Summary
A hallmark of Gram-negative bacteria is an asymmetric outer membrane containing lipopolysaccharides (LPS) in the extracellular leaflet. LPS molecules consist of lipid-A that is connected to the inner and outer core oligosaccharides. This LPS core structure is extended in the periplasm by the O antigen, a variable and serotype-defining polysaccharide. In the ABC transporter-dependent LPS biosynthesis pathway, the WzmWzt transporter secretes the complete O antigen across the inner membrane for ligation to the LPS core. In some O antigen transporters, the nucleotide binding domain (NBD) of Wzt is fused C terminally to a carbohydrate-binding domain (CBD) that interacts with the O antigen chain. Here, we present the crystal structure of the Aquifex aeolicus CBD that reveals a conserved flat and variable twisted jelly-roll surface. The CBD dimer is stabilized by mutual β-strand exchange. Microbial glycan array binding studies with the isolated CBD provide insights into its interaction with complex carbohydrates.
eTOC
Bi and Zimmer determined the crystal structure of the isolated carbohydrate-binding domain of the Aquifex aeolicus O antigen ABC transporter WzmWzt. The domain is essential for O antigen translocation by interacting with the O antigen polysaccharide. The structure allows modeling the architecture of the full length O antigen ABC transporter.
Graphical Abstract
Introduction
A ubiquitous architectural feature of microbial cells is the production of cell surface polysaccharides as an outermost shell. These complex carbohydrates protect many pathogens by reducing the efficacy of the host’s innate immune response, encapsulating the cell in a protective coat to outlast detrimental environmental conditions, or mediating host-microbe interactions. Lipopolysaccharides (LPS) of the outer membrane (OM) in Gram-negative bacteria represent such complex carbohydrates. The molecules consist of the lipid A membrane anchor that is extended by conserved inner and outer core oligosaccharides as well as a variable high molecular weight polysaccharide, called the O antigen (Raetz and Whitfield, 2002).
O antigens are serotype specific and consist of a characteristic repeat unit structure that is 3–5 sugars long (Greenfield and Whitfield, 2012). The polymer is attached to the conserved outer core oligosaccharide in the periplasm before the mature LPS molecule is shuttled to the outer membrane by the Lpt pathway (May et al., 2015). O antigens are synthesized by two different yet equally abundant mechanisms (Raetz and Whitfield, 2002). In the Wzx dependent pathway, the individual O antigen repeat units are synthesized intracellularly on an undecaprenyl anchor (Und-P) and flipped to the periplasmic side of the inner membrane (IM) by the flippase Wzx. Following transport, the individual repeat units are ligated into a high molecular weight polymer, before being transferred to the outer core oligosaccharide by WaaL (Islam and Lam, 2014). In the ABC transporter-dependent pathway, however, the entire O antigen polymer is synthesized on the cytosolic side of the inner membrane, also on an Und-P carrier, and translocated to the membrane’s periplasmic side by the WzmWzt ABC transporter. Following reorientation, WaaL also transfers this polysaccharide to the lipid-A core to complete an LPS molecule (Greenfield and Whitfield, 2012).
Some bacteria signal completion of O antigen biosynthesis by chemically modifying its terminal non-reducing end sugar (Clarke et al., 2004). These modifications, as observed for example in several E. coli and Klebsiella species, include methylation, phospho-methylation, or attachment of non-canonical sugar units, such as β-Kdo (Clarke et al., 2011; Cuthbertson et al., 2005). When these modifications occur, the corresponding ABC transporter contains a CBD covalently fused to the C-terminus of its NBD. It has been demonstrated that the CBD specifically recognizes the O antigen’s terminal modification in the context of the repeat unit structure, and this interaction is necessary for secretion in vivo (Cuthbertson et al., 2005; Mann et al., 2016). Specifically, O antigens with different repeat unit structures but chemically identical modifications are only recognized by the CBD of the corresponding transporter, suggesting that the O antigen repeat also contributes to recognition (Mann et al., 2019). However, the transporter and CBD can function as separated proteins and co-expression of the CBD together with a CBD-truncated ABC transporter restores O antigen translocation in vivo (Cuthbertson et al., 2005). Furthermore, the isolated CBD and corresponding ABC transporter interact in the absence of an O antigen translocation substrate, albeit with weak affinity (Bi et al., 2018).
The CBD likely functions to increase the local O antigen concentration near the ABC transporter (Bi et al., 2018), which is expected to facilitate initiation of O antigen transport. Accordingly, the biosynthesis and membrane translocation of capped O antigens can be uncoupled, i.e. translocation is possible after accumulation of O antigens at the inner membrane (Kos et al., 2009). In contrast, ABC transporter-mediated translocation of uncapped O antigens, as exemplified by Klebsiella pneumoniae O2a, must occur concomitant with biosynthesis, perhaps because of direct interactions of the transporter with the synthesizing enzymes (Kos et al., 2009).
Biopolymer translocating ABC transporters employ a different transport mechanism compared to classical ABC exporters. Because the lengths of the translocated substrates substantially exceed the dimensions of the transporter itself, polypeptide and polysaccharide ABC transporters likely use a processive transport mechanism in which the polymer is pushed through a channel formed by the transporters’ transmembrane segments (Bi et al., 2018; Morgan et al., 2017).
Recent crystal structures of the O antigen ABC transporter WzmWzt from Aquifex aeolicus (Aa) provided the first structural insights into the O antigen translocation mechanism (Bi et al., 2018; Caffalette, 2019). In nucleotide-free and ATP-bound states, the transporter forms a large transmembrane channel sufficiently wide to accommodate a polysaccharide chain 8–10 sugar units long. The channel’s interior surface is decorated with aromatic residues that could interact with the translocating O antigen via CH-π stacking interactions, as observed for cellulose synthase (Morgan et al., 2013).
The Aa O antigen transporter contains a CBD at the C-terminus of its NBD, which was removed to facilitate crystallization (Bi et al., 2018). Further, the exact Aa O antigen structure as well as the chemical nature of its non-reducing end cap is unknown to date (Mamat et al., 2009). Hence, the truncated Aa-WzmWzt structure provided little information on the crucial steps required to initiate O antigen translocation in vivo.
To fill this void, we determined the crystal structure of the isolated Aa CBD and performed microbial glycan array binding assays to determine its substrate specificity. We speculate that cap binding occurs at a conserved CBD surface that also faces the NBD dimer, such that ligand binding could affect the transporter’s hydrolytic activity.
Results
Structure of the Wzt Carbohydrate Binding Domain
The Aa CBD (residues 235–395) was expressed in E. coli with a C-terminal hexa-histidine tag to facilitate purification by metal-affinity chromatography (see Supplemental Experimental Procedures). The domain purifies as a stable dimer (Figure S1A) and was crystallized by standard techniques, yielding small needle-shaped crystals that diffracted to ~3.6 Å resolution. Following structure determination by molecular replacement using the corresponding domain from the E. coli O9a ABC transporter as a search model (Cuthbertson et al., 2007), we identified 19 poorly ordered N-terminal residues potentially interfering with crystal packing. A second CBD construct lacking eight N terminal residues crystallized in a different crystal form that diffracted X-rays to 2.65 Å resolution. With the exception of the deleted N-terminal residues, the structure of this construct is identical to the slightly longer version and was used for refinement and coordinate deposition.
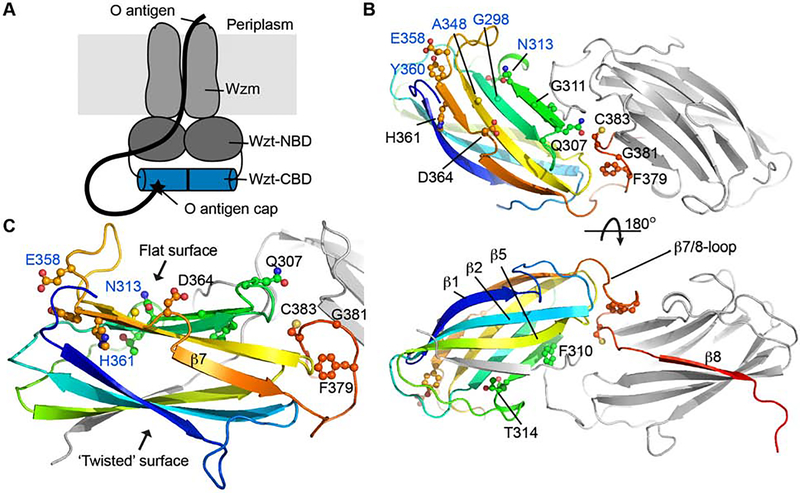
Aa CBD forms a partially SDS-resistant dimer in which the two protomers are positioned side-by-side (Figures 1 and S1A). Each protomer adopts a jelly-roll fold with extended loops connecting the β-strands at the edges. The CBD dimer is stabilized by β-strand exchange between the protomers: The C-terminal residues of protomer ‘A’ (387–394) complete the jelly-roll fold of protomer ‘B’ by inserting between its β-strands 4 and 5. The completed jelly-roll contains a twisted and a rather flat surface, with the twisted surface formed by β-strands 1, 2 and 5 as well as the C terminus of the second protomer (Figure 1).
Figure 1. Crystal structure of the Aquifex aeolicus O antigen ABC transporter’s Carbohydrate Binding Domain.
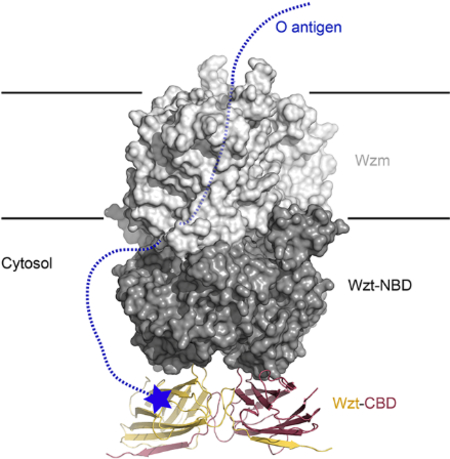
(A) The heterodimeric O antigen ABC transporter consists of the Wzt and Wzm subunits. Wzt contains a nucleotide binding domain (NBD) as well a C terminal carbohydrate binding domain (CBD, blue cylinder). The CBD binds the O antigen’s modified non-reducing end (black star).
(B) Cartoon representation of the isolated CBD. One subunit is colored in rainbow color from the N (blue) to the C (red) terminus. Conserved residues are shown as ‘ball and sticks’ and labeled.
(C) Close up view on one of the CBD subunits. (See also Figure S1)
The dimer interface is further fortified by residues preceding the exchanged β-strand, which form a loop sandwiched between the subunits (Figure 1). A similar domain organization has been observed for the homologous transporter CBDs from Raoultella terrigena and E. coli (Cuthbertson et al., 2007; Mann et al., 2016). Of note, the loop region at the dimer interface includes a Cys residue (Cys383) whose sulfhydryl group is within 4.5 Å to its symmetry mate. The cysteines appear to be reduced based on the experimental electron density map (Figure S1B) and because the purified CBD dimer migrates as a monomer after heat treatment during non-reducing SDS-PAGE (Figure S1A). However, we cannot exclude that the full-length ABC transporter is stabilized by a disulfide bond in this region under the hyper-thermophilic growth conditions of Aquifex aeolicus.
CBDs from O antigen-translocating ABC transporters share an overall low level of sequence identity. However, several conserved and solvent exposed residues surround the flat jelly-roll surface, including Gln307, Phe379 and Gly381 near the dimer interface and Thr314, Glu358, and Tyr360 at the CBD’s opposite end (Figure 1). Among these, residues equivalent to Gly298, Asn313, Ala348, and Glu358 in the E. coli O9a CBD have been implicated in interacting with the O antigen ligand (Cuthbertson et al., 2007), and single point mutants thereof severely affect in vitro LPS binding as well as O antigen translocation in vivo.
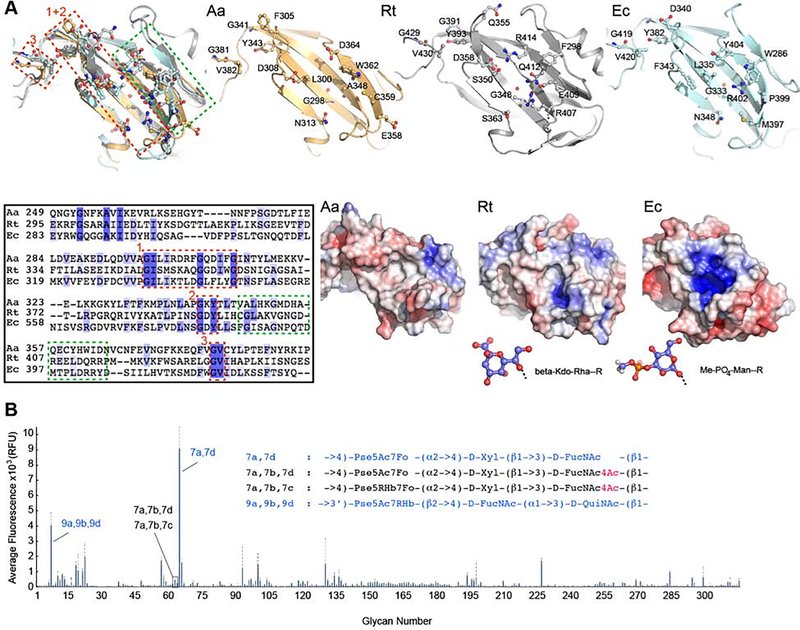
A sequence comparison of the three structurally characterized Wzt-CBDs reveals conserved GIxI/MxxxxGxxxxG and GxY motifs located within β-strand 3 and 6, respectively, as well as a GV motif preceding the exchanged C-terminal β-strand at the dimer interface. The motifs cluster to one side of the flat CBD surface (Figure 2A). The surface’s other half is formed from less conserved residues, perhaps accounting for the chemical diversity of O antigen end modifications recognized by the respective CBDs (Figure 2A). These sequence variations give rise to profound differences in surface properties. While Aa CBD forms a predominantly neutral and polar ligand binding surface, a small electropositive cavity is formed by the R. terrigena CBD, which recognizes a β-Kdo moiety (Cuthbertson et al., 2007). Further, an even larger positively charge pocket is evident on the surface of the E. coli O9a CBD, which binds a methyl-phosphate-derivatized mannosyl unit (Mann et al., 2019) (Figure 2A).
Figure 2. CBD-ligand interaction.
(A) Superimposition of the Aa CBD (orange) with the corresponding domains from R. terrigena (gray, PDB entry 5HNO) and E. coli O9a (pale cyan, PDB entry 2R5O) based on secondary structure matching. Residues lining the putative ligand-binding pocket are shown as ‘ball-and-sticks’ and labeled. Lower panel (left): Sequence alignment of the shown domains, boxed sequences are highlighted and numbered accordingly in the structure overlay; (right) electrostatic potential calculated in Pymol with the APBS plugin from red (−5k) to blue (+5k). Stick models of the known ligands for the 5HNO and 2R5O domains are shown in blue for carbon atoms.
(B) Microbial glycan array studies. Aa CBD was incubated with a printed array of 313 different LPS structures and binding was detected immunologically. The averaged fluorescence is shown for 4 replicas. O antigen structures recognized by the CBD are shown in blue, selected structures not recognized are shown in black. Pse: Pseudaminic acid, Xyl: Xylose, FucN: Fucoseamine, Qui: Quinovose, Rha: rhamnose. Sugar modifications: Ac: acetyl, Fo: formyl, Hb: hydroxybutanoyl. (See also Figure S1)
Glycan microarray binding assays identify a putative CBD substrate homolog
The presence of a WecA homolog in the Aa VF5 genome suggests that the O antigen polymer is assembled on an Und-PP-N-acetylglucosamine (Und-PP-GlcNAc) anchor because WecA catalyzes the transfer of N-acetyl-glucosamine-1-phosphate to Und-P (Al-Dabbagh et al., 2008). However, with the exception of one predicted mannosyltransferase located in proximity to the wzm and wzt genes, no homologs of other O antigen biosynthetic enzymes have been identified in the Aquifex genome thus far.
Therefore, we tested whether carbohydrate-binding studies using a microbial glycan array could provide additional information on Aa CBD’s binding specificity. To this end, the purified CBD was incubated with a printed array of 313 LPS or O antigen structures and CBD binding was determined immunologically using a fluorescently labeled antibody against the protein’s hexa-His tag (Stowell et al., 2014). Binding studies were performed at 5 and 50 μg/ml protein concentration and revealed significant binding to Pseudomonas aeruginosa 7a,7d and 9a,9b,9d O antigens with binding to 7a,7d being most prominent (Figures 2B and S1C). While both O antigen structures contain derivatives of pseudaminic acid, a comparison of the bound 7a,7d structure with the 7a,7b,7d O antigen, which the CBD does not recognize, is most informative. The O antigen structures are essentially identical, with the exception that 7a,7b,7d contains a 4-acetylated N-acetylfucosamine (FucNAc) while this sugar unit is unmodified in the recognized 7a,7d structure (Knirel et al., 2006) (Figure 2). The binding profiles are essentially identical for assays performed at 5 or 50 μg/ml protein concentration and were repeated six-times. Thus, the Aa CBD discriminates between 4-acetylated and unmodified FucNAc in the context of the 7a,7d O antigen structure, which could reflect its specificity towards the natural O antigen cap.
However, it should be noted that the identified glycans represent O antigen repeat unit structures and not end modifications, therefore information on the CBDs specificity towards its native substrate is indirect. Yet, because the identified Pseudomonas O antigen structures are synthesized by the Wzx-dependent pathway and thus not end-modified (Islam and Lam, 2014), we can assume that the detected selectivity is indeed due to variations of the O antigen repeat unit structures (Figure 2B).
Putative CBD-NBD interaction in the full length ABC transporter
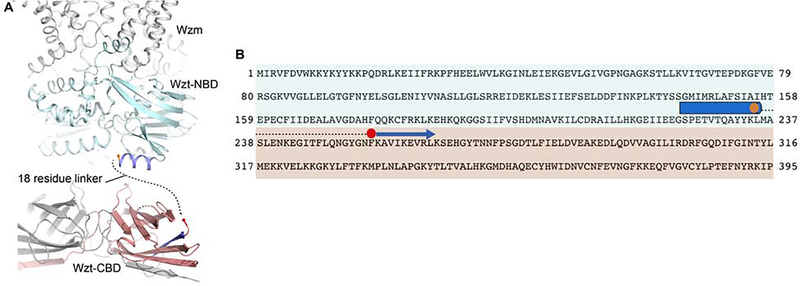
In the full-length ABC transporter, the CBD forms a C-terminal extension of Wzt’s NBD (Figure 1). The crystal structures of the NBD-truncated WzmWzt transporter reveal a well-ordered C-terminal α-helix underneath the NBD with Leu235 as the terminal residue (Bi et al., 2018; Caffalette, 2019) (Figure 3). Based on our CBD structure, residues Asn254 to Pro395 are also well ordered and form a stable tertiary and quaternary structure. Hence, the nucleotide- and carbohydrate-binding domains are connected by an approximately 20 residue long linker (Figure 3).
Figure 3. Domain organization of Wzt.
(A) The structure of ATP-bound WztWzm is shown (PDB entry 6M96) with the NBDs and TM regions colored cyan and gray, respectively. The C-terminal helix of the truncated Wzt is colored dark blue with Leu235 at the C-terminus shown as an orange sphere. The protomers of the Aa CBD dimer are colored gray and red with the N-terminal β-strand colored blue. Phe255 at the N-terminus is shown as a red sphere.
(B) Wzt primary sequence with structurally resolved regions near the NBD-CBD interface shown as cylinders and arrows for α-helices and β-strands, respectively. Aa Wzt is colored according to the domains shown in panel A. The missing linker region is shown as a dashed line.
Because O antigen repeat unit and cap chemical structures differ substantially between species and serotypes, it is likely that CBD sequence conservation is primarily due to interactions with the transporter’s conserved NBD (Figure 4). Accordingly, we recently proposed a model in which the NBD and CBD dimers in the full-length transporter are rotate approximately 90 degree with respect to each other (Caffalette, 2019), which maximizes their interface. Our dimeric CBD structure is consistent with such an ‘antiparallel’ orientation.
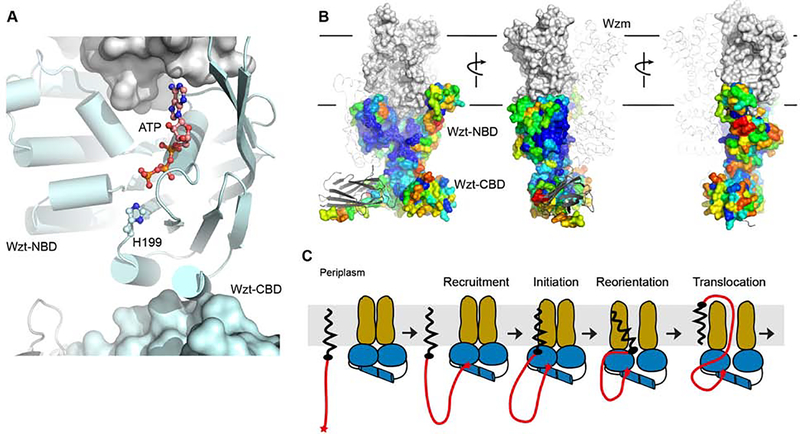
Figure 4. Impact of the CBD on the full-length WztWzm structure and function.
(A) Wzt’s H-loop connects to a peripheral α-helix at the putative NBD-CBD interface. His199 of the H-loop and ATP are shown as ‘ball-and-sticks’ (PDB entry 6M96). The CBD, shown as a surface, was manually placed underneath the NBD to maximize the interface.
(B) Proposed position of Wzt’s CBD and NBD domains. The ABC transporter is shown as a ribbon and surface for the individual heterodimers, respectively. The NBD surface is colored according to sequence conservation from blue (highest) to red (lowest) conservation (generated in CONSURF (Ashkenazy et al., 2010)). The CBD dimer is shown as for the transporter.
(C) Proposed function of the CBD as a substrate-recruiting domain. Initial interactions of the O antigen’s non-reducing end cap with the CBD tethers the polymeric substrate to the transporter where it could alter the transporter’s hydrolytic activity. Substrate insertion into the transporter likely occurs via interactions of the substrate’s lipid anchor with the transporter’s lateral gate.
Due to the short distance of the NBD-CBD linker (~20 residues), both Wzt domains are likely located within the same half transporter and not diagonally connected. The domains must be separated by only a short distance facilitating direct CBD-NBD contacts. This explains the weak association of the isolated domains in vitro and in vivo. (Bi et al., 2018; Cuthbertson et al., 2005). Strikingly, the most likely domain of the NBD to interact with the CBD is a short α-helix that follows the conserved H-loop (Figure 4A). Histidine 199 within this loop interacts with the γ-phosphate of ATP and helps to stabilize the catalytic water molecule for hydrolysis (ter Beek et al., 2014). Thus CBD-induced perturbations of the H-loop could have a profound effect on the transporter’s hydrolytic activity, as observed in vitro.
Discussion
LPS molecules are synthesized and integrated into the bacterial OM in a multi-step process (Raetz and Whitfield, 2002). While export of LPS molecules from the IM’s periplasmic leaflet to the extracellular side of the OM is fairly well characterized biochemically and structurally (Botos et al., 2016; Freinkman et al., 2011; May et al., 2015; Okuda et al., 2016), the biosynthesis of the O antigen polysaccharide and its transport from the IM’s cytosolic to the periplasmic side remain poorly understood to date. For the ABC transporter-dependent biosynthesis pathway, an important unresolved question is the mechanism of transport initiation.
A defining feature of O antigen-translocating ABC transporters is the presence of a ‘gate helix’ within the NBD (Bi et al., 2018). This helix is formed by an extended loop connecting the first two conserved β-strands of the NBD core and packs against the transporter’s putative cytosolic substrate entrance. Because the helix is conserved among ABC transporters recognizing substrates attached to an Und-PP-GlcNAc anchor, such as O antigens and teichoic acids (van der Es et al., 2017), it has been speculated that it mediates the insertion of the substrate into the ABC transporter starting with the Und-PP-GlcNAc head group (Bi et al., 2018) (Figure 4). Thus, the lipid anchor would establish critical interactions with the transporter required to initiate the transport process.
This initiation model likely requires an elevated local substrate concentration, achievable by associating the ABC transporter with the polymer-producing glycosyltransferases, as suggested for the terminally unmodified Klebsiella pneumoniae O2a antigen (Kos et al., 2009). Conversely, if O antigen biosynthesis is terminated by end modification, the corresponding ABC transporter’s CBD binds this cap, thereby also leading to locally increased substrate concentrations. In this case, close proximity of the transporter and biosynthetic machinery may not be required, as suggested for the E. coli O9a transporter (Kos et al., 2009).
Biochemical analyses of isolated CBDs recognizing either modified mannosyl or β-Kdo caps suggest that substrate interaction occurs on the CBD’s flat conserved surface (Cuthbertson et al., 2007; Greenfield and Whitfield, 2012) (Figure 1). We propose that this surface also faces the NBDs, thereby sandwiching the bound O antigen cap between the carbohydrate- and nucleotide-binding domains. Interestingly, previously reported interaction studies of the isolated CBD and CBD-truncated WzmWzt transporter demonstrated significantly increased ATP hydrolysis rates upon CBD-NBD interaction (Bi et al., 2018), perhaps due to conformational changes of the transporter’s H-loop. It is possible that ligand binding to the CBD-NBD interface modifies (perhaps further increase) the transporter’s hydrolytic rate, thereby initiating transport (Figure 4B). Yet, the precise CBD-NBD interactions and the mode of O antigen cap recognition have to await further structural analyses of the full-length WzmWzt transporter.
STAR Methods
LEAD CONTACT AND DATA AVAILABILITY
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Jochen Zimmer, (jochen zimmer@virginia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Both Aa VF5 Wzt-CBD constructs (residues 235–395 and 254–395) were cloned into the pET28a vector (Novagen) with NdeI and HindIII restriction sites encoding a C-terminal hexa-histidine tag.
METHOD DETAILS
Constructs
Both Aa VF5 Wzt-CBD constructs (residues 235–395 and 254–395) were cloned into the pET28a vector (Novagen) with NdeI and HindIII restriction sites, thereby generating C-terminally hexa-His tagged proteins.
Protein Expression
Both constructs were expressed in E. coli BL21 (DE3) cells (Invitrogen). The E. coli cells were cultured in LB medium at 37 °C and protein expression was induced at an optical density at 600 nm of 0.6 with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG). The cells were harvested by centrifugation after 4 hrs of incubation at 37 °C.
Protein Purification
The harvested cell paste was resuspended in RB buffer containing 20 mM sodium phosphate pH7.5, 0.05 M NaCl and 5 mM β-mercaptoethanol (β-ME) and then lysed in a microfluidizer. The insoluble material was removed by centrifugation for 30 min at 200,000xg in a Beckman Ti45 rotor. The supernatant was batch incubated with Ni-NTA agarose (Qiagen) for 60 min at 4 °C. The protein was then purified by Ni-NTA affinity chromatography at 4 °C. After washing with 50 ml WB1 buffer containing 25 mM Tris HCl pH8.5, 0.5 M NaCl, 30 mM imidazole, and 5 mM β-ME, 50 ml WB2 buffer containing 25 mM Tris HCl pH8.5, 50 mM NaCl, 50 mM imidazole, and 5 mM β-ME, the protein was eluted with EB buffer, consisting of 25 mM Tris HCl pH8.5, 50 mM NaCl, 300 mM imidazole, and 5 mM β-ME. The protein was further purified by gel filtration chromatography using a Superdex-200 column (GE) in 25 mM Tris HCl pH8.5, 50 mM NaCl, and 5 mM β-ME.
Crystallization
AaCBD crystals were obtained by sitting-drop vapor diffusion after mixing 2 μL of protein at 7.6 mg/ml with 1 μL of well solution containing 0.1 M citric acid pH 5.5, 0.5 M tri-sodium citrate in the presence of at 30 °C. The shorter CBD construct containing residues 245–395 was crystallized at 3.7 mg/ml by combining 1 μl of well solution (0.1 M MOPS pH 7, 1.5 M sodium acetate) with 1 μl of protein solution and sitting-drop vapor diffusion at 27 °C.
Initial crystals were observed after approximately three days and reached their final size within up to three weeks for both constructs. Crystals were cryo-protected in the presence of 25% glycerol and 3 M sodium acetate for the long and short constructs, respectively.
Data Collection and Structure Determination
Diffraction data were collected at the Argonne National Laboratory beamline SER-CAT as well as the AMX beamline at the Brookhaven National Laboratories (NSLS-II) for the long and short CBD constructs, respectively. Data were integrated in XDS (Kabsch, 2010) and reduced in Aimless, as part of the CCP4 program suite (CCP4, 1994). The structure of the long CBD construct was determined by molecular replacement phasing using Phaser (McCoy et al., 2007) with a poly-alanine model of the E. coli O9a CBD dimer structure (PDB ID: 2R5O) as search model with all flexible loops removed. This crystal form contained four CBD dimers per asymmetric unit, which were placed by Phaser in consecutive rounds. Phases were improved significantly by density modification in PARROT (Cowtan, 2010) including non-crystallographic symmetry (NCS) averaging. The model was manually built in COOT (Emsley and Cowtan, 2004) and refined in Phenix (Adams et al., 2010) to an Rfree of about 33% including NCS averaging. The obtained dimeric CBD model was used for MR replacement phasing of the truncated CBD structure, which contained one CBD dimer in the asymmetric unit. This model was completed and refined as described above. Validation of the final model was assessed by MolProbity as part of the Phenix.refine package, and the PDB Validation Service. Coordinates have been deposited at the Protein Data Bank under accession code 6O14. Figures were prepared using Pymol (PyMol, 2002).
Microbial Glycan Microarray Binding Studies
The purified long Wzt-CBD construct was used for microbial glycan array binding studies, performed by the National Center for Functional Glycomics. The array consists of LPS molecules from 313 different Gram-negative bacteria. Detailed information on the array and LPS structures are available at http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8p.shtml and http://www.ncbi.nlm.nih.gov/pubmed/24814672. Binding studies were performed in 6 replicas and at 5 and 50 μg/ml CBD concentration with identical results. Bound CBD was detected immunologically using a Fluor488-conjugated antibody (Qiagen) against CBD’s C terminal poly-histidine tag. The highest and lowest points from each set of six replicates have been removed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data collection and refinement statistics for X-ray crystallography can be found in Supplemental Table S1.
DATA AND CODE AVAILABILITY
The atomic coordinates and structure factors of EcBcsC2 have been deposited into the PDB with accession code PDB ID: 6O14.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| E. coli C41(λDE3) | Lucigen | Cat# 60442 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Nis-Pur Ni-NTA Resin | Thermo-Fisher | Cat # 88221 |
| Superdex S200 10/30 | GE | Cat# 17-1043-10 |
| Critical Commercial Assays | ||
| MemGold | Molecular Dimensions | Cat# MD-139 |
| Deposited Data | ||
| Aa Wzt-CBD | This study | PDB ID: 6O14 |
| R. terrigena Wzt-CBD | (Mann, E., et al., JBC 2016) | PDB ID: 5HNO |
| K. pneumoniae Wzt-CBD | (Cuthbertson, L., et al., PNAS 2007) | PDB ID: 2R5O |
| Oligonucleotides | ||
| AaCBD_NdeI_F ATATACATATGAACTTCAAAGCTGTC | IDT | N/A |
| AaCBD_HindIII_R (short) TATTAAAGCTTAGGAATTTTTCGGTAATTG | IDT | N/A |
| AaCBD_HindIII_R (long) TATAAAGCttAGGAATTTTTCGGTAATTGAAC | IDT | N/A |
| Recombinant DNA | ||
| pET28_AaCBD-His | This paper | N/A |
| Software and Algorithms | ||
| HKL3000 | (Minor et al, 2006) | https://sbgrid.org/ |
| Phenix 1.9 | (Adams et al., 2010) | https://sbgrid.org/ |
| Coot | (Emsley and Cowtan 2004) | https://sbgrid.org/ |
| Pymol 2.2 | Schrodinger, LLC | https://sbgrid.org/ |
| CCP4 7.0 | Winn et al., 2011 | https://sbgrid.org/ |
Highlights.
Crystal structure of the O antigen transporter carbohydrate binding domain
Microbial glycan array binding studies provide insights into ligand specificity
A model of the full length O antigen ABC transporter
Acknowledgements
We thank the beamline staffs at the Brookhaven and Argonne National Laboratories, beamlines AMX and SER-CAT, respectively. The AMX beamline of the National Synchrotron Light Source II is a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. Data for this research was also in part collected at the Southeast Regional (SER) Collaborative Access Team beamline at the Advanced Photon Source, Argonne National Laboratory. J.Z. is supported by NIH grant 1R01GM129666. We appreciate the participation of the Protein-Glycan Interaction Resource of the Center for Functional Glycomics, supporting grant R24 GM098791, and the National Center for Functional Glycomics, supporting grant P41 GM103694, for microbial glycan array assays. We would like to thank David Cafiso, David Nyenhuis and Sarah Nyenhuis for discussions.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Dabbagh B, Mengin-Lecreulx D, and Bouhss A (2008). Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J Bacteriol 190, 7141–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, and Ben-Tal N (2010). ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38, W529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Mann E, Whitfield C, and Zimmer J (2018). Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature 553, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botos I, Majdalani N, Mayclin SJ, McCarthy JG, Lundquist K, Wojtowicz D, Barnard TJ, Gumbart JC, and Buchanan SK (2016). Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens. Structure 24, 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffalette C, Corey R, Sansom MSP, Stansfeld PJ, Zimmer J (2019). A lipid gating mechanism for the O antigen ABC transporter. Nature Comm 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BR, Cuthbertson L, and Whitfield C (2004). Nonreducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J Biol Chem 279, 35709–35718. [DOI] [PubMed] [Google Scholar]
- Clarke BR, Richards MR, Greenfield LK, Hou D, Lowary TL, and Whitfield C (2011). In vitro reconstruction of the chain termination reaction in biosynthesis of the Escherichia coli O9a O-polysaccharide: the chain-length regulator, WbdD, catalyzes the addition of methyl phosphate to the non-reducing terminus of the growing glycan. J Biol Chem 286, 41391–41401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Kimber MS, and Whitfield C (2007). Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc Natl Acad Sci U S A 104, 19529–19534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Powers J, and Whitfield C (2005). The C-terminal domain of the nucleotide-binding domain protein Wzt determines substrate specificity in the ATP-binding cassette transporter for the lipopolysaccharide O-antigens in Escherichia coli serotypes O8 and O9a. J Biol Chem 280, 30310–30319. [DOI] [PubMed] [Google Scholar]
- Freinkman E, Chng S-S, and Kahne D (2011). The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci U S A 108, 2486–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield LK, and Whitfield C (2012). Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr Res 356, 12–24. [DOI] [PubMed] [Google Scholar]
- Islam ST, and Lam JS (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol 60, 697–716. [DOI] [PubMed] [Google Scholar]
- Karplus PA, and Diederichs K (2012). Linking Crystallographic Model and Data Quality. Science 336, 1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel YA, Bystrova OV, Kocharova NA, Zahringer U, and Pier GB (2006). Conserved and variable structural features in the lipopolysaccharide of Pseudomonas aeruginosa. J Endotoxin Res 12, 324–336. [DOI] [PubMed] [Google Scholar]
- Kos V, Cuthbertson L, and Whitfield C (2009). The Klebsiella pneumoniae O2a antigen defines a second mechanism for O antigen ATP-binding cassette transporters. J Biol Chem 284, 2947–2956. [DOI] [PubMed] [Google Scholar]
- Mamat U, Schmidt H, Munoz E, Lindner B, Fukase K, Hanuszkiewicz A, Wu J, Meredith TC, Woodard RW, Hilgenfeld R, et al. (2009). WaaA of the hyperthermophilic bacterium Aquifex aeolicus is a monofunctional 3-deoxy-D-manno-oct-2-ulosonic acid transferase involved in lipopolysaccharide biosynthesis. J Biol Chem 284, 22248–22262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E, Kelly SD, Al-Abdul-Wahid MS, Clarke BR, Ovchinnikova OG, Liu B, and Whitfield C (2019). Substrate recognition by a carbohydrate-binding module in the prototypical ABC transporter for lipopolysaccharide O antigen from Escherichia coli O9a. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E, Mallette E, Clarke BR, Kimber MS, and Whitfield C (2016). The Klebsiella pneumoniae O12 ATP-binding Cassette (ABC) Transporter Recognizes the Terminal Residue of Its O-antigen Polysaccharide Substrate. J Biol Chem 291, 9748–9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JM, Sherman DJ, Simpson BW, Ruiz N, and Kahne D (2015). Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Philos Trans R Soc Lond B Biol Sci 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J, Strumillo J, and Zimmer J (2013). Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Acheson JF, and Zimmer J (2017). Structure of a Type-1 Secretion System ABC Transporter. Structure 25, 522–529. [DOI] [PubMed] [Google Scholar]
- Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, and Kahne D (2016). Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, and Whitfield C (2002). Lipopolysaccharide endotoxins. Annu Rev Biochem 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, McBride R, Berger O, Razi N, Heimburg-Molinaro J, Rodrigues LC, Gourdine JP, Noll AJ, von Gunten S, et al. (2014). Microbial glycan microarrays define key features of host-microbial interactions. Nat Chem Biol 10, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Beek J, Guskov A, and Slotboom DJ (2014). Structural diversity of ABC transporters. J Gen Physiol 143, 419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Es D, Hogendorf WF, Overkleeft HS, van der Marel GA, and Codee JD (2017). Teichoic acids: synthesis and applications. Chem Soc Rev 46, 1464–1482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of EcBcsC2 have been deposited into the PDB with accession code PDB ID: 6O14.