Summary
Networks of transcription factors regulate diverse physiological processes in plants to ensure that plants respond to abiotic stresses rapidly and efficiently. In this study, expression of two DREB/CBF genes, TaDREB3 and TaCBF5L, was modulated in transgenic wheat and barley, by using stress‐responsive promoters HDZI‐3 and HDZI‐4. The promoters were derived from the durum wheat genes encoding the γ‐clade TFs of the HD‐Zip class I subfamily. The activities of tested promoters were induced by drought and cold in leaves of both transgenic species. Differences in sensitivity of promoters to drought strength were dependent on drought tolerance levels of cultivars used for generation of transgenic lines. Expression of the DREB/CBF genes under both promoters improved drought and frost tolerance of transgenic barley, and frost tolerance of transgenic wheat seedlings. Expression levels of the putative TaCBF5L downstream genes in leaves of transgenic wheat seedlings were up‐regulated under severe drought, and up‐ or down‐regulated under frost, compared to those of control seedlings. The application of TaCBF5L driven by the HDZI‐4 promoter led to the significant increase of the grain yield of transgenic wheat, compared to that of the control wild‐type plants, when severe drought was applied during flowering; although no yield improvements were observed when plants grew under well‐watered conditions or moderate drought. Our findings suggest that the studied HDZI promoters combined with the DREB/CBF factors could be used in transgenic cereal plants for improvement of abiotic stress tolerance, and the reduction of negative influence of transgenes on plant development and grain yields.
Keywords: Phenotype, HDZI‐4 and HDZI‐3 promoters, transcription factors, transgenic barley and wheat, C‐repeat binding factor 5‐like protein
Introduction
Drought and low temperature are two significant abiotic stress factors limiting the yields of staple crops globally. To survive under harsh environments, plants need to provide rapid responses to these stress factors. The environmental stimuli are perceived by receptors and sensors such as cytoskeleton and hydroxyproline‐rich and arabinogalactan glycoproteins (Humphrey et al., 2007; Luan, 2002; Śniegowska‐Świerk et al., 2015; Thion et al., 1996). These stimuli are converted into intracellular signals by second messengers such as Ca2+ (Cao et al., 2017; Cheong et al., 2003; Klimecka and Muszynska, 2007; Knight et al., 1997; Sanders et al., 2002; Urao et al., 1994) that trigger regulatory networks through abscisic acid (ABA)‐dependent and ABA‐independent pathways, which guide diverse physiological changes in metabolism to provide plant adaptation and/or tolerance to detrimental influences of stresses (Heidarvand and Amiri, 2010; Kidokoro et al., 2017; Shinozaki et al., 2003; Todaka et al., 2017; Yang et al., 2011).
Two groups of genes involved in abiotic stress regulatory networks have been identified (Gong et al., 2015; Hu et al., 2007; Sazegari et al., 2015; Yang et al., 2016). The first group is represented by functional genes, whose expression is initiated or altered by stress‐related transcription factors (TFs), and the final products of these genes are directly involved in biochemical and physiological changes required for stress acclimations (Nakashima et al., 2014; Novillo et al., 2011; Shinozaki et al., 2003). The second group comprises regulatory genes, which include numerous genes encoding TFs that carry out up‐ or down‐regulation of downstream cascades of regulatory and functional genes (Harris et al., 2011; Pujol and Galaud, 2013; Raza et al., 2016; Smith, 2000). Significant changes in transcriptomes in response to environmental stresses and hence use of stress‐related TFs are among effective strategies adopted by plants to deal with unfavourable growth conditions.
The APETALA2 (AP2)/ethylene‐responsive element‐binding (ERF) group is a superfamily of TFs with the majority of members involved in abiotic and/or biotic stress responses. The superfamily of AP2/ERF is classified into five groups, which are represented by the following subfamilies: AP2, ERF, RAV, DREB/CBF, and the subfamily of other TFs (Agarwal et al., 2017; Sakuma et al., 2002). The drought‐responsive element binding (DREB) factors comprise a subfamily of the AP2/ERF family of TFs containing single AP2 DNA‐binding domains, which recognize six nucleotides (A/G)CCGAC of the dehydration‐responsive element/C‐repeat (DRE/CRT). These cis‐elements are located on promoter regions of target stress‐responsive genes and play an important role in regulation of stress‐inducible transcription (Agarwal et al., 2017; Bouaziz et al., 2015; Hrmova and Lopato, 2014; Sakuma et al., 2002).
Numerous TFs belonging to the DREB/CBF subfamily have been reported to enhance the stress durability of transgenic plants by regulating stress‐responsive downstream genes, if overexpressed under the control of strong constitutive promoters (Agarwal et al., 2017; Ban et al., 2011; Chen et al., 2007; Sarkar et al., 2014; Xianjun et al., 2011). However, constitutive overexpression of stress‐related regulatory genes often leads to severe growth retardation and/or a grain yield decrease under normal growth conditions (Agarwal et al., 2017; Kasuga et al., 1999; Lopato and Langridge, 2011; Morran et al., 2011). Several promoters of stress‐inducible functional genes such as rd29A (Kasuga et al., 2004; Mallikarjuna et al., 2011), HVA22 (Lee et al., 2003), ZmRab17 (Morran et al., 2011) and TdCor39 (Kovalchuk et al., 2013), and promoters of stress‐inducible regulatory genes such as LIP19 (Nakashima et al., 2007), OsNAC6 (Nakashima et al., 2007) and OsWRKY71 (Kovalchuk et al., 2013), including the promoter of the rice γ‐clade HD‐Zip I gene Oshox24 (Nakashima et al., 2013), reduce the negative effects of overexpressed TFs on plant growth and/or yield. Therefore, finding and testing novel stress‐inducible promoters for optimization of expression levels of transgenes is one of the critical methodologies to improve plant developmental phenotypes and yields (Agarwal et al., 2017; Hrmova and Lopato, 2014).
It was demonstrated that HD‐Zip I genes from wheat, TaHDZipI‐3 and TaHDZipI‐4, are stress‐responsive and hence their promoters can be potentially used for moderate stress‐inducible transgene expression in transgenic plants. It was shown that the TaHDZipI‐4 gene can be induced by ABA, drought and cold, while the TaHDZipI‐3 gene was induced by drought, but no significant responses of this gene on the elevated levels of ABA or cold were detected (Harris et al., 2016). Therefore, the promoters of TaHDZipI‐3 and TaHDZipI‐4 were expected to have different properties and hence could serve as the candidates of the moderate strength stress‐inducible promoters for molecular breeding.
In this work, the promoters of the wheat γ‐clade of HD‐Zip I genes, TdHDZipI‐3 and TdHDZipI‐4, were isolated from durum wheat (Triticum turgidum ssp. durum) and are designated HDZI‐3 and HDZI‐4, respectively. HDZI‐3 and HDZI‐4 promoters were used to optimize TaCBF5L expression in transgenic wheat, and TaDREB3 in transgenic barley, under two abiotic stresses drought and cold. We demonstrate that in contrast to the findings on the expression levels of TaHDZipI‐3 and TaHDZipI‐4 genes from Triticum aestivum (Harris et al., 2016), both HDZI‐3 and HDZI‐4 promoters from T. turgidum ssp. durum were induced by drought and cold. Furthermore, these two promoters had low levels of expression in unstressed wheat. Based on our study, DREB/CBF transgene expression under HDZI‐3 and HDZI‐4 promoters led to the improvement of drought and/or frost tolerance of transgenic barley and wheat. Aberrant development was observed in some transgenic lines, but it did not correlate with transgene expression levels. The use of the HDZI‐4 promoter in combination with the TaCBF5L gene significantly increased the grain yield of transgenic wheat under severe drought during flowering.
Results
Isolation of the TaCBF5L and TaDREB3 genes and phylogenetic relationships of their products to other DREB/CBF TFs
A 687‐bp long cDNA of TaCBF5L containing a full‐length coding region was isolated from roots of drought‐stressed bread wheat (T. aestivum L. genotype RAC875), using a yeast‐one‐hybrid (Y1H) screen with the drought‐responsive element (DRE) sequence as a bait. TaDREB3 was isolated from the developing grain of the same wheat genotype as a bait sequence (Lopato et al., 2006). The phylogenetic reconstruction of phylogeny of DREB TFs at protein levels was performed with the neighbour joining algorithm in MEGA 6.06 (Tamura et al., 2013). The reconstruction of phylogeny showed clear subdivisions among different groups of DREB TFs from wheat, maize, rice, barley and Arabidopsis (Figure S1), whereby the TaCBF5L and TaDREB3 proteins allocated to the same subclade of subgroup C.
Based on the reconstruction of phylogeny (Figure S1), TaCBF5L shows a closer evolutionary relationship with TaCBF5 (84% sequence identity‐SI), TdDREB3 (77% SI), HvCBF5 (76% SI), TaDREB3 (74% SI), TmCBF5 (74% SI) and ZmDBP4 (64% SI), compared to other entries in the tree (Figure S1, Table S1). The analysis of the multiple sequence alignment of six close homologous proteins with TaCBF5L revealed that both TaCBF5L and TaDREB3 contained an APETALA 2 (AP2) DNA‐binding domain of 35 amino acid residues and the well‐conserved PKKPAGR motifs (PKK/RPAGRxKFxETRHP), positioned at the N‐termini of proteins. Additionally, the LWSY motif was identified to be a conserved motif that was positioned at the C‐termini of proteins (Figure S2).
Expression levels of TaHDZipI‐3 and TaHDZipI‐4 in different wheat tissues during different stages of plant development
The spatial expression patterns of two wheat γ‐clade HD‐Zip I genes, TaHDZipI‐3 and TaHDZipI‐4, were investigated in a variety of wheat tissues (Figure 1a). The highest expression levels of both genes were seen in bract and pistil tissues suggesting that they play roles during floral development. In other tissues under well‐watered conditions, the two TaHDZipI‐3 and TaHDZipI‐4 genes demonstrated a relatively low level of basal expression (Figure 1a).
Figure 1.
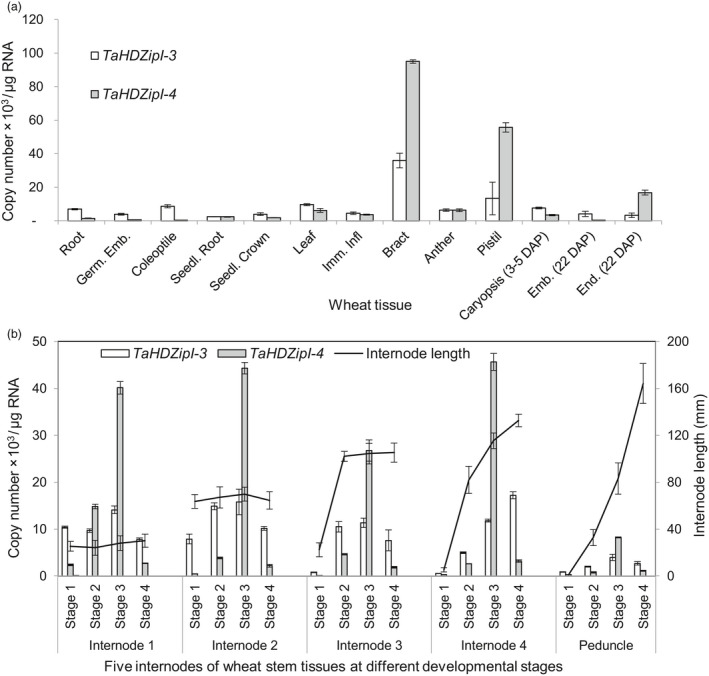
Levels of TaHDZipI‐3 and TaHDZipI‐4 expression in different wheat tissues and at different stages of stem development. (a) Levels of TaHDZipI‐3 and TaHDZipI‐4 expression in different tissues of wheat cv. Chinese Spring. ERF—ethylene‐responsive element‐binding; Emb. (22DAP)—Embryo 22 days after pollination; End. (22 DAP)—Endosperm 22 days after pollination; Germ. Emb.—Embryo in germinating seed; Imm.Infl—Immature inflorescence; Int—Internode; Seedl. crown—Seedling crown; Seedl. Root—Seedling root. (b) Spatial expression patterns of TaHDZipI‐3 and TaHDZipI‐4 expression in four stem internodes (Int) below peduncle (Ped) at four different stages of wheat (T. aestivum cv. RAC875) development. Internode length parameters are plotted against the secondary vertical axis. Stem stages are as follows: Stage 1 (100 mm); Stage 2 (300 mm) awns emerging; Stage 3 (400 mm) head emerging; Stage 4 (500 mm) at anthesis; peduncle emerged. Error bars represent the standard deviation of three biological replicates (a) and three technical replicates (a).
It is considered that the γ‐clade HD‐Zip I TFs contribute to growth modulation under water deficit and that in Arabidopsis this translates to reduced stem elongation (Harris et al., 2011). As DREB/CBF TFs also contribute to growth modulation under water deficit, expression of the two TaHDZipI‐3 and TaHDZipI‐4 γ‐clade genes was characterized in the internodes of wheat stem at different stages of development to explain any differences in the stem length that may have been observed in transgenic plants, where the expression level of DREB/CBF TFs was controlled by HDZI promoters. Expression was investigated in stem internodes over four different developmental stages: Stage 1 (100 mm); Stage 2 (300 mm)—awns emerging; Stage 3 (400 mm)—head emerging; Stage 4 (500 mm)—anthesis and peduncle emergence. Internodes 1–4 and the peduncle were at different stages of elongation/maturation at each stem stage development, enabling us to establish correlations between the expression levels of the two γ‐clade HD‐Zip I genes, and internode elongation and maturation. The analyses of stem developmental series revealed that the two wheat γ‐clade HD‐Zip I TFs were expressed differentially, both spatially and temporally, during normal stem development (Figure 1b).
The level of TaHDZipI‐3 expression was associated with the maturity of any given internode. At stage 1, internodes 1 and 2 have reached their final length and expressed TaHDZipI‐3 to relatively high levels, compared to internodes 3, 4 and the peduncle, which just started to elongate. Likewise, internode 3 reached its final length by awns emerging (stage 2) and TaHDZipI‐3 expression remained steady through stage 3 and stage 4. However, the four‐time harvest points were not sufficient to determine the final length of internode 4 and the peduncle, however, internode 4 showed increases in TaHDZipI‐3 expression as length of stem increased, whereas expression in the peduncle remained at lower levels (Figure 1b).
Expression of TaHDZipI‐4 showed a steady pattern in all five internodes, although there was an increase of gene expression from stage 1 to reach maximal expression at stage 3, followed by a dramatic decrease at stage 4 (Figure 1b).
Expression of TaCBF5L and TaDREB3 driven by HDZI‐3 or HDZI‐4 promoters in transgenic wheat and barley sublines, under rapid dehydration and under various drought conditions
Expression levels of TaCBF5L were detected in leaves of T2 lines of transgenic wheat under well‐watered and after 6‐h dehydration treatments; these data were compared with those of control wild‐type (WT) plants, using northern blot hybridization (Figure 2a). Expression levels of TaDREB3 were assessed in leaves of transgenic T1 barley lines grown under hydroponic conditions, before application of stress conditions and after 7‐h incubation of seedlings without growth media components, and compared with those of control WT plants, using northern blot hybridization (Figure 2b). The results of these comparisons showed that expression of transgenes TaCBF5L and TaDREB3, controlled by either of the tested HDZI‐3 or HDZI‐4 promoters, was much stronger under dehydration than the expression levels under well‐watered conditions in both wheat and barley (Figure 2). In contrast, both the TaCBF5L endogenous gene of WT wheat plants and HvDREB3 endogenous gene of WT barley plants showed either very weak or undetectable hybridization signal under applied experimental conditions independently of whether RNA was isolated from leaves collected before or after dehydration.
Figure 2.
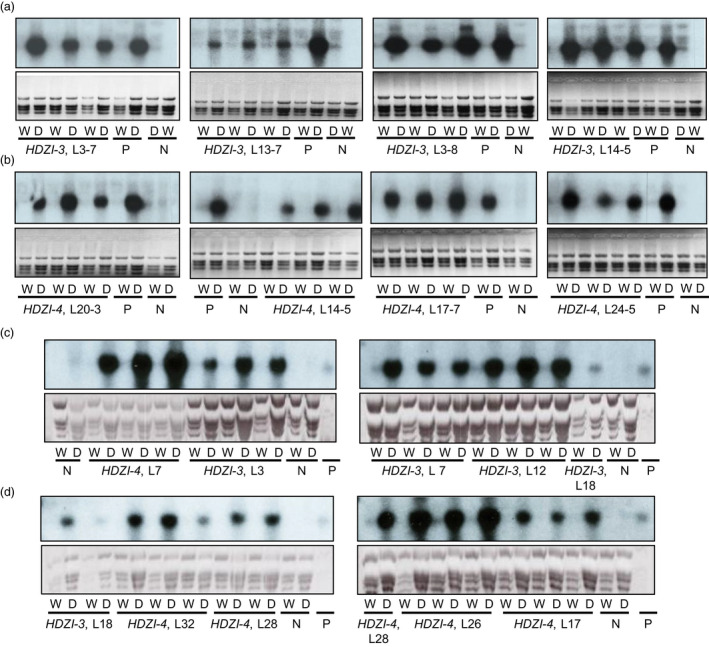
Induction of wheat HDZI‐3 and HDZI‐4 promoters in leaves of 3‐week‐old control and transgenic T2 wheat seedlings (a) and in control and transgenic T1 barley seedlings of the same age (b) before (W) and after 6 h of dehydration (d). N: WT plants with the endogenous TaCBF5L or HvDREB3 genes either cannot be seen or seen as a weak band under both well‐watered and dehydration conditions, and therefore were used as negative control; P: transgenic wheat plants with TaCBF5L transgene showing a strong band under dehydration conditions (a and b), and/or a 1000‐fold diluted purified DNA fragment of the TaDREB3 coding region (c and d) were used as positive controls; W: well‐watered; D: drought.
The expression levels of TaCBF5L in T4 transgenic sublines under four different drought stages were determined using the Q‐PCR method. Evaluation of the data showed that the expression levels of TaCBF5L transgene in the drought‐tolerant wheat cultivar, controlled by the HDZI‐3 or HDZI‐4 promoter, showed no or a little increase during the leaf wilting point (−1.5 to −2 MPa) or moderate drought (−2 to −3 MPa), compared to those with the basal level of expression under well‐watered (−1.2 to −1.5 MPa) conditions (Figure 3a,b). However, the TaCBF5L expression levels were obviously up‐regulated by a severe drought stress stage >−4 MPa). In contrast, in the drought‐sensitive barley cultivar, the expression levels of TaDREB3, controlled by HDZI‐3 or HDZI‐4 promoter, increased several folds already at wilting point (−0.7 to −1.2 MPa), and in most of the tested lines expression decreased during a more severe drought stress stage >−3 MPa; Figure 3c,d).
Figure 3.
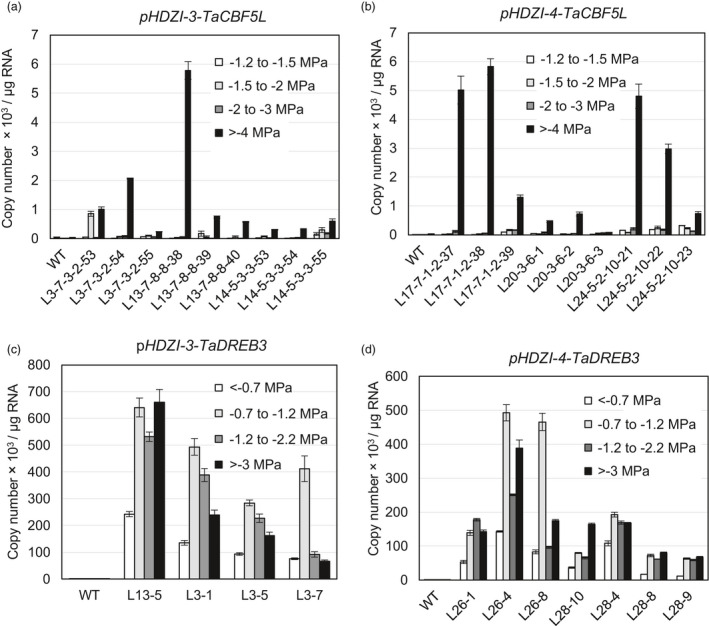
TaCBF5L or TaDREB3 transgene expression in wheat (a and b) or barley (c and d) plants controlled by the promoter HDZI‐3 (a and c) and the promoter HDZI‐4 (b and d) under various drought stages: well‐watered condition (leaf water potential with −1.2 to −1.5 MPa for wheat or 0 to −0.7 MPa for barley), the leaf wilting point (leaf water potential with −1.5 to −2 MPa for wheat or −0.7 to −1.2 MPa for barley), moderate drought (leaf water potential with −2 to −3 MPa for wheat or −1.2 to −1.5 MPa for barley) and drought condition (leaf water potential >−4 MPa for wheat or >−3 MPa for barley). The error bars represent ±SD of three technical replicates.
Comparison of growth and yield characteristics of T1 transgenic and control WT barley plants grown under well‐watered conditions
Comparisons of growth and yield characteristics of selected T1 transgenic (Figure S3) and control WT barley grown under well‐watered conditions revealed that the most transgenic lines at the beginning of their reproductive stages appeared to be similar as the control WT and null‐segregant plants (Figures S4 and S5). The most of transgenic lines showed similar height, number of tillers, flowering time and yield as control plants, although size and yield of a few lines had significantly decreased compared to WT plants (Figure S5). According to the northern blot hybridization data both types of lines expressed transgene, although the levels of transgene expression were not precisely quantified. Null segregants identified by PCR were removed from the experiment.
Evaluation of phenotypes of T3 transgenic wheat lines grown under well‐watered conditions and under moderate drought applied during the flowering stage
Four sublines of T3 transgenic and WT wheat plants were planted in two deep containers and subjected to constant well‐watered conditions in the first container, and to moderate drought during the flowering stage in the second container. Plant growth characteristics and yield components of these plants were evaluated and compared at the end of their reproduction stages.
Transgenic sublines L13‐7‐8 and L14‐5‐3 transformed with the pHDZI‐3‐TaCBF5L construct showed similar phenotypic features such as tiller, spike, seed number, single grain weight, plant height, grain weight per plant and total dry biomass, compared to those of WT plants (Figure 4a). However, two other transgenic sublines L3‐7‐3 and L3‐8‐8, derived from the same L3 line, showed significantly smaller sizes of plants, fewer seeds, less biomass and grain yield than those of WT under moderate drought (Figure 4a). In addition, all sublines were subjected to well‐watered conditions, and the subline L14‐5‐3 that was exposed to moderate drought conditions, flowered between 2 and 3 days earlier than WT plants.
Figure 4.
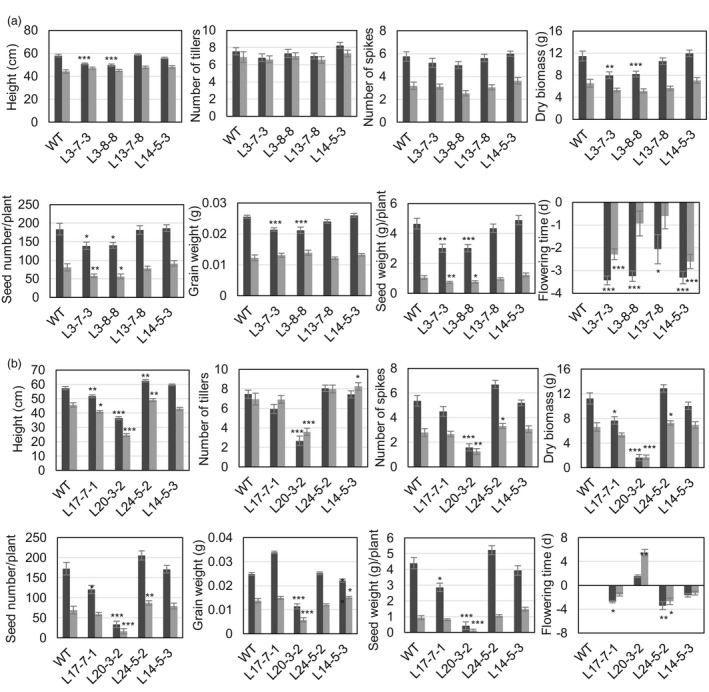
Growth characteristics and yield components of control wild‐type (WT) and transgenic wheat (Triticum aestivum cv. Gladius) transformed with pHDZI‐3‐TaCBF5L (a) and pHDZI‐4‐TaCBF5L (b) under well‐watered (black boxes) and moderate drought (grey boxes) conditions. Flowering time of transgenic plants was compared with the average flowering time of 16 control WT plants, which is represented as day 0. Values represent means ± SE (n varies for each column and is shown in each case directly on the graphs) at ‘*’ P < 0.05, ‘**’ for P < 0.01 and ‘***’ for P < 0.001, which were calculated by the Student's t‐test (unpaired, two‐tailed).
From the four transgenic sublines transformed with the pHDZI‐4‐TaCBF5L construct, two sublines grown under well‐watered conditions, and one subline exposed to mild drought, showed lower spike numbers and grain yields than WT plants. Three sublines of transgenic plants transformed with the pHDZI‐4‐TaCBF5L construct flowered 2–3 days earlier than WT plants, although the L20‐3‐2 subline was significantly delayed in growth and flowered 5 days later than WT plants (Figure 4b). In addition, L20‐3‐2 subline had a lower plant height, lower tiller and seed numbers, and produced less biomass compared to the WT plants. However, the rest of sublines had similar number of tillers than the WT plants (Figure 4b).
Evaluation of phenotypes of T4 transgenic wheat lines grown under severe drought during the flowering stage
Two independent lines of transgenic wheat transformed with the pHDZI‐3‐TaCBF5L or pHDZI‐4‐TaCBF5L constructs were grown alongside WT plants in pots with water‐saturated soil for 3–4 weeks, then the plant watering was withheld, and the phenotypic evaluation was performed at the end of the reproductive stage. Over 95% of the transgenic and WT wheat plants survived seedling stages and proceeded to reproductive stages. The soil water content curves indicated that the plants were exposed to severe drought (25%–35% of soil water content) during flowering time (Figures S6 and S7).
Transgenic sublines L14‐5‐3‐3 and L13‐7‐8‐11 with transgene driven by the HDZI‐3 promoter had similar numbers of spikes and tillers compared to those of WT plants (Figure 5a). However, both sublines showed slightly delayed flowering, in addition, the L13‐7‐8‐11 subline plants had smaller size, fewer seeds, less biomass and lower grain yield than the control WT plants (Figure 5a). Consistently, transgenic L14‐5‐3‐1 and L24‐5‐2‐1 sublines with a transgene driven by HDZI‐4 promoter showed similar spike and tiller numbers as the control WT plants (Figure 5b). However, both types of transgenic lines had significantly larger size, higher biomass and seed numbers, and, therefore, higher grain yields compared to the control WT plants (Figure 5b). In addition, both transgenic sublines with TaCBF5L driven by the HDZI‐4 promoter flowered 3–4 days earlier than the control WT plants (Figure 5b).
Figure 5.
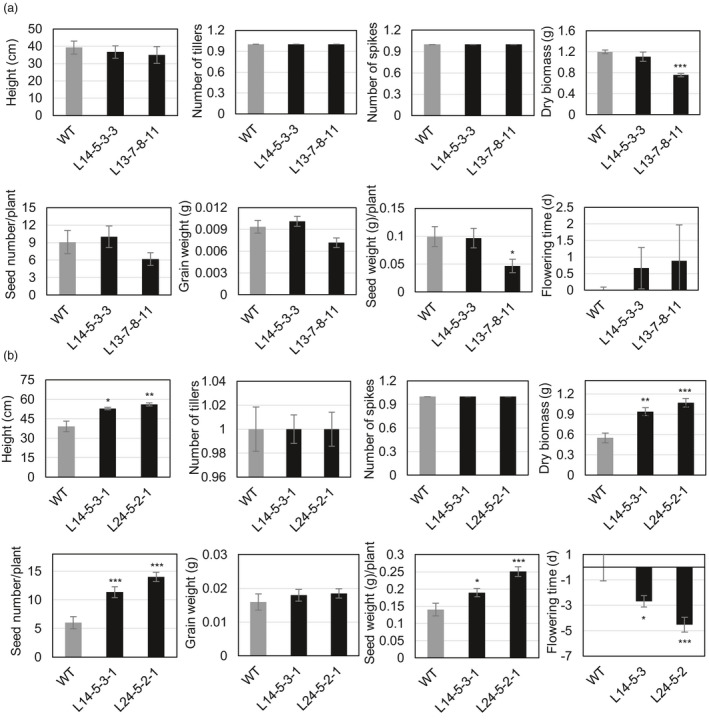
Growth characteristics and yield components of control wild‐type (WT) and transgenic wheat (T. aestivum cv. Gladius) transformed with pHDZI‐3‐TaCBF5L (a), and pHDZI‐4‐TaCBF5L (b) under severe drought. Flowering time of transgenic plants was compared to average flowering times of 16 control WT plants, which is represented as day 0. Values represent means ± SE (n varies for each column and is shown in each case directly on the graphs) at *P < 0.05, ** for P < 0.01 and *** for P < 0.001, which were calculated by Student's t‐test (unpaired, two‐tailed).
Stress‐inducible expression of TaDREB3 gene driven by the HDZI‐3 or HDZI‐4 promoters improves drought tolerance of transgenic barley seedlings
The comparison of drought tolerance of transgenic wheat and barley was performed at the vegetative stage of plant development. It was measured as a recovery rate of seedlings subjected to stringent (lethal effect for the most control plants) drought conditions. Control and transgenic plants of the similar size were selected for the experiment. Three consecutive experiments using wheat seedlings revealed no significant improvement of transgenic seedlings’ survival rates compared to the control WT seedlings for both promoter‐transgene constructs (data not shown). In contrast, improvement of drought tolerance of transgenic barley seedlings was obvious in every experiment, where the transgene driven by either HDZI‐3 or HDZI‐4 promoter in barley yielded positive results (Figure S8).
Expression levels of putative downstream genes of TaCBF5L in leaves in control WT and transgenic wheat plants under well‐watered conditions and under drought
The levels of expression of the TaCBF5L transgene and those of stress‐inducible LEA⁄COR⁄DHN genes, TaRab17, TaCor410, TaCor18, TaRab15 and Wlt10, in control WT and transgenic wheat plants were very different under well‐watered condition (leaf water potential −1.2 to −1.5 MPa) and severe drought (leaf water potential >−4 MPa; Figure 6). In all cases except Wlt10, the levels of transgene expression under severe drought increased compared to those under well‐watered conditions with the HDZI‐3 promoter application. In most cases, the increase of expression levels of most tested genes was higher in transgenic than in the control WT lines. As an exception, TaRab15 gene expression in most transgenic lines was lower than that of the control WT plants.
Figure 6.
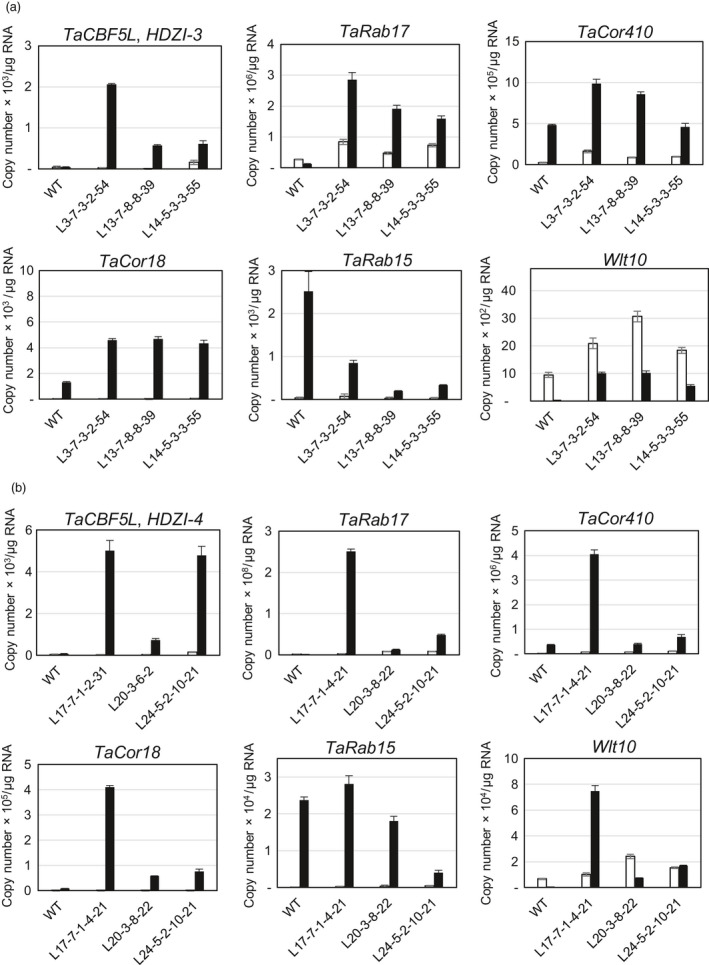
Expression of the TaCBF5L transgene and stress‐inducible LEA⁄COR⁄DHN genes in control WT and transgenic wheat plants with inducible overexpression of TaCBF5L controlled by HDZI‐3 (a) and HDZI‐4 (b) promoters. Expression levels of the TaCBF5L transgene and selected stress‐inducible genes were estimated under well‐watered conditions (white boxes) and severe drought (leaf water potential >−4 MPa; black boxes).
Stress‐inducible expression of DREB/CBF genes driven by HDZI‐3 or HDZI‐4 promoter improves frost tolerance of transgenic wheat and barley seedlings
Wheat seedling frost tolerance of three T4 lines transformed with the pHDZI‐3‐TaCBF5L construct was compared with that of control WT plants. Based on the evaluation of survival rates (Figure 7a), control WT plants did not grow well and only no more than 6% of them survived the harsh conditions of frost. However, all examined transgenic lines showed strong tolerance to frost, with a survival rate that was three‐ to fourfold higher than that of the control WT plants (Figure 7a). Moreover, survival rates of each two transgenic L3‐8‐8‐11 and L14‐5‐3‐3 lines were significantly higher than those of the control WT plants (Figure 7a).
Figure 7.
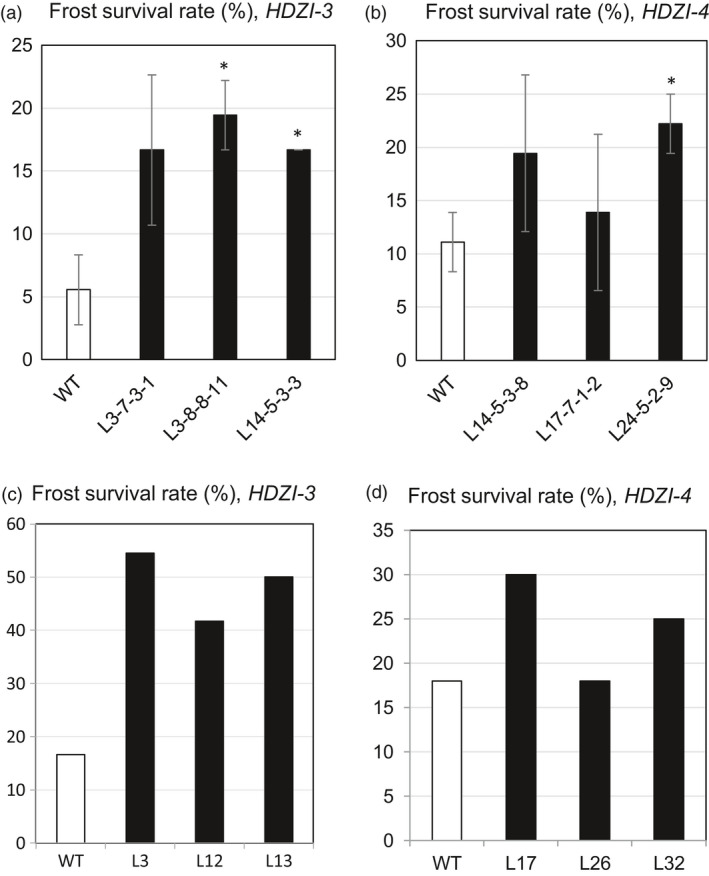
Frost survival rates of control WT and transgenic T3 wheat plants transformed with pHDZI‐3‐TaCBF5L (a) and pHDZI‐4‐TaCBF5L (b) constructs. Error bars represent ±SD of three technical replicates. Differences between transgenic and WT plants were tested in the unpaired Student's t‐test (*P < 0.05). Frost survival rate of control WT and transgenic T1 barley seedlings transformed with pHDZI‐3‐TaDREB3 (c) and pHDZI‐4‐TaDREB3 (d); data in panels (c) and (d) are based on a single experiment, thus no ±SD values are included.
Three T4 lines transformed with the pHDZI‐4‐TaCBF5L construct after frost treatment showed a tendency to recover stronger than the control WT plants. Survival rates of transgenic wheat plants were 1.2‐ to 2.0‐fold higher than that of the control WT plants, suggesting that the TaCBF5L under HDZI‐4 promoter provides a bit lower enhancement of the wheat frost tolerance than the HDZI‐3 promoter (Figure 7b). Frost tolerance data obtained in similar experiments for T1 barley seedlings revealed a similar picture (Figure 7c,d). Frost tolerance improvement was delivered by both pHDZI‐3‐TaDREB3 and pHDZI‐4‐TaDREB3 constructs; however, in the case of the HDZI‐3 promoter, the frost tolerance enhancement was clearly stronger than that of the HDZI‐4 promoter (Figure 7c,d).
The expression levels of the TaCBF5L transgene in most wheat sublines with HDZI‐3 or HDZI‐4 promoters increased up to several folds after cold treatment. In some plants, however, no significant activation of the promoter was observed, although the basal levels of the promoter activity were high (Figure 8a,b). In contrast, when examining barley transgenic plants, the picture was slightly different. Firstly, the basal activities of both HDZI‐3 and HDZI‐4 promoters (relatively to stress‐induced activities) were overall stronger in transgenic barley lines than those in transgenic wheat plants, and, secondly, the activation of the HDZI‐4 promoter under the low temperature of 4 °C was in general stronger than the activation of the HDZI‐3 promoter (Figure 8c,d).
Figure 8.
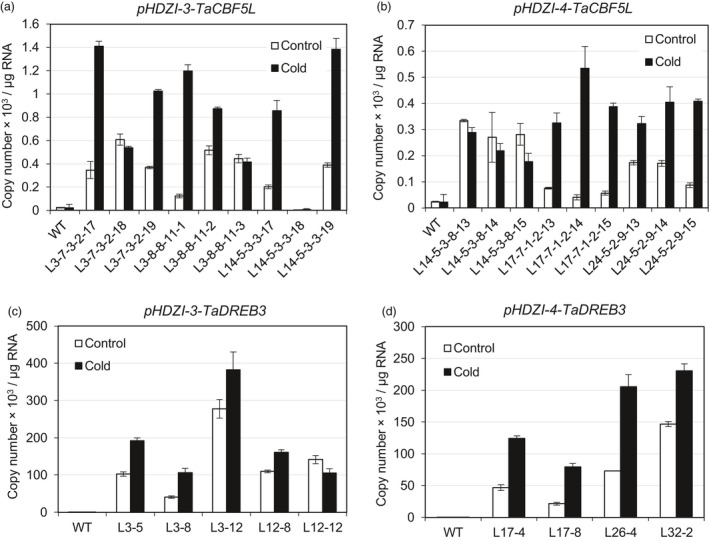
TaCBF5L (a and b) or TaDREB3 (c and d) transgene expression levels controlled by the HDZI‐3 (a and c) and HDZI‐4 (b and d) promoters in leaves of control WT and transgenic T4 wheat (a and b) or T1 barley plants (c and d) grown at 24 °C (control) and subjected to cold treatment at 4 °C. The error bars represent ±SD of three technical replicates.
Activation of stress‐inducible genes by overexpression of TaCBF5L under low temperature
Expression of five LEA⁄COR⁄DHN genes, TaRab17, TaCor410, TaCor18, TaRab15 and Wlt10, as the putative downstream stress‐inducible genes of TaCBF5L, was examined in the leaves of transgenic wheat and control WT plants in the absence of stress and after the exposure to 4 °C (Figure 9). Nearly all tested genes in all transgenic lines transformed with either pHDZI‐3‐TaCBF5L or pHDZI‐4‐TaCBF5L constructs demonstrated stronger expression than the control WT plants under normal growth temperatures. Some of the tested downstream genes in the transgenic lines were up‐regulated, while the others were clearly repressed, and other genes kept their expression levels unchanged, when compared to the expression levels of the same genes in the WT plants. Overall the expression patterns of downstream genes in HDZI‐3 and HDZI‐4 wheat transgenic lines were comparable. However, in HDZI‐3 transgenic lines these patterns were more consistent than those in HDZI‐4 transgenic lines.
Figure 9.
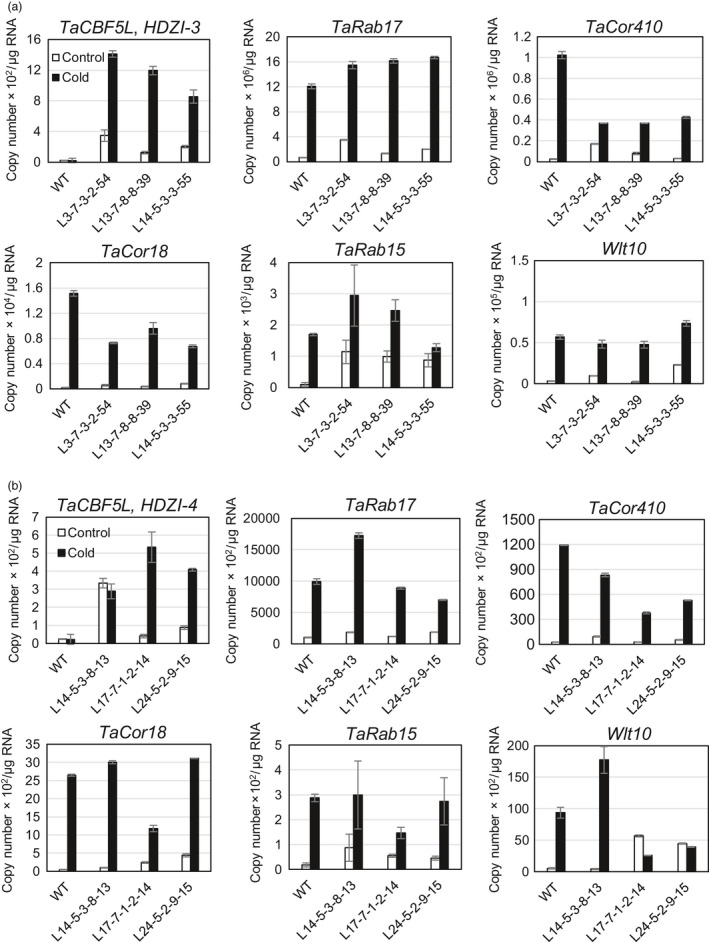
Expression of the TaCBF5L transgene and stress‐inducible LEA⁄COR⁄DHN genes in transgenic wheat plants with overexpression of TaCBF5L controlled by HDZI‐3 (a) and HDZI‐4 (b) promoters in control WT and transgenic T4 lines at 23 °C (Control) and under the cold treatment at 4 °C (Cold).
GUS expression pattern under different stresses in the T1 transgenic wheat transformed with the pHDZI‐3‐GUS or HDZI‐4‐GUS construct
To analyse the spatial and temporal activity of the pHDZI‐3 and pHDZI‐4 promoters, wheat was transformed with pHDZI‐3‐GUS and pHDZI‐4‐GUS fusion constructs. Twenty‐two independent T1 transgenic wheat lines (six lines with the pHDZI‐3‐GUS reporter construct and sixteen lines with the pHDZI‐4‐GUS reporter construct; Tables S2 and S3) were generated and analysed in the pilot experiment using hydroponic conditions. Plants from each subline were treated with cold, high salinity, increased ABA levels and dehydration, respectively (Figure S9). All analysed transgenic wheat plants transformed with either pHDZI‐3‐GUS or pHDZI‐4‐GUS constructs showed no GUS expression under salinity and ABA. Three transgenic T1 pHDZI‐3‐GUS lines (Lines 3, 5 and 7) showed GUS expression in the coleoptiles and roots of seedlings under cold stress, and one line (Line 3) showed a weak HDZI‐3 promoter activity in the roots under dehydration (Figure S9a,b). Weak staining of coleoptiles was observed in eight T1 pHDZI‐4‐GUS transgenic lines (Lines 1a, 2, 4, 6, 8, 10, 12 and 13) after cold treatment (Figure S9a,c). However, the detected GUS activity was too weak to proceed with histochemical analysis of the spatial pattern of the promoter activity. No GUS activity was detected in other plant tissues.
Discussion
Drought and frost may impair plant growth and development at any time point of a plant life cycle. However, the sensitivity to drought and frost is especially acute during reproductive stages. In the case of drought this is because of the plant–water status changes, leading to a high transpiration rate and the declining reserves of soil moisture towards the end of a vegetation season (Saini and Westgate, 1999). The exact reasons of high sensitivity of wheat and barley to night frosts at flowering are unknown. It is noteworthy that a particularly strong sensitivity of one or both gametophytes to below‐zero temperatures occurs a short time before, during and/or short time after fertilization.
In this work, we used two representatives of the wheat DREB/CBF family of TFs to investigate stress‐inducible expression in transgenic wheat and barley, and to study the impact of transgenes driven by two distinct stress‐inducible durum wheat promoters on growth characteristics, yield components and tolerance of transgenic plants to drought and frost at reproductive and/or vegetative stages of plant development. Additional details regarding the selection of donor plants and transgenes can be found in Supporting Discussion.
Q‐PCR analyses of TaHDZipI‐3 and TaHDZipI‐4 expression in a variety of plant tissues in the absence of stress revealed relatively low levels of expression of both genes in all examined tissues except for the floral tissues, suggesting that the TaHDZipI‐3 and TaHDZipI‐4 promoters could elevate expression of target genes in frost vulnerable florets and initiate accumulation of protective proteins before stress (Figure 1a). The analysis of TaHDZipI‐3 and TaHDZipI‐4 expression in expanding parts of the stem at different stages of development revealed that expression of the TaHDZipI‐3 gene was low, while the expression levels of TaHDZipI‐4 were more variable and relatively high during head emergence. Although the DREB/CBF proteins may suppress the growth of transgenic plants (Kasuga et al., 1999; Kovalchuk et al., 2013; Morran et al., 2011), relatively low basal levels of both HDZI promoters (particularly low in wheat) applied through expression of the DREB/CBF transgenes may not significantly affect stem elongation under the optimal growth conditions, except during transitioning to flowering. Based on TaHDZipI‐3 and TaHDZipI‐4 expression data under stress (Harris et al., 2016), we expected that the promoters would exert different properties: the HDZI‐3 would be induced only by drought/dehydration, while HDZI‐4 would be induced by both drought/dehydration and low temperatures. However, our study shows that both promoters in wheat and barley were induced by drought and cold, albeit with different strength. The reason for this is unclear, although we suggest that the minor differences in promoter sequences of genes from bread and durum wheat, or the absence of the distal repressor sequences from the HDZI‐3 promoter fragment controlling cold response, could play roles (Figures S10–S13). In addition to different strength of promoters, we identified promoter‐dependent differences in phenotypes, stress tolerance and downstream gene expression in both transgenic wheat and barley plants. Further discussion of the promoter activity studies using transgenic plants transformed with promoter‐GUS fusion constructs can be found in Supporting Discussion.
Based on the statement above and the results of our previous works (Kovalchuk et al., 2013; Morran et al., 2011; Shavrukov et al., 2016), we conclude that: (i) accurate selection of lines; (ii) use of untransformed donor plants and two or three backcrosses of selected homozygous transgenic lines with acceptable phenotypes and (iii) accurate selection of backcrossed plants for transgene presence and/or expression could provide stable lines with low detrimental effects on genomic DNA (that occur during the process of plant transformation) and enhance stress tolerance and decrease or abolish the negative influence of the transgene on plant development.
The analysis of plant phenotypes and yield was performed under well‐watered and drought conditions. In addition, the molecular analysis of regulation of several stress‐responsive genes, which are potential downstream genes of DREB/CBF TFs showed that this analysis (Figures 7 and 8) supported the observed enhancement of stress tolerance.
Drought tolerance improvement was not observed in transgenic wheat seedlings in three consecutive experiments. This result was not unexpected if one considered the relatively high tolerance of the wheat Gladius cultivar to drought, achieved through breeding programmes for the Australian environment. Our previous attempts to enhance drought tolerance in the drought‐tolerant Gladius cultivar by overexpression of the DREB/CBF and bZIP encoding genes resulted in minor or no improvements of tolerance (Amalraj et al., 2016; Luang et al., 2018). However, the significant improvement was achieved by using HD‐Zip I and ERF‐like (SHN1) TFs (Bi et al., 2018; Yang et al., 2018), which most likely regulate different aspects of drought response. These improvements suggested the reasons for a high tolerance of the Gladius cultivar to drought, which was introgressed during breeding of this cultivar. In contrast, the drought tolerance of all the tested transgenic barley lines was higher than that of the control WT plants. Thus, the sensitivity of the barley Golden Promise cultivar to drought provided us with the opportunity to improve its drought tolerance through DREB/CBF overexpression.
The analysis of transgenic wheat and barley growth characteristics, and yield components under well‐watered conditions and mild drought (wheat only) revealed some lines had the same or very similar phenotypes as the control WT plants and some lines had worsened. However, both types of lines expressed transgenes and demonstrated their function of the significant improvements of frost tolerance. It is notable that all except one of the tested transgenic lines flowered a few days earlier than the control WT plants; this is very unusual for overexpression DREB/CBF TFs, which typically lead to significant delays in flowering due to a slower growth of transgenic plants.
Taking into consideration that both tested promoters in transgenic wheat were activated only under strong drought, we performed the ‘drought‐during‐flowering’ experiment under harsh drought conditions (Figures S6 and S7). In this case, the behaviour of transgenic wheat lines was dependent on the HDZI‐3 and HDZI‐4 promoters used in each transgenic plant. The pHDZI‐3‐TaCBF5L lines showed a decline in most yield components. In contrast, both transgenic wheat lines transformed with the pHDZI‐4‐TaCBF5L construct significantly increased plant biomass and seed number per spike, resulting in the significant increase of the grain weight per plant (yield). Transition to flowering in these two lines occurred 3–5 days earlier than that in the control WT plants. We have no explanation for differences in yield and their dependence upon the promoter used except a possibility that the differences in the drought‐induced spatial expression pattern could be attributed to each promoter regulating TaCBF5L overexpression.
The analysis of potential downstream stress‐responsive genes directly or indirectly regulated by TaCBF5L in transgenic wheat plants revealed that all five tested genes were up‐regulated in transgenic wheat lines compared to the control WT plants under well‐watered conditions (obviously because of the basal levels of transgene expression), while the behaviour of the tested genes under strong drought was different (Figure 6). The results obtained for the HDZI‐3 promoter were more consistent than those for the HDZI‐4 promoter, which likely point out to differences in spatial patterns of promoter activities.
Vegetative frost tolerance enhancement was observed in both transgenic species and through the application of both HDZI promoters. However, while with the HDZI‐3 promoter a significant improvement of frost tolerance was achieved in both transgenic wheat and barley compared to control WT plants, the HDZI‐4 promoter performance in both transgenic plants was less convincing. The possible explanation could be the higher overall basal activity level of the HDZI‐3 promoter. This could provide slightly higher transgene expression, and hence higher basal levels of the target stress‐responsive genes and their products prior to stress, which could lead to better pre‐adaptation of HDZI‐3 transgenic plants to cold. The other explanation could be in the differences of spatial expression of transgenes under two tested promoters. These differences could lead to the diverse levels of transgene product accumulation in the most vulnerable to stress plant tissues that in turn could provide various levels of the transgene‐produced advantages under stress.
The analysis of expression of downstream genes perhaps confirms the role of overall higher basal levels of transgene expression in transgenic lines, when the HDZI‐3 promoter was applied, and consequently a better preparation of plants to a cold stress during growth under the optimal for plant temperatures. Notably, similar downstream genes may be regulated by the same TaCBF5L transgene under drought and cold conditions in different ways. For instance, the stress‐inducible TaCor410 gene was up‐regulated under drought and down‐regulated under low temperature independently of whether either HDZI‐3 or HDZI‐4 promoters were used. On the other hand, TaCor18 gene expression was up‐regulated by drought but down‐regulated by cold, only when the HDZI‐3 promoter was used. The Wlt10 expression level was up‐regulated by drought and but it was not affected by cold by the HDZI‐3 promoter application. Notably, in the absence of stress, all tested downstream genes were up‐regulated by the basal levels of the TaCBF5L transgene.
By summarizing our data, we conclude that the application of each of two tested HDZI‐3 and HDZI‐4 promoters has its own advantages and disadvantages. Transgenic lines with developmental phenotypes similar to those of the WT donor plants can be selected for both promoters. In barley, both promoters were effective tools to increase drought tolerance at a vegetative stage by overexpression of the DREB/CBF TFs, and therefore could be used for drought tolerance enhancements of the drought‐sensitive crop species. However, both promoters in combination with TaCBF5L failed to improve the survival rates of the drought‐tolerant wheat under water deficit. The HDZI‐3 promoter provided better frost tolerance than the HDZI‐4 promoter in both wheat and barley, most likely due to higher basal activity levels, which lead to a better provision for upcoming stress. On the contrary, the application of HDZI‐4 promoter delivered yield improvements in wheat, providing flowering occurred under strong drought, while the application of the HDZI‐3 promoter provided no gains in a grain yield under the same conditions.
In conclusion, we suggest that both tested wheat HDZI‐3 and HDZI‐4 promoters could be used in transgenic crop plants in combination with DREB/CBF TFs for the improvement of the abiotic stress tolerance, and for the concurrent retention of original phenotypes and yields.
Experimental procedures
Isolation and identification of the TaCBF5L and TaDREB3 genes
A full‐length cDNA of TaCBF5L was isolated from roots of the drought‐stressed T. aestivum L. genotype RAC875, using a modified yeast‐one hybrid approach (Lopato et al., 2006; Pyvovarenko and Lopato, 2011) with DRE cis‐element TACCGAC as a bait. Isolation of TaDREB3 cDNA and characterization of the gene in transgenic wheat and barley was described earlier (Kovalchuk et al., 2013; Lopato et al., 2006; Morran et al., 2011). The homologous to TaCBF5L and TaDREB3 proteins from a variety of species such as Arabidopsis, wheat, rice, maize and barley were found using the Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990), and a non‐redundant protein sequence database of the National Center for Biotechnology Information (NCBI). The multiple protein sequence alignment of the homologous proteins to TaCBF5L and TaDREB5 was conducted using MAFFT version 7 (Katoh and Standley, 2013). A phylogenetic tree was reconstructed based on the alignment results using Molecular Evolutionary Genetics Analysis (MEGA 6.06; Tamura et al., 2013) with neighbour joining and p‐distance specifications and 1000 bootstrap replications.
Supporting Experimental procedures contain protocols for plasmid construction and plant transformation, determination of transgene copy number and expression levels by quantitative real‐time PCR (Q‐PCR) and northern blot hybridization, selection of transgenic wheat and barley sublines, comparison of growth and yield components of selected sublines with control WT wheat plants grown under different drought conditions and well‐watered conditions, survival rates comparison of wheat and barley seedlings under terminal drought and frost and analysis of promoter activation in transgenic wheat seedlings by checking GUS expression.
GenBank accession numbers
TaCBF5L—MF406152, TdHDZipI‐3 promoter (HDZI‐3)—MG063277, TdHDZipI‐4 promoter (HDZI‐4)—MG063278.
Conflict of interest
Authors declare no conflict of interest.
Author contributions
Y.Y. performed experiments and analysed data. H.H.J.B., J.H., M.R., Y.L., I.M., N.B., L.C. and S.S.H. assisted with experiments and analysed data. N.K., S.L. and S.H. conceived the project, designed experiments and analysed data. Y.Y., S.H., N.K. and S.L. wrote the manuscript. H.H.J.B. and M.H. contributed to writing. M.H. and S.L. edited manuscript. All authors commented on the manuscript.
Supporting information
Figure S1 The phylogenetic tree of DREB TFs from a representative dicot Arabidopsis and monocots wheat and barley.
Figure S2 Multiple sequence alignment of TaCBF5L and six homologous proteins with a close evolutionary relationship to TaCBF5L from wheat (Ta—T. aestivum; Tm—T. monoccocum), maize (Zm—Zea mais), and barley (Hv—Hordeum vulgare).
Figure S3 Copy numbers of the TaCBF5L transgene estimated by Q‐PCR in T1 transgenic wheat (a) and barley (b).
Figure S4 Control WT barley (H. vulgare cv. Golden promise) and transgenic T1 barley lines transformed with the pHDZI‐3‐TaDREB3 and pHDZI‐4‐TaDREB3 constructs.
Figure S5 Growth characteristics and yield components of control WT and transgenic barley transformed with pHDZI‐3‐TaDREB3 (a), and pHDZI‐4‐TaDREB3 (b) constructs grown under well‐watered conditions.
Figure S6 Details of the flowering‐under‐severe‐drought experiment of wheat plants.
Figure S7 Pot soil water content for plant flowering experiment under severe drought.
Figure S8 Drought survival rates of control WT and transgenic T1 barley seedlings transformed with the pHDZI‐3‐TaDREB3 (a) and pHDZI‐4‐TaDREB3 (b) constructs, subjected to severe drought.
Figure S9 The results of the promoter‐GUS activity studies using T1 transgenic wheat plants transformed with pHDZI‐3‐GUS and pHDZI‐4‐GUS constructs.
Figure S10 Soil water tension monitored at 10 cm and 30 cm depths in large containers used for wheat growth under well‐watered conditions or gradually increasing drought.
Figure S11 Details of drought tolerance experiments.
Figure S12 Details of frost tolerance experiments.
Figure S13 Alignments of TdHDZipI‐3 and TdHDZipI‐4 promoter sequences and sequences of corresponding genes of Triticum aestivum cv. Chinese Spring, identified in the Whole Genome Reference Assembly Pseudomolecules v1.0 databases of the International Wheat Genome Sequencing Consortium, using the BLAST software (Altschul et al., 1997).
Table S1 List of PCR primers, and Q‐PCR and Northern hybridisation primers and probes, used in this study for investigated genes
Table S2 Proteins, homologous to TaCBF5L from wheat, maize, rice, barley and Arabidopsis were searched using the BLAST tool (Altschul et al., 1990)
Table S3 The list of T1 transgenic wheat lines transformed with promoter‐GUS constructs and tested for the GUS activity
Data S1 Supporting Experimental procedures, Supporting Discussion, Supporting References
Acknowledgements
We acknowledge contributions of Ainur Ismagul in plant transformation. We thank Ursula Langridge, Yuriy Onyskiv, Alex Kovalchuk and Yongle Li for technical support, Mario Fruzangohar for bioinformatic analyses, Margaret Pallotta for the assistance with the BAC library screen and Carl Simmons for critically reading the manuscript. YY is thankful to the University of Adelaide for the postgraduate scholarship and HHJA‐B for the Master Biotechnology scholarship. SH and YL contributions were supported by the Australian Research Council Industrial Transforming Research Hub (IH130200027). This work was supported by the Australian Research Council Linkage Project (LP120100201 to MH and SL), Australian Government: Grains Research and Development Corporation, the Government of South Australia and Pioneer Hi Bred International.
References
- Agarwal, P.K. , Gupta, K. , Lopato, S. and Agarwal, P. (2017) Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 68, 2135–2148. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalraj, A. , Luang, S. , Kumar, M.Y. , Sornaraj, P. , Eini, O. , Kovalchuk, N. , Bazanova, N. et al. (2016) Change of function of the wheat stress‐responsive transcriptional repressor TaRAP2.1L by repressor motif modification. Plant Biotechnol. J. 14, 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, Q. , Liu, G. and Wang, Y. (2011) A DREB gene from Limonium bicolor mediates molecular and physiological responses to copper stress in transgenic tobacco. J. Plant Physiol. 168, 449–458. [DOI] [PubMed] [Google Scholar]
- Bi, H. , Shi, J. , Kovalchuk, N. , Luang, S. , Bazanova, N. , Chirkova, L. , Zhang, D. et al. (2018) Overexpression of the TaSHN1 transcription factor in bread wheat leads to leaf surface modifications, improved drought tolerance and no yield penalty under controlled growth conditions. Plant Cell Environ. 41, 2549–2566. [DOI] [PubMed] [Google Scholar]
- Bouaziz, D. , Charfeddine, M. , Jbir, R. , Saidi, M.N. , Pirrello, J. , Charfeddine, S. , Bouzayen, M. et al. (2015) Identification and functional characterization of ten AP2/ERF genes in potato. Plant Cell, Tissue Organ Cult. 123, 155–172. [Google Scholar]
- Cao, X.Q. , Jiang, Z.H. , Yi, Y.Y. , Yang, Y. , Ke, L.P. , Pei, Z.M. and Zhu, S. (2017) Biotic and abiotic stresses activate different Ca2 + permeable channels in Arabidopsis . Front. Plant Sci. 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. , Wang, Q.Y. , Cheng, X.G. , Xu, Z.S. , Li, L.C. , Ye, X.G. , Xia, L.Q. et al. (2007) GmDREB2, a soybean DRE‐binding transcription factor, conferred drought and high‐salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353, 299–305. [DOI] [PubMed] [Google Scholar]
- Cheong, Y.H. , Kim, K.N. , Pandey, G.K. , Gupta, R. , Grant, J.J. and Luan, S. (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis . Plant Cell, 15, 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, L. , Zhang, H. , Gan, X. , Zhang, L. , Chen, Y. , Nie, F. , Shi, L. et al. (2015) Transcriptome profiling of the potato (Solanum tuberosum L.) plant under drought stress and water‐stimulus conditions. PLoS ONE, 10, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, J.C. , Hrmova, M. , Lopato, S. and Langridge, P. (2011) Modulation of plant growth by HD‐Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 190, 823–837. [DOI] [PubMed] [Google Scholar]
- Harris, J.C. , Sornaraj, P. , Taylor, M. , Bazanova, N. , Baumann, U. , Lovell, B. , Langridge, P. et al. (2016) Molecular interactions of the γ‐clade homeodomain‐leucine zipper class I transcription factors during the wheat response to water deficit. Plant Mol. Biol. 90, 435–452. [DOI] [PubMed] [Google Scholar]
- Heidarvand, L. and Amiri, R.M. (2010) What happens in plant molecular responses to cold stress?. Acta Physiol. Plant. 32, 419–431. [Google Scholar]
- Hrmova, M. and Lopato, S. (2014) Enhancing abiotic stress tolerance in plants by modulating properties of stress responsive transcription factors. In Genomics of Plant Genetic Resources ( Tuberosa, R. , Graner, A. and Frison, E. , eds), pp. 291–316. Netherlands: Springer. [Google Scholar]
- Hu, Z. , Killion, P.J. and Iyer, V.R. (2007) Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 39, 683–687. [DOI] [PubMed] [Google Scholar]
- Humphrey, T.V. , Bonetta, D.T. and Goring, D.R. (2007) Sentinels at the wall: cell wall receptors and sensors. New Phytol. 176, 7–21. [DOI] [PubMed] [Google Scholar]
- Kasuga, M. , Liu, Q. , Miura, S. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress‐inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Kasuga, M. , Miura, S. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2004) A combination of the Arabidopsis DREB1A gene and stress‐inducible rd29A promoter improved drought‐and low‐temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. [DOI] [PubMed] [Google Scholar]
- Katoh, K. and Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro, S. , Yoneda, K. , Takasaki, H. , Takahashi, F. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2017) Different cold‐signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell, 29, 760–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimecka, M. and Muszynska, G. (2007) Structure and functions of plant calcium‐dependent protein kinases. Acta Biochim. Pol. 54, 219–233. [PubMed] [Google Scholar]
- Knight, H. , Trewavas, A.J. and Knight, M.R. (1997) Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, N. , Jia, W. , Eini, O. , Morran, S. , Pyvovarenko, T. , Fletcher, S. , Bazanova, N. et al. (2013) Optimization of TaDREB3 gene expression in transgenic barley using cold‐inducible promoters. Plant Biotechnol. J. 11, 659–670. [DOI] [PubMed] [Google Scholar]
- Lee, J.T. , Prasad, V. , Yang, P.T. , Wu, J. , David Ho, T.H. , Charng, Y.Y. and Chan, M.T. (2003) Expression of Arabidopsis CBF1 regulated by an ABA/stress inducible promoter in transgenic tomato confers stress tolerance without affecting yield. Plant Cell Environ. 26, 1181–1190. [Google Scholar]
- Lopato, S. and Langridge, P. (2011) Engineering Stress Tolerance in Cereals Using DREB/CBF Genes: Outcomes, Problems and Perspectives. ISB News Report. [Google Scholar]
- Lopato, S. , Bazanova, N. , Morran, S. , Milligan, A.S. , Shirley, N. and Langridge, P. (2006) Isolation of plant transcription factors using a modified yeast one‐hybrid system. Plant Methods, 2, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, S. (2002) Signalling drought in guard cells. Plant Cell Environ. 25, 229–237. [DOI] [PubMed] [Google Scholar]
- Luang, S. , Sornaraj, P. , Bazanova, B. , Jia, W. , Eini, O. , Hussain, S.S. , Kovalchuk, N. et al. (2018) TabZIP2 from wheat is a part of the signalling pathway that mobilises plants to respond to nutrient starvation under drought. Plant Mol. Biol. 96, 543–561. [DOI] [PubMed] [Google Scholar]
- Mallikarjuna, G. , Mallikarjuna, K. , Reddy, M. and Kaul, T. (2011) Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol. Lett. 33, 1689–1697. [DOI] [PubMed] [Google Scholar]
- Morran, S. , Eini, O. , Pyvovarenko, T. , Parent, B. , Singh, R. , Ismagul, A. , Eliby, S. et al. (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. [DOI] [PubMed] [Google Scholar]
- Nakashima, K. , Tran, L.S. , Van Nguyen, D. , Fujita, M. , Maruyama, K. , Todaka, D. , Ito, Y. et al. (2007) Functional analysis of a NAC‐type transcription factor OsNAC6 involved in abiotic and biotic stress‐responsive gene expression in rice. Plant J. 51, 617–630. [DOI] [PubMed] [Google Scholar]
- Nakashima, K. , Jan, A. , Todaka, D. , Maruyama, K. , Goto, S. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2013) Comparative functional analysis of six drought‐responsive promoters in transgenic rice. Planta, 239, 1–14. [DOI] [PubMed] [Google Scholar]
- Nakashima, K. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo, F. , Medina, J. , Rodríguez‐Franco, M. , Neuhaus, G. and Salinas, J. (2011) Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. J. Exp. Bot. 63, 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol, B. and Galaud, J.P. (2013) A practical guide to quantifying the effect of genes underlying adaptation in a mixed genomics and evolutionary ecology approach. Acta Bot. Gallica. 160, 197–204. [Google Scholar]
- Pyvovarenko, T. and Lopato, S. (2011) Isolation of plant transcription factors using a yeast one‐hybrid system. Methods Mol. Biol. 754, 45–66. [DOI] [PubMed] [Google Scholar]
- Raza, G. , Ahmad, N. , Hussain, M. , Zafar, Y. and Rahman, M. (2016) Role of genetics and genomics in mitigating abiotic stresses in soybeans. In Environmental Stresses in Soybean Production: Soybean Production ( Miransari, M. , ed), pp. 205–228. New York, NY: Academic Press, Elsevier. [Google Scholar]
- Saini, H.S. and Westgate, M.E. (1999) Reproductive development in grain crops during drought. Adv. Agron. 68, 59–96. [Google Scholar]
- Sakuma, Y. , Liu, Q. , Dubouzet, J.G. , Abe, H. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2002) DNA‐binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration‐ and cold‐inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. [DOI] [PubMed] [Google Scholar]
- Sanders, D. , Pelloux, J. , Brownlee, C. and Harper, J.F. (2002) Calcium at the crossroads of signaling. Plant Cell, 14, S401–S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, T. , Thankappan, R. , Kumar, A. , Mishra, G.P. and Dobaria, J.R. (2014) Heterologous expression of the AtDREB1A gene in transgenic peanut‐conferred tolerance to drought and salinity stresses. PLoS ONE, 9, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazegari, S. , Niazi, A. and Ahmadi, F.S. (2015) A study on the regulatory network with promoter analysis for Arabidopsis DREB genes. Bioinformation, 11, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavrukov, Y. , Baho, M. , Lopato, S. and Langridge, P. (2016) The TaDREB3 transgene transferred by conventional crossings to different genetic backgrounds of bread wheat improves drought tolerance. Plant Biotechnol. J. 14, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K. , Yamaguchi‐Shinozaki, K. and Seki, M. (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. [DOI] [PubMed] [Google Scholar]
- Smith, H. (2000) Phytochromes and light signal perception by plants–an emerging synthesis. Nature, 407, 585–591. [DOI] [PubMed] [Google Scholar]
- Śniegowska‐Świerk, K. , Dubas, E. and Rapacz, M. (2015) Drought‐induced changes in the actin cytoskeleton of barley (Hordeum vulgare L.) leaves. Acta Physiol. Plant. 37, 1–13. [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion, L. , Mazars, C. , Thuleau, P. , Graziana, A. , Rossignol, M. , Moreau, M. and Ranjeva, R. (1996) Activation of plasma membrane voltage‐dependent calcium‐permeable channels by disruption of microtubules in carrot cells. FEBS Lett. 393, 13–18. [DOI] [PubMed] [Google Scholar]
- Todaka, D. , Zhao, Y. , Yoshida, T. , Kudo, M. , Kidokoro, S. , Mizoi, J. , Kodaira, K.S. et al. (2017) Temporal and spatial changes in gene expression, metabolite accumulation and phytohormone content in rice seedlings grown under drought stress conditions. Plant J. 90, 61–78. [DOI] [PubMed] [Google Scholar]
- Urao, T. , Katagiri, T. , Mizoguchi, T. , Yamaguchi‐Shinozaki, K. , Hayashida, N. and Shinozaki, K. (1994) Two genes that encode Ca2+‐dependent protein kinases are induced by drought and high‐salt stresses in Arabidopsis thaliana . Mol. Gen. Genet. 244, 331–340. [DOI] [PubMed] [Google Scholar]
- Xianjun, P. , Xingyong, M. , Weihong, F. , Man, S. , Liqin, C. , Alam, I. , Lee, B.H. et al. (2011) Improved drought and salt tolerance of Arabidopsis thaliana by transgenic expression of a novel DREB gene from Leymuschinensis. Plant Cell Rep. 30, 1493–1502. [DOI] [PubMed] [Google Scholar]
- Yang, W. , Liu, X.D. , Chi, X.J. , Wu, C.A. , Li, Y.Z. , Song, L.L. , Liu, X.M. et al. (2011) Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA‐dependent and ABA‐independent pathways. Planta, 233, 219–229. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Sornaraj, P. , Borisjuk, N. , Kovalchuk, N. and Haefele, S.M. (2016) Transcriptional network involved in drought response and adaptation in cereals. In Abiotic and Biotic Stress in Plants ‐ Recent Advances and Future Perspectives ( Shanker, A.K. and Shanker, C. , eds), pp. 3–29. Rijeka: InTech. [Google Scholar]
- Yang, Y. , Luang, S. , Harris, J. , Riboni, M. , Li, Y. , Bazanova, N. , Hrmova, M. et al. (2018) Overexpression of the class I homeodomain transcription factor TaHD‐ZipI‐5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol. J. 16, 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The phylogenetic tree of DREB TFs from a representative dicot Arabidopsis and monocots wheat and barley.
Figure S2 Multiple sequence alignment of TaCBF5L and six homologous proteins with a close evolutionary relationship to TaCBF5L from wheat (Ta—T. aestivum; Tm—T. monoccocum), maize (Zm—Zea mais), and barley (Hv—Hordeum vulgare).
Figure S3 Copy numbers of the TaCBF5L transgene estimated by Q‐PCR in T1 transgenic wheat (a) and barley (b).
Figure S4 Control WT barley (H. vulgare cv. Golden promise) and transgenic T1 barley lines transformed with the pHDZI‐3‐TaDREB3 and pHDZI‐4‐TaDREB3 constructs.
Figure S5 Growth characteristics and yield components of control WT and transgenic barley transformed with pHDZI‐3‐TaDREB3 (a), and pHDZI‐4‐TaDREB3 (b) constructs grown under well‐watered conditions.
Figure S6 Details of the flowering‐under‐severe‐drought experiment of wheat plants.
Figure S7 Pot soil water content for plant flowering experiment under severe drought.
Figure S8 Drought survival rates of control WT and transgenic T1 barley seedlings transformed with the pHDZI‐3‐TaDREB3 (a) and pHDZI‐4‐TaDREB3 (b) constructs, subjected to severe drought.
Figure S9 The results of the promoter‐GUS activity studies using T1 transgenic wheat plants transformed with pHDZI‐3‐GUS and pHDZI‐4‐GUS constructs.
Figure S10 Soil water tension monitored at 10 cm and 30 cm depths in large containers used for wheat growth under well‐watered conditions or gradually increasing drought.
Figure S11 Details of drought tolerance experiments.
Figure S12 Details of frost tolerance experiments.
Figure S13 Alignments of TdHDZipI‐3 and TdHDZipI‐4 promoter sequences and sequences of corresponding genes of Triticum aestivum cv. Chinese Spring, identified in the Whole Genome Reference Assembly Pseudomolecules v1.0 databases of the International Wheat Genome Sequencing Consortium, using the BLAST software (Altschul et al., 1997).
Table S1 List of PCR primers, and Q‐PCR and Northern hybridisation primers and probes, used in this study for investigated genes
Table S2 Proteins, homologous to TaCBF5L from wheat, maize, rice, barley and Arabidopsis were searched using the BLAST tool (Altschul et al., 1990)
Table S3 The list of T1 transgenic wheat lines transformed with promoter‐GUS constructs and tested for the GUS activity
Data S1 Supporting Experimental procedures, Supporting Discussion, Supporting References


