Abstract
Drug development is currently hampered by the inability of animal experiments to accurately predict human response. While emerging organ on chip technology offers to reduce risk using microfluidic models of human tissues, the technology still mostly relies on end-point assays and biomarker measurements to assess tissue damage resulting in limited mechanistic information and difficulties to detect adverse effects occurring below the threshold of cellular damage. Here we present a sensor-integrated liver on chip array in which oxygen is monitored using two-frequency phase modulation of tissue-embedded microprobes, while glucose, lactate and temperature are measured in real time using microfluidic electrochemical sensors. Our microphysiological platform permits the calculation of dynamic changes in metabolic fluxes around central carbon metabolism, producing a unique metabolic fingerprint of the liver's response to stimuli. Using our platform, we studied the dynamics of human liver response to the epilepsy drug Valproate (Depakine™) and the antiretroviral medication Stavudine (Zerit™). Using E6/E7LOW hepatocytes, we show TC50 of 2.5 and 0.8 mM, respectively, coupled with a significant induction of steatosis in 2D and 3D cultures. Time to onset analysis showed slow progressive damage starting only 15–20 hours post-exposure. However, flux analysis showed a rapid disruption of metabolic homeostasis occurring below the threshold of cellular damage. While Valproate exposure led to a sustained 15% increase in lipogenesis followed by mitochondrial stress, Stavudine exposure showed only a transient increase in lipogenesis suggesting disruption of β-oxidation. Our data demonstrates the importance of tracking metabolic stress as a predictor of clinical outcome.
Introduction
Drug development is currently hindered by the inability of animal experiments to predict human response,1,2 leading to clinical failures and post-market withdrawals costing the industry an estimated $2B annually. In addition, animal testing for cosmetics is no longer possible in Europe (EC no. 1223/2009) limiting development. While emerging organ on chip technology offers to reduce risk using microfluidic models of human tissues, it has thus far failed to show concrete advantages over traditional methods. One of the main challenges lies in detecting idiosyncratic or “unexplained” toxicity occurring due to physiological stress developing below the threshold of cellular damage.3
Recently, we demonstrated the real-time measurement of oxygen, glucose and lactate in 3D liver organoids maintained under physiological conditions using tissue-embedded microsensors.4 Oxygen was measured by phase shift that is independent of signal intensity, and thus unaffected by tissue growth, compaction or death,5,6 but amperometric sensors for glucose and lactate had to be microfluidically addressed in a sequential manner, limiting dynamic resolution to once every 80 minutes and overall measurement to 24 hours.4 This design allowed us to detect relatively rapid changes in mitochondrial function showing the CYP450-independent stress induced by sub-toxic concentration of troglitazone (Rezulin™). Other microphysiological platforms similarly fall short when it comes to sampling sensitivity, total period and frequency,7,8 limiting measurement resolution needed to examine sub-toxic effects.
In this work, we chose to focus on fatty liver disease, steatosis, a medical condition affecting over 25% of the global population9–12 and a common adverse event reported in drug induced liver injury13–15 and during prescription drug use. While fatty liver disease can range from simple steatosis to nonalcoholic steatohepatitis (NASH), cirrhosis, and hepatocellular carcinoma, one of the early and more severe forms of the disease is microvesicular steatosis.14 Microvesicular steatosis is often associated with mitochondrial dysfunction and can be life threatening when long lasting.16–18 Drugs associated with microvesicular steatosis include Valproate, Aspirin, glucocorticoids, anti-retroviral drugs (e.g. Stavudine), nonsteroidal anti-inflammatory drugs (e.g. Bromfenac) and cocaine.19–23
Valproate (2-n-propylpentanoic acid) is a widely used antiepileptic drug. Over 40% of the patients on Valproate therapy show elevated liver enzymes, with 61% showing evidence of steatosis based on abdominal ultrasound.24 Idiosyncratic injury was reported in about 1/20 000 patients and is characterized by microvesicular steatosis developing over weeks of continuous use.25–27 Valproate toxicity was primarily studied in rodents, showing suppression of β-oxidation and mitochondrial dysfunction as mechanism of action,28 at least in high doses.29,30 Stavudine (d4T) is a nucleoside reverse transcriptase inhibitor used in the treatment of HIV/AIDS. Elevation of liver enzymes was reported in 30% of the patients, with a similar number showing steatosis in ultrasound.31,32 Stavudine toxicity was similarly studied in rodents and is thought to similarly suppress β-oxidation and induce mitochondrial dysfunction, possibly due to inhibition of mitochondrial mDNA synthesis.33,34
To elucidate the mechanism of both Valproate and Stavudine we designed a 6-unit microphysiological flux balance platform using a microfluidic in-line biosensor array that permits metabolic measurements at a rate of 1 per 25 seconds for 3–4 days. This design allows us to calculate dynamic changes in metabolic fluxes around central carbon metabolism (Fig. 1) producing a unique metabolic fingerprint of the liver response to stimuli. Our metabolic analysis points to lipid accumulation as the main mechanism of damage in both drugs. However, we show that at sub-toxic concentrations Valproate exposure shifts pyruvate toward citrate and fatty acid production in minutes, suggesting direct metabolic rerouting of glucose, rather than the β-oxidation disruption reported in rodents. In contrast, sub-toxic concentrations of Stavudine do not appear to increase lipogenesis and its metabolic effects develop over hours, suggesting a transcriptional suppression of β-oxidation as a possible mode of action. Interestingly, analytical derivation of lowest exposure levels (LEL) were not significantly different than clinically observed Cmax suggesting the safety margin of both drugs should be reconsidered.
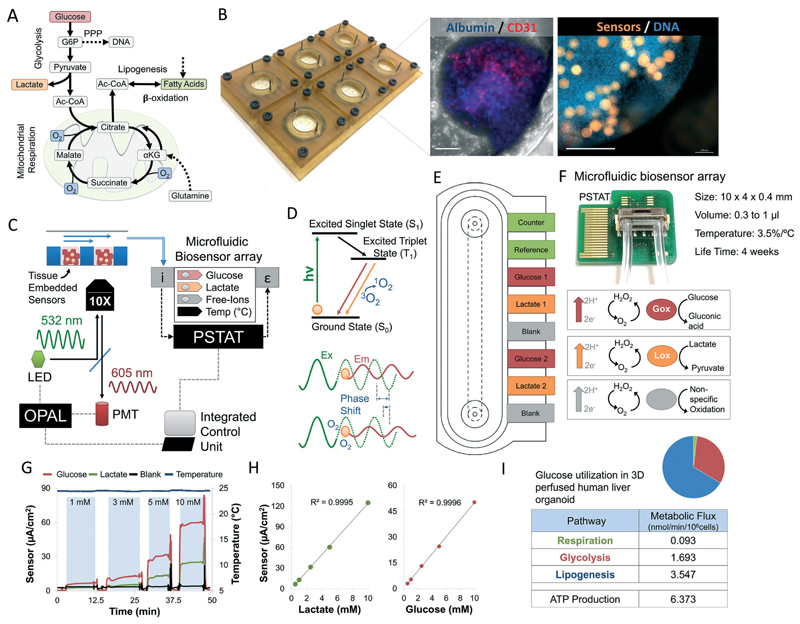
Fig. 1. Design of a microphysiological flux balance platform.
(A) Metabolic pathways of glucose utilization in human hepatocytes. Flux balance analysis permits the calculation of intracellular fluxes using extracellular oxygen, glucose, and lactate measurements. Dotted arrows note experimentally-limited fluxes. (B) CNC-fabricated 6-unit bioreactor plate. Laser-cut disposable microwell chips containing 9 organoids are seeded with microsensors in an open configuration and then perfused until metabolic stabilization achieved. Immunofluorescent staining shows a human liver organoid composed of albumin-positive E6/E7LOW hepatocytes (blue) and CD31-positive endothelial cells (red). Oxygen sensors (orange) are embedded inside the microtissue (blue) during seeding. Scale bar = 250 μm (C) platform schematics. Bioreactor is loaded with tissue-embedded oxygen sensors and mounted on an Olympus IX83. OPAL-controlled modulation LED signal excites the embedded oxygen sensors. Phase shift is measured through a hardware-filtered photomultiplier (PMT). Bioreactor outflow is connected to a microfluidic biosensor array containing electrochemical sensors for glucose and lactate, continuously adjusted according to non-specific oxidation events and changes in the ambient temperature. Sensors are connected to an on-chip potentiostat (PSTAT). All measurements (optical and electronic) are processed in real-time by single microprocessor, synchronizing the signal continuously. (D) Jablonski diagram describing the generation of phosphorescence with Ru-CPOx beads under the influence of oxygen. The quenching of the phosphorescence by triplet oxygen leads to a decrease in signal intensity and phosphorescence decay time T1. The effect induces a phase shift between the intensity-modulated excitation and emission light, proportional to oxygen concentration. Two-superimposed frequencies are used to screen out background interference. (E) Low volume microfluidic amperometric, 8-electrode, biosensor array. Anodic oxidation of H2O2 on platinum produces a current rapidly (t90 < 25 s), while embedded catalase activity prevents cross-contamination. A 450 mV potential between the working and counter electrodes is monitored against a reference electrode to minimize background noise caused by reversible electrolysis events. (F) Photo of microfluidic biosensor array with total internal volume of 0.3–1 μL and integrated temperature sensors and PSTAT. (G) Raw measurements of glucose, lactate, blank and temperature sensors of calibration measurements for different analyte concentrations. Measurements were carried automatically out under continuous flow of 2 μL min−1. Air gap between samples ensure a sharp change in chemical gradient on the sensor during in calibration. (H) Amperometric calibration curves of glucose and lactate concentrations in bioreactor outflow. (I) Intracellular metabolic fluxes for polarized HepG2/C3A organoid under steady state conditions. Glucose utilization in each pathway is shown as nmol min−1 per 106 cells as well as calculated ATP production (methods). Relative glucose utilization is shown as pie chart.
Materials and methods
Bioreactor design and fabrication
Bioreactor manifold and disposable PDMS microwell insert design was carried out using AutoCAD® (Autodesk, USA), and adapted for computer numerical control (CNC) using SolidWorks® (SolidWorks, USA). Design and dimensions are presented in Supplement S1A-C. Bioreactor manifold was machined from biocompatible polyetherimide (ULTEM) blocks using Haas VF-2SSYT (Hass Automations, USA) machining (Fig. 1B). Each unit was composed of two 50.8 mm circular support structures that fit standard 2 inch inserts, imbedded with a biocompatible epoxy-glued glass windows for efficient light transmission and a stainless-steel needle connection for perfusion.
PDMS microwell inserts were fabricated using laser cutting. Briefly, a thin sheet of PDMS (Dow Corning) was cast to 0.7 mm height using a motorized film applicator (Erichsen) and cured at 70 °C for 1 h. Microwells were cut to 1.5 mm diameter, and a center-to-center distance of 3 mm using a 355 nm pulsed Nd-YAG laser (3D-Micromac). PDMS inserts were washed with 70% (vol/vol) Ethanol (EtOH), nitrogen dried, and covalently bound to clean 0.5 mm thick glass coverslips (Schott) using oxygen plasma activation.
Bioreactor assembly
Each bioreactor was housed with a removable polydimethylsiloxane (PDMS) microwell insert in which cells are protected from the negative effect of shear. Sealing around the microwells was realized with a rubber gasket creating a perfusion chamber with an internal volume of 200 μL. Pressure and sealing was maintained using 4 stainless steel screws. Each bioreactor was perfused separately via 0.03″ Tygon® low adhesion tubing (Saint-Gobain, France) fitted onto the steel needles (S1†).
Cell culture
All cells were cultured under standard conditions in a humidified incubator at 37 °C, under 5% CO2. E6/E7LOW human hepatocyte were expanded and differentiated as previously described.35 Differentiation and maintenance medium is composed of William's E basal medium supplemented with dexamethasone (Sigma-Aldrich, USA), Bovine serum albumin (Fraction V, MP, USA), insulin, transferrin and selenium (ITS, Gibco, USA), l-alanyl-l-glutamine (BI, Israel), 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin (BI, Israel).
HepG2/C3A cells were obtained from the American Type Culture Collection (ATCC, USA). Cells were cultured in Dulbecco's Modified Eagle Medium basal medium (DMEM) supplemented with fetal bovine serum (BI, Israel), Eagle-MEM non-essential amino acids (BI, Israel), l-alanyl-l-alutamine (BI, Israel) or l-glutamine (BI, Israel), 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin (BI, Israel).
Microvascular cardiac endothelial cells (VEC Technologies, USA) were cultured in PeproGrow™ MicroV (Microvascular Endothelial Cell Media, Peprotech, USA) supplemented with 10% fetal bovine serum, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin (BI, Israel).
Organoid seeding
A suspension of HepG2/C3A cells and CPOx-50-RuP beads (Colibri Photonics, Germany) were suspended in 2.5 mg ml−1 rat tail collagen type I (Corning, USA) at a cell density of 7 × 105 cells per μl. A volume of 1.1 μl of the gel-imbedded mixture was injected into each microwell and left to form spontaneously until their metabolic activity stabilized around day 4.
E6/E7LOW human hepatocyte and microvascular endothelial cells counted and mixed in a 1 : 1 ratio in 2.5 mg ml−1 rat tail collagen type I (Corning, USA) at a cell density of 4.6 × 105 cells per μl. Organoids were seeded in a similar manner as mentioned above.
E6/E7LOW human hepatocyte viability assessment
E6/E7LOW human hepatocyte were cultured with different concentrations of compounds dissolved in culture medium for 24 hours. Cell viability was subsequently determined using LIVE/DEAD Cytotoxicity kit (Molecular Probes, USA) according to manufacturer directions. Briefly, cultures were incubated for 30 min with 2 μM calcein AM and 3 μM ethidium homodimer-1. Live cells were positive for green fluorescence due to hydrolysis of the acetoxymethyl ester group by intracellular esterases. Dead cells were positive for red fluorescence due to ethidium homodimer-1 binding intracellular DNA. Cellular viability was expressed as live over dead ratio and normalized based on negative (DMSO/DDW/EtOH) and positive (0.1% Saponin) controls. Fluorescence micrographs were analyzed for total fluorescence using ImageJ for 12 repeats for each sample. TC50 and TC20 concentrations were calculated using MATLAB® Curve Fitting Tool. Results were fitted to a sigmoidal curve.36,37 Error bars indicate ± S. E.
Adverse outcome pathway analysis
Quantification of Apoptotic index was performed using DeadEnd™ fluorometric TUNEL System (Promega) according to manufacturer directions. Briefly, cells were treated with TC20 concentrations of compounds dissolved in culture medium for 24 hours, and subsequently fixed in 4% Paraformaldehyde (PFA). Cell were then permeabilized and exposed to fluorescein-12-dUTP and terminal deoxynucleotidyl transferase (TdT). The reaction was subsequently stopped and the cells counterstained with 1 μg mL−1 Hoechst 33258. Apoptotic cells were positive for green fluorescence in the nucleus. Percentage apoptosis was defined as the number of TUNEL positive nuclei normalized to Hoechst 33258 positive nuclei. Quantification of Steatosis was performed using HCS LipidTOX™ Phospholipidosis and Steatosis Detection Kit (ThermoFisher). Briefly, differentiated cells were incubated with different concentrations of compounds dissolved in culture medium and 1× Phospholipidosis Detection Reagent for 48 h, subsequently fixed in 4% PFA. Cells were then stained with 1× LipidTOX™ for 20 min and counterstained with 1 μg mL−1 Hoechst 33258. Staining intensity was normalized to number of Hoechst 33258 positive nuclei. Quantification of Bile Secretion was performed using 5(6)-carboxy-2′,7′-dichlorofluorescein diacetate (CDFDA) staining. Briefly, differentiated cells were incubated with different concentrations of compounds dissolved in culture medium for 24 h. Cells were then treated with 10 μg mL−1 CDFDA and 1 μg mL−1 Hoechst 33342 for 30 min. Incubation medium was removed and cultures washed with ice-cold PBS containing calcium and magnesium. Staining intensity was normalized to number of Hoechst 33342 positive nuclei. Analysis excluded Intracellular CDFDA staining based on intensity and phase images. All quantifications were made in comparison to cells treated with melatonin as negative control.
Real-time oxygen measurement
Real-time oxygen measurements were performed optically using on-chip lifetime-based luminescence quenching (LBLQ).5,6,38 Fifty-micrometer-diameter polystyrene microbeads were loaded with ruthenium-phenanthroline-based phosphorescence dye (CPOx-50-RuP). RuP phosphorescence signal shows a characteristic delay given by the lifetime of its excited triplet state (Fig. 1D). Oxygen acts as a quencher, leading to a decrease in decay time and signal intensity with increasing concentration. We chose to measure decay time, rather than signal intensity, as it is not sensitive to changes in probe concentration or excitation intensity over the course of the experiment. The signal was measured using the OPAL system (Colibri Photonics, Germany) that comprises of a control module, 532 nm LED excitation source, and a photomultiplier (PMT) detector mounted on the ocular of an IX81 Olympus microscope (Olympus, Japan). A filter cube with 531/40 (Ex), 555, 607/70 (Em) was inserted in the optical light path during measurements (Fig. 1C). To accurately measure decay time, we chose phase modulation in which sinusoidal amplitude-modulated light is shifted in phase due to oxygen quenching (Fig. 1D). To overcome the superposition of inphase background fluorescence that alters the phase of the detected signal, we used a novel 53.5 and 31.3 kHz two-frequency phase modulation that allowed us to screen out the interference.4,5,38–40 Measurements were carried out by averaging five consecutive 4 s exposures. Measurements were taken every 15 min. Under similar conditions, 28 days of measurement of organoids was done with no apparent phototoxicity, signal drift, or relevant loss of signal intensity.4
Assessment of cellular toxicity and time to onset
Bioreactors were perfused with different concentrations of compounds dissolved in culture medium. Cell viability was determined by oxygen uptake following 24 hours of exposure unless otherwise noted. TC50 concentrations were determined using MATLAB by sigmoidal curve fitting.36,37 Time to onset was analyzed by MATLAB based on LPF and trend assessment.
Assessment of a lowest exposure level
To find the asymptotic concentration for which damage occurs only at infinite exposure time, we used a previously developed model for accumulative drug exposure.41,42 The model is based on the steady-state dynamics of flux accumulation, using a linear first-order approximation of the cell membrane. The flux behavior is described by
| (1) |
where J(C) is the concentration dependent flux, Jmin is the minimal flux that will cause an effect, t is time of exposure and τ is a drug-cell specific constant accounting reaction dynamics.43,44 In our case, t is the time to onset (TTO) of damage creating a bi-asymptotic behavior. This adapted model is used to describe lowest exposure level (LEL) under first order, steady state assumptions, as
| (2) |
| (3) |
Real-time glucose and lactate measurements
Amperometric glucose and lactate sensors were purchased from Innovative Sensor Technology (IST, Switzerland). The sensors are based on the enzymatic reactions of glucose oxidase, with a linear range of 0.5 mM to 30 mM, and lactate oxidase, with a linear range of 0.5 mM to 20 mM. Both sensors produce H2O2 in amounts proportional to the measured metabolite, which is detected with platinum electrodes under polarized condition. Measurements were made continuously over the whole experiment, 8–24 hours prior to exposure and until respiratory response was confirmed. Measurements were carried out and calibrated to sensitivity decrease by onchip potentiostat (IST, Switzerland).
Metabolic pathways and ATP production
Glucose uptake, oxygen uptake, and lactate production rates were measured by calculating the change in metabolite concentration between the bioreactor in- and outflow as a function of perfusion rate and cell number. Metabolic rates were calculated assuming negligible contribution to oxygen uptake by fatty acid oxidation and enzymatic activity. Low level of lipids in our culture medium ensured that fatty acid uptake was more than 50-fold lower than glucose, whereas glutamine contribution to the Krebs cycle was minor and glycogen content following 12 hour exposure to the drug showed no significant change (ESI† S4).
Based on these assumptions, oxidative phosphorylation flux was calculated by dividing the oxygen uptake rate by six. We estimated 32 ATP molecules generated by complete oxidation of one molecule of glucose. Glycolysis flux was calculated by dividing lactate production rate by two, with maximal rate defined by glucose uptake rate minus the oxidative phosphorylation flux. ATP production in glycolysis was estimated to be two molecules per molecule of glucose. We assumed that any glucose left over was directed toward lipogenesis, because the contribution of pentose phosphate pathway in non-proliferating cells is minor.45 Finally, we assumed that excess lactate was produced by glutaminolysis, an assumption confirmed by off-chip measurement of glutamine uptake.4 ATP production in glutaminolysis was estimated to be three molecules per molecule of lactate generated.
Quantitative RT-PCR
RNA was isolated and purified using Macherey–Nagel NucleoSpin RNA II kit according to manufacturer instructions. RNA concentration and purity were determined using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from 1 μg RNA sample using qScript cDNA SuperMix (Quanta BioSciences) according to the manufacturer's protocol. Gene expression analysis was carried out using KAPA SYBR FAST Universal 2× qPCR Master Mix (Kapa Biosystems) on BioRad CFX96 Real-Time System, according to manufacturer's directions. Gene transcription was evaluated using the ΔΔCt method normalized to 60S ribosomal protein L32 (RPL32) or ubiquitin C (UBC).
Statistical analysis
Experiments were repeated 2 or 3 times with triplicate samples for each experimental condition, unless stated otherwise. Data from representative experiments are presented, and similar trends were seen in multiple trials. A parametric two-tailed Student's t-test was used for calculating significant differences between groups. All error bars represent ± standard error, unless otherwise noted. TC50 and LEL error indicate the standard error calculated based on the 95% confidence bound calculated by the curve-fitting tool.
Results & discussion
Design of a microphysiological flux balance platform
Changes in central carbon metabolism and fuel utilization are sensitive markers of physiological stress. Flux balance analysis is a computational method to derive these intracellular fluxes by measuring changes in extracellular fluxes.4,45 Interestingly, for non-proliferating cells growing in lipid and glutamine poor medium (methods), central carbon fluxes can be estimated by measuring glucose, lactate and oxygen fluxes alone (Fig. 1A). To monitor the dynamic transition between these metabolic pathways, we designed a microfluidic system that maintains hepatic organoids under physiological conditions mimicking zonation4 while dynamically measuring oxygen, glucose, and lactate concentrations (Fig. 1B and C). 6-Unit bioreactor platform manifold was fabricated from biocompatible polyetherimide (ULTEM™) using CNC, while disposable multi-well microchips were fabricated using laser cutting (Fig. 1B, ESI† S1). Oxygen is measured using tissue-embedded microsensors loaded with a ruthenium-based dye, whose phosphorescence is quenched in the presence of oxygen, decreasing decay time (Fig. 1C). In contrast to intensity measurement, decay time is insensitive to probe concentration or excitation intensity. We use sinusoidal intensity-modulated light; resulting in an oxygen-dependent phase shift in the 605 nm emission (Fig. 1D) that is stable down to three particles, and 1.5 mm away from the focus permitting accurate measurement even during toxic damage and subsequent tissue disintegration.40 We have recently coupled the bioreactor to off-chip electrochemical sensors for glucose and lactate, addressed using a complex microfluidic switchboard.4 However, sensor crosstalk, environmental noise (temperature, salts) and switchboard complexity prevented large scale integration.
To address this problem, we integrated a microfluidic biosensor array in our platform with an on-chip temperature sensor, and a three-electrode design in which the counter and reference electrodes are separated (Fig. 1E, ESI† S2). The reference electrode is used to measure the working electrode potential without passing current through it, while the counter electrode closes a circuit, allowing current to pass. This circuit is not possible in a two-electrode system. Anodic oxidation of H2O2 on platinum produces a current rapidly (t90 < 25 s), while embedded catalase activity prevents cross-contamination. A 450 mV potential between the working and counter electrodes is monitored against a reference electrode to minimize background noise caused by reversible electrolysis events. Finally, an on-chip potentiostat (PSTAT) monitors an 8-electrode array is integrated in the 10 × 4 × 0.4 mm microchip (Fig. 1F) with a total volume of 0.3 to 1 μL suitable to be connected directly to the bioreactor outflow. A single central processing unit (CPU) controls the entire system, simplifying synchronization (Fig. 1C). Sensors shows linear range from 0.05 mM to 15 mM lactate and 25 mM glucose (Fig. 1G and H). Using our microphysiological platform we calculated the intracellular metabolic fluxes of polarized HepG2/C3A organoids under steady state condition (Fig. 1I). Glucose utilization in each pathway is shown as nmol min−1 per 106 cells as well as calculated ATP production (methods). Relative glucose utilization is shown as pie chart. To validate our model, we carried out a complete HPLC-based metabolic flux balance analysis46 on HepG2/C3A cells and primary human hepatocytes showing that glutamine uptake and lipid secretion are less than 0.3 nmol min−1 per 106 cells (ESI† S3 and S4).
Valproate and Stavudine induced steatosis
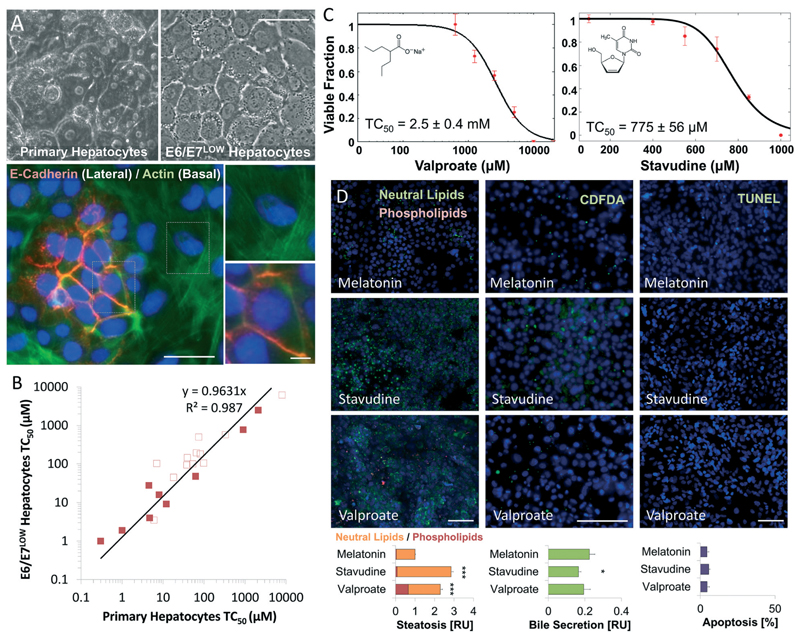
The broad-spectrum antiepileptic drug Valproate and antiretroviral drugs such as Stavudine (d4T) are known to induce hepatic steatosis in patients and animal models. We therefore assessed their effect on differentiated, polarized cultures of E6/E7LOW human hepatocytes (Fig. 2A). Recently published E6/E7LOW hepatocytes show equivalent toxicity profile to primary human hepatocytes,35 but less batch-to-batch variability (Fig. 2B). Analysis of Valproate and Stavudine toxicity showed TC50 of 2.5 ± 0.4 and 0.8 ± 0.06 mM, (Fig. 2C), 5-times and 100-times higher than reported Cmax, respectively. Adverse outcome analysis showed no evidence of apoptosis following 24 hours exposure (Fig. 2D), and only mild inhibition of bile secretion for Stavudine (p < 0.05, n = 4). However, both compounds show strong and significant increase in neutral lipids (p < 0.001, n = 4). Evidence of phospholipidosis was seen following Valproate but not Stavudine exposure.
Fig. 2. Valproate and Stavudine show drug-induced steatotic injury following 24 hours exposure in E6/E7LOW hepatocytes.
(A) Phase micrographs showing cuboidal morphology of differentiated E6/E7LOW hepatocytes compared to primary cells. Immunofluorescence staining of E-cadherin and actin shows nodales of epitelial polarization. Scale bar = 50 μm. (B) Graph showing the linear correlation in TC50 values of 21 compounds between E6/E7LOW and primary human hepatocytes. Steatosis-inducing drugs (full squares) show R2 > 0.99 correlation to primary hepatocytes compared with R2 > 0.97 for other drugs (empty squares). In general, differentiated E6/E7LOW hepatocytes show slightly elevated toxicity than cryopreserved cells. (C) Dose-dependent toxicity curves of differentiated E6/E7LOW hepatocytes treated with Valproate or Stavudine for 24 hours in standard 2D cell culture. TC50 values were 2.5 and 0.8 mM, respectively. (D) Fluorescence micrographs and total quantification of adverse outcome pathway in differentiated E6/E7LOW hepatocytes. Disruption of bile acid secretion, cholestasis, was evaluated by CDFDA staining. Lipid accumulation (steatosis) by LipidTOX assay and apoptosis by TUNEL assay (methods). Measurements were normalized to the number of nuclei. (***P < 0.001, *P < 0.05; n = 12). Data are from donor 653. All n values represent the number of experimental/biological repeats. Error bars indicate ± S.E. Scale bar = 200 μm.
Real time analysis of respiratory dynamics suggests slow accumulative damage induced by Valproate and Stavudine
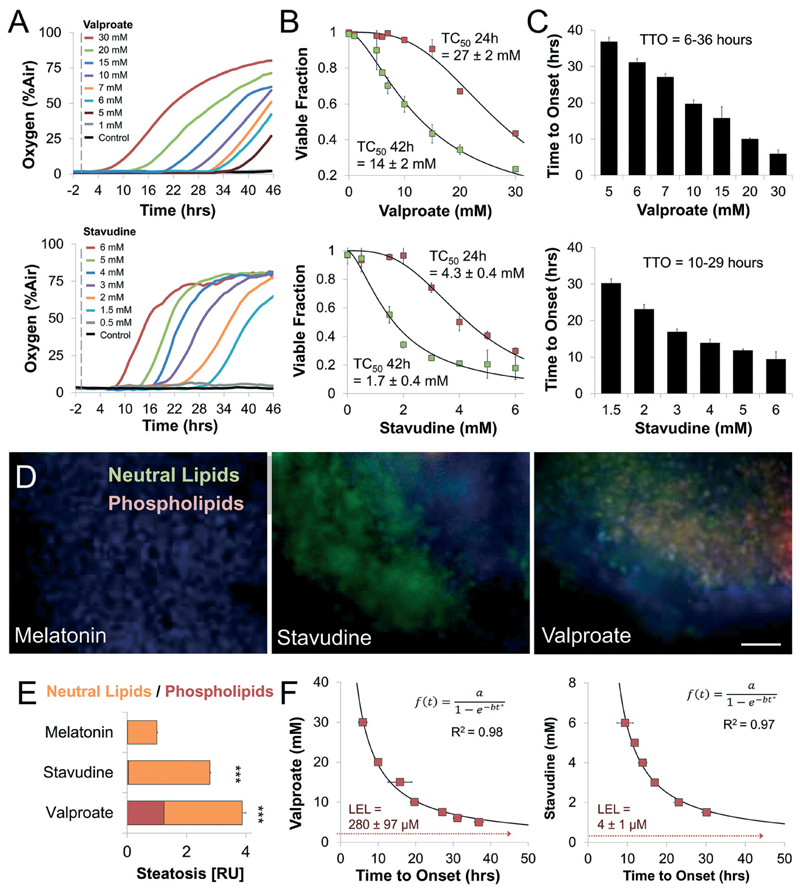
We have recently shown that time resolved information can offer insight into mechanism of action as direct damage (e.g. rotenone) occurs in minutes while indirect accumulative damage progresses over hours (e.g. amiodarone).4,40 To study the respiratory dynamics of Valproate and Stavudine we exposed our chip to each drug tracking effects for 46 hours (Fig. 3A). Both drugs showed progressive accumulation of damage, reaching TC50 of 14 ± 2 mM for Valproate and 1.7 ± 0.4 mM for Stavudine at 42 hours (Fig. 3B). Differences in TC50 between E6/E7LOW hepatocytes (Fig. 2C) and HepG2/C3A cells suggest metabolic activation is required for toxicity. In both cases, time to onset of damage (TTO) was dose dependent, ranging from 6–36 hours for Valproate to 10–29 hours for Stavudine (Fig. 3C). These results suggest a clear secondary mechanism of damage, possibly due to excess lipid accumulation. To confirm drug-induced steatosis on our chip, we stained the cells for neutral lipids and phospholipids (Fig. 3D). HepG2/C3A organoids showed strong 4-fold and 3-fold increase (p < 0.001, n = 9) in the amount of intracellular lipids for Valproate and Stavudine, respectively (Fig. 3E).
Fig. 3. Tissue-embedded microsensors show prolonged accumulative damage and analytical derivation of no observed effect levels in steatosis-inducing drugs.
(A) Representative oxygen uptake over time response of differentiated HepG2/C3A organoids exposed to increasing concentrations of Valproate and Stavudine. Dotted line notes exposure onset. (B) Dose-dependent toxicity curves of differentiated HepG2/C3A organoids treated with Valproate and Stavudine. TC50 for Valproate ranged from 27 mM at 24 hours to 14 mM at 42 hours. TC50 for Stavudine ranged from 4.3 mM at 24 hours to 1.7 mM at 42 hours. (C) Time to onset (TTO) of response of differentiated HepG2/C3A organoids to Valproate and Stavudine. Both drugs showed a dose-dependent decrease in TTO ranging from 6–36 hours in Valproate to 10–29 hours for Stavudine suggesting slowly accumulative steatotic damage. (D) Fluorescence micrographs and (E) quantification of steatosis and phospholipidosis in differentiated HepG2/C3A organoids as a result of exposure to different drugs. (***P < 0.001; n = 9). n represent the number of experimental repeats. Scale bar = 200 μm. (F) Analytical derivation of lowest exposure level (LEL) using the time to onset-dependent flux accumulation equation. LEL was defined as the horizontal asymptote, concentration for which onset of damage is at infinite time (methods). Valproate and Stavudine showed LEL of 280 ± 97 and 4 ± 1 μM, respectively, close to clinically reported Cmax. All error bars indicate ± standard error. TC50 and LEL error calculated by curve fitting (methods).
Interestingly, our data describes a relationship between drug concentration and the time it takes for damage to develop upon chronic exposure. Thus, we can fit the behavior on a flux accumulation model (methods) and extract the asymptotic concentration for which damage would occur only at infinite exposure time (Fig. 3F). Our analysis suggests that the lowest exposure levels (LEL) for Valproate and Stavudine are 280 ± 97 and 4 ± 1 μM, respectively. Alarmingly these calculated LEL levels fall within the observed Cmax in patients47–50 suggesting that the clinically observed steatosis for both drugs might be due to improper safety margins and should be revaluated.
Valproate induces a rapid shift from glycolysis to lipogenesis
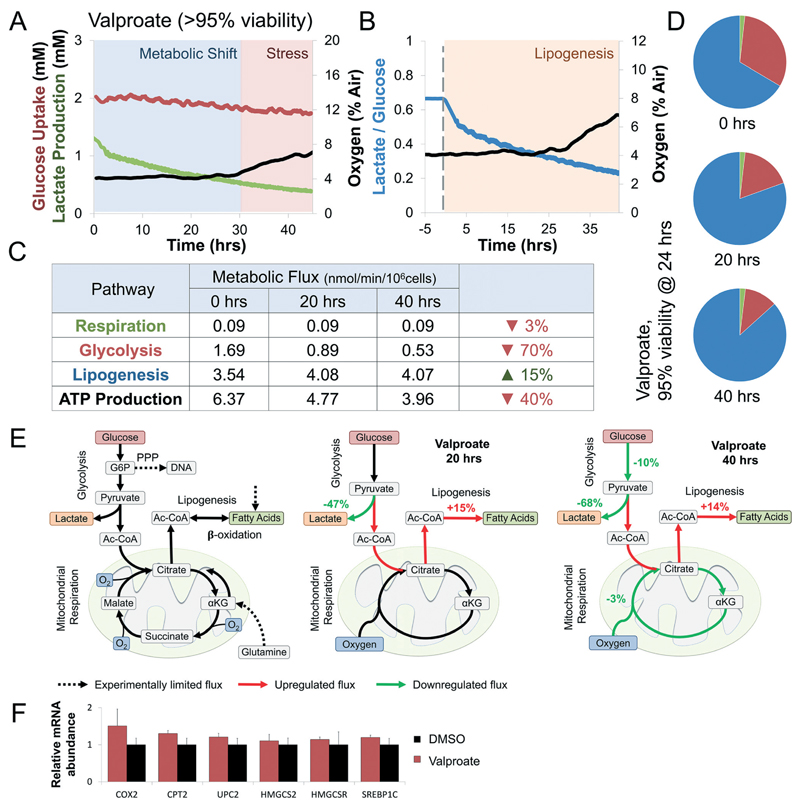
To study the metabolic effects of chronic Valproate exposure we chose a sub-toxic concentration correlating to 95% viability (Fig. 3B). Surprisingly, while oxygen consumption was only affected after 29 hours of exposure, lactate production dropped by 25% within minutes and continued to decrease throughout (Fig. 4A and B) suggesting direct modulation of cellular metabolism by Valproate. Intracellular metabolic fluxes were calculated following 0, 20, and 40 hours exposure to sub-toxic concentrations of Valproate (Fig. 3C). Lipogenesis increased by 15% while glycolysis and ATP production drop by 40% and 70%, respectively. The metabolic shift translated into an increasing percentage of glucose utilized for lipogenesis over other pathways as a function of time (Fig. 4D). Mapping metabolic fluxes at 20 and 40 hour time points illuminates Valproate mechanism of action. Valproate shifts pyruvate from lactate to citrate production, increasing lipogenesis through the first 30 hours of exposure (Fig. 4E). Lipid accumulation affects glucose uptake and mitochondrial function, with prolonged exposure leading to metabolic stress (Fig. 4E). These results suggest that disruption of β-oxidation may not play a major role in Valproate toxicity, and indeed gene expression analysis of E6/E7LOW hepatocytes shows no significant evidence of β-oxidation suppression (Fig. 4F), supporting enzymatic driven mechanism in sub-toxic exposure of human livers.
Fig. 4. Valproate induces a rapid shift from glycolysis to lipogenesis at sub-toxic concentrations.
(A) Curves of oxygen, glucose, and lactate fluxes during continuous perfusion with 5 mM Valproate. Oxygen uptake (black) drops by 3% only after 29 hours of continuous exposure. In contrast, glucose uptake (red) and lactate production (green) drop immediately after exposure. Determination of metabolic shift and stress phases are interpretation of the experimental results against trends measured simultaneously of a control bioreactor (Fig. S3A†). Metabolic shift was determined at the onset of a significant change in glucose uptake or lactate production, while stress was determined at the onset of a significant change in oxygen uptake. (B) Changes in lactate over glucose ratio following exposure to Valproate (blue line). Ratio drops by 25% immediately upon exposure, suggesting enzymatic rather than transcriptional effect. Lipogenesis was determined at the onset of a significant shift in the metabolic fluxes towered lipid synthesis (methods) (C) intracellular metabolic fluxes calculated following 0, 20, and 40 hours exposure to sub-toxic concentration Valproate (>95% viability). Glucose utilization in each pathway is shown as nmol min−1 per 106 cells as well as calculated ATP production (methods). Lipogenesis increases by 15% while glycolysis and ATP production drop by 40% and 70%, respectively. (D) Relative glucose utilization is shown as pie chart. Lipogenesis utilizes an increasing percentage of available glucose during Valproate exposure. (E) Schematics depicting the metabolic response of liver cells to Valproate. Dotted arrows note experimentally-limited fluxes, red and green arrows note up- and down-regulated fluxes, respectively. Valproate exposure shift glucose from lactate to citrate production, increasing lipogenesis through the first 30 hours of exposure. Continued exposure suppressed glucose uptake and oxidative respiration, hallmarks of metabolic stress. (F) Gene expression analysis in E6/E7LOW hepatocytes shows metabolic changes but no significant evidence of β-oxidation suppression, supporting enzymatic driven mechanism in sub-toxic exposure.
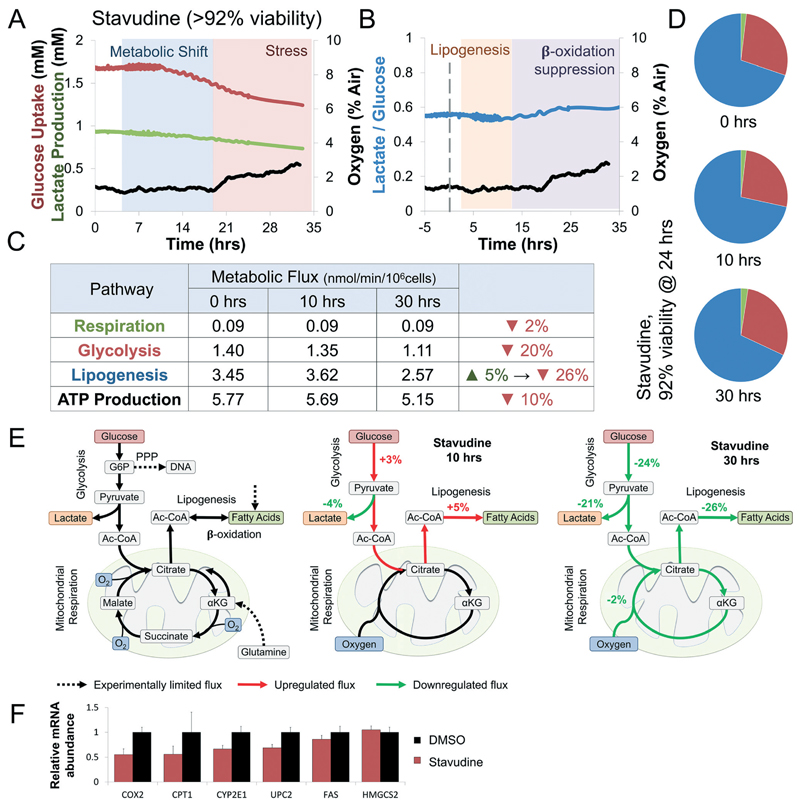
Stavudine (d4T) shows a transient lipogenesis and global metabolic suppression
To study the metabolic effects of chronic Stavudine exposure we chose a sub-toxic concentration correlating to 92% viability (Fig. 5B). As expected oxygen consumption was only affected after 20 hours of exposure suggesting slow accumulation of damage. Metabolic changes were only observed after 2–3 hours following exposure, suggesting transcriptional effects of Stavudine. Glucose uptake was upregulated during the first 10 hours (Fig. 5B), followed by gradual down-regulation. Lactate over glucose ratio dropped by 7% in the first 10 hours, suggesting a shift from lactate to citrate production, similar to Valproate (Fig. 4B and 5B), however, the ratio increase by 13% after 11 hours suggesting mild mitochondrial stress (Fig. 5B). Intracellular metabolic fluxes were calculated following 0, 10, and 30 hours exposure to sub-toxic concentrations of Stavudine (Fig. 5C). Lipogenesis increases by 5% with the first 10 hours. In contrast, both glycolysis and lipogenesis were suppressed at 30 hours, while ATP production was only marginally affected. Interestingly, no significant changes in the percentage of glucose utilization can be seen (Fig. 5D). Mapping metabolic fluxes at 10 and 30 hour time points illuminates Stavudine mechanism of action. Stavudine exposure causes a minor shift of pyruvate from lactate to citrate production, increasing lipogenesis by only 5% through the first 10 hours of exposure (Fig. 5E). However, prolonged exposure, coupled with accumulation of triglycerides, suppressed glucose uptake and all subsequent pathways suggesting mild mitochondrial stress.4,51 Our data suggest that lipid accumulation is not a result of increased production but rather suppression of fatty acid oxidation. Indeed, gene expression analysis of E6/E7LOW hepatocytes shows Stavudine-induced down-regulation of β-oxidation genes CPT1, COX2, UPC2, and CYP2E1 (Fig. 5F), supporting transcriptionally driven β-oxidation suppression in sub-toxic exposure of human livers.
Fig. 5. Stavudine (d4T) shows a transient increase in lipogenesis followed by global suppression of glucose utilization at sub-toxic concentrations.
(A) Curves of oxygen, glucose, and lactate fluxes during continuous perfusion with 1.5 mM Stavudine. Oxygen uptake (black) drops by 2% only after 20 hours of continuous exposure. Glucose uptake (red) and lactate production (green) drop 11 hours following exposure. Determination of metabolic shift and stress phases are interpretation of the experimental results against trends measured simultaneously of a control bioreactor (Fig. S3A†). Metabolic shift was determined at the onset of a significant change in glucose uptake or lactate production, while stress was determined at the onset of a significant change in oxygen uptake. (B) Changes in lactate over glucose ratio following exposure to Stavudine (blue line). Ratio drops by 7% in the first 10 hours, suggesting a shift from lactate to citrate production. Ratio increase by 13% after 11 hours suggesting mild mitochondrial stress. Lipogenesis or β-oxidation were determined at the onset of a significant shift in the metabolic fluxes towards either phase (methods) (C) intracellular metabolic fluxes calculated following 0, 10, and 30 hours exposure to sub-toxic concentration Stavudine (>92% viability). Glucose utilization in each pathway is shown as nmol min−1 per 106 cells as well as calculated ATP production (methods). Lipogenesis increases by 5% with the first 10 hours, primarily due to increase glucose uptake and a shift from lactate to citrate production. In contrast, both glycolysis and lipogenesis were suppressed at 30 hours, while ATP production was only marginally affected. (D) Relative glucose utilization is shown as pie chart. No significant changes in the percentage of glucose utilization is seen. (E) Schematics depicting the metabolic response of liver cells to Stavudine. Dotted arrows note experimentally-limited fluxes, red and green arrows note up- and down-regulated fluxes, respectively. Stavudine exposure shift glucose from lactate to citrate production, marginally increasing lipogenesis through the first 10 hours of exposure. Continued exposure suppressed glucose uptake and oxidative respiration, and lipogenesis hallmarks of mild mitochondrial stress. (F) Gene expression analysis in E6/E7LOW hepatocytes shows β-oxidation genes CPT1, COX2, UPC2, and CYP2E1 are inhibited by Stavudine suggesting a slow transcriptional suppression of lipid oxidation.
Conclusions
In this work we established a robust microphysiological flux analysis platform with streamlined microfluidic analysis of central carbon metabolism and glucose utilization (Fig. 1). Our on-chip CPU-synchronized sensors permit unparalleled measurements at a rate of 1 per 25 seconds for 3–4 days, allowing us to discover rapid changes in cellular metabolism such as the quick shift from lactate to citrate induced by within minutes of Valproate exposure (Fig. 4). In addition, integrated noise reduction, and stabilized electrochemical measurements, allowed us to run 60 hour experiments with continuous metabolic measurement, about 20 000 data points, uncovering the dynamics of both fast and slow metabolic events (Fig. 4 and 5). In fact, though metabolic adaptation was detectable within minutes to hours, mitochondrial stress took more than a day to develop (Fig. 4 and 5) and would have been missed using standard assays.
Non-alcoholic fatty liver disease is a growing epidemic affecting over 25% of the global population.9,12,52 Driven by life style choices and prescription drug use, the disease can range from steatosis to steatohepatitis, cirrhosis, and hepatocellular carcinoma. Interestingly, in spite of fundamental differences between human and rodent in the regulation of lipid metabolism,53–56 rodents are still predominantly being used to study the disease. Importantly, while in human metabolism glucose serves as the predominant precursor for lipogenesis, rodents utilize acetic acid in de novo lipogenesis, circumventing mitochondrial pathways. In fact, most drug candidates identified in rodent models of fatty liver injury have not proven effective in clinical studies.57,58 This suggests the development of a robust microphysiological model of human fatty liver disease could prove useful in drug development and safety assessment.
In current understanding, Valproate-induced steatosis is caused by β-oxidation impairment,26 due to the formation of valproyl-CoA conjugate in the mouse liver. Interestingly, valproyl-CoA is virtually undetectable in Valproate treated patients serum or urine sample59,60 and in a sub-toxic concentration its formation should be negligible.26,61 However, in mouse models exposed to high doses of Valproate, valproyl-CoA leads to mitochondrial CoA depletion, coupled with CPT1 inhibition, blocks β-oxidation and impairs ATP production.62,63 In rat hepatocytes, toxic concentration of Valproate induces mitochondrial swelling, increase ROS production and led to cytochrome C release, suggesting direct mitochondrial damage and apoptosis.29,64 These findings, however, do no correlate to development of Valproate-induced liver injury in the clinical settings, nor with our findings (Fig. 3 and 4). Valproate induces only a mild elevation of liver enzymes in patients suggesting minimal cellular damage,65 developing damage only month to years following initial exposure.25–27
Our findings demonstrate that sub-toxic levels of Valproate show no indication of direct mitochondrial damage in human hepatocytes (Fig. 4). Analysis of metabolic fluxes following Valproate exposure shows a shift from pyruvate production of lactate to citrate, occurring in minutes following exposure (Fig. 4A and B) and leading to increased lipogenesis. Mild mitochondrial stress occurs, but only after prolonged exposure to microvesicular steatosis (Fig. 4E) and we find no evidence of apoptosis (Fig. 2D). Surprisingly, β-oxidation impairment does not appear to play a major role in Valproate induced steatosis in human hepatocytes (Fig. 4F), at least not in sub-toxic concentration. Importantly, recent studies exposing mice and rats to sub-toxic levels of Valproate similarly showed no indication of β-oxidation impairment confirming our findings.66,67 These results demonstrate that mechanism of acute toxicity differ from chronic exposure to sub-toxic concentrations of the drug, and that dynamic assessment of metabolic stress could accurately replicate clinically observed manifestations of damage in human patients.
Stavudine (d4T) related hepatotoxicity is suggested to occur by inhibition of DNA polymerase γ, the enzyme responsible for the replication of mitochondrial DNA.33,34 Early studies showed that toxic concentration of Stavudine led to mitochondrial dysfunctional, ROS production, and apoptosis.68 Recently, it has also been suggested that intracellular lipid accumulation may be the result of interactions with nuclear receptors and subsequent transcriptional changes.69–72
Our findings show that sub-toxic levels of Stavudine show no indication of direct mitochondrial damage, with ATP production remaining relatively unchanged throughout the exposure (Fig. 5C and D). Analysis of the metabolic fluxes following Stavudine exposure suggests that sub-toxic concentrations cause a transient increase in lipogenesis followed by global suppression of both glycolysis and lipogenesis (Fig. 5). Indeed, gene expression analysis suggests that in this case, it does seem that transcriptional suppression of β-oxidation leads to lipid accumulation in human hepatocytes (Fig. 5F). Our data fits recent findings regarding the ability of Stavudine to bind LXR and ER, suppressing PPAR-induced β-oxidation. Transcriptional modulation seems like a prominent lead to understand and prevent steatosis caused by sub-toxic exposure to Stavudine and other antiretroviral drugs.
In conclusion, our work demonstrates the real-time measurement of glucose, lactate, and oxygen in a liver-on-chip bioreactor perfused under physiological conditions. The platform is uniquely able to monitor metabolic changes indicating metabolic shifts and phenomena occurring at drug concentrations previously regarded as safe, revealing mechanisms relevant to the clinical manifestations of chronic exposure to drug-induced liver injury.
Supplementary Material
† Electronic supplementary information (ESI) available. See DOI: 10.1039/c8lc00357b
Acknowledgements
This research was carried out at the Alexander Grass Center for Bioengineering at the Hebrew University of Jerusalem. Funding was provided by European Research Council Consolidator Grant OCLD (project no. 681870), by grant no. 3-13971 from the Chief Scientist Office of the Ministry of Health, Israel and L'Oréal Research and Innovation. The authors thank Mrs. Sheila Dunphy, Mr. Steven Van Put (CMST, IMEC) and Mr. Gerhard Jobst (Jobst Technologies GmbH) for technical support.
Footnotes
Avner Ehrlich: 0000-0002-6339-1665
Muneef Ayyash: 0000-0001-7842-6501
Merav Cohen: 0000-0001-7802-0499
Yaakov Nahmias: 0000-0002-6051-616X
Conflicts of interest
Y. N. holds equity in Tissue Dynamics Ltd. and chairs its scientific advisory board.
References
- 1.Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, et al. Regul Toxicol Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien PJ, Irwin W, Diaz D, Howard-Cofield E, Krejsa CM, Slaughter MR, Gao B, Kaludercic N, Angeline A, Bernardi P, Brain P, et al. Arch Toxicol. 2006;80:580–604. doi: 10.1007/s00204-006-0091-3. [DOI] [PubMed] [Google Scholar]
- 3.Bhatia SN, Ingber DE. Nat Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 4.Bavli D, Prill S, Ezra E, Levy G, Cohen M, Vinken M, Vanfleteren J, Jaeger M, Nahmias Y. Proc Natl Acad Sci U S A. 2016;113:E2231–2240. doi: 10.1073/pnas.1522556113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmalzlin E, van Dongen JT, Klimant I, Marmodee B, Steup M, Fisahn J, Geigenberger P, Lohmannsroben HG. Biophys J. 2005;89:1339–1345. doi: 10.1529/biophysj.105.063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papkovsky DB. Methods Enzymol. 2004;381:715–735. doi: 10.1016/S0076-6879(04)81046-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, Massa S, Riahi R, Chae S, Hu N, Avci H, et al. Proc Natl Acad Sci U S A. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weltin A, Slotwinski K, Kieninger J, Moser I, Jobst G, Wego M, Ehret R, Urban GA. Lab Chip. 2014;14:138–146. doi: 10.1039/c3lc50759a. [DOI] [PubMed] [Google Scholar]
- 9.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 10.Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, Landt CL, Harrison SA. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 11.Vernon G, Baranova A, Younossi ZM. Aliment Pharmacol Ther. 2011;34:274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 12.Lazo M, Hernaez R, Eberhardt MS, Bonekamp S, Kamel I, Guallar E, Koteish A, Brancati FL, Clark JM. Am J Epidemiol. 2013;178:38–45. doi: 10.1093/aje/kws448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrade RJ, Lucena MI, Fernandez MC, Pelaez G, Pachkoria K, Garcia-Ruiz E, Garcia-Munoz B, Gonzalez-Grande R, Pizarro A, Duran JA, Jimenez M, et al. Gastroenterology. 2005;129:512–521. doi: 10.1016/j.gastro.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Kleiner DE, Chalasani NP, Lee WM, Fontana RJ, Bonkovsky HL, Watkins PB, Hayashi PH, Davern TJ, Navarro V, Reddy R, Talwalkar JA, et al. Hepatology. 2014;59:661–670. doi: 10.1002/hep.26709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massart J, Begriche K, Moreau C, Fromenty B. J Clin Transl Res. 2017;3:212–232. doi: 10.18053/jctres.03.2017S1.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hautekeete ML, Degott C, Benhamou JP. Acta Clin Belg. 1990;45:311–326. doi: 10.1080/17843286.1990.11718105. [DOI] [PubMed] [Google Scholar]
- 17.Tandra S, Yeh MM, Brunt EM, Vuppalanchi R, Cummings OW, Unalp-Arida A, Wilson LA, Chalasani N. J Hepatol. 2011;55:654–659. doi: 10.1016/j.jhep.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benedict M, Zhang X. World J Hepatol. 2017;9:715–732. doi: 10.4254/wjh.v9.i16.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wanless IR, Dore S, Gopinath N, Tan J, Cameron R, Heathcote EJ, Blendis LM, Levy G. Gastroenterology. 1990;98:497–501. doi: 10.1016/0016-5085(90)90845-r. [DOI] [PubMed] [Google Scholar]
- 20.Freneaux E, Fromenty B, Berson A, Labbe G, Degott C, Letteron Larrey PD, Pessayre D. J Pharmacol Exp Ther. 1990;255:529–535. [PubMed] [Google Scholar]
- 21.Letteron P, Brahimi-Bourouina N, Robin MA, Moreau A, Feldmann G, Pessayre D. Am J Physiol. 1997;272:G1141–1150. doi: 10.1152/ajpgi.1997.272.5.G1141. [DOI] [PubMed] [Google Scholar]
- 22.Kimura S, Kobayashi T, Tanaka Y, Sasaki Y. Brain Dev. 1991;13:95–100. doi: 10.1016/s0387-7604(12)80114-8. [DOI] [PubMed] [Google Scholar]
- 23.Freneaux E, Labbe G, Letteron P, The Le D, Degott C, Geneve J, Larrey D, Pessayre D. Hepatology. 1988;8:1056–1062. doi: 10.1002/hep.1840080513. [DOI] [PubMed] [Google Scholar]
- 24.Luef GJ, Waldmann M, Sturm W, Naser A, Trinka E, Unterberger I, Bauer G, Lechleitner M. Ann Neurol. 2004;55:729–732. doi: 10.1002/ana.20074. [DOI] [PubMed] [Google Scholar]
- 25.Scheffner D, Konig S, Rauterberg-Ruland I, Kochen W, Hofmann WJ, Unkelbach S. Epilepsia. 1988;29:530–542. doi: 10.1111/j.1528-1157.1988.tb03757.x. [DOI] [PubMed] [Google Scholar]
- 26.Silva MF, Aires CC, Luis PB, Ruiter JP, L IJ, Duran M, Wanders RJ, Tavares de Almeida I. J Inherited Metab Dis. 2008;31:205–216. doi: 10.1007/s10545-008-0841-x. [DOI] [PubMed] [Google Scholar]
- 27.Zimmerman HJ, Ishak KG. Hepatology. 1982;2:591–597. doi: 10.1002/hep.1840020513. [DOI] [PubMed] [Google Scholar]
- 28.Zhang LF, Liu LS, Chu XM, Xie H, Cao LJ, Guo C, A JY, Cao B, Li MJ, Wang GJ, Hao HP. Acta Pharmacol Sin. 2014;35:363–372. doi: 10.1038/aps.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong V, Teng XW, Chang TK, Abbott FS. Toxicol Sci. 2005;86:436–443. doi: 10.1093/toxsci/kfi185. [DOI] [PubMed] [Google Scholar]
- 30.Eyer F, Felgenhauer N, Gempel K, Steimer W, Gerbitz KD, Zilker T. J Clin Psychopharmacol. 2005;25:376–380. doi: 10.1097/01.jcp.0000168485.76397.5c. [DOI] [PubMed] [Google Scholar]
- 31.Guaraldi G, Squillace N, Stentarelli C, Orlando G, D'Amico R, Ligabue G, Fiocchi F, Zona S, Loria P, Esposito R, Palella F. Clin Infect Dis. 2008;47:250–257. doi: 10.1086/589294. [DOI] [PubMed] [Google Scholar]
- 32.Crum-Cianflone N, Dilay A, Collins G, Asher D, Campin R, Medina S, Goodman Z, Parker R, Lifson A, Capozza T, Bavaro M, et al. J Acquired Immune Defic Syndr. 2009;50:464–473. doi: 10.1097/QAI.0b013e318198a88a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundar K, Suarez M, Banogon PE, Shapiro JM. Crit Care Med. 1997;25:1425–1430. doi: 10.1097/00003246-199708000-00034. [DOI] [PubMed] [Google Scholar]
- 34.Akhtar MA, Mathieson K, Arey B, Post J, Prevette R, Hillier A, Patel P, Ram LJ, Van Thiel DH, Nadir A. Eur J Gastroenterol. Hepatol. 2008;20:1194–1204. doi: 10.1097/MEG.0b013e328305b9e0. [DOI] [PubMed] [Google Scholar]
- 35.Levy G, Bomze D, Heinz S, Ramachandran SD, Noerenberg A, Cohen M, Shibolet O, Sklan E, Braspenning J, Nahmias Y. Nat Biotechnol. 2015;33:1264–1271. doi: 10.1038/nbt.3377. [DOI] [PubMed] [Google Scholar]
- 36.Wenner MM, Wilson TE, Davis SL, Stachenfeld NS. J Appl Physiol. 2011;111:1703–1709. doi: 10.1152/japplphysiol.00780.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prentice RL. Biometrics. 1976;32:761–768. [PubMed] [Google Scholar]
- 38.Ast C, Schmalzlin E, Lohmannsroben HG, van Dongen JT. Sensors. 2012;12:7015–7032. doi: 10.3390/s120607015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelhard S, Kumke MU, Lohmannsroben HG. Anal Bioanal Chem. 2006;384:1107–1112. doi: 10.1007/s00216-005-3364-4. [DOI] [PubMed] [Google Scholar]
- 40.Prill S, Bavli D, Levy G, Ezra E, Schmalzlin E, Jaeger MS, Schwarz M, Duschl C, Cohen M, Nahmias Y. Arch Toxicol. 2016;90:1181–1191. doi: 10.1007/s00204-015-1537-2. [DOI] [PubMed] [Google Scholar]
- 41.El-Kareh AW, Secomb TW. Neoplasia. 2000;2:325–338. doi: 10.1038/sj.neo.7900096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Currie GM. J Nucl Med Technol. 2018;46(2):81–86. doi: 10.2967/jnmt.117.199588. [DOI] [PubMed] [Google Scholar]
- 43.Gowrishankar TR, Weaver JC. Proc Natl Acad Sci U S A. 2003;100:3203–3208. doi: 10.1073/pnas.0636434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotnik T, Miklavcic D. IEEE Trans Biomed Eng. 2000;47:1074–1081. doi: 10.1109/10.855935. [DOI] [PubMed] [Google Scholar]
- 45.Ouattara DA, Prot JM, Bunescu A, Dumas ME, Elena-Herrmann B, Leclerc E, Brochot C. Mol BioSyst. 2012;8:1908–1920. doi: 10.1039/c2mb25049g. [DOI] [PubMed] [Google Scholar]
- 46.Levy G, Habib N, Guzzardi MA, Kitsberg D, Bomze D, Ezra E, Uygun BE, Uygun K, Trippler M, Schlaak JF, Shibolet O, et al. Nat Chem Biol. 2016;12:1037–1045. doi: 10.1038/nchembio.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sussman NM, McLain LW., Jr JAMA. 1979;242:1173–1174. [PubMed] [Google Scholar]
- 48.Klotz U, Antonin KH. Clin Pharmacol Ther. 1977;21:736–743. doi: 10.1002/cpt1977216736. [DOI] [PubMed] [Google Scholar]
- 49.Rezk NL, Tidwell RR, Kashuba AD. J Chromatogr B: Anal Technol Biomed Life Sci. 2003;791:137–147. doi: 10.1016/s1570-0232(03)00224-1. [DOI] [PubMed] [Google Scholar]
- 50.Verweij-van Wissen CP, Aarnoutse RE, Burger DM. J Chromatogr B: Anal Technol Biomed Life Sci. 2005;816:121–129. doi: 10.1016/j.jchromb.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Bournat JC, Brown CW. Curr Opin Endocrinol, Diabetes Obes. 2010;17:446–452. doi: 10.1097/MED.0b013e32833c3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bedogni G, Miglioli L, Masutti F, Tiribelli C, Marchesini G, Bellentani S. Hepatology. 2005;42:44–52. doi: 10.1002/hep.20734. [DOI] [PubMed] [Google Scholar]
- 53.Di Girolamo M, Rudman D. Am J Physiol. 1966;210:721–727. doi: 10.1152/ajplegacy.1966.210.4.721. [DOI] [PubMed] [Google Scholar]
- 54.Bergen WG, Mersmann HJ. J Nutr. 2005;135:2499–2502. doi: 10.1093/jn/135.11.2499. [DOI] [PubMed] [Google Scholar]
- 55.Chandrasekera PC, Pippin JJ. ALTEX. 2014;31:157–176. doi: 10.14573/altex.1309231. [DOI] [PubMed] [Google Scholar]
- 56.Eizirik DL, Pipeleers DG, Ling Z, Welsh N, Hellerstrom C, Andersson A. Proc Natl Acad Sci U S A. 1994;91:9253–9256. doi: 10.1073/pnas.91.20.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cole BK, Feaver RE, Wamhoff BR, Dash A. Expert Opin Drug Discovery. 2018;13:193–205. doi: 10.1080/17460441.2018.1410135. [DOI] [PubMed] [Google Scholar]
- 58.Hansen HH, Feigh M, Veidal SS, Rigbolt KT, Vrang N, Fosgerau K. Drug Discovery Today. 2017;22:1707–1718. doi: 10.1016/j.drudis.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 59.McLaughlin DB, Andrews JA, Hooper WD, Cannell GR, Eadie MJ, Dickinson RG. Br J Clin Pharmacol. 2000;49:409–415. doi: 10.1046/j.1365-2125.2000.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghodke-Puranik Y, Thorn CF, Lamba JK, Leeder JS, Song W, Birnbaum AK, Altman RB, Klein TE. Pharmacogenet Genomics. 2013;23:236–241. doi: 10.1097/FPC.0b013e32835ea0b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aires CC, Ruiter JP, Luis PB, ten Brink HJ, Ijlst L, de Almeida IT, Duran M, Wanders RJ, Silva MF. Biochim Biophys Acta. 2007;1771:533–543. doi: 10.1016/j.bbalip.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Aires CC, Ijlst L, Stet F, Prip-Buus C, de Almeida IT, Duran M, Wanders RJ, Silva MF. Biochem Pharmacol. 2010;79:792–799. doi: 10.1016/j.bcp.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 63.Silva MF, Ruiter JP, L IJ, Allers P, ten Brink HJ, Jakobs C, Duran M, Tavares de Almeida I, Wanders RJ. Anal Biochem. 2001;290:60–67. doi: 10.1006/abio.2000.4947. [DOI] [PubMed] [Google Scholar]
- 64.Jafarian I, Eskandari MR, Mashayekhi V, Ahadpour M, Hosseini MJ. Toxicol Mech Methods. 2013;23:617–623. doi: 10.3109/15376516.2013.821567. [DOI] [PubMed] [Google Scholar]
- 65.Powell-Jackson PR, Tredger JM, Williams R. Gut. 1984;25:673–681. doi: 10.1136/gut.25.6.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seckin S, Basaran-Kucukgergin C, Uysal M. Pharmacol Toxicol. 1999;85:294–298. doi: 10.1111/j.1600-0773.1999.tb02025.x. [DOI] [PubMed] [Google Scholar]
- 67.Punapart M, Eltermaa M, Oflijan J, Sutt S, Must A, Koks S, Schalkwyk LC, Fernandes C, Vasar E, Soomets U, Terasmaa A. PPAR Res. 2014;2014 doi: 10.1155/2014/349525. 349525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stankov MV, Panayotova-Dimitrova D, Leverkus M, Vondran FW, Bauerfeind R, Binz A, Behrens GM. AIDS. 2012;26:1995–2006. doi: 10.1097/QAD.0b013e32835804f9. [DOI] [PubMed] [Google Scholar]
- 69.da Cunha J, Maselli LM, Stern AC, Spada C, Bydlowski SP. World J Virol. 2015;4:56–77. doi: 10.5501/wjv.v4.i2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Svard J, Blanco F, Nevin D, Fayne D, Mulcahy F, Hennessy M, Spiers JP. Br J Pharmacol. 2014;171:480–497. doi: 10.1111/bph.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang ZX, Shen W, Sun H. Hepatol Int. 2010;4:741–748. doi: 10.1007/s12072-010-9202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viengchareun S, Caron M, Auclair M, Kim MJ, Frachon P, Capeau J, Lombes M, Lombes A. Antiviral Ther. 2007;12:919–929. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.