Abstract
Existing long-read assemblers require thousands of CPU hours to assemble a human genome and are being outpaced by sequencing technologies in terms of both throughput and cost. We developed a long-read assembler wtdbg2 (https://github.com/ruanjue/wtdbg2) that is 2–17 times as fast as published tools while achieving comparable contiguity and accuracy. It paves the way for population-scale long-read assembly in future.
De novo sequence assembly reconstructs a sample genome from relatively short sequence reads. It is essential to the study of new species and structural genomic changes that often fail mapping-based analysis as the reference genome may lack the regions of interest. With the rapid advances in single-molecule sequencing technologies by Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT), we are able to sequence reads of 10–100 kilobases (kb) at low cost. Such long reads resolve major repeat classes in primates and help to improve the contiguity of assemblies. Long-read assembly has become a routine for bacteria and small genomes, thanks to the development of several high-quality assemblers1–5. For mammalian genomes, however, existing assemblers may require significant computing resources. The computing cost with commercial cloud services is comparable to the sequencing cost with one ONT’s PromethION machine, which is capable of sequencing a human genome at 30-fold coverage in two days6. To address this issue, we developed wtdbg2, a new long-read assembler that is times faster for large genomes with little compromise on the assembly quality.
Wtdbg2 broadly follows the overlap-layout-consensus paradigm. It advances the existing assemblers with a fast all-vs-all read alignment implementation and a novel layout algorithm based on fuzzy-Bruijn graph (FBG), a new data structure for sequence assembly that is related to sparse de Bruijn graphs and A-Bruijn graphs.
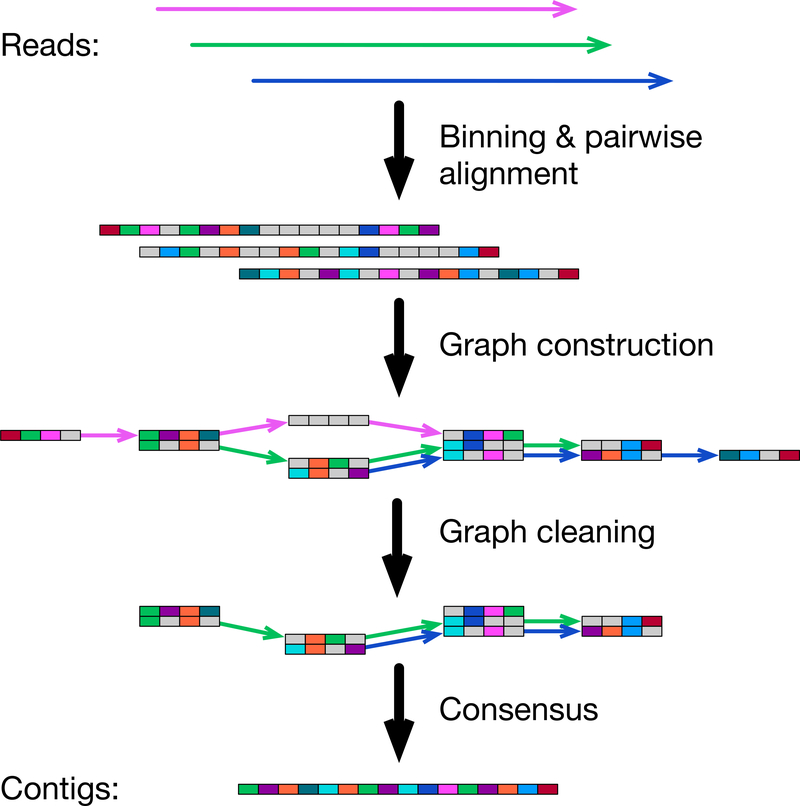
For mammalian genomes, current read overlappers7–9 split input reads into many smaller batches and perform all-vs-all alignment between batches. This strategy wastes compute time on repeated file I/O and on indexing and querying non-informative k-mers. These overlappers do not build a single hash table as they worry the hash table may take too much memory. Interestingly, this should not be a major concern. Wtdbg2 first loads all reads into memory and counts k-mer occurrences. It then takes each tiling 256bp subsequence on reads as one unit, defined as a bin (each small box in Figure 1), and builds a hash table with keys being k-mers occurring ≥2 times in reads, and values being locations of associated bins on reads. For example, among PacBio reads sequenced from the CHM1 human genome to 60-fold coverage10, there are only 1.5 billion non-unique homopolymer-compressed 21-mers9. Staging raw read sequences in memory and constructing the hash table takes 250GB at the peak, which is comparable to the memory usage of short-read assemblers.
Fig. 1.
Outline of the wtdbg2 algorithm. Wtdbg2 groups 256 base pairs into a bin, a small box in the figure. Bins/boxes with the same color suggest they share k-mers, except that a gray bin doesn’t match other bins due to sequencing errors. Wtdbg2 performs all-vs-all alignment between binned reads and constructs the fuzzy-Bruijn assembly graph, where a vertex is a 4-bin segment and an edge connects two vertices if they are both present on a read. Wtdbg2 then trims tips and pops bubbles and produces the final contig sequences from the consensus of read subsequences attached to each edge.
Sequence binning described above aims to speed up pairwise alignment with dynamic programming (DP) between binned sequences. With 256bp binning, the DP matrix is 65536 (=256×256) times smaller than a per-base DP matrix as is used by the Smith-Waterman algorithm11. This reduces DP to a much smaller scale in comparison to k-mer based8, 9 or base-level DP7.
FBG extends the basic ideas behind de Bruijn graph (DBG) to work with long noisy reads. In analogy to DBG, a “base” in FBG is a 256bp bin and a “K-mer” or K-bin in FBG consists of K consecutive bins on reads. A vertex in FBG is a K-bin and an edge between two vertices indicates their adjacency on a read. Unlike DBG, different K-bins may be represented by a single vertex if they are aligned together based on all-vs-all read alignment. This treatment tolerates errors in noisy long reads. FBG is closer to sparse DBG12 than standard DBG in that it does not inspect every K-bin on reads. The sparsity reduces the memory to construct FBG. Furthermore, FBG explicitly keeps the read names and the offsets of bins going through each edge to retain long-range information without a separate “read threading” step as with standard DBG assembly. After graph simplification4, 13, wtdbg2 writes the final FBG to disk with read sequences on edges contained in the file. Wtdbg2 constructs the final consensus with partial order alignment14 over edge sequences.
We evaluated wtdbg2 v2.5 on four datasets along with CANU-1.83, FALCON-1808311, Flye-2.3.62, MECAT-1803145 and Ra-190327 (Table 1; see Supplementary Table 1 for more datasets). We used minimap2 to align assembled contigs to the reference genome and to collect metrics. Depending on datasets, wtdbg2 is 2–17 times as fast as the closest competitors. Its contiguity and assembly accuracy are generally comparable to other assemblers. Wtdbg2 assemblies sometimes cover less reference genomes, which is a weakness of wtdbg2, but its contigs tend to have fewer duplicates (metric “% genome covered more than once” in the table). The low redundancy rate is particularly evident for the Col-0/Cvi-0 A. thaliana dataset that has a relatively high heterozygosity of ~1%. On a M. schizocarpa (banana) ONT dataset sequenced to 45-fold coverage15, wtdbg2 delivers a 507Mb assembly with 1.0Mb N50. While this is not as good as the published result, it is larger and more contiguous than the Flye and Ra assemblies (Online Methods).
Table 1. Evaluating long-read assemblies.
FALCON requires PacBio-style read names and does not work with ONT data or the A4 strain of D. melanogaster which was downloaded from SRA. The A. thaliana assembly by FALCON is acquired from PacBio website as our assembly is fragmented. MECAT produces fragmented assemblies for the ONT dataset. Human assemblies were performed by the developers of each assembler. Base-level evaluations and NGA50 are only reported when the sequenced strain or individual is close to the reference genome. BUSCO scores are computed for genomes sequenced to 50-fold coverage or higher.
| Dataset | Metric | CANU | FALCON | Flye | MECAT | Ra | Wtdbg2 |
|---|---|---|---|---|---|---|---|
| C. elegans Bristo ref. strain PacBio x80 | Total length (>= 50kbp) | 106.5Mb | 100.8Mb | 102.0Mb | 102.1Mb | 108.1Mb | 104.8Mb |
| % reference genome covered | 99.58 | 99.16 | 99.29 | 99.51 | 99.55 | 99.37 | |
| % genome covered more than once | 0.33 | 0.25 | 0.15 | 0.35 | 0.69 | 0.13 | |
| NG75 (75% ref. in contigs longer than NG75) | 1,884,280 | 935,802 | 1,275,590 | 1,424,674 | 1,320,829 | 2,255,274 | |
| NG50 (50% ref. in contigs longer than NG50) | 2,677,990 | 1,629,544 | 1,926,198 | 2,113,456 | 2,047,105 | 3,596,268 | |
| NGA50 (50% ref in alignments longer than NGA50) | 1,283,814 | 980,062 | 1,087,075 | 1,119,713 | 1,019,386 | 1,365,602 | |
| # alignment breakpoints | 681 | 192 | 284 | 278 | 724 | 177 | |
| BUSCO (% complete single-copy genes) | 98.2% | 88.1% | 98.4% | 97.0% | 90.9% | 97.5% | |
| # substitutions/1Mb (pre-/post-polish) | 64.1 / 62.2 | 233.2 / 50.1 | 61.6 / 57.6 | 65.9 / 62.8 | 309.9 / 66.8 | 83.8 / 60.3 | |
| # insertions/1Mb (pre-/post-polish) | 31.1 / 22.4 | 592.7 / 19.4 | 29.8 / 21.8 | 43.9 / 21.9 | 3011.2 / 24.3 | 110.6 / 20.8 | |
| # deletions/1Mb (pre-/post-polish) | 152.8 / 55.1 | 1822.7 / 56.7 | 381.4 / 56.9 | 366.0 / 57.9 | 144.1 / 53.1 | 343.0 / 57.7 | |
| Wall-clock time over 32 CPUs (pre-polish) | 9h30m | 2h06m | 2h58m | 3h08m | 2h23m | 26m | |
| D. melanogaster ISO1 ref. strain ONT x32 | Total length (>= 50kbp) | 135.0Mb | 130.7Mb | 126.5Mb | 127.4Mb | ||
| % reference genome covered | 91.74 | 89.40 | 86.35 | 89.34 | |||
| % genome covered more than once | 1.19 | 0.14 | 0.68 | 0.22 | |||
| NG75 | 714,013 | 1,367,004 | 685,943 | 1,752,322 | |||
| NG50 | 4,298,595 | 6,016,667 | 1,898,336 | 10,631,323 | |||
| NGA50 | 1,837,928 | 2,210,468 | 1,700,400 | 2,989,107 | |||
| # alignment breakpoints | 823 | 248 | 225 | 276 | |||
| # substitutions per 1Mb (pre-polish) | 847.6 | 1318 | 1976.2 | 1109.2 | |||
| # insertions per 1Mb (pre-polish) | 255.9 | 10669.9 | 4388.7 | 371.2 | |||
| # deletions per 1Mb (pre-polish) | 7168.2 | 1901.3 | 2324.6 | 9746.3 | |||
| Wall-clock time over 32 CPUs (pre-polish) | 22h23m | 1h41m | 2h10m | 50m | |||
| A. thaliana F1 generation of Col-0 and Cvi-0 strains (~1% heterozygosity) PacBio x185 | Total length (>= 50kbp) | 196.5Mb | 138.1Mb | 122.3Mb | 188.4Mb | 133.3Mb | 125.0Mb |
| % reference genome covered | 99.04 | 97.03 | 93.55 | 97.47 | 92.52 | 92.66 | |
| % genome covered more than once | 47.61 | 11.35 | 3.72 | 51.46 | 3.38 | 1.08 | |
| NG75 | 460,325 | 4,810,976 | 180,227 | 1,096,121 | 404,218 | 2,182,254 | |
| NG50 | 873,036 | 7,979,657 | 370,306 | 3,525,236 | 1,210,836 | 8,707,235 | |
| # alignment breakpoints | 3,059 | 2,102 | 1,674 | 2,573 | 2,078 | 1,777 | |
| BUSCO (% complete single-copy genes) | 43.8% | 91.9% | 93.1% | 49.2% | 87.8% | 90.3% | |
| Wall-clock time over 32 CPUs (pre-polish) | 30h42m | (by PacBio) | 20h3m | 11h33m | 18h33m | 1h12m | |
| Human CHM1 cell line PacBio x100 | Total length (>= 50kbp) | 2,837Mb | 2,938Mb | 2,712Mb | |||
| % reference genome covered | 89.33 | 90.13 | 86.03 | ||||
| % genome covered more than once | 0.53 | 0.72 | 0.02 | ||||
| NG75 | 3,793,440 | 7,726,658 | 4,387,668 | ||||
| NG50 | 17,570,750 | 26,132,317 | 18,220,221 | ||||
| NGA50 | 7,128,216 | 9,262,902 | 8,017,241 | ||||
| # alignment breakpoints | 1,795 | 7,966 | 1,619 | ||||
| BUSCO (% complete single-copy genes) | 91.3% | 91.5% | 90.5% | ||||
| # substitutions per 1Mb (post-polish) | 961.5 | 966.6 | 963.6 | ||||
| # insertions per 1Mb (post-polish) | 142.8 | 140.1 | 140.2 | ||||
| # deletions per 1Mb (post-polish) | 140.0 | 137.6 | 141.1 | ||||
| Total CPU hours (pre-polish CPU hours) | 22,750 | 68,789 | 2,506 (632) | ||||
For samples close to the reference genome, we also compared the consensus accuracy before and after signal-based polishing16 when applicable. Without polishing, CANU, Flye and MECAT tend to produce better consensus sequences. This is probably because they perform at least two rounds of error correction or the consensus step, while wtdbg2 applies one round of consensus only. After Quiver polishing, the consensus accuracy of all assemblers is very close and significantly higher than the accuracy of consensus without polishing. This observation reconfirms that polishing consensus is still necessary17 and suggests that the pre-polishing consensus accuracy is not obviously correlated with post-polishing accuracy. In the past, Quiver was taking a small fraction of total assembly time, but it is now several times slower than wtdbg2 (7 wall-clock hours for C. elegans and 37 wall-clock hours for CHM1) and becomes the new bottleneck. This calls for future improvement to the polishing step.
We assembled four additional human datasets (Table 2). Wtdbg2 finishes each assembly in <2 days on a single computer. This performance broadly matches the throughput of a PromethION machine. In comparison, Flye and CANU required ~5,000 and ~40,000 CPU hours, respectively, to assemble NA128782,18. For this sample, wtdbg2 uses 235GB memory, less than half of memory used by Flye. Partly due to the relatively low memory footprint, wtdbg2 is scalable to huge non-human genomes. It can assemble axolotl, with a 32Gb genome, in two days using 1.2TB memory. The NG50 is 392kb, longer than the published assembly19.
Table 2.
Wtdbg2 performance on other human genomes. Performance metrics were obtained on a machine with 96 CPU cores. G. size: size of the reference genome; Cov.: sequencing coverage; NG50: 50% of the reference genome are in contigs longer than this length.
| Data set | Technology | Cov. | CPU hour | Real hour | Peak RAM (GB) | NG50 (Mb) |
|---|---|---|---|---|---|---|
| NA12878 | Nanopore | 36 | 1513 | 26 | 235 | 10.3 |
| NA19240 | Nanopore | 35 | 1197 | 19 | 226 | 4.4 |
| NA24385 | PacBio CCS | 28 | 410 | 6 | 108 | 11.8 |
| HG00733 | PacBio Sequel | 93 | 1906 | 37 | 338 | 29.2 |
Ten years ago when the Illumina sequencing technology entered the market, the sheer volume of data effectively decommissioned all aligners and assemblers developed earlier. History repeats itself. Affordable population-scale long-read sequencing is on the horizon. Wtdbg2 is an assembler that is able to keep up with the throughput and the cost. With heterozygote-aware consensus algorithms and phased assembly planned for future, wtdbg2 and upcoming tools might fundamentally change the current practices on sequence data analysis.
Online methods
The wtdbg2 algorithm
Wtdbg2 reads all input sequences into memory and encodes each base with 2 bits. By default, it selects a quarter of k-mers based on their hash code and counts their occurrences using a hash table with 46-bit key to store a k-mer and 17-bit value to store its count. Wtdbg2 filters out k-mer occurring once or over 1000 times in reads, and then scans reads again to build a hash table for the remaining k-mers and their positions in bins.
For all-vs-all read alignment, wtdbg2 traverses each read, from the longest to the shortest, and uses the hash table to retrieve the reads that share k-mers with the read in query. It takes each bin as a basepair and applies Smith-Waterman-like DP between binned sequences, penalizing gaps and mismatching bins that do not share k-mers. Wtdbg2 retains alignments no shorter than 8×256bp. After finishing alignments for all reads, wtdbg2 frees the hash table but keeps the all-vs-all alignments in memory (alignments are also written to disk as intermediate results).
At this step, wtdbg2 drops base sequences. It only sees binned sequences and the alignments between them. On an L-long binned sequence B = b1 b2 … bL, a K-bin BKi = bi bi+1 … bi+K−1 is a K-long subsequence starting at the i-th position on B. If binned sequences B and B’ can be aligned, we can infer the overlap length between K-bins BKi and B′Ki′ by lifting their coordinates between the two sequences based on the alignment. We say two K-bins BKi and b′Ki′ are equivalent if the overlap length between them is K (i.e. the two bins are completely aligned). Using the all-vs-all alignment, wtdbg2 collects a maximal non-redundant set Ω of K-bins such that no K-bin in Ω is equivalent to others. For each K-bin in Ω, its coverage is defined as the number of equivalent K-bins in all reads. Wtdbg2 records the locations and coverage of each K-bin.
Two K-bins in Ω may have an overlap up to K-1 bins. The vertex set V of FBG is intended to be an Ω’s subset in which no K-bins overlap with each other. To construct V, wtdbg2 traverses each non-redundant K-bin in the descending order of their initial coverage. Given a K-bin BK, wtdbg2 reduces its coverage by deducting the number of K-bins already in V that overlap with BK. If the reduced coverage is ≥3 and higher than half of the initial coverage, BK will be added to V; otherwise it will be ignored. After the construction of V, wtdbg2 adds an edge between two K-bins if they are located on the same read. There are often multiple edges between two K-bins. Wtdbg2 retains one edge and keeps the count. An edge covered by <3 reads are discarded. This generates FBG. The coverage thresholds can be adjusted on the wtdbg2 command line.
Assembling evaluation datasets
With wtdbg2, we specified the genome size and sequence technology on the command line, which automatically applies multiple options. Specifically, we used “-xrs -g100m” for C. elegans, “-xsq -g125m” for A. thaliana, “-xrs -g144m” for D. melanogaster A4 strain, “-xont -g144m” for the ISO1 strain, “-xrs -g3g” for CHM1, “-xont -g3g” for human NA12878 and NA19240 ONT reads, “-xsq -g3g” for HG00733, “-xccs -g3g” for NA24385 and “-xrs -g3g” for the axolotl dataset. Here, option “-x” specifies the preset. “rs” uses homopolymer-compressed9 (HPC) 21-mer. Both “sq” and “ont” apply 15-mer to genomes smaller than 1Gb but use HPC 19-mer for larger genomes. Note that 415=1GB. We change the type of k-mers for larger genomes to avoid non-specific seed hits, which reduce the performance. We use shorter k-mers for Nanopore data due to their higher error rates and relatively low coverage in our evaluation. Increasing k-mer length for Nanopore helps to resolve paralogous regions but reduces alignment sensitivity and leads to more fragmented assemblies for data at ~30-fold coverage.
For CANU, Flye and MECAT, we similarly specified the genome size and the sequencing technology only. The FALCON configure file for assembling C. elegans is provided as supplementary data. The FALCON A. thaliana assembly was downloaded at http://bit.ly/pbpubdat. We are using AC:GCA_000983455.1 for the CANU CHM1 assembly and AC:GCA_001297185.1 for the FALCON CHM1 assembly.
Assembling the M. schizocarpa (banana) dataset
The authors who produced the dataset failed to run CANU, so we skipped CANU and MECAT (which is based on CANU). This is a nanopore dataset to which FALCON is not applicable. We used wtdbg2’s nanopore preset for large genome for assembly (“-xont -g600m -k0 -p19”) and got an 507Mb assembly with N50=1.0Mb for contigs longer than 10kb. Flye assembled a 505Mb genome with N50=300kb. The authors of the dataset managed to get N50=2.1Mb with Ra on all raw reads. However, with Ra, we could only produce a small assembly of 490Mb at 643kb N50. Instead, we get the best contiguity with miniasm, which generated a 520Mb assembly with N50=1.9Mb. Wtdbg2 is ~10 times as fast as Flye and Ra.
Evaluating assemblies
To count alignment breakpoints, we mapped all assemblies to the corresponding reference genomes with minimap2 under the option “--paf-no-hit -cxasm20 -r2k -z1000,500”. We used the companion script paftools.js to collect various metrics (command line: “paftools.js asmstat -q50000 -d.1”, where “-q” sets the minimum contig length and “-d” sets the max sequence divergence). To count substitutions and gaps, we applied a different minimap2 setting “-cxasm5 --cs -r2k”. This setting introduces more alignment breakpoints but avoids poorly aligned regions harboring spuriously high number of differences that are likely caused by large-scale variations and skew the counts. We used “paftools.js call” to call variations.
Data availability
C. elegans and A. thaliana Ler-0 reads are available at the PacBio public datasets portal: http://bit.ly/pbpubdat. We downloaded SRR5439404 for the D. melanogaster A4 strain, SRR6702603 for the D. melanogaster reference ISO1 strain, ERR2571284 through ERR2571302 for M. schizocarpa (banana; MinION reads only), PRJNA378970 for axolotl, SRR7615963 for HG00733, and ERR2631600 and ERR2631601 for NA19240. CHM1 reads were acquired from SRP044331 (http://bit.ly/chm1p6c4 for raw signals), NA12878 reads from http://bit.ly/na12878ont (release 5) and NA24385 from http://bit.ly/NA24385ccs. For the A. thaliana Col-0/Cvi-0 dataset, the FASTQ files at SRA (AC: PRJNA314706) were not processed properly. Jason Chin, the first author of the paper1 describing the dataset, provided us with reprocessed raw reads, which are now hosted at public ftp site: ftp://ftp.dfci.harvard.edu/pub/hli/col0-cvi0/. The CHM1 CANU and FALCON assemblies and the axolotl assembly are available at NCBI (GCA_000983455.1, GCA_001297185.1 and GCA_002915635.1, respectively). All the evaluated assemblies generated by us can be obtained at ftp://ftp.dfci.harvard.edu/pub/hli/wtdbg/. The FTP site also provides the detailed command lines and the FALCON configuration files.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
The wtdbg2 source code is hosted by GitHub at: https://github.com/ruanjue/wtdbg2.
Supplementary Material
Acknowledgements
We are grateful to Jason Chin for providing the properly processed raw reads for the A. thaliana Col-0/Cvi-0 dataset. We would like to thank Chengxi Ye from University of Maryland for frequent and fruitful discussion in the development of wtdbg and thank Alun Li and Shigang Wu from CAAS for the help in polishing assemblies. We also thank the reviewers whose comments have helped us to improve wtdbg2. This study was supported by Natural Science Foundation of China (NSFC; grant 31571353 and 31822029 to J.R.) and by US National Institutes Health (NIH; grant R01-HG010040 to H.L.).
Footnotes
Competing interests. The authors declare no competing interests.
Reference
- 1.Chin CS et al. Nat Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolmogorov M, Yuan J, Lin Y & Pevzner PA Nat Biotechnol 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Koren S et al. Genome Res 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H Bioinformatics 32, 2103–2110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao CL et al. Nat Methods 14, 1072–1074 (2017). [DOI] [PubMed] [Google Scholar]
- 6.De Coster W et al. Genome Res 29, 1178–1187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers G in WABI, Vol. 8701 (eds. Brown DG & Morgenstern B) 52–67 (Springer, Wroclaw, Poland; 2014). [Google Scholar]
- 8.Berlin K et al. Nat Biotechnol 33, 623–630 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Li H Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaisson MJ, Wilson RK & Eichler EE Nat Rev Genet 16, 627–640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith TF & Waterman MS J Mol Biol 147, 195–197 (1981). [DOI] [PubMed] [Google Scholar]
- 12.Ye C, Ma ZS, Cannon CH, Pop M & Yu DW BMC Bioinformatics 13 Suppl 6, S1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerbino DR & Birney E Genome Res 18, 821–829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C, Grasso C & Sharlow MF Bioinformatics 18, 452–464 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Belser C et al. Nat Plants 4, 879–887 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Chin CS et al. Nat Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Watson M & Warr A Nat Biotech (2019). [DOI] [PubMed] [Google Scholar]
- 18.Jain M et al. Nat Biotechnol 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowoshilow S et al. Nature 554, 50–55 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
C. elegans and A. thaliana Ler-0 reads are available at the PacBio public datasets portal: http://bit.ly/pbpubdat. We downloaded SRR5439404 for the D. melanogaster A4 strain, SRR6702603 for the D. melanogaster reference ISO1 strain, ERR2571284 through ERR2571302 for M. schizocarpa (banana; MinION reads only), PRJNA378970 for axolotl, SRR7615963 for HG00733, and ERR2631600 and ERR2631601 for NA19240. CHM1 reads were acquired from SRP044331 (http://bit.ly/chm1p6c4 for raw signals), NA12878 reads from http://bit.ly/na12878ont (release 5) and NA24385 from http://bit.ly/NA24385ccs. For the A. thaliana Col-0/Cvi-0 dataset, the FASTQ files at SRA (AC: PRJNA314706) were not processed properly. Jason Chin, the first author of the paper1 describing the dataset, provided us with reprocessed raw reads, which are now hosted at public ftp site: ftp://ftp.dfci.harvard.edu/pub/hli/col0-cvi0/. The CHM1 CANU and FALCON assemblies and the axolotl assembly are available at NCBI (GCA_000983455.1, GCA_001297185.1 and GCA_002915635.1, respectively). All the evaluated assemblies generated by us can be obtained at ftp://ftp.dfci.harvard.edu/pub/hli/wtdbg/. The FTP site also provides the detailed command lines and the FALCON configuration files.