Alfalfa (Medicago sativa L.) is one of the most important forage legume crops for hay and silage production worldwide (Li and Brummer, 2012). Understanding the genetic basis underlying its agronomic performance will provide critical molecular insights in support of alfalfa breeding. Alfalfa is a tetraploid, perennial, open pollinated legume with a high level of heterozygosity, which increases the complexity of the genetics required to support plant breeding. Genotyping‐by‐sequencing (GBS)‐based genome‐wide association studies (GWAS), which utilize genetically diverse accessions, offer an alternative to QTL mapping with biparental mapping populations (Flint‐Garcia et al., 2003) and are particularly suitable for orphan crop species, even those without reference genomes. In the present study, we applied the GWAS approach to a collection of 322 alfalfa genotypes of diverse geographic origin to investigate marker‐trait associations for nine agronomic traits characterized across three consecutive years. Phenotypic data analysis revealed that all nine traits were significantly influenced by genotype and were of varying degrees of heritability.
A total of 115 654 high‐quality single nucleotide polymorphisms (SNPs) were identified, and 44 757 SNPs (38.7%) were uniquely and physically mapped onto the M. truncatula reference genome with an average map density of 8.8 kilobases. Population structure on 322 alfalfa genotypes based on these SNPs with a minor allele frequency > 5% in multiple analyses (STRUCTURE, PCoA and phylogenetic trees) showed that the genotypes from China were distinct from those collected from the other regions of the world.
The best linear unbiased prediction (BLUP) values of each genotype for nine agronomic traits were used as phenotypic values for GWAS. GWAS were conducted in the mixed linear model using TASSEL 5.0. For the nine agronomic traits, plant height, plant branching, number of stem nodes, first inflorescence position, biomass yield, leaf to stem ratio, plant regrowth, flowering date and plant height in the fall, a total of 42 putative significant marker‐trait associations (MTAs) were detected (P < 1/44757 ≈ 2.23 × 10−5) with at least one MTA identified for each trait except for biomass yield.
To identify the candidate genes associated with the significant loci based on a GWAS, we performed a pairwise alignment using the flanking sequences of the significant SNP loci against the M. truncatula reference genome sequences (Mt4.0 V1) and NCBI nucleotide acid databases.
Plant height (PH) is considered a critical indicator of biomass yield in alfalfa (Santos et al., 2018). Although this trait is desirable in alfalfa cultivars, its genetic characterization has not been well documented. In our study, a significant SNP (rs12428), which explained 12.6% of the phenotypic variation in PH, was located in a gene (MTR_2 g105090) on chromosome 2 (Fig. 1b1). The gene containing the SNP encodes a protein that harbours an ACT domain and shares high protein sequence similarity with the Arabidopsis thaliana ACR11 gene and was labelled MsACR11. Recent research has reported that ACR11 is an activator of a glutamine synthetase (GS) 2, giving it a mechanistic role in nitrogen assimilation in A. thaliana (Sung et al., 2011). Nitrogen is one of the limiting factors for growth and development in crops. In higher plants, inorganic nitrogen cannot be absorbed and utilized unless it is assimilated into organic matter such as glutamine or glutamic acid. GS is a key enzyme which plays an important role in nitrogen metabolism in higher plants. GS2, encoded by GLN2, is the only plastid‐type GS in A. thaliana (Osanai et al., 2017). Until now, the regulatory mechanism of nitrogen metabolism‐related genes in determining the PH trait of the legume forage crop has remained unelucidated. Therefore, this SNP might represent an important locus controlling plant height in alfalfa.
Figure 1.
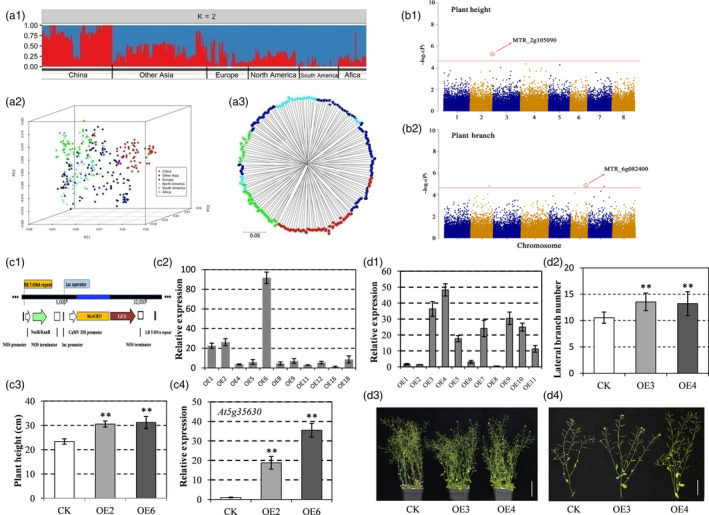
(a) Structure (a1), principal coordinate analysis (a2) and NJ tree (a3) of the alfalfa association panel using the marker data generated by GBS. (b) Manhattan plot showing significant alfalfa locus associated with plant height (b1), and plant branching (b2). Negative log10 (P) values from a genome‐wide scan are plotted against position on each of eight chromosomes. (c) Functional characterization of MsACR11 as an ACR‐domain protein in Arabidopsis thaliana. (c1) Schematic showing the transfer DNA of the vector. (c2) expression analysis of MSACR11 in A. thaliana overexpression lines by qRT‐PCR. Total RNA was extracted from 3‐week‐old A. thaliana transgenic lines. The data are shown as the means ± SD of three biological replicates. (c3) the plant height of WT and transgenic A. thaliana plants. (c4) the transcription level of AtGLN2 (At5g35630) in transgenic lines and WT plants (data are the means ± SD of three biological replicates). (d) Functional characterization of MsPB1 in A. thaliana. (d1) the expression analysis of MsPB1 in A. thaliana transgenic lines by qRT‐PCR. Total RNA was extracted from 3‐week‐old A. thaliana transgenic lines (data are the means ± SD of the biological replicates). (d2) the plant branch number of transgenic A. thaliana and WT plants (means ± SD, n = 6). (d3) phenotypes of plants of control (wild type col‐0) and transgenic lines. (d4) phenotypes of lateral branches in control and transgenic plants. CK, Control (Col‐0); OE, Overexpression. Bars = 5 cm in d3 and d4. ‘**’ in c3, c4 and d2 indicated P < 0.01.
We next examined the function of MsACR11 by constructing the 35S::MsACR11 binary plant transformation vector and generating transgenic A. thaliana lines (Fig. 1c1, c2). Overexpression of MsACR11 in the two highest expressing transgenic A. thaliana lines (OE2 and OE6) resulted in a significant increase in PH (P‐value < 0.01) of approximately 30%, on average, compared to WT (Fig. 1c3). This is the first report of a biological function for the ACT‐domain protein.
Furthermore, the transcription level of GLN2 was measured by qRT‐PCR. The transgenic lines displayed a significant increase, 13‐ to 23‐fold on average, compared with WT plants (Fig. 1c4), indicating a positive regulation of GS2 by MsACR11, which is consistent with the results of a previous study (Osanai et al., 2017). Thus, this research demonstrated that MsACR11 is probably an activator of GS2 and plays a key role in nitrogen assimilation in alfalfa. Overall, these data contribute to our knowledge on the evolution of nitrogen assimilation in higher plants, especially in the forage legume alfalfa.
One MTA, rs33833 (Fig. 1b2) that explained 7.7% of the phenotypic variation was defined as residing in the most significant possible candidate gene related to plant branching (PB), MTR_6 g082400. MTR_6 g082400 encodes an octicosapeptide/phox/Bem1p family protein, these proteins contain PB1 domains that act as universal structural modules that use surfaces of different charges for protein–protein interactions (Korasick et al., 2015). Recent structural studies revealed the presence of a Phox/Bem1p domain in the C‐terminal III/IV interaction sequence motif present in ARF and Aux/IAA proteins, which are master regulators of plant growth and development as well as branch numbers (Han et al., 2014; Liu et al., 2015; Shinohara et al., 2013). Using the sequence of MTR_6 g082400, a 1,870‐bp full‐length cDNA sequence, of a gene which is closely related to an octicosapeptide/phox/Bem1p family protein and contains a Phox/Bem1 (PB1) domain, was obtained from alfalfa leaves, termed MsPB1. The 35S::MsPB1 overexpression binary vector was constructed, transformed into A. thaliana and transgenic lines created (Fig. 1d1). The branch number of selected transgenic lines (OE3 and OE4) and WT plants was determined at the initial flowering stage. The overexpression of MsPB1 resulted in a significant increase in branch number, especially lateral branches, in transgenic lines compared with WT (Fig. 1d2–d4). Furthermore, the transgenic lines showed a disordered lateral branch launching (Fig. 1d4).
It has been reported that PB1 domains adopt a ubiquitin‐like β‐grasp fold that can present two oppositely charged faces on the protein surface, the negative interface of one PB1 domain generally binds to the positive interface of another PB1 domain protein, to mediate the protein–protein interaction (Sumimoto et al., 2007). Accordingly, the PB1 domains are classified into three types. Type I contains an OPCA motif, type II contains an invariant Lys residue on the first β strand, and type I/II contain both motifs. Type I/II PB1 domain proteins will allow higher‐order protein multimerization. A lysine is present in the first β‐strand of the identified PB1 domain protein, which also harbours an OPCA motif. Therefore, we surmise that this protein belongs to the type I/II PB1 protein group (Sumimoto et al., 2007). These findings demonstrate that the MsPB1 gene is probably involved in facilitating interactions between various members of auxin‐related protein families to mediate branch number in plants.
In conclusion, our study demonstrates that integrating GBS and GWAS can be a powerful approach for dissecting agronomic traits in alfalfa, and the results will be valuable for further characterization of candidate genes and to assist in alfalfa breeding.
Conflict of interest
The authors declare no conflict of interest.
Author contribution
ZW conceived and designed the study. HZ, HMZ, LM and GBL collected and analysed the phenotypic data. ZW conducted the GWAS analyses, HZ and JC performed the functional verification. ZW and XMW drafted the manuscript. ZW and CSJ revised the manuscript.
Acknowledgements
This work was supported by National Natural Science Foundation of China (31761143013, 31872410), Earmarked Fund for China Agriculture Research System (CARS34) and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS‐XTCX2018020).
References
- Flint‐Garcia, S.A. , Thornsberry, J.M. and Buckler, E.S. IV . (2003) Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54, 357–374. [DOI] [PubMed] [Google Scholar]
- Han, M. , Park, Y. , Kim, I. , Kim, E.H. , Yu, T.K. , Rhee, S. and Suh, J.Y. (2014) Structural basis for the auxin‐induced transcriptional regulation by Aux/IAA17. Proc. Natl. Acad. Sci. 111, 18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick, D.A. , Chatterjee, S. , Tonelli, M. , Dashti, H. , Lee, S.G. , Westfall, C.S. and Jez, J.M. (2015) Defining a two‐pronged structural model for PB1 (Phox/Bem1p) domain interaction in plant auxin responses. J. Biol. Chem. 290, 12868–12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.H. and Brummer, E.C. (2012) Applied genetics and genomics in Alfalfa breeding. Agronomy 2, 40–61. [Google Scholar]
- Liu, S. , Hu, Q. , Luo, S. , Li, Q. , Yang, X. , Wang, X. and Wang, S. (2015) Expression of wild‐type PtrIAA14. 1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 6, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai, T. , Kuwahara, A. , Otsuki, H. , Saito, K. and Yokota Hirai, M. (2017) ACR6 is an activator of plastid‐type glutamine synthetase GS2 in Arabidopsis thaliana . Plant Cell Physiol. 58, 650–657. [DOI] [PubMed] [Google Scholar]
- Santos, I.G.D. , Cruz, C.D. , Nascimento, M. , Rosado, R.D.S. and Ferreira, R.D.P. (2018) Direct, indirect and simultaneous selection as strategies for alfalfa breeding on forage yield and nutritive value. Pesqui. Agropecu. Trop. 48, 178–189. [Google Scholar]
- Shinohara, N. , Taylor, C. and Leyser, O. (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11, e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto, H. , Kamakura, S. and Ito, T. (2007) Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci. STKE 2007, re6. [DOI] [PubMed] [Google Scholar]
- Sung, T.Y. , Chung, T.Y. , Hsu, C.P. and Hsieh, M.H. (2011) The ACR10 encodes a novel type of chloroplastic ACT domain repeat protein that is coordinately expressed with GLN2 in Arabidopsis. BMC Plant Biol. 11, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]


