osteolysis disorder MCTO is known to be caused by a dominant missense mutation in the transactivation domain. A mutation in the zinc finger DNA-binding domain of MafB may also cause DRS.
DRS is another disorder related to a mutation in MafB that differs from MCTO at the site of the mutation along the MafB gene [59]. DRS is caused by a mutation in the DNA-binding domain of MafB, with patients suffering from defects in the abducens nerves, FSGS, and dysacusis but exhibiting no major abnormalities in hematological signatures (Fig. 3) [59, 66]. Interestingly, homozygous mice with the Duane syndrome mutation and ENU-induced DNA-binding domain-mutated mice are incapable of surviving soon after birth [63]. Since this lethal phenotype is similar to that of Mafb-deficient mice, mutations in the DNA-binding domain may reflect the loss of MafB function.
It is possible that more patients with Mafb mutations in the DNA-binding domain will be diagnosed with symptoms that could be different from those in previous reports. Therefore, it is important to deeply analyze our mutant mice to uncover the possible symptoms that DRS patients could show.
Conclusion
The functions of MafB in macrophages have been slowly revealed for the past 20 years. These functions can be summarized as a regulator of specific genes in various types of macrophages, such as tissue-specific macrophages, tissue repairing macrophages at pathological lesions, and macrophages exposed to lipid/cholesterol-rich environments. The studies discussed in this review illustrate that MafB may function particularly in macrophages that restrain the immune system and maintain homeostasis.
In addition, whole genome sequencing has been applied to diseases with unknown causes, showing mutation of MAFB as the reason for DRS and MCTO. It is possible that accumulation of such findings may clarify and increase the number of patients diagnosed with a mutation in the MAFB gene. Therefore, precise analysis of Mafb-mutated mice may help to predicting other phenotypes of patients to build the basis for both prevention and treatment.
Acknowledgments
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan through Grants-in-Aid for Scientific Research (16K18398 and 19K07499). It was also supported by the Takamatsunomiya Cancer Foundation (15-24724; M. Hamada) and grants from the Uehara Memorial Foundation and Takeda Science Foundation.
References
- 1.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., Butte A.J., Bhattacharya M.2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20: 163–172. doi: 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auffray C., Fogg D., Garfa M., Elain G., Join-Lambert O., Kayal S., Sarnacki S., Cumano A., Lauvau G., Geissmann F.2007. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317: 666–670. doi: 10.1126/science.1142883 [DOI] [PubMed] [Google Scholar]
- 3.Aziz A., Soucie E., Sarrazin S., Sieweke M.H.2009. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science 326: 867–871. doi: 10.1126/science.1176056 [DOI] [PubMed] [Google Scholar]
- 4.Aziz A., Vanhille L., Mohideen P., Kelly L.M., Otto C., Bakri Y., Mossadegh N., Sarrazin S., Sieweke M.H.2006. Development of macrophages with altered actin organization in the absence of MafB. Mol. Cell. Biol. 26: 6808–6818. doi: 10.1128/MCB.00245-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boersma-Vreugdenhil G.R., Kuipers J., Van Stralen E., Peeters T., Michaux L., Hagemeijer A., Pearson P.L., Clevers H.C., Bast B.J.2004. The recurrent translocation t(14;20)(q32;q12) in multiple myeloma results in aberrant expression of MAFB: a molecular and genetic analysis of the chromosomal breakpoint. Br. J. Haematol. 126: 355–363. doi: 10.1111/j.1365-2141.2004.05050.x [DOI] [PubMed] [Google Scholar]
- 6.Botto M., Dell’Agnola C., Bygrave A.E., Thompson E.M., Cook H.T., Petry F., Loos M., Pandolfi P.P., Walport M.J.1998. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 19: 56–59. doi: 10.1038/ng0598-56 [DOI] [PubMed] [Google Scholar]
- 7.Cao S., Liu J., Song L., Ma X.2005. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J. Immunol. 174: 3484–3492. doi: 10.4049/jimmunol.174.6.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemente C., Rius C., Alonso-Herranz L., Martín-Alonso M., Pollán Á., Camafeita E., Martínez F., Mota R.A., Núñez V., Rodríguez C., Seiki M., Martínez-González J., Andrés V., Ricote M., Arroyo A.G.2018. MT4-MMP deficiency increases patrolling monocyte recruitment to early lesions and accelerates atherosclerosis. Nat. Commun. 9: 910. doi: 10.1038/s41467-018-03351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochain C., Vafadarnejad E., Arampatzi P., Pelisek J., Winkels H., Ley K., Wolf D., Saliba A.E., Zernecke A.2018. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ. Res. 122: 1661–1674. doi: 10.1161/CIRCRESAHA.117.312509 [DOI] [PubMed] [Google Scholar]
- 10.Cordes S.P., Barsh G.S.1994. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79: 1025–1034. doi: 10.1016/0092-8674(94)90033-7 [DOI] [PubMed] [Google Scholar]
- 11.Cuevas V.D., Anta L., Samaniego R., Orta-Zavalza E., Vladimir de la Rosa J., Baujat G., Domínguez-Soto Á., Sánchez-Mateos P., Escribese M.M., Castrillo A., Cormier-Daire V., Vega M.A., Corbí A.L.2017. MAFB determines human macrophage anti-inflammatory polarization: relevance for the pathogenic mechanisms operating in multicentric carpotarsal osteolysis. J. Immunol. 198: 2070–2081. doi: 10.4049/jimmunol.1601667 [DOI] [PubMed] [Google Scholar]
- 12.Daassi D., Hamada M., Jeon H., Imamura Y., Nhu Tran M.T., Takahashi S.2016. Differential expression patterns of MafB and c-Maf in macrophages in vivo and in vitro. Biochem. Biophys. Res. Commun. 473: 118–124. doi: 10.1016/j.bbrc.2016.03.063 [DOI] [PubMed] [Google Scholar]
- 13.Dick S.A., Macklin J.A., Nejat S., Momen A., Clemente-Casares X., Althagafi M.G., Chen J., Kantores C., Hosseinzadeh S., Aronoff L., Wong A., Zaman R., Barbu I., Besla R., Lavine K.J., Razani B., Ginhoux F., Husain M., Cybulsky M.I., Robbins C.S., Epelman S.2019. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20: 29–39. doi: 10.1038/s41590-018-0272-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K., Pamer E.G., Li M.O.2014. The cellular and molecular origin of tumor-associated macrophages. Science 344: 921–925. doi: 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., Mazloom A.R., Ma’ayan A., Chua W.J., Hansen T.H., Turley S.J., Merad M., Randolph G.J., Immunological Genome Consortium.2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13: 1118–1128. doi: 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gemelli C., Orlandi C., Zanocco Marani T., Martello A., Vignudelli T., Ferrari F., Montanari M., Parenti S., Testa A., Grande A., Ferrari S.2008. The vitamin D3/Hox-A10 pathway supports MafB function during the monocyte differentiation of human CD34+ hemopoietic progenitors. J. Immunol. 181: 5660–5672. doi: 10.4049/jimmunol.181.8.5660 [DOI] [PubMed] [Google Scholar]
- 17.Gemelli C., Zanocco Marani T., Bicciato S., Mazza E.M., Boraschi D., Salsi V., Zappavigna V., Parenti S., Selmi T., Tagliafico E., Ferrari S., Grande A.2014. MafB is a downstream target of the IL-10/STAT3 signaling pathway, involved in the regulation of macrophage de-activation. Biochim. Biophys. Acta 1843: 955–964. doi: 10.1016/j.bbamcr.2014.01.021 [DOI] [PubMed] [Google Scholar]
- 18.Ginhoux F., Schultze J.L., Murray P.J., Ochando J., Biswas S.K.2016. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 17: 34–40. doi: 10.1038/ni.3324 [DOI] [PubMed] [Google Scholar]
- 19.Goudot C., Coillard A., Villani A.C., Gueguen P., Cros A., Sarkizova S., Tang-Huau T.L., Bohec M., Baulande S., Hacohen N., Amigorena S., Segura E.2017. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity 47: 582–596.e6. doi: 10.1016/j.immuni.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 20.Grapin-Botton A., Bonnin M.A., Sieweke M., Le Douarin N.M.1998. Defined concentrations of a posteriorizing signal are critical for MafB/Kreisler segmental expression in the hindbrain. Development 125: 1173–1181. [DOI] [PubMed] [Google Scholar]
- 21.Hamada M., Moriguchi T., Yokomizo T., Morito N., Zhang C., Takahashi S.2003. The mouse mafB 5’-upstream fragment directs gene expression in myelomonocytic cells, differentiated macrophages and the ventral spinal cord in transgenic mice. J. Biochem. 134: 203–210. doi: 10.1093/jb/mvg130 [DOI] [PubMed] [Google Scholar]
- 22.Hamada M., Nakamura M., Tran M.T., Moriguchi T., Hong C., Ohsumi T., Dinh T.T., Kusakabe M., Hattori M., Katsumata T., Arai S., Nakashima K., Kudo T., Kuroda E., Wu C.H., Kao P.H., Sakai M., Shimano H., Miyazaki T., Tontonoz P., Takahashi S.2014. MafB promotes atherosclerosis by inhibiting foam-cell apoptosis. Nat. Commun. 5: 3147. doi: 10.1038/ncomms4147 [DOI] [PubMed] [Google Scholar]
- 23.Havixbeck J.J., Rieger A.M., Wong M.E., Wilkie M.P., Barreda D.R.2014. Evolutionary conservation of divergent pro-inflammatory and homeostatic responses in Lamprey phagocytes. PLoS One 9: e86255. doi: 10.1371/journal.pone.0086255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herath N.I., Rocques N., Garancher A., Eychène A., Pouponnot C.2014. GSK3-mediated MAF phosphorylation in multiple myeloma as a potential therapeutic target. Blood Cancer J. 4: e175. doi: 10.1038/bcj.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hume D.A.2008. Macrophages as APC and the dendritic cell myth. J. Immunol. 181: 5829–5835. doi: 10.4049/jimmunol.181.9.5829 [DOI] [PubMed] [Google Scholar]
- 26.Hume D.A., Summers K.M., Rehli M.2016. Transcriptional regulation and macrophage differentiation. Microbiol. Spectr. 4: 1–19. [DOI] [PubMed] [Google Scholar]
- 27.Iacovazzo D., Flanagan S.E., Walker E., Quezado R., de Sousa Barros F.A., Caswell R., Johnson M.B., Wakeling M., Brändle M., Guo M., Dang M.N., Gabrovska P., Niederle B., Christ E., Jenni S., Sipos B., Nieser M., Frilling A., Dhatariya K., Chanson P., de Herder W.W., Konukiewitz B., Klöppel G., Stein R., Korbonits M., Ellard S.2018. MAFA missense mutation causes familial insulinomatosis and diabetes mellitus. Proc. Natl. Acad. Sci. USA 115: 1027–1032. doi: 10.1073/pnas.1712262115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igarashi K., Kataoka K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M.1994. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367: 568–572. doi: 10.1038/367568a0 [DOI] [PubMed] [Google Scholar]
- 29.Jablonski K.A., Gaudet A.D., Amici S.A., Popovich P.G., Guerau-de-Arellano M.2016. Control of the Inflammatory Macrophage Transcriptional Signature by miR-155. PLoS One 11: e0159724. doi: 10.1371/journal.pone.0159724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamitani-Kawamoto A., Hamada M., Moriguchi T., Miyai M., Saji F., Hatamura I., Nishikawa K., Takayanagi H., Hitoshi S., Ikenaka K., Hosoya T., Hotta Y., Takahashi S., Kataoka K.2011. MafB interacts with Gcm2 and regulates parathyroid hormone expression and parathyroid development. J. Bone Miner. Res. 26: 2463–2472. doi: 10.1002/jbmr.458 [DOI] [PubMed] [Google Scholar]
- 31.Kataoka K., Fujiwara K.T., Noda M., Nishizawa M.1994. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol. 14: 7581–7591. doi: 10.1128/MCB.14.11.7581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kataoka K., Noda M., Nishizawa M.1994. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 14: 700–712. doi: 10.1128/MCB.14.1.700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh M.C., Jung Y., Ugboma C.M., Shimbo M., Kuno A., Basha W.A., Kudo T., Oishi H., Takahashi S.2018. MafB Is Critical for Glucagon Production and Secretion in Mouse Pancreatic α Cells In Vivo. Mol. Cell. Biol. 38: e00504-17. doi: 10.1128/MCB.00504-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawauchi S., Takahashi S., Nakajima O., Ogino H., Morita M., Nishizawa M., Yasuda K., Yamamoto M.1999. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem. 274: 19254–19260. doi: 10.1074/jbc.274.27.19254 [DOI] [PubMed] [Google Scholar]
- 35.Kelly L.M., Englmeier U., Lafon I., Sieweke M.H., Graf T.2000. MafB is an inducer of monocytic differentiation. EMBO J. 19: 1987–1997. doi: 10.1093/emboj/19.9.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H.2017. The transcription factor MafB promotes anti-inflammatory M2 polarization and cholesterol efflux in macrophages. Sci. Rep. 7: 7591. doi: 10.1038/s41598-017-07381-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J.W., Yang H.J., Oel A.P., Brooks M.J., Jia L., Plachetzki D.C., Li W., Allison W.T., Swaroop A.2016. Recruitment of Rod Photoreceptors from Short-Wavelength-Sensitive Cones during the Evolution of Nocturnal Vision in Mammals. Dev. Cell 37: 520–532. doi: 10.1016/j.devcel.2016.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.I., Ho I.C., Grusby M.J., Glimcher L.H.1999. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity 10: 745–751. doi: 10.1016/S1074-7613(00)80073-4 [DOI] [PubMed] [Google Scholar]
- 39.Kim K., Kim J.H., Lee J., Jin H.M., Kook H., Kim K.K., Lee S.Y., Kim N.2007. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood 109: 3253–3259. doi: 10.1182/blood-2006-09-048249 [DOI] [PubMed] [Google Scholar]
- 40.Klein C., Bellity J., Finidori G., Glorion C., Pannier S.2018. Multicentric carpotarsal osteolysis syndrome: long-term follow-up of three patients. Skeletal Radiol. 47: 1015–1019. doi: 10.1007/s00256-018-2874-z [DOI] [PubMed] [Google Scholar]
- 41.Koshida R., Oishi H., Hamada M., Takei Y., Takahashi S.2017. MafB is required for development of the hindbrain choroid plexus. Biochem. Biophys. Res. Commun. 483: 288–293. doi: 10.1016/j.bbrc.2016.12.150 [DOI] [PubMed] [Google Scholar]
- 42.Krishnasamy K., Limbourg A., Kapanadze T., Gamrekelashvili J., Beger C., Häger C., Lozanovski V.J., Falk C.S., Napp L.C., Bauersachs J., Mack M., Haller H., Weber C., Adams R.H., Limbourg F.P.2017. Blood vessel control of macrophage maturation promotes arteriogenesis in ischemia. Nat. Commun. 8: 952. doi: 10.1038/s41467-017-00953-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I.2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159: 1312–1326. doi: 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao C.T., Andrews R., Wallace L.E., Khan M.W., Kift-Morgan A., Topley N., Fraser D.J., Taylor P.R.2017. Peritoneal macrophage heterogeneity is associated with different peritoneal dialysis outcomes. Kidney Int. 91: 1088–1103. doi: 10.1016/j.kint.2016.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Littink K.W., Stappers P.T.Y., Riemslag F.C.C., Talsma H.E., van Genderen M.M., Cremers F.P.M., Collin R.W.J., van den Born L.I.2018. Autosomal recessive NRL mutations in patients with enhanced S-Cone syndrome. Genes (Basel) 9: E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M.2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25: 677–686. doi: 10.1016/j.it.2004.09.015 [DOI] [PubMed] [Google Scholar]
- 47.Matcovitch-Natan O., Winter D.R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada González F., Perrin P., Keren-Shaul H., Gury M., Lara-Astaiso D., Thaiss C.A., Cohen M., Bahar Halpern K., Baruch K., Deczkowska A., Lorenzo-Vivas E., Itzkovitz S., Elinav E., Sieweke M.H., Schwartz M., Amit I.2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353: aad8670. doi: 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- 48.Matsushita M., Matsushita A., Endo Y., Nakata M., Kojima N., Mizuochi T., Fujita T.2004. Origin of the classical complement pathway: Lamprey orthologue of mammalian C1q acts as a lectin. Proc. Natl. Acad. Sci. USA 101: 10127–10131. doi: 10.1073/pnas.0402180101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A.2001. Nrl is required for rod photoreceptor development. Nat. Genet. 29: 447–452. doi: 10.1038/ng774 [DOI] [PubMed] [Google Scholar]
- 50.Menéndez-Gutiérrez M.P., Rőszer T., Fuentes L., Núñez V., Escolano A., Redondo J.M., De Clerck N., Metzger D., Valledor A.F., Ricote M.2015. Retinoid X receptors orchestrate osteoclast differentiation and postnatal bone remodeling. J. Clin. Invest. 125: 809–823. doi: 10.1172/JCI77186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyai M., Hamada M., Moriguchi T., Hiruma J., Kamitani-Kawamoto A., Watanabe H., Hara-Chikuma M., Takahashi K., Takahashi S., Kataoka K.2016. Transcription Factor MafB Coordinates Epidermal Keratinocyte Differentiation. J. Invest. Dermatol. 136: 1848–1857. doi: 10.1016/j.jid.2016.05.088 [DOI] [PubMed] [Google Scholar]
- 52.Miyai M., Tanaka Y.G., Kamitani A., Hamada M., Takahashi S., Kataoka K.2010. c-Maf and MafB transcription factors are differentially expressed in Huxley’s and Henle’s layers of the inner root sheath of the hair follicle and regulate cuticle formation. J. Dermatol. Sci. 57: 178–182. doi: 10.1016/j.jdermsci.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 53.Moriguchi T., Hamada M., Morito N., Terunuma T., Hasegawa K., Zhang C., Yokomizo T., Esaki R., Kuroda E., Yoh K., Kudo T., Nagata M., Greaves D.R., Engel J.D., Yamamoto M., Takahashi S.2006. MafB is essential for renal development and F4/80 expression in macrophages. Mol. Cell. Biol. 26: 5715–5727. doi: 10.1128/MCB.00001-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukundan L., Odegaard J.I., Morel C.R., Heredia J.E., Mwangi J.W., Ricardo-Gonzalez R.R., Goh Y.P., Eagle A.R., Dunn S.E., Awakuni J.U., Nguyen K.D., Steinman L., Michie S.A., Chawla A.2009. PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 15: 1266–1272. doi: 10.1038/nm.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mumm S., Huskey M., Duan S., Wenkert D., Madson K.L., Gottesman G.S., Nenninger A.R., Laxer R.M., McAlister W.H., Whyte M.P.2014. Multicentric carpotarsal osteolysis syndrome is caused by only a few domain-specific mutations in MAFB, a negative regulator of RANKL-induced osteoclastogenesis. Am. J. Med. Genet. A. 164A: 2287–2293. doi: 10.1002/ajmg.a.36641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., Locati M., Mantovani A., Martinez F.O., Mege J.L., Mosser D.M., Natoli G., Saeij J.P., Schultze J.L., Shirey K.A., Sica A., Suttles J., Udalova I., van Ginderachter J.A., Vogel S.N., Wynn T.A.2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41: 14–20. doi: 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A., Caputo V., Cecchetti S., Prudente S., Fiorenza M.T., Boitani C., Philip N., Niyazov D., Leoni C., Nakane T., Keppler-Noreuil K., Braddock S.R., Gillessen-Kaesbach G., Palleschi A., Campeau P.M., Lee B.H., Pouponnot C., Stella L., Bocchinfuso G., Katsanis N., Sol-Church K., Tartaglia M.2015. Mutations Impairing GSK3-Mediated MAF Phosphorylation Cause Cataract, Deafness, Intellectual Disability, Seizures, and a Down Syndrome-like Facies. Am. J. Hum. Genet. 96: 816–825. doi: 10.1016/j.ajhg.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishizawa M., Kataoka K., Goto N., Fujiwara K.T., Kawai S.1989. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc. Natl. Acad. Sci. USA 86: 7711–7715. doi: 10.1073/pnas.86.20.7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J.G., Tischfield M.A., Nugent A.A., Cheng L., Di Gioia S.A., Chan W.M., Maconachie G., Bosley T.M., Summers C.G., Hunter D.G., Robson C.D., Gottlob I., Engle E.C.2016. Loss of MAFB Function in Humans and Mice Causes Duane Syndrome, Aberrant Extraocular Muscle Innervation, and Inner-Ear Defects. Am. J. Hum. Genet. 98: 1220–1227. doi: 10.1016/j.ajhg.2016.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perveen R., Favor J., Jamieson R.V., Ray D.W., Black G.C.2007. A heterozygous c-Maf transactivation domain mutation causes congenital cataract and enhances target gene activation. Hum. Mol. Genet. 16: 1030–1038. doi: 10.1093/hmg/ddm048 [DOI] [PubMed] [Google Scholar]
- 61.Puranik A.S., Leaf I.A., Jensen M.A., Hedayat A.F., Saad A., Kim K.W., Saadalla A.M., Woollard J.R., Kashyap S., Textor S.C., Grande J.P., Lerman A., Simari R.D., Randolph G.J., Duffield J.S., Lerman L.O.2018. Kidney-resident macrophages promote a proangiogenic environment in the normal and chronically ischemic mouse kidney. Sci. Rep. 8: 13948. doi: 10.1038/s41598-018-31887-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rőszer T., Menéndez-Gutiérrez M.P., Lefterova M.I., Alameda D., Núñez V., Lazar M.A., Fischer T., Ricote M.2011. Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator-activated receptor gamma or retinoid X receptor alpha deficiency. J. Immunol. 186: 621–631. doi: 10.4049/jimmunol.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadl V., Jin F., Yu J., Cui S., Holmyard D., Quaggin S., Barsh G., Cordes S.2002. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev. Biol. 249: 16–29. doi: 10.1006/dbio.2002.0751 [DOI] [PubMed] [Google Scholar]
- 64.Sanjurjo L., Aran G., Roher N., Valledor A.F., Sarrias M.R.2015. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J. Leukoc. Biol. 98: 173–184. doi: 10.1189/jlb.3RU0215-074R [DOI] [PubMed] [Google Scholar]
- 65.Sarrazin S., Mossadegh-Keller N., Fukao T., Aziz A., Mourcin F., Vanhille L., Kelly Modis L., Kastner P., Chan S., Duprez E., Otto C., Sieweke M.H.2009. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell 138: 300–313. doi: 10.1016/j.cell.2009.04.057 [DOI] [PubMed] [Google Scholar]
- 66.Sato Y., Tsukaguchi H., Morita H., Higasa K., Tran M.T.N., Hamada M., Usui T., Morito N., Horita S., Hayashi T., Takagi J., Yamaguchi I., Nguyen H.T., Harada M., Inui K., Maruta Y., Inoue Y., Koiwa F., Sato H., Matsuda F., Ayabe S., Mizuno S., Sugiyama F., Takahashi S., Yoshimura A.2018. A mutation in transcription factor MAFB causes Focal Segmental Glomerulosclerosis with Duane Retraction Syndrome. Kidney Int. 94: 396–407. doi: 10.1016/j.kint.2018.02.025 [DOI] [PubMed] [Google Scholar]
- 67.Satpathy A.T., Wu X., Albring J.C., Murphy K.M.2012. Re(de)fining the dendritic cell lineage. Nat. Immunol. 13: 1145–1154. doi: 10.1038/ni.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schejbel L., Skattum L., Hagelberg S., Åhlin A., Schiller B., Berg S., Genel F., Truedsson L., Garred P.2011. Molecular basis of hereditary C1q deficiency--revisited: identification of several novel disease-causing mutations. Genes Immun. 12: 626–634. doi: 10.1038/gene.2011.39 [DOI] [PubMed] [Google Scholar]
- 69.Shichita T., Ito M., Morita R., Komai K., Noguchi Y., Ooboshi H., Koshida R., Takahashi S., Kodama T., Yoshimura A.2017. MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat. Med. 23: 723–732. doi: 10.1038/nm.4312 [DOI] [PubMed] [Google Scholar]
- 70.Sieweke M.H., Tekotte H., Frampton J., Graf T.1996. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell 85: 49–60. doi: 10.1016/S0092-8674(00)81081-8 [DOI] [PubMed] [Google Scholar]
- 71.Suzuki K., Numata T., Suzuki H., Raga D.D., Ipulan L.A., Yokoyama C., Matsushita S., Hamada M., Nakagata N., Nishinakamura R., Kume S., Takahashi S., Yamada G.2014. Sexually dimorphic expression of Mafb regulates masculinization of the embryonic urethral formation. Proc. Natl. Acad. Sci. USA 111: 16407–16412. doi: 10.1073/pnas.1413273111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tozaki-Saitoh H., Masuda J., Kawada R., Kojima C., Yoneda S., Masuda T., Inoue K., Tsuda M.2019. Transcription factor MafB contributes to the activation of spinal microglia underlying neuropathic pain development. Glia 67: 729–740. doi: 10.1002/glia.23570 [DOI] [PubMed] [Google Scholar]
- 73.Tran M.T., Hamada M., Nakamura M., Jeon H., Kamei R., Tsunakawa Y., Kulathunga K., Lin Y.Y., Fujisawa K., Kudo T., Takahashi S.2016. MafB deficiency accelerates the development of obesity in mice. FEBS Open Bio 6: 540–547. doi: 10.1002/2211-5463.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran M.T.N., Hamada M., Jeon H., Shiraishi R., Asano K., Hattori M., Nakamura M., Imamura Y., Tsunakawa Y., Fujii R., Usui T., Kulathunga K., Andrea C.S., Koshida R., Kamei R., Matsunaga Y., Kobayashi M., Oishi H., Kudo T., Takahashi S.2017. MafB is a critical regulator of complement component C1q. Nat. Commun. 8: 1700. doi: 10.1038/s41467-017-01711-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsunakawa Y., Hamada M., Matsunaga Y., Fuseya S., Jeon H., Wakimoto Y., Usui T., Kanai M., Mizuno S., Morito N., Takahashi S.2019. Mice harboring an MCTO mutation exhibit renal failure resembling nephropathy in human patients. Exp. Anim. 68: 103–111. doi: 10.1538/expanim.18-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wende H., Lechner S.G., Cheret C., Bourane S., Kolanczyk M.E., Pattyn A., Reuter K., Munier F.L., Carroll P., Lewin G.R., Birchmeier C.2012. The transcription factor c-Maf controls touch receptor development and function. Science 335: 1373–1376. doi: 10.1126/science.1214314 [DOI] [PubMed] [Google Scholar]
- 77.Wu X., Briseño C.G., Durai V., Albring J.C., Haldar M., Bagadia P., Kim K.W., Randolph G.J., Murphy T.L., Murphy K.M.2016. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J. Exp. Med. 213: 2553–2565. doi: 10.1084/jem.20160600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wynn T.A., Chawla A., Pollard J.W.2013. Macrophage biology in development, homeostasis and disease. Nature 496: 445–455. doi: 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yap N.V., Whelan F.J., Bowdish D.M., Golding G.B.2015. The Evolution of the Scavenger Receptor Cysteine-Rich Domain of the Class A Scavenger Receptors. Front. Immunol. 6: 342. doi: 10.3389/fimmu.2015.00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida T., Ohkumo T., Ishibashi S., Yasuda K.2005. The 5’-AT-rich half-site of Maf recognition element: a functional target for bZIP transcription factor Maf. Nucleic Acids Res. 33: 3465–3478. doi: 10.1093/nar/gki653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zankl A., Duncan E.L., Leo P.J., Clark G.R., Glazov E.A., Addor M.C., Herlin T., Kim C.A., Leheup B.P., McGill J., McTaggart S., Mittas S., Mitchell A.L., Mortier G.R., Robertson S.P., Schroeder M., Terhal P., Brown M.A.2012. Multicentric carpotarsal osteolysis is caused by mutations clustering in the amino-terminal transcriptional activation domain of MAFB. Am. J. Hum. Genet. 90: 494–501. doi: 10.1016/j.ajhg.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang C., Moriguchi T., Kajihara M., Esaki R., Harada A., Shimohata H., Oishi H., Hamada M., Morito N., Hasegawa K., Kudo T., Engel J.D., Yamamoto M., Takahashi S.2005. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 25: 4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Chen Q., Ross A.C.2012. Retinoic acid and tumor necrosis factor-α induced monocytic cell gene expression is regulated in part by induction of transcription factor MafB. Exp. Cell Res. 318: 2407–2416. doi: 10.1016/j.yexcr.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhuang L., Adler S., Aeberli D., Villiger P.M., Trueb B.2017. Identification of a MAFB mutation in a patient with multicentric carpotarsal osteolysis. Swiss Med. Wkly. 147: w14529. [DOI] [PubMed] [Google Scholar]