Abstract
Despite the importance of magnetic properties of biological samples for biomagnetism and related fields, the exact magnetic susceptibilities of most biological samples in their physiological conditions are still unknown. Here we used superconducting quantum interferometer device to detect the magnetic properties of nonfixed, nondehydrated live cell and cellular fractions at a physiological temperature of 37°C (310 K). It is obvious that there are paramagnetic components within human nasopharyngeal carcinoma CNE-2Z cells. More importantly, the magnetic properties of the cytoplasm and nucleus are different. Although within a single cell, the magnetic susceptibility difference between cellular fractions (nucleus and cytoplasm) could only cause ∼41–130 pN forces to the nucleus by gradient ultrahigh magnetic fields of 13.1–23.5 T (92–160 T/m), these forces are enough to cause a relative position shift of the nucleus within the cell. This not only demonstrates the importance of magnetic susceptibility in the biological effects of magnetic field but also illustrates the potential application of high magnetic fields in biomedicine.
Significance
The magnetic properties of biological samples are the basis for understanding various biomagnetism phenomena and biomedical applications of magnetic fields. Theoretically, magnetic susceptibility of living biological samples could allow quantifications of the magnetic forces and torques acting on them by magnetic fields. However, the magnetic susceptibilities of most biological samples in physiological conditions are still unknown. We used SQUID to detect the magnetic properties of live cells in their native state. We found that even in a micrometer-range human cell, the force generated by a high-gradient magnetic field and resulting from the magnetic susceptibility difference between the nucleus and cytoplasm could lead to nucleus repositioning within the cell, which could potentially change cell polarity and cell division, as well as stem cell differentiation.
Introduction
In recent years, the development of an SQUID (superconducting quantum interferometer device) has enabled detection of extremely weak biomagnetic signals in magnetocardiography and magnetoencephalography for high-sensitivity and real-time signal detection, which enables early diagnosis of epilepsy and other diseases (1). Although the magnetic signals of biological samples are generally very weak, the differences between various samples and their different conditions could be used in medical diagnosis. For example, deoxyhemoglobin is paramagnetic, but oxyhemoglobin is diamagnetic; therefore, various states of red blood cells have different magnetism. This finding has been used in cerebral hemorrhage diagnosis to judge the timing of hemorrhage (2). In addition, paramagnetic products are produced in the erythrocyte (red blood cell) infected by Plasmodium, and gradient magnetic field has been used to diagnose malaria and isolate the Plasmodium-infected erythrocyte (3,4).
In the meantime, magnetic fields have various effects on living organisms (5, 6, 7), but the underlying mechanisms still remain incompletely understood. Similar to other nonliving materials, biological samples may be affected by magnetic torque and/or force in magnetic fields, and one of the most important physical quantities is their magnetic susceptibility, the degree of magnetization in response to an applied magnetic field (5,8). Mathematically, it can be calculated as the ratio of magnetization M (magnetic moment per unit) to the applied magnetizing field intensity H. However, depending on parameter measured, people use various ways to present the magnetic susceptibility, such as volume magnetic susceptibility, molar magnetic susceptibility, mass magnetic susceptibility, etc. For example, the volume susceptibility (χ) of pure water was shown to be −9.05 × 10−6 (international system of units) (9), and the theoretical molar magnetic susceptibility of a peptide bond is −5.36 × 10−6 emu (CGS (centimeter-gram-second) units) (10), which are the basis for later calculations. However, very limited magnetic susceptibility information is available for the biological samples at their physiological conditions, which means in solution and at body temperature for human samples. For example, Senftle and Thorpe (11) measured the mass susceptibility of a transplanted hepatoma as (−0.688 ± 0.0046) × 10−6 emu/g and (−0.670 ± 0.0012) × 10−6 emu/g and (−0.637 ± 0.0059) × 10−6 emu/g for the liver tissues of a tumor-bearing rat and normal rat, respectively, at 263 K. However, the magnetic susceptibility information of these tissues at 37°C (310 K) is unknown.
It is well known that temperature is critical for the physiological functions and structures of biological samples. Moreover, Nakamae et al. (12) found that λ-DNA was paramagnetic below 20 K in the B-DNA state in an applied magnetic field, but it was diamagnetic down to 2 K in the A-DNA state. There are a few studies about human blood samples at their physiological conditions. For example, in 2001, Spees et al. (13) made use of an SQUID to measure the magnetic susceptibility of human red blood cells at the physiological temperature of 37°C (310 K) and found that the χ of carbon-monoxygenated and deoxygenated erythrocytes are (−0.749 ± 0.010) × 10−6 and (−0.483 ± 0.013) × 10−6, respectively, which has served as the foundation for numerous calculations and medical applications (14,15). In 2012, Jain et al. (16) used magnetic resonance susceptometry to detect the χ of fresh human blood at 37°C (310 K), and they discovered that the difference between fully oxygenated and fully deoxygenated blood (Δχdo) was 0.273 ± 0.006 ppm (centimeter-gram-second units), but the susceptibility of oxygenated blood relative to water was −0.008 ± 0.003 ppm (centimeter-gram-second units).
However, despite the importance of magnetic property characterization of biological samples, the exact magnetic susceptibilities of most live biological samples in their physiological conditions are unknown, which prevents people from getting an accurate understanding of the magnetic properties of biological organisms. The metabolic state and composition, as well as many other aspects, could be altered if the biological samples are fixed or not examined right away. Therefore, in this study, we made use of DC-SQUID mode (MPMS3; Quantum Design, San Diego, CA) to measure live biological samples, including human cells and cellular fractions, in their native state (at a body temperature of 37°C (310 K)) to provide some basic information for future investigations.
Materials and Methods
Reagents and materials
Antibodies of COX IV (1124-1-AP; Proteintech, Chicago, Illinois), β-tubulin (HC101; TransGen Biotech, Beijing, China), and histone H3 (ab1791; Abcam, Cambridge, MA) were used. Horseradish-peroxidase-linked secondary anti-rabbit (7074P2; CST, Danvers, MA) and anti-mouse (7076S; CST) antibodies were also used, along with phalloidin 594 (AB-2315633; Invitrogen, Carlsbad, CA) and Alexa Fluor 488-conjugated secondary antibody (AB-2315147; Invitrogen). Mammalian protein extraction reagent (M-PER) lysis buffer (78501; Thermo Fisher Scientific, Waltham, MA), protease (04639116; Roche Diagnostics, Basel, Switzerland) and phosphatase (04906837; Roche Diagnostics) inhibitor cocktail, polyvinylidene fluoride (PVDF) membranes (MB0323; Millipore, Burlington, MA), nonfat dried milk (A600669; Sangon Biotech, Shanghai, China), and Enhanced Chemiluminescence Kits (P90720; Millipore) were used. Sterilized nonmagnetic materials were used throughout the sample preparation and testing process.
SQUID measurement
In this research, we made use of DC-SQUID mode (MPMS3; Quantum Design) to measure human cells and cellular fractions. We loaded samples into the sealed liquid sample holders (C130D; Quantum Design) and tightened it to avoid leakage in the vacuum environment. For the magnetization (M) versus magnetic field (H) test at 310 K, the field was swept as 0 Oe (oersted, the centimeter-gram-second unit of magnetic intensity),→ 30,000 Oe→ -30,000 Oe,→ 30,000 Oe → 0 Oe and one measurement point per 5000 Oe. For the magnetization (M) versus temperature (T) test, the magnetic field was set as 1000 or 50,000 Oe, and the temperature climbed from 2 to 310 K at a speed of 4 K/min. Magnetization of sample is measured every 4 K when temperature is below 102 K but 8 K when above 102 K.
The mass susceptibility (χ) was calculated as
| (1) |
in which χ is the mass susceptibility, M is the magnetic moment, m is the sample mass, and H is the magnetic field strength.
Cell culture for SQUID test
CNE-2Z cells (Research Resource Identifier: CVCL 6890) were cultured in RPMI (Roswell Park Memorial Institute)-1640 (10–040-CVR; Corning Life Sciences, Acton, MA) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin under 310 K in an incubator. During their exponential growth phase, CNE-2Z cells were harvested by a scrapper and gently washed by phosphate-buffered saline (PBS) three times through centrifugation at 2000 rotations per minute (rpm). The pellets were then sent for M versus H or M versus T test.
Cell nuclei and cytoplasm sample preparation
20 million exponential growth phase CNE-2Z cells were harvested by a scrapper and washed by phosphate-buffered saline buffer for three times through centrifugation at 2000 rpm. Then, the pellets were suspended into 2 mL of reagent A of the Cytoplasm and Nuclear Protein Extraction Kit (P0027; Beyotime Biotechnology, Shanghai, China) supplied with 1 mM protease inhibitor phenylmethylsulphonyl fluoride (PMSF) in the final concentration. After being bathed in an ice-water mixture for 15 min, 100 μl of reagent B were added into the suspension. After vortex at the highest speed, the suspension was rebathed in an ice-water mixture for 1 min. We centrifuged the lysed cell suspension for 5 min at 13,000 rpm under 4°C in a centrifuge (Legend Micro 17R; Thermo Fisher Scientific). The supernatant was a cytoplasm in the lysis buffer, and the precipitation in the tube was a nucleus.
For immunofluorescence (IF) characterization, intact CNE-2Z cells and isolated CNE-2Z nuclei were fixed by 4% (vol/vol) formaldehyde at room temperature for 20 min. After perforating and blocking by AbDil-Tx (Tris-buffered saline (TBS) supplemented with 0.1% (vol/vol) Triton X-100, 2% (wt/vol) bovine serum albumin, and 0.05% (vol/vol) sodium azide) at room temperature for at least 30 min, samples were stained with anti-COX IV antibody at 4°C for overnight, followed by fluorescently conjugated secondary antibody Alexa Fluor 488. As for IF characterization of water-cooled magnet #4 (WM4)-treated CNE-2Z cells, microfilaments (actin filament) were stained by fluorescently labeled phalloidin at room temperature for 30 min. Both the in situ and isolated nuclei were stained with 1 μg/mL 4′, 6-diamidino-2-phenylindole at room temperature for half an hour. Fluorescent images were acquired by DMI4000B microscope (SCR-000011; Leica, Wetzlar, Germany).
For the immunoblotting test, the proper amount of cytoplasm in lysis buffer and nuclei precipitation were further lysed on ice with the M-PER lysis buffer supplemented with a protease and phosphatase inhibitor cocktail at 4°C for 30 min. Mixed with 5× loading buffer, thermal denatured protein samples were electrophoresed on SDS-PAGE gels and transferred onto PVDF membranes. Then the PVDF membrane was blocked with 5% nonfat dried milk at room temperature for 30 min and followed by incubating with corresponding primary antibodies and horseradish-peroxidase-conjugated secondary antibodies. Visualization was performed using Enhanced Chemiluminescence Kits, and the blots were analyzed using Tanon Fine-do X6 (Tanon, Shanghai, China).
Statistics
Analysis and presentation of the SQUID data were performed using Origin 9 software. Analysis and comparisons between groups were performed using Graphpad 5 software and unpaired two-tailed Student’s t-test. Data were considered significantly different at p < 0.05 or below. Results were presented as mean ± SEM.
Results and Discussion
Mass susceptibilities of CNE-2Z cells
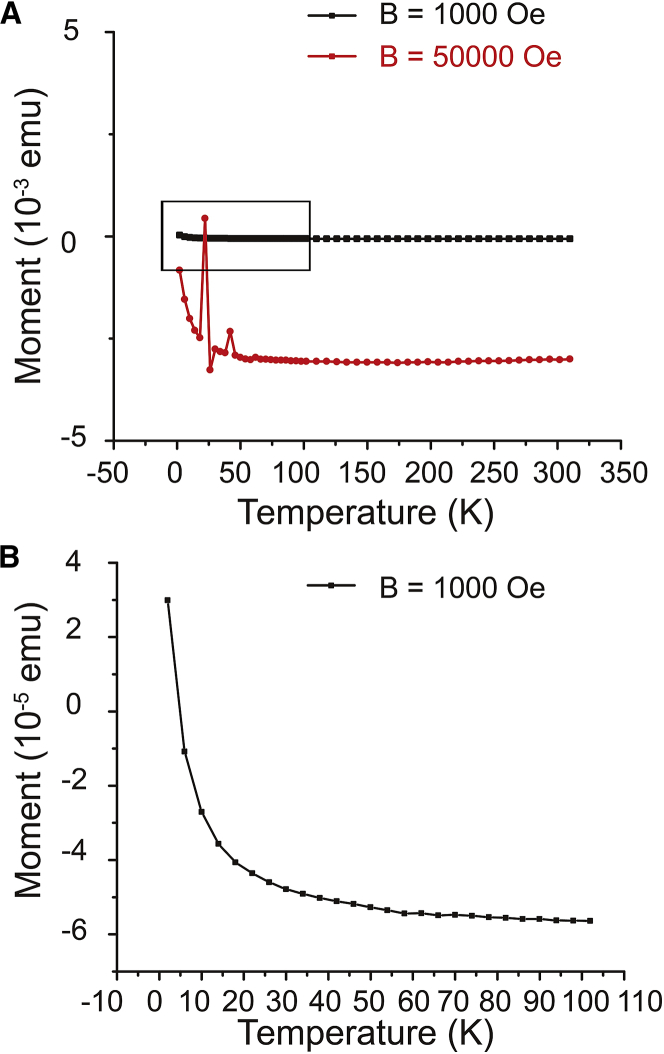
We first examined the mass susceptibility of human nasopharyngeal carcinoma CNE-2Z cells, which has been used in a few previous studies for the effects of static magnetic fields (6,17, 18, 19). Magnetization of 40 million human CNE-2Z cells as a function of temperature at both 1000 and 50,000 Oe were performed by SQUID measurement. It is obvious from the M-H curves that there are paramagnetic upturns at low temperature (Fig. 1), which indicate that there are some paramagnetic components existing in CNE-2Z cells. This is confirmed by both 50,000 Oe (Fig. 1 A) and 1000 Oe (Fig. 1 B).
Figure 1.
M-T curves of human CNE-2Z cells by SQUID measurement show that there are paramagnetic components in CNE-2Z cells. (A) Magnetization of 40 million CNE-2Z cells as a function of temperature at 1000 or 50,000 Oe is shown here. The box area in (A) was enlarged in (B). Magnetization changes of 40 million CNE-2Z cells at 1000 Oe between 2 and 102 K are shown here. To see this figure in color, go online.
The susceptibility difference between the cytoplasm and nucleus of CNE-2Z cells
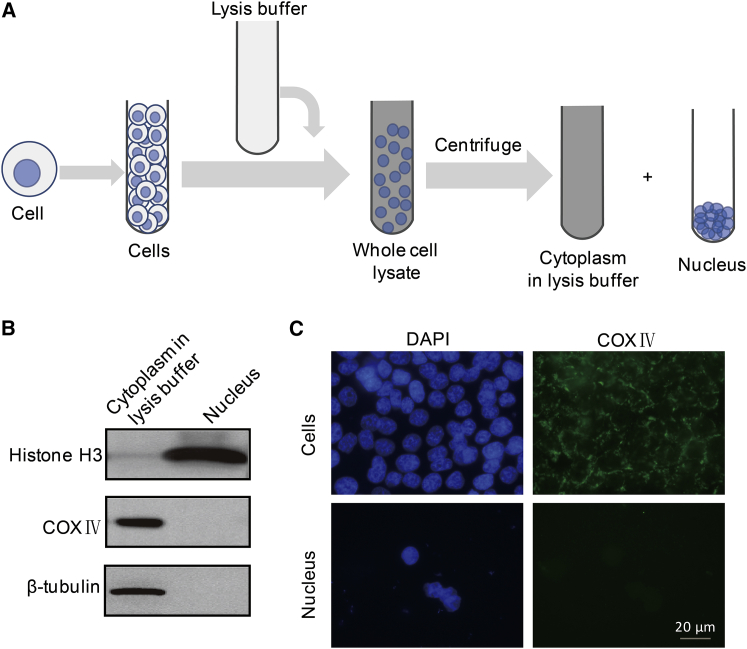
Next, we further explored the magnetic properties of organelles within cells. Because the nucleus is the largest component of a cell, which exhibits vital roles both in cells’ genetic and physical behaviors (20), it is essential to investigate its magnetic property. Here, we chose nondividing CNE-2Z cells, which mainly contain a nucleus and the rest as cytoplasm (Fig. 2 A) and are much simpler than dividing cells.
Figure 2.
Nucleus isolation and purity verification. (A) An illustration of nuclei isolation procedure is shown here. (B) Immunoblotting and (C) immunofluorescence test were used to verify the purity of the isolated nuclei and cytoplasm. The scale bar represents 20 μm. To see this figure in color, go online.
Fig. 2 B displayed the nucleus preparation processes through centrifugation. To check the sample purity, we characterized the isolated nucleus using immunoblotting (Fig. 2 B) and IF (Fig. 2 C). COX IV and β-tubulin are cytoplasm markers, whereas histone H3 is normally retained only in nucleus (21). As in Fig. 2 B, COX IV and β-tubulin were only present in cytoplasm in lysis buffer, whereas histone H3 only appeared in nucleus, which verified that there was no contamination between isolated cytoplasm and nucleus. The IF result in Fig. 2 C further proved the purity of our samples.
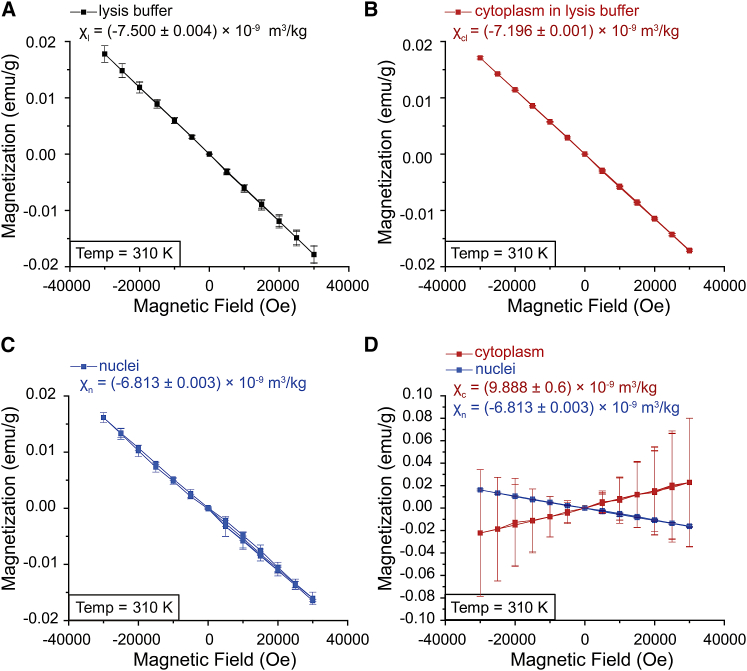
Fig. 3, A–C and Fig S1, A–C showed that the mass susceptibilities of lysis buffer χl, cytoplasm in lysis buffer χcl, and nuclei χn were (−7.500 ± 0.004) × 10−9 m3/kg, (−7.196 ± 0.001) × 10−9 m3/kg, and (−6.813 ± 0.003) × 10−9 m3/kg, respectively. The mass susceptibility of cytoplasm itself χc was calculated as
| (2) |
in which χc is the mass susceptibility of cytoplasm; Mcl and Ml are the magnetic moments of the cytoplasm in the lysis buffer or the lysis buffer, respectively; m is the sample mass of the cytoplasm in the lysis buffer; and H is the magnetic field strength. ξl and ξc are the mass ratios of the lysis buffer or the cytoplasm in the lysis buffer, respectively, and the mass of the cytoplasm was obtained through subtracting the masses of nuclei from the masses of CNE-2Z cells. After three independent experiments, we found that the χc was calculated to be (9.888 ± 0.6) × 10−9 m3/kg, which means that the cytoplasm of CNE-2Z cells is paramagnetic (Fig. 3 D and Document S1. Supporting Materials and Methods and Fig. S1, Document S2. Article plus Supporting Material). Because the cytoplasm contains a large number of mitochondria and peroxisomes, which are the major organelles to generate ROS (reactive oxygen species), we speculate that the paramagnetic ROS that harbor unpaired electrons may be one of the reasons for the paramagnetic property of cytoplasm (22). However, the nucleus is diamagnetic, which is probably due to its composition of DNA, nuclear proteins, and lipids.
Figure 3.
Magnetization of isolated cytoplasm and nuclei from 20 million CNE-2Z cells. (A) Magnetization versus magnetic field strength of lysis buffer, (B) cytoplasm in lysis buffer, and (C) nuclei at 310 K are shown here. (D) Magnetization versus magnetic field strength of cytoplasm and nuclei at 310 K are shown here. Data are presented as mean ± standard error of the mean deviation (SD). For nuclei and cytoplasm, n = 3; for lysis buffer, n = 4. To see this figure in color, go online.
Force analysis induced by the susceptibility difference
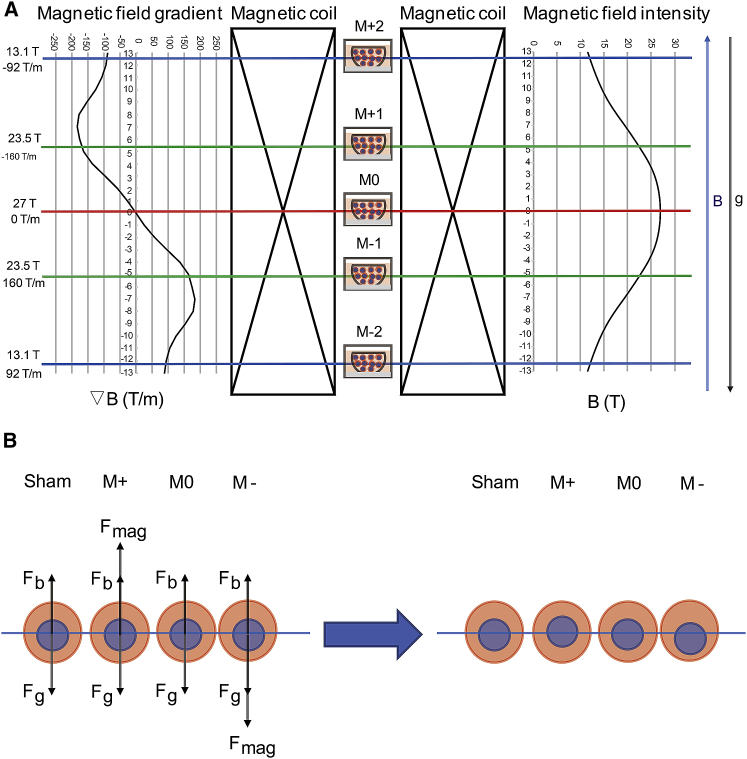
Because even the nonuniform intrabead distribution of nanoparticles could contribute to their anisotropy (23) as the smallest living biological unit, we hypothesize that the intracellular fractions may also be nonuniform. Because a cell can be roughly divided into two parts (the nucleus and the cytoplasm), nuclear positioning is a prerequisite for the correct execution of centered mitosis (24), cell differentiation (25), cell migration (26), and neural system development (27). Because of its importance for correct cell function and tissue development, the position of the cell nucleus is tightly controlled by cytoskeletal elements, and both actin (28) and microtubules (25) can exert pulling or pushing forces on nuclei through a variety of mechanisms. For example, in the cytoplasm of J774 macrophages, the local intracellular transport can be driven by local active forces on the order of 50–460 pN (29). Here, we aim to see whether the difference between the magnetic susceptibilities of the nucleus and the cytoplasm within a single cell could generate cellular effect in a large gradient magnetic field. We utilized the WM4 in the Chinese High Magnetic Field Laboratory (Hefei, China), which can generate an upward-directed high magnetic field (6). At the center of the magnet, it is 27 T with no gradient (referred to as M0). For the upper part (referred to as M+) and lower part (referred to as M−) of the magnet, the absolute value of gradient ranges from 160 to 92 T/m (Fig. 4 A).
Figure 4.
Illustration of the magnetic field parameters and predicted relative position changes of the nucleus in gradient high magnetic field in cells. (A) A diagram of the magnetic field intensity and gradient distribution is shown here. (B) An illustration of forces on the nuclei in different magnetic fields is shown here. “M+” includes cells from M + 1 and M + 2. “M−” includes cells from M − 1 to M − 2. Fb, Fbuoyancy; Fg, Fgravity; Fmag, Fmagnetic. To see this figure in color, go online.
Because the cell nucleus is suspended in the cytoplasm, based on the formulas of gravity (Eq. 3), buoyance force (Eq. 4), and magnetic force with the gradient of the magnetic field (Eq. 5; 30,31), we could estimate the force acted on the nucleus and infer the relative positions of the nucleus and the cytoplasm:
| (3) |
| (4) |
| (5) |
Here, mn is the nuclei mass, g is gravitational acceleration, Vn represents the average volume of CNE-2Z nuclei, Δχ is the difference of value between the volume susceptibility of the nuclei and the volume susceptibility of the cytoplasm, μ0 is the permeability of free space equal to 4π × 107 N/A2, B is the intensity of magnetic field, and ▿B represents the gradient magnitude of the magnetic field.
To simplify the model, we assumed that the cell nucleus is suspended in the cytoplasm, and both the density of the nuclei ρn and the density of the cytoplasm ρc are same, with the density of water ρw as 1.0 × 103 kg/m3.
For cells at a specific position in the WM4, the resultant force acted on cell nucleus can be calculated as
| (6) |
At the M0 position (27 T, 0 T/m), the resultant force acted on the cell nucleus is FM0 = 0 N. We predict that the cell nucleus would not shift relative to the cytoplasm.
At the M + 1 position (23.5 T, −160 T/m), the resultant force is FM + 1 = +128.54 pN, and at the M + 2 position (13.1 T, −92 T/m), the resultant force is FM + 2 = +41.2 pN, with the plus sign representing an upward direction.
However, at the M − 1 position (23.5 T, 160 T/m), the resultant force is FM − 1 = −128.54 pN, and the resultant force is FM − 2 = −41.2 pN at the M − 2 position (13.1 T, 92 T/m), with the minus sign representing a downward direction (calculation details are in Supporting Materials and Methods).
Although the forces seem not to be strong, it should be mentioned that in a well-defined in vitro model cytoskeleton, Brangwynne et al. (32) calculated that the order of magnitude of the actin tension force is only ∼10 pN, and the measured mitotic forces in vivo were on the order of 10–100 pN (33,34). Comparing the calculated forces FM with the intracellular forces generated by the cytoskeleton, we predict that the relative position of the nucleus toward the cytoplasm at the M+ position (M + 1 and M + 2 ) would be higher than the nucleus in the cytoplasm at the M− position (M − 1 and M − 2) (Fig. 4 B).
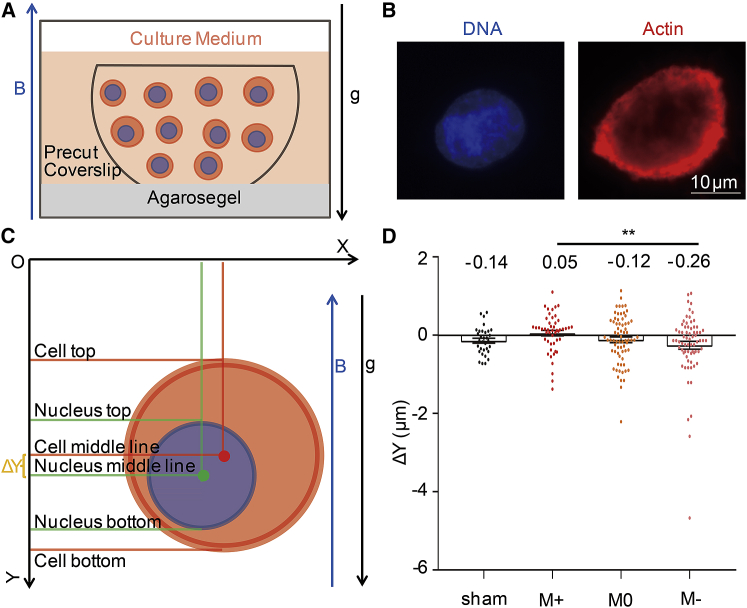
Experimental observation of nuclei localization
To test our hypothesis, we put CNE-2Z cells in the magnet and compared the relative position of the nucleus in cells at positions with no gradient or ±92 or ±160 T/m gradient with opposite gradient directions. We also prepared the sham control group of CNE-2Z, which was maintained in the same condition with cells in 27 T magnet except magnetic field exposure. Fig. 5 A showed the magnetic-exposed cells, which were described previously (6). Briefly, cells were seeded on precut coverslips one night before magnetic exposure, and then coverslips were vertically inserted on agarose gel so that they are parallel with the magnetic field direction (upward) and gravity. After 4 h of magnetic treatment, cells were fixed immediately, and microfilaments (actin filament) were marked by fluorescently labeled phalloidin (red) (Fig. 5 B). We used microfilaments to determine the edge of the cell and measured the region stained with 4′, 6-diamidino-2-phenylindole, which stains DNA, to represent the nucleus area. The relative positions of the nucleus and the cytoplasm along the magnetic-field-changing direction can be measured (Fig. 5 C). We only selected round-interphase cells. To increase sample size, we combined cells from the M + 1 and M + 2 positions as M+-treated cells (in the upper part of the magnet) and cells from the M − 1 and M − 2 positions as M−-treated cells (in the lower part of the magnet). Consistent with our theoretical prediction (Fig. 4 B), we found that CNE-2Z cells in the M+ position intend to have a relatively higher nucleus position compared to the cells in the M− position (Fig. 5 D). The relative positions of nuclei in the sham control and the M0 position where there is no magnetic field gradient are similar. However, the nuclei of M+ cells are ∼0.17 μm higher than the sham control and M0 position, whereas the nuclei of M− cells are ∼0.14 μm lower than the sham control and M0 positions, which is perfectly consistent with the calculation results based on the SQUID test for the nucleus and cytoplasm in vitro. Therefore, the magnetic susceptibility difference in different cellular fractions within a single cell could cause their relative location changes in a high-gradient magnetic field.
Figure 5.
Statistical results of relative position changes of the nucleus in gradient high magnetic field. (A) An illustration of experimental set-up for CNE-2Z cells exposed to magnetic field is shown here. (B) Immunofluorescence images of a CNE-2Z cell are shown here. The scale bar represents 10 μm. (C) A measurement illustration is shown here. (D) Quantification of the relative position of the nucleus in cells in sham control-, M+-, M0-, and M−-treated cells is shown. A total of 32–70 round interphase CNE-2Z cells from three to four independent coverslips were examined for each condition. Data are presented as mean ± standard error of the mean (SEM). Student’s t-test was used to determine the difference. ∗∗p < 0.01. To see this figure in color, go online.
Conclusions
Our study not only measured the magnetic susceptibilities of cells and cellular components but also provided evidence that the magnetic susceptibilities of living biological samples could allow quantifications of the magnetic forces and torques acting on them by high magnetic fields. More specifically, we found that in round-interphase human CNE-2Z cells in the micrometer range, the 41–130 pN force generated by a high-gradient magnetic field and resulting from the magnetic susceptibility differences of cell nuclei and cytoplasm can lead to nucleus position changes, which could potentially change cell polarity, cell division, and stem cell differentiation. Therefore, a systematic study of the magnetic properties of different biological samples in their physiological conditions may reveal in-depth mechanisms of magnetic effects, which may provide mechanistic foundations for the application of magnetic fields in biological research and medicine.
Author Contributions
X.Z. designed the study. Q.T. and L.Z. performed experiments and analyzed data. X.H., H.C., and X.J. performed cellular experiments. Q.T., L.Z., and X.Z. prepared the manuscript. All authors approved the final version of the article.
Acknowledgments
We thank the High Magnetic Field Laboratory Facility, Chinese Academy of Sciences, Hefei, China.
This work was supported by the National Key Research and Development Program of China (2016YFA0400900), National Natural Science Foundation of China (U1532151 and 31900506), the CASHIPS (Chinese Academy of Sciences Hefei Institutes of Physical Science) Director’s Fund (YZJJ201704 and KP-2017-26), Heye Health Technology Foundation (HYJJ20170811), the National Science Foundation for Young Scientists of China (31900506), the National Science Foundation of Anhui Province (1908085MA11), and China Postdoctoral Science Foundation Funded Project (2019M652222). A portion of this work was supported by the High Magnetic Field Laboratory of Anhui Province.
Editor: Philip LeDuc.
Footnotes
Qingping Tao and Lei Zhang contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.12.020.
Contributor Information
Lei Zhang, Email: leizhang@hmfl.ac.cn.
Xin Zhang, Email: xinzhang@hmfl.ac.cn.
Supporting Material
References
- 1.Stefan H., Trinka E. Magnetoencephalography (MEG): past, current and future perspectives for improved differentiation and treatment of epilepsies. Seizure. 2017;44:121–124. doi: 10.1016/j.seizure.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 2.Deistung A., Mentzel H.J., Reichenbach J.R. Demonstration of paramagnetic and diamagnetic cerebral lesions by using susceptibility weighted phase imaging (SWI) Z. Med. Phys. 2006;16:261–267. doi: 10.1078/0939-3889-00324. [DOI] [PubMed] [Google Scholar]
- 3.Paul F., Roath S., Osisanya J.O. Separation of malaria-infected erythrocytes from whole blood: use of a selective high-gradient magnetic separation technique. Lancet. 1981;2:70–71. doi: 10.1016/s0140-6736(81)90414-1. [DOI] [PubMed] [Google Scholar]
- 4.Kasetsirikul S., Buranapong J., Pimpin A. The development of malaria diagnostic techniques: a review of the approaches with focus on dielectrophoretic and magnetophoretic methods. Malar. J. 2016;15:358. doi: 10.1186/s12936-016-1400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zablotskii V., Lunov O., Dejneka A. Effects of high-gradient magnetic fields on living cell machinery. J. Phys. D Appl. Phys. 2016;49:493003. [Google Scholar]
- 6.Zhang L., Hou Y., Zhang X. 27 T ultra-high static magnetic field changes orientation and morphology of mitotic spindles in human cells. eLife. 2017;6:e22911. doi: 10.7554/eLife.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi-Sekino S., Kira T., Akahane M. Effects of 7 T static magnetic fields on the expression of biological markers and the formation of bone in rats. Bioelectromagnetics. 2019;40:16–26. doi: 10.1002/bem.22161. [DOI] [PubMed] [Google Scholar]
- 8.Lipfert J., Kerssemakers J.W., Dekker N.H. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat. Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 9.Schenck J.F. The role of magnetic susceptibility in magnetic resonance imaging: MRI magnetic compatibility of the first and second kinds. Med. Phys. 1996;23:815–850. doi: 10.1118/1.597854. [DOI] [PubMed] [Google Scholar]
- 10.Pauling L. Diamagnetic anisotropy of the peptide group. Proc. Natl. Acad. Sci. USA. 1979;76:2293–2294. doi: 10.1073/pnas.76.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Senftle F.E., Thorpe A. Magnetic susceptibility of normal liver and transplantable hepatoma tissue. Nature. 1961;190:410–413. doi: 10.1038/190410a0. [DOI] [PubMed] [Google Scholar]
- 12.Nakamae S., Cazayous M., Bouchiat H. Intrinsic low temperature paramagnetism in B-DNA. Phys. Rev. Lett. 2005;94:248102. doi: 10.1103/PhysRevLett.94.248102. [DOI] [PubMed] [Google Scholar]
- 13.Spees W.M., Yablonskiy D.A., Ackerman J.J. Water proton MR properties of human blood at 1.5 Tesla: magnetic susceptibility, T(1), T(2), T∗(2), and non-Lorentzian signal behavior. Magn. Reson. Med. 2001;45:533–542. doi: 10.1002/mrm.1072. [DOI] [PubMed] [Google Scholar]
- 14.Wehrli F.W., Fan A.P., Langham M.C. Susceptibility-based time-resolved whole-organ and regional tissue oximetry. NMR Biomed. 2017;30:e3495. doi: 10.1002/nbm.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biondetti E., Rojas-Villabona A., Shmueli K. Investigating the oxygenation of brain arteriovenous malformations using quantitative susceptibility mapping. Neuroimage. 2019;199:440–453. doi: 10.1016/j.neuroimage.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Jain V., Abdulmalik O., Wehrli F.W. Investigating the magnetic susceptibility properties of fresh human blood for noninvasive oxygen saturation quantification. Magn. Reson. Med. 2012;68:863–867. doi: 10.1002/mrm.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Ji X., Zhang X. Cell type- and density-dependent effect of 1 T static magnetic field on cell proliferation. Oncotarget. 2017;8:13126–13141. doi: 10.18632/oncotarget.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., Wang J., Zhang X. Moderate and strong static magnetic fields directly affect EGFR kinase domain orientation to inhibit cancer cell proliferation. Oncotarget. 2016;7:41527–41539. doi: 10.18632/oncotarget.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Yang X.X., Zhang X. 1 T moderate intensity static magnetic field affects Akt/mTOR pathway and increases the antitumor efficacy of mTOR inhibitors in CNE-2Z cells. Sci. Bull. (Beijing) 2015;60:2120–2128. [Google Scholar]
- 20.Kirby T.J., Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 2018;20:373–381. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y.M., Du J.Y., Lau A.T.Y. Posttranslational modifications of human histone H3: an update. Proteomics. 2014;14:2047–2060. doi: 10.1002/pmic.201300435. [DOI] [PubMed] [Google Scholar]
- 22.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 23.van Oene M.M., Dickinson L.E., Dekker N.H. Biological magnetometry: torque on superparamagnetic beads in magnetic fields. Phys. Rev. Lett. 2015;114:218301. doi: 10.1103/PhysRevLett.114.218301. [DOI] [PubMed] [Google Scholar]
- 24.Tran P.T., Marsh L., Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fridolfsson H.N., Starr D.A. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fruleux A., Hawkins R.J. Physical role for the nucleus in cell migration. J. Phys. Condens. Matter. 2016;28:363002. doi: 10.1088/0953-8984/28/36/363002. [DOI] [PubMed] [Google Scholar]
- 27.Tsai J.W., Bremner K.H., Vallee R.B. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 28.Gomes E.R., Jani S., Gundersen G.G. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Bausch A.R., Möller W., Sackmann E. Measurement of local viscoelasticity and forces in living cells by magnetic tweezers. Biophys. J. 1999;76:573–579. doi: 10.1016/S0006-3495(99)77225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durmus N.G., Tekin H.C., Demirci U. Magnetic levitation of single cells. Proc. Natl. Acad. Sci. USA. 2015;112:E3661–E3668. doi: 10.1073/pnas.1509250112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wosik J., Chen W., Kloc M. Magnetic field changes macrophage phenotype. Biophys. J. 2018;114:2001–2013. doi: 10.1016/j.bpj.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brangwynne C.P., Koenderink G.H., Weitz D.A. Nonequilibrium microtubule fluctuations in a model cytoskeleton. Phys. Rev. Lett. 2008;100:118104. doi: 10.1103/PhysRevLett.100.118104. [DOI] [PubMed] [Google Scholar]
- 33.Garzon-Coral C., Fantana H.A., Howard J. A force-generating machinery maintains the spindle at the cell center during mitosis. Science. 2016;352:1124–1127. doi: 10.1126/science.aad9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicklas R.B. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.