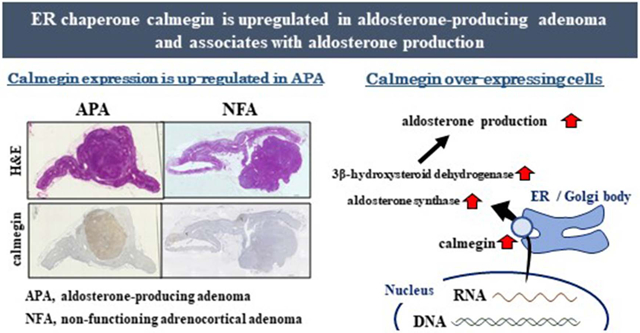
Abstract
The endoplasmic reticulum (ER) plays a pivotal role in syntheses of proteins and steroid hormones, and regulation of intracellular Ca2+ level. We aimed to investigate ER-associated genes in aldosterone-producing adenomas (APAs) and clarify their effect on aldosterone production. Microarray analysis targeting 288 ER-associated genes was conducted using non-functioning adrenocortical adenomas (NFA, n = 5) and APAs (n = 19). Immunohistochemistry (IHC) and qPCR analyses were performed with 13 NFA and 48 APA samples. Functional studies were performed with human adrenocortical carcinoma (HAC15) cells, some of which were genetically modified using lentiviruses. The ER chaperone calmegin (CLGN) was the most highly expressed ER-associated gene in APAs relative to NFAs. Analysis with qPCR revealed CLGN to be 9.5-fold upregulated in APAs relative to NFAs. There were no differences among different APA genotypes affecting aldosterone production. IHC analysis revealed that CLGN was strongly expressed in APAs and aldosterone-producing cell clusters. Angiotensin II stimulation or KCNJ5 T158A overexpression in HAC15 cells did not affect CLGN mRNA levels. CLGN overexpression in HAC15 cells increased aldosterone levels, but did not stimulate CYP11B2 mRNA levels. Pathway and gene ontology analyses using RNA sequencing results showed that tRNA aminoacyl metabolism was the most enriched pathway in CLGN-overexpressing cells. CYP11B2 and HSD3B2 protein expression were more abundant in CLGN-overexpressing cells. CLGN knockdown using CRISPR/Cas9 method in HAC15 cells that carry the KCNJ5 mutation did not affect aldosterone production. To summarize, CLGN was upregulated and associated with aldosterone production via translational regulation of CYP11B2 in APAs.
Keywords: aldosterone-producing adenoma, CYP11B2, translation, CLGN, aldosterone
Graphical Abstract
Introduction
Aldosterone is an important hormone that regulates blood pressure by promoting sodium reabsorption in the distal tube of the kidney. Primary aldosteronism (PA), which is the most common form of secondary hypertension, leads to cardiovascular disease and renal dysfunction by excess aldosterone secretion.1, 2 The rates of these complications are higher than the rates in essential hypertension patients with similar risk profiles.1, 2 There are two main subtypes of PA: aldosterone-producing adenoma (APA) and bilateral idiopathic hyperaldosteronism.3 In APA, CYP11B2 (aldosterone synthase) transcription is activated by intracellular Ca2+ signaling, resulting in autonomous over-production of aldosterone.4-7 However, the mechanisms by which translation mediated aldosterone production have not been elucidated.
The endoplasmic reticulum (ER) is a major site of steroid hormone synthesis along with mitochondria, and plays a pivotal role in modulating intracellular Ca2+ level via regulation of Ca2+ storage. Sustained Ca2+ signaling caused by mutated ion channels or pumps is a key factor for autonomous over-production of aldosterone in APA. Additionally, protein synthesis, quality control, and transport are one of the most important functions of the ER. The ER is recognized as a hub of post-transcriptional gene regulation for cellular signaling, and a recent study has suggested that cytosolic protein-encoding mRNA, as well as secretory and membrane proteins, are also translated on ER-bound ribosomes.8 Taken together, the ER is a crucial subcellular organelle for aldosterone synthesis, thus its function is presumed to be essential for aldosterone production in APA. However, the specific function of ER in APA has never been reported.
First, we sought a key molecule that regulates aldosterone production among ER-associated genes in APA, using a transcriptome-analysis-based approach. This analysis showed that the gene CLGN, encoding an ER chaperone protein called calmegin, had the highest expression in APA. We hypothesized that CLGN was associated with aldosterone production in APA, and it had a crucial role in aldosterone synthesis. Accordingly, we confirmed the regulatory function of CLGN in aldosterone production in adrenal cells.
Methods
The data that support the findings of this study are available from the corresponding author on reasonable request.
Patients and tissue samples
The diagnosis of PA was performed based on the guidelines of the Japan Endocrine Society,9 and the details were provided in our previous report and online-only Data Supplement.10 Non-functioning adrenocortical adenoma (NFA) was diagnosed based on radiological findings and endocrinological results, which did not show cortisol or aldosterone elevation, also as previously reported.11 Unilateral laparoscopic adrenalectomy was performed to remove the APA or NFA. The diagnosis of an APA was confirmed by detecting the expression of CYP11B2 by immunohistochemistry of the resected adrenal.12 The clinical characteristics of the patients with APA (n = 48) and NFA (n = 13) are shown in Table S1. The genotypes of APAs were ATP1A1 mutation (n = 5), KCNJ5 mutation (n = 27), and no mutation (n = 16). Our study was approved by the ethics committee of Hiroshima University, and a written informed consent was obtained from all the patients.
Cell culture and materials
The HAC15 cell line, a subclone of the H295R human adrenocortical carcinoma cells, was provided by W. E. Rainey (University of Michigan, USA).13 The cell culture was performed as previously reported.14 Angiotensin II (A-II) and forskolin were purchased from Sigma Aldrich Co. Ltd. (St. Louis, MO, USA).
Transcriptome analysis
Among the samples, 5 of 13 NFAs and 19 of 48 APAs were subjected to transcriptome analysis, and the data are the same as that previously reported.15 SurePrint G3 Human Gene Expression 8 × 60K v2, which included approximately 50,000 unique probes (Agilent Technologies Inc., Santa Clara, CA, USA), was used for the detection of gene expression. The gene set for ER-localized proteins in humans was retrieved from the AmiGO 2 website (http://amigo2.geneontology.org/amigo).
RNA extraction and qPCR assay
Total RNA extraction, reverse transcription, and qPCR assay of CYP11B2 and GAPDH were performed as previously reported.14 The oligonucleotide primers for CLGN were 5′-ACGACTTGGACATCTGAGCTGTC-3′ and 5′-TTCCGTCTCAACATCATCATCC-3′.
DNA extraction, genotyping, and methylation analysis
Genomic DNA was extracted from NFAs and APAs, and genotyping was performed as previously reported.16 In brief, genomic DNA from frozen tissue were genotyped in KCNJ5, ATP1A1, ATP2B3, CACNA1D, and CTNNB1 by PCR-based direct sequencing. The details of methylation analysis and clinical characteristics were described in our previous report.10 Briefly, genomic DNA from 12 NFA and 35 APA samples were analyzed by Infinium HumanMethylation450 BeadChip kit (Illumina, San Diego, CA) after bisulfite conversion. Methylation levels for each CpG residue were presented as β-values, which were expressed as 0 to 1, representing completely nonmethylated to completely methylated values, respectively.
Lentiviral production and infection
The lentiviral vector plasmid, pCDH-CMV-MCS-EF1-Puro-GFP, was purchased from System Bioscience (Palo Alto, CA, USA). Full length cDNA of CLGN was generated by PCR using an APA sample and ligated into the multiple cloning site of the plasmid. The open reading frame of the ATP1A1 gene with the L104R mutant was obtained from GenScript (Piscataway, NJ) and ligated into the multiple cloning site of the plasmid. For KCNJ5 mutation, KCNJ5-T158A plasmid was used as reported previously.7
To investigate the effect of silencing the CLGN gene in HAC15 cells, CRISPR/Cas9 vectors targeting specific sites and regions within the CLGN gene were constructed. The sgRNA (TAGTTAGGATTATCGACCAG) was designed using CHOPCHOP (http://chopchop.cbu.uib.no/index.php), and the gRNA (GCACTACCAGAGCTAACTCA) was used for the negative control. The sgRNA oligomers for CLGN and negative control were synthesized and cloned into the pLentiCRISPR v2 vector (GenScript Co. Piscataway, NJ), and denoted sgCLGN and sgControl, respectively.
To assess the effects of CLGN overexpression and KCNJ5 and ATP1A1 mutation, lentivirus production and infection in HAC15 cells were conducted as reported previously.7, 17 For the investigation of CLGN knock-down, the cells were stably infected with control or sgCLGN lentivirus as previously reported.7 Briefly, 72 h after infection, the cells stably expressing the transgene were subjected to 10 μg/ml puromycin selection for more than one week.
Western blot analysis
Cell lysis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transfer, and blot processing were performed as previously reported.15 Briefly, cells were lysed with RIPA buffer supplemented with a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, USA). Thereafter, 240 μg of total protein was separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. The blot was probed using antibodies against CLGN (Abcam, #EPR11832-11, Cambridge, UK) and β-actin (Cell Signaling, #4967, Danvers, MA, USA). Antibodies against human CYP11B2, StAR, and 3βHSD were applied as we previously reported.18, 19 Protein levels were determined as arbitrary units normalized against β-actin expression.
ER calcium levels
The Ca2+ concentration in the ER was measured by CEPIA (calcium-measuring organelle-entrapped protein indicator 1) as we previously reported.15 Briefly, HAC15 cells were first transfected with pCMV R-CEPIA1er plasmid (Addgene, Cambridge, MA) and 48 h later infected with CLGN or control lentivirus. After the incubation with Hanks Balanced Salt Solution without phenol red for 30 m, the protein was detected at excitation wavelength of 562 nm and an emission wavelength of 641 nm.
RNA Sequencing analysis
Total RNA from control and CLGN-overexpressing HAC15 cells was extracted as previously reported.14 For next generation sequencing, a paired-end RNA library was prepared from 1.0 μg total RNA according to the manufacturer’s instructions (Illumina Truseq Library Construction, Illumina Inc., San Diego, CA), and then sequenced on an Illumina Hiseq 2500 platform.
Animals
Dahl salt-sensitive (SSjr) rats were housed at University of Mississippi Medical Center. The detailed methods were previously reported.15 At 8 weeks of age, they were fed with low-salt containing (0.02% NaCl) but otherwise complete chow, or with complete chow in addition to high-salt water (0.9% NaCl) for 23 days before euthanasia and tissue removal for analysis. This protocol was approved from the Institutional animal review board.
Aldosterone, cortisol and protein assay
Aldosterone and cortisol levels in cell culture medium were measured using an ELISA kit as previously described.7 Cellular protein levels were measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The aldosterone and cortisol levels were normalized by cellular protein levels.
Statistical Analysis
Results were expressed as mean ± S.E. of at least three separate experiments in which each sample was assayed in triplicates. Data obtained from adrenal tumor were analyzed by Mann-Whitney U test or Kruskal-Wallis test. In vitro study, differences between groups were analyzed for statistical significance by t-test, and multiple groups were analyzed by one-way ANOVA followed by Bonferroni comparisons. P values < 0.05 from each analysis were considered statistically significant. The analyses were performed using SPSS for Windows (release 24.0; SPSS Inc., Chicago, IL, USA). R software package (https://www.r-project.org/) was used for the depiction of the heat map, and pathway or gene ontology (GO) analysis.
Results
CLGN is the highest differentially expressed gene in APA
We conducted a transcriptomic analysis of 288 ER-expressed genes in 19 APA and 5 NFA samples. The results are depicted as a heat map in Figure S1. All the differentially expressed genes were shown in our previous report.15 Among 288 ER-expressed genes, 16 genes showed a more than 2-fold expression difference between APA and NFA samples (Table S2). Of these differentially expressed genes, 5 were upregulated and 11 were downregulated in APA.
APA has higher CLGN expression
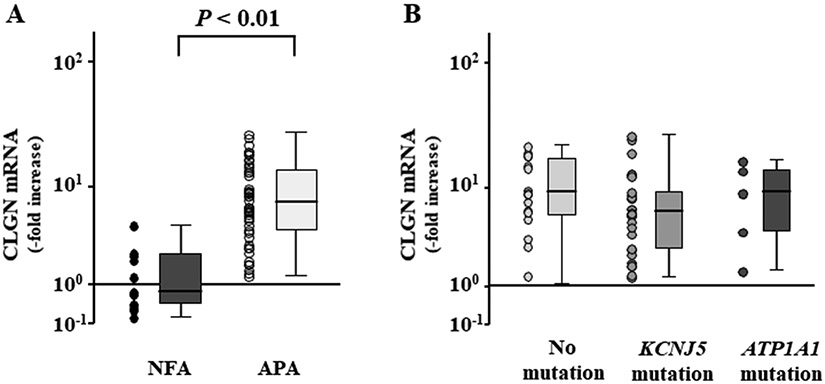
Although CLGN has two transcript variants, only the transcript variant 1 of CLGN was detected in the adrenal tissues by RNA sequence analysis (Figure S2A). We created the qPCR primers only for CLGN transcript variant 1 (Figure S2B), and qPCR assay revealed CLGN to be 9.5-fold upregulated in APAs (n = 48) relative to NFAs (n = 13) (Figure 1A). There were no differences of CLGN expression levels among the APA genotypes which drive aldosterone production (Figure 1B). As we previously reported, CYP11B2 expression was 535.1-fold higher in APAs than in NFAs according to qPCR analysis.15 The CLGN expression levels did not correlate with the CYP11B2 expression levels in the APAs (r = −0.063, P = 0.668).
Figure 1.
CLGN expression in aldosterone-producing adenoma (APA) (A) Comparison of CLGN mRNA expression levels between non-functioning adrenocortical adenoma (NFA) and APA. (B) CLGN mRNA expression levels among the APA genotypes including no known mutation, KCNJ5 mutation, and ATP1A1 mutation.
The diagnosis of adrenal overt Cushing’s syndrome was performed based on the guidelines of the Endocrine Society,20 and the details were provided in online-only Data Supplement and Table S3. There were no differences of CLGN expression between NFA and cortisol-producing adenoma (CPA) (Figure S3).
CLGN expression in APA and adrenal cells
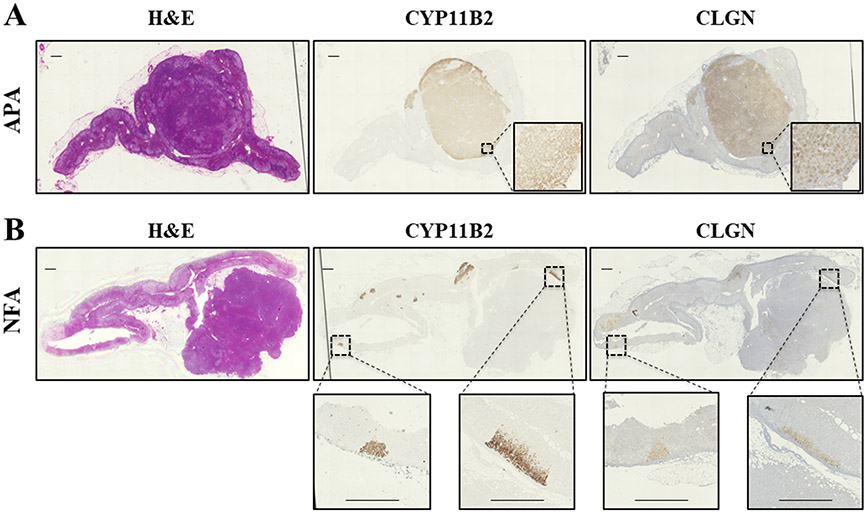
CLGN immunolocalization in APA and NFA are shown in Figure 2A and 2B, respectively. CLGN protein was found to be expressed in the APAs, but not in the NFA. CLGN expression is strong and clearly noticeable in aldosterone producing cell clusters (APCC) of the adrenal gland tissue adjacent to the APAs with no staining in the zona fasciculata or reticularis (Figure 2B).
Figure 2.
Immunohistochemistry analyses of CYP11B2 and CLGN in an aldosterone-producing adenoma (APA) and non-functioning adrenocortical adenoma (NFA). Hematoxylin and Eosin staining (left), and immunohistochemistry using antibodies against CYP11B2 (middle) and CLGN (right) in an APA (A) and NFA (B). Scale bar indicates 1 mm.
Adrenal glands of the rats fed with a high- or low-salt diet were immunostained with anti-CYP11B2 and anti-CLGN antibodies. As we previously reported in detail, adrenal ZG of the low-salt–fed rats was strongly immunoreactive for CYP11B2 and had a wider area than the ZG of the high-salt–fed rat (Figure S5). CLGN protein was not detected in adrenal glands of either low-salt– or high-salt–fed rats (Figure S5).
Effects of A-II, forskolin, and a KCNJ5 mutation on CLGN expression in HAC15 cells
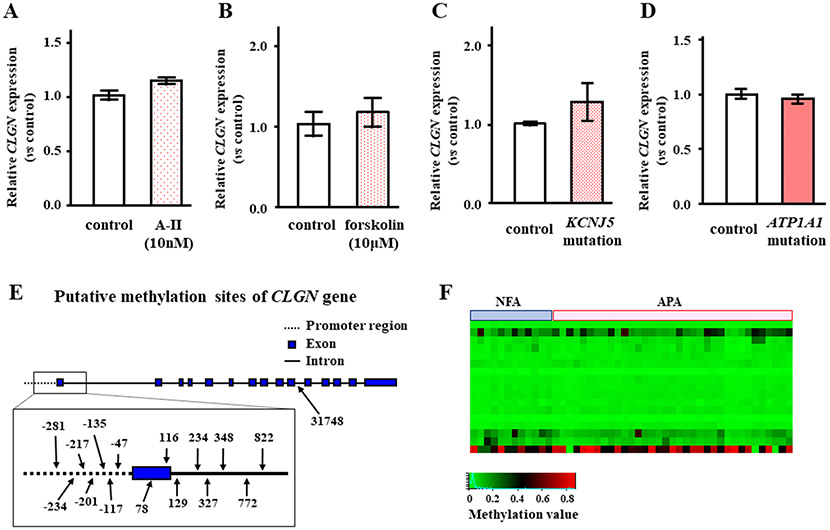
To investigate the regulation of CLGN expression, we tested A-II and forskolin stimulation in HAC15 cells. There were no effects of A-II or forskolin stimulation on CLGN mRNA expression in HAC15 cells (Figure 3A and 3B). The KCNJ5 or ATP1A1 mutation did not influence the transcription of CLGN in HAC15 cells (Figure 3C and 3D). Since major intracellular signaling to stimulate aldosterone production had no effect on CLGN expression, we analyzed the DNA methylation levels of CLGN between the NFAs and APAs. Sixteen DNA methylation sites were found on CLGN (Figure 3E). There was no difference in CLGN promoter methylation levels between the NFAs and APAs (Figure 3F).
Figure 3.
Effect of angiotensin II (A-II), forskolin, and KCNJ5 mutation in HAC15 cells, and DNA methylation on mRNA expression of CLGN in human adrenocortical tumors. Confluent HAC15 cells were serum deprived for 24 h, then incubated with A-II (A), forskolin (B), or no stimulant for 3 h. After aspiration of the media, the cells were harvested, and CLGN and GAPDH mRNA levels were measured (n = 3). (C) Serum-deprived HAC15 cells with or without a KCNJ5 mutation were harvested, and CLGN and GAPDH mRNA levels were measured (n = 3). (D) Serum-deprived HAC15 cells with or without a ATP1A1 mutation were harvested, and CLGN and GAPDH mRNA levels were measured (n = 3). (E) Putative methylation sites numbered from the transcription start site of CLGN gene are shown. (F) A heat map of CLGN methylation levels between NFA (n = 12) and APA (n = 35). There were no differences of CLGN methylation levels between NFA and APA.
Effects of CLGN overexpression in HAC15 cells
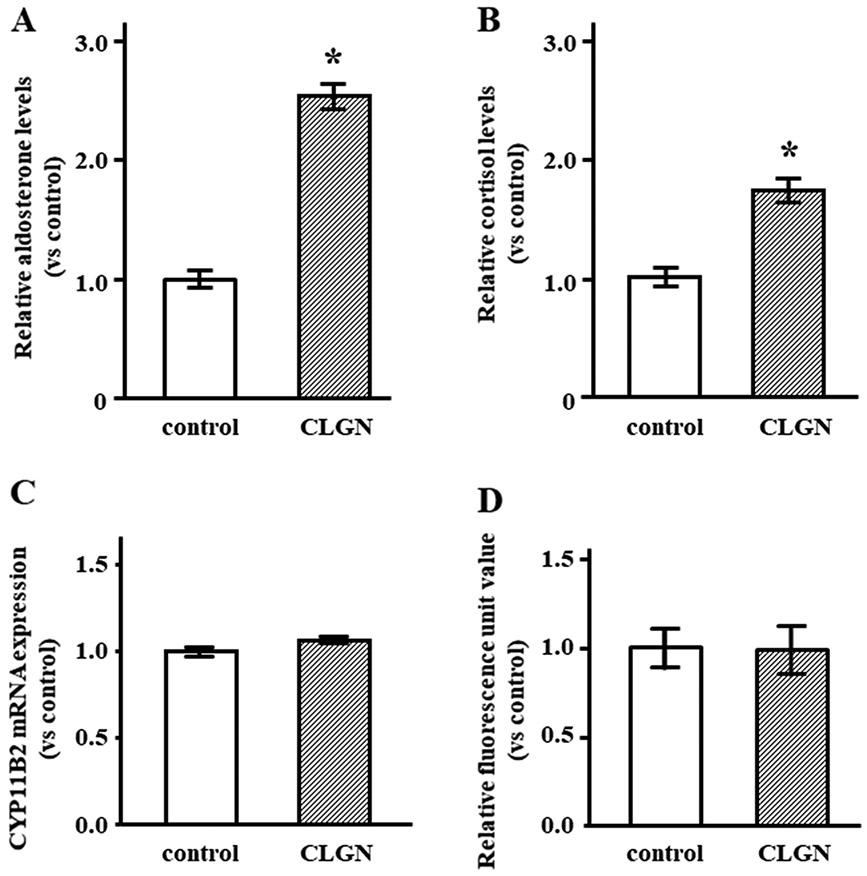
CLGN was delivered into HAC15 cells by lentiviruses. We measured aldosterone concentration in the medium of HAC15 cells with the overexpression of CLGN or control cDNA. HAC15 cells infected with CLGN had 2.5-fold higher aldosterone level relative to control cells (Figure 4A). Cortisol levels in HAC15 cells were slightly higher than those in control cells (Figure 4B). CYP11B2 mRNA expression levels were not different between CLGN-overexpressing and control cells (Figure 4C). We measured ER calcium levels using the calcium indicator CEPIA; no difference between CLGN-overexpressing and control cells was observed (Figure 4D).
Figure 4.
CLGN overexpression increased aldosterone production in HAC15 cells. (A, B) After confluent HAC15 cells with control or CLGN were serum deprived for 24 h, cells were incubated with fresh media with 0.1% serum for 24 h. Relative aldosterone and cortisol levels in the media are compared between HAC15 cells with control and CLGN. *P < 0.05 vs. control (n = 3). (C) Confluent HAC15 cells with control or CLGN were serum deprived for 24 h, and the serum-deprived cells were harvested for CYP11B2 and GAPDH mRNA levels were measured. *P < 0.05 vs. each control (n = 3). (D) After transduction of control or CLGN in HAC15 cells, R-CEPIAer was applied to compare intra-endoplasmic reticulum Ca2+ levels between control and CLGN cells (n = 3).
RNA sequence analysis in CLGN-overexpressing cells
The mRNA expressions of steroidogenic enzymes in CLGN-overexpressing cells did not change relative to those in control cells (Figure S6). To investigate the relationship between CLGN and aldosterone production, we performed pathway and GO analyses using the RNA sequencing data. tRNA metabolic process, aminoacyl tRNA biosynthesis, and cytosolic tRNA aminoacylation were the most enriched pathways identified using bioinformatics data such as GO (http://geneontology.org/), KEGG (https://www.genome.jp/kegg/), and REACTOME (https://reactome.org/), respectively. (Table S4). Expression levels of the genes related to these pathways in the CLGN-overexpressing cells are shown in Figure S7 with respect to the expression levels in native HAC15 cells
Protein levels of steroidogenic enzymes in CLGN-overexpressing cells
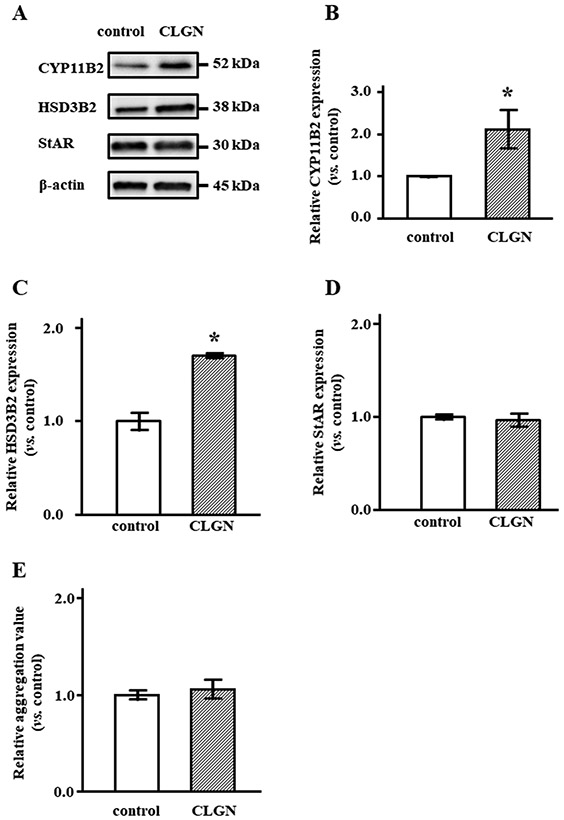
CYP11B2 and HSD3B2 protein levels were significantly increased in CLGN-overexpressing cells, by 2.1- and 1.7-fold, respectively (Figure 5A-C). StAR protein levels were not different between CLGN and control cells (Figure 5A, D). We examined whether CLGN overexpression affected protein aggregation because CLGN may act as a molecular chaperon. The aggregation levels in CLGN-overexpressing cells did not differ from those in control cells (Figure 5E), although protein production, including CYP11B2 was increased in the CLGN-overexpressing cells.
Figure 5.
Steroidogenic enzymes and StAR protein expression in HAC15 cells with control or CLGN. After confluent HAC15 cells with control or CLGN were serum deprived for 24 h, the cells were harvested for SDS-PAGE and immunoblotting analysis. (A) The band intensities of CYP11B2, HSD3B2, StAR and β-actin were depicted. (B-D) CYP11B2, HSD3B2 and StAR protein levels normalized by β-actin levels were compared in between control and CLGN over-expressing cells. *P < 0.05 vs. control (n = 3). (E) Protein aggregation measured using the Proteostat Protein Aggregation Assay Kit (Enzo Life Sciences, Farmingdale, NY, USA) following the manufacturer's instructions (n = 3).
Effects of CLGN suppression on aldosterone production
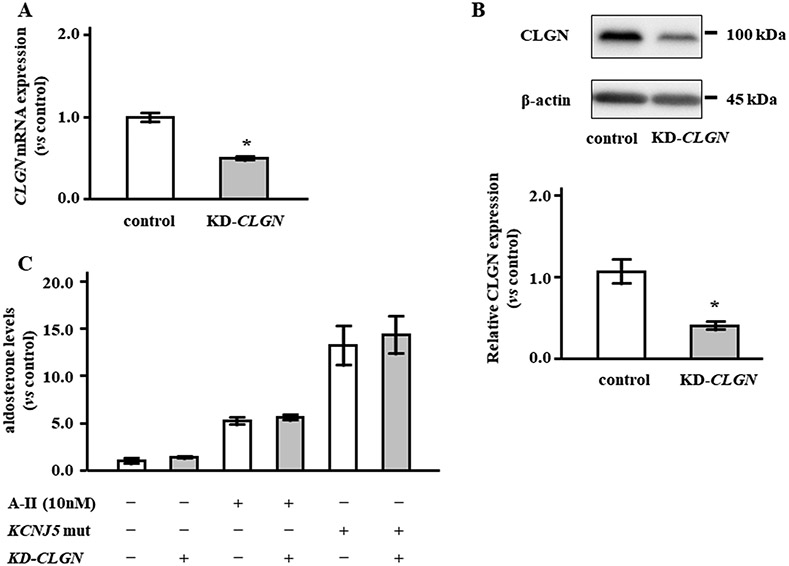
Infection of HAC15 cells with sgRNA-CLGN-carrying lentivirus significantly decreased mRNA levels by 49.8% and protein levels by 62.0% (Figure 6A and 6B). We investigated the effects of CLGN suppression on the aldosterone levels stimulated by A-II and KCNJ5 mutations. Knockdown of CLGN did not decrease aldosterone production in HAC15 cells stimulated by either A-II or overexpression of KCNJ5 mutation (Figure 6C).
Figure 6.
CLGN silencing by CRISPR/Cas9 technology had no effect on aldosterone production in HAC15 cells. (A) CLGN mRNA expression was analyzed in HAC15 cells with control (sg-Control) or knock-down CLGN (KD-CLGN). (B) CLGN protein levels were evaluated by western blot in control and KD-CLGN cells. CLGN protein levels were adjusted by β-actin, and the protein levels in KD-CLGN cells were expressed as fold change vs. control cells (C) Relative aldosterone levels between control and KD-CLGN cells +/− angiotensin II (A-II) stimulation or KCNJ5 mutation (KCNJ5 mut) transduction. Results are shown as fold change vs. control cells. *P < 0.05 vs. control (n = 3).
Discussion
We demonstrated that CLGN is highly expressed in APA and APCC, but not in CPA or ZG of rat adrenal. HAC15 cells infected with CLGN had higher aldosterone levels relative to control cells, however mRNA expressions of steroidogenic enzymes were not increased in HAC15 cells with CLGN. RNA sequencing analysis indicated that CLGN has a role in aminoacyl tRNA biosynthesis. CLGN overexpression in HAC15 cells increased CYP11B2 and HSD3B2 protein expression. Knockdown of CLGN did not affect aldosterone production in HAC15 cells stimulated by A-II or overexpression of KCNJ5 T158A.
We showed that overexpression of CLGN potentiated aldosterone production by enhancing CYP11B2 protein, but not mRNA, level in HAC15 cells. CLGN is a testis-specific molecular chaperone that disappears along with the elimination of the ER during spermatogenesis.21 Male CLGN knockout mice are infertile because their sperm are unable to bind the zona pellucida.22 However detailed functions of CLGN are still unknown. Our pathway analysis using RNA sequencing of CLGN-overexpressing cells indicated an increase in the pathway related to tRNA aminoacylation. tRNA is essential for accurate translation, and its modification by aminoacylation has been reported to ensure tRNA stability and translational efficiency.23 tRNA hypo-modification results in translational inaccuracy and accumulation of misfolded proteins.23 Taken together, CLGN appears to participate in aldosterone production by the translational regulation of CYP11B2.
CYP11B2 mRNA expression did not correlated with CLGN mRNA expression in APA, and CLGN protein was not likely to be co-localize with CYP11B2 protein by immunohistochemical analysis (Figure 2). CLGN expression showed a large spread of values in APA and a partial overlapping between APA and NFA (Figure 1A). Considering with the results, the proposed mechanisms by which CLGN mediated CYP11B2 expression in adrenal cells are that CLGN basically require abundant CYP11B2 mRNA expression, wherein it enhance the CYP11B2 protein production in APA.
CLGN transcription is unlikely to be regulated by acute activation of intracellular signaling for aldosterone production, because CLGN expression was not altered by A-II, forskolin, or KCNJ5 T158A expression in HAC15 cells. Remarkably, CLGN was not expressed in the rat zona glomerulosa even after chronic stimulation by sodium deficiency although CLGN mRNA is weakly expressed in rat adrenal.24 Therefore, CLGN may be involved in autonomous and persistent aldosterone production because of no difference of CLGN expression among genotype. In considering the pathogenesis of APA, APA may be developed from the precursor cells with CLGN-expression.
According to the bioinformatics analyses (AmiGO 2; http://amigo.geneontology.org/amigo), CLGN has calcium ion binding sites, thus may have a function in calcium ion storage. Aldosterone production by A-II and the mutations described thus far in APA involve activation of intracellular calcium signaling.7, 25, 26 As we previously reported, the ER in APA has abundant calcium storage partly by virtue of CALN1.15 Therefore, we investigated the calcium storage of ER in HAC15 cells with CLGN overexpression. CLGN overexpression did not influence the calcium storage (Figure 4E). We concluded that enhanced aldosterone production by CLGN overexpression was not associated with calcium storage in HAC15 cells.
CLGN mRNA and protein expression were not enhanced in CPA. Additinally, the effects of CLGN on cortisol production were not identical to those on aldosterone production in HAC15 cells. The machineries of cortisol production including intracellular signaling in CPA are different from those of aldosterone production in APA.4, 27, 28 Translational regulation via CLGN should be distinctive aspect for aldosterone production in APA. It is suggested that CLGN have pivotal roles for aldosterone production in APA, but not cortisol production in CPA.
Silencing of CLGN expression did not suppress A-II or KCNJ5-mutation stimulated aldosterone production in HAC15 cells. The HAC15 were derived from a human adrenocortical cancer, and aminoacyl tRNA biosynthesis is highly upregulated in cancer.29 A recent report indicated that aminoacyl-tRNA synthetases (ARSs) have various functions beyond ensuring accurate protein synthesis, including their promotion of tumorigenesis.30 Bioinformatics studies demonstrate that ARSs may interact with the members of the PI3K/AKT/mTOR signaling pathway.31 Therefore, suppression of ARSs function would be a novel therapeutic approach to decrease aldosterone production in patients with APA.
This study has some limitations. First, the mechanism of interaction between CLGN, ARSs and CYP11B2 is not clarified. Although it is unclear whether the enhancement of CYP11B2 protein expression by CLGN overexpression is a direct or indirect effect, aldosterone production machinery via a key molecule of translation is novel findings. Second, pathological findings in some APAs are heterogeneous, and thus results might be variable according to the site of biopsy. Further cases are required to determine the association of CLGN and CYP11B2 in APA.
Perspectives
We demonstrated that CLGN, which is highly expressed in APA, potentiated aldosterone production by enhancing CYP11B2 protein expression. The translational regulation of CYP11B2 through ARSs by CLGN would be a novel mechanism of aldosterone production in APA. Although silencing of CLGN did not suppress A-II or KCNJ5 mutation-mediated aldosterone production, overall suppression of ARSs function or genes is a potential therapeutic approach in patients with APA and perhaps other hormone-producing endocrine neoplasia. Importantly, there are no therapeutic agents to regulate aldosterone production in APA. Taken together, this study might be useful to clarify the entire molecular mechanisms of aldosterone production in APA and the sequential development of agents to target aldosterone production.
Supplementary Material
Novelty and Significance.
1). What Is New?
Since the endoplasmic reticulum (ER) is a crucial subcellular organelle for protein synthesis, we targeted 288 ER-expressed genes in aldosterone producing adenoma (APA).
CLGN is highly expressed in APA and aldosterone producing cell clusters, but not in zona glomerulosa of rat adrenal.
CLGN did not potentiate CYP11B2 transcription, but increased aldosterone production and CYP11B2 protein expression.
2). What Is Relevant?
Results of RNA sequencing analysis using CLGN over-expressing cells indicated that CLGN has a role in aminoacyl tRNA biosynthesis.
Translational regulation of CYP11B2 by CLGN provide a foundation to seek a potential therapy to suppress aldosterone production in APA.
3). Summary
We demonstrated that CLGN was upregulated and associated with aldosterone production via translational regulation of CYP11B2 in APAs. The translational regulation of CYP11B2 through aminoacyl-tRNA synthetases by CLGN would be a novel mechanism of aldosterone production in APA.
Acknowledgement
This work was partly carried out with the kind cooperation of the Analysis Center of Life Science, Hiroshima University and the Program of the network-type joint Usage/Research Center for Radiation Disaster Medical Science of Hiroshima University, Nagasaki University and Fukushima Medical University.
Sources of Funding
This study was financially supported by JSPS KAKENHI Grant Number JP19K17964 (K.I.), JSPS KAKENHI Grant Number JP17K09883 (K.O.), SENSHIN Medical Research Foundation (K.O.), Okinaka Memorial Institute for Medical Research Grant (K.O.), JSPS KAKENHI Grant Number JP19K17600 (K.K.), JSPS KAKENHI Grant Number JP19K17972 (Y.Y.), National Heart, Lung and Blood Institute grant R01 HL144847 (C.-E.GS.), and the National Institute of General Medical Sciences under Award Number 1U54GM115428 (C.-E.GS.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure statements
The authors have nothing to disclose.
References
- 1.Milliez P, Girerd X, Plouin PF, Blacher J, Safar ME, Mourad JJ. Evidence for an increased rate of cardiovascular events in patients with primary aldosteronism. Journal of the American College of Cardiology. 2005;45:1243–1248. [DOI] [PubMed] [Google Scholar]
- 2.Mulatero P, Monticone S, Bertello C, Viola A, Tizzani D, Iannaccone A, Crudo V, Burrello J, Milan A, Rabbia F, Veglio F. Long-term cardio- and cerebrovascular events in patients with primary aldosteronism. J Clin Endocrinol Metab. 2013;98:4826–4833. [DOI] [PubMed] [Google Scholar]
- 3.Funder JW, Carey RM, Mantero F, Murad MH, Reincke M, Shibata H, Stowasser M, Young WF, Jr. The management of primary aldosteronism: Case detection, diagnosis, and treatment: An endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2016;101:1889–1916. [DOI] [PubMed] [Google Scholar]
- 4.Choi M, Scholl UI, Yue P, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331:768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azizan EA, Poulsen H, Tuluc P, et al. Somatic mutations in atp1a1 and cacna1d underlie a common subtype of adrenal hypertension. Nat Genet. 2013;45:1055–1060. [DOI] [PubMed] [Google Scholar]
- 6.Beuschlein F, Boulkroun S, Osswald A, et al. Somatic mutations in atp1a1 and atp2b3 lead to aldosterone-producing adenomas and secondary hypertension. Nat Genet. 2013;45:440–444, 444e441-442. [DOI] [PubMed] [Google Scholar]
- 7.Oki K, Plonczynski MW, Luis Lam M, Gomez-Sanchez EP, Gomez-Sanchez CE. Potassium channel mutant kcnj5 t158a expression in hac-15 cells increases aldosterone synthesis. Endocrinology. 2012;153:1774–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reid DW, Nicchitta CV. Diversity and selectivity in mrna translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2015;16:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishikawa T, Omura M, Satoh F, Shibata H, Takahashi K, Tamura N, Tanabe A, Task Force Committee on Primary Aldosteronism TeJES. Guidelines for the diagnosis and treatment of primary aldosteronism--the japan endocrine society 2009. Endocr J. 2011;58:711–721. [DOI] [PubMed] [Google Scholar]
- 10.Yoshii Y, Oki K, Gomez-Sanchez CE, Ohno H, Itcho K, Kobuke K, Yoneda M. Hypomethylation of cyp11b2 in aldosterone-producing adenoma. Hypertension. 2016;68:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oki K, Yamane K, Nakanishi S, Shiwa T, Kohno N. Influence of adrenal subclinical hypercortisolism on hypertension in patients with adrenal incidentaloma. Exp Clin Endocrinol Diabetes. 2012;120:244–247. [DOI] [PubMed] [Google Scholar]
- 12.Seccia TM, Caroccia B, Gomez-Sanchez EP, Gomez-Sanchez CE, Rossi GP. The biology of normal zona glomerulosa and aldosterone-producing adenoma: Pathological implications. Endocr Rev. 2018;39:1029–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parmar J, Key RE, Rainey WE. Development of an adrenocorticotropin-responsive human adrenocortical carcinoma cell line. J Clin Endocrinol Metab. 2008;93:4542–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishimoto R, Oki K, Yoneda M, Gomez-Sanchez CE, Ohno H, Kobuke K, Itcho K, Kohno N. Gonadotropin-releasing hormone stimulate aldosterone production in a subset of aldosterone-producing adenoma. Medicine (Baltimore). 2016;95:e3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobuke K, Oki K, Gomez-Sanchez CE, Gomez-Sanchez EP, Ohno H, Itcho K, Yoshii Y, Yoneda M, Hattori N. Calneuron 1 increased ca(2+) in the endoplasmic reticulum and aldosterone production in aldosterone-producing adenoma. Hypertension. 2018;71:125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itcho K, Oki K, Kobuke K, Yoshii Y, Ohno H, Yoneda M, Hattori N. Aberrant g protein-receptor expression is associated with DNA methylation in aldosterone-producing adenoma. Mol Cell Endocrinol. 2018;461:100–104. [DOI] [PubMed] [Google Scholar]
- 17.Oki K, Plonczynski MW, Gomez-Sanchez EP, Gomez-Sanchez CE. Ypel4 modulates hac15 adrenal cell proliferation and is associated with tumor diameter. Mol Cell Endocrinol. 2016;434:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez-Sanchez CE, Qi X, Velarde-Miranda C, Plonczynski MW, Parker CR, Rainey W, Satoh F, Maekawa T, Nakamura Y, Sasano H, Gomez-Sanchez EP. Development of monoclonal antibodies against human cyp11b1 and cyp11b2. Mol Cell Endocrinol. 2014;383:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oki K, Kopf PG, Campbell WB, Luis Lam M, Yamazaki T, Gomez-Sanchez CE, Gomez-Sanchez EP. Angiotensin ii and iii metabolism and effects on steroid production in the hac15 human adrenocortical cell line. Endocrinology. 2013;154:214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieman LK, Biller BM, Findling JW, Newell-Price J, Savage MO, Stewart PM, Montori VM. The diagnosis of cushing's syndrome: An endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2008;93:1526–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe D, Yamada K, Nishina Y, Tajima Y, Koshimizu U, Nagata A, Nishimune Y. Molecular cloning of a novel ca(2+)-binding protein (calmegin) specifically expressed during male meiotic germ cell development. J Biol Chem. 1994;269:7744–7749. [PubMed] [Google Scholar]
- 22.Ikawa M, Wada I, Kominami K, Watanabe D, Toshimori K, Nishimune Y, Okabe M. The putative chaperone calmegin is required for sperm fertility. Nature. 1997;387:607–611. [DOI] [PubMed] [Google Scholar]
- 23.Pereira M, Francisco S, Varanda AS, Santos M, Santos MAS, Soares AR. Impact of trna modifications and trna-modifying enzymes on proteostasis and human disease. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y, Fuscoe JC, Zhao C, et al. A rat rna-seq transcriptomic bodymap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sackmann S, Lichtenauer U, Shapiro I, Reincke M, Beuschlein F. Aldosterone producing adrenal adenomas are characterized by activation of calcium/calmodulin-dependent protein kinase (camk) dependent pathways. Horm Metab Res. 2011;43:106–111. [DOI] [PubMed] [Google Scholar]
- 26.Condon JC, Pezzi V, Drummond BM, Yin S, Rainey WE. Calmodulin-dependent kinase i regulates adrenal cell expression of aldosterone synthase. Endocrinology. 2002;143:3651–3657. [DOI] [PubMed] [Google Scholar]
- 27.Zennaro MC, Boulkroun S, Fernandes-Rosa F. Genetic causes of functional adrenocortical adenomas. Endocr Rev. 2017;38:516–537. [DOI] [PubMed] [Google Scholar]
- 28.Beuschlein F, Fassnacht M, Assie G, et al. Constitutive activation of pka catalytic subunit in adrenal cushing's syndrome. N Engl J Med. 2014;370:1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J, Locasale JW, Bielas JH, O'Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31:522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, You S, Hwang D. Aminoacyl-trna synthetases and tumorigenesis: More than housekeeping. Nat Rev Cancer. 2011;11:708–718. [DOI] [PubMed] [Google Scholar]
- 31.Hyeon DY, Kim JH, Ahn TJ, Cho Y, Hwang D, Kim S. Evolution of the multi-trna synthetase complex and its role in cancer. J Biol Chem. 2019;294:5340–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.