Summary
Adenine base editors (ABEs) have been exploited to introduce targeted adenine (A) to guanine (G) base conversions in various plant genomes, including rice, wheat and Arabidopsis. However, the ABEs reported thus far are all quite inefficient at many target sites in rice, which hampers their applications in plant genome engineering and crop breeding. Here, we show that unlike in the mammalian system, a simplified base editor ABE‐P1S (Adenine Base Editor‐Plant version 1 Simplified) containing the ecTadA*7.10‐nSpCas9 (D10A) fusion has much higher editing efficiency in rice compared to the widely used ABE‐P1 consisting of the ecTadA‐ecTadA*7.10‐nSpCas9 (D10A) fusion. We found that the protein expression level of ABE‐P1S is higher than that of ABE‐P1 in rice calli and protoplasts, which may explain the higher editing efficiency of ABE‐P1S in different rice varieties. Moreover, we demonstrate that the ecTadA*7.10‐nCas9 fusion can be used to improve the editing efficiency of other ABEs containing SaCas9 or the engineered SaKKH‐Cas9 variant. These more efficient ABEs will help advance trait improvements in rice and other crops.
Keywords: ABE‐P1S, editing efficiency, ecTadA deaminase, Cas9 variants, rice
Introduction
The clustered regularly interspaced short palindromic repeat (CRISPR)‐CRISPR‐associated protein (Cas) is an immune system of bacteria and archaea that can protect them from infectious nucleic acids or other foreign genetic elements (Hille et al., 2018). Owing to its simplicity, the class 2 CRISPR‐Cas system has been repurposed for genome editing in a wide range of organisms, including many plant species (Chen et al., 2019; Komor et al., 2017; Mahfouz et al., 2014; Mao et al., 2017). The endonuclease Cas9 (or Cas12a or Cas12b) proteins from class 2 type II (or V) CRISPR‐Cas systems can target a specific DNA sequence under the guidance of an sgRNA (or crRNA), where they generate a DNA double‐stranded break (DSB) (Cong et al., 2013; Mali et al., 2013; Teng et al., 2018; Zetsche et al., 2015). The DSB is mainly repaired through the non‐homologous end joining (NHEJ) pathway in most eukaryotic cells, which can induce random insertions or deletions (indels) at the DNA cleavage site (Carroll, 2014). In contrast, base editing, a newly developed genome editing tool based on the CRISPR‐Cas9 system, enables targeted base conversions without inducing DSBs or requiring exogenous repair template DNA (Gaudelli et al., 2017; Komor et al., 2016; Rees and Liu, 2018). Therefore, base editing is a promising tool for precision molecular breeding because the varietal differences in many important agronomic traits in crops are determined by one or a few base changes (Zhang et al., 2018). Cytosine base editor (CBE), the first member in the base editing toolbox, can convert cytidine (C) to thymidine (T) (or G to A) at specific genomic sites in an sgRNA‐dependent manner (Komor et al., 2016). The most commonly used CBEs in plants consist of a Cas9 nickase (nSpCas9 (D10A)) fused with a cytidine deaminase and a uracil glycosylase inhibitor (UGI) (Li et al., 2017; Lu and Zhu, 2017; Qin et al., 2019a; Shimatani et al., 2017; Zong et al., 2017). Although the CBEs have been shown to be functional in many plant species, the narrow target range and low editing efficiency considerably limit their application in crop breeding. Recently, the target scope of CBEs in plants was expanded by constructing new CBEs with different Cas9 variants (Hua et al., 2019; Qin et al., 2019b). Moreover, the editing efficiency of CBEs in plants was improved by exploiting different cytidine deaminases, such as human AID and APOBEC3A (Ren et al., 2018; Zong et al., 2018).
In addition to the CBEs, adenine base editors (ABEs) were recently developed that can introduce targeted A to G (or T to C) substitutions in a programmable manner (Gaudelli et al., 2017). The ABEs consist of a Cas9 nickase (or nuclease dead Cas9, dCas9) fused with an evolved adenine deaminase TadA from E. coli (Gaudelli et al., 2017). Of note, the adenine base editors used for genome editing in bacteria and mammalian cells are different. The ABEs that work efficiently in bacteria contain the evolved TadA variant ecTadA* fused to the dCas9 protein. However, in mammalian cells, providing the ecTadA*‐nSpCas9 (D10A) fusion with a wild‐type ecTadA or an additional evolved ecTadA* monomer is necessary for efficient base editing (Gaudelli et al., 2017). This difference is attributed to the biochemical property of tRNA deaminase TadA as a homodimer (Losey et al., 2006). The E.coli genome encodes a wild‐type ecTadA that can assist adenine base editing of the ecTadA*‐dCas9 fusion. But the human genome lacks an ecTadA homolog, thus necessitating the in‐cis tethering of a wild‐type ecTadA to support base editing of the ecTadA*‐nSpCas9 (D10A) fusion (Gaudelli et al., 2017). The ABE architecture used for human genome editing has since been adopted to perform adenine base editing in other eukaryotes (Liang et al., 2018; Liu et al., 2018a,2018b; Ma et al., 2018; Ryu et al., 2018; Yang et al., 2018), including various plant species (Hua et al., 2018; Kang et al., 2018; Li et al., 2018; Negishi et al., 2019; Wang et al., 2019; Yan et al., 2018). However, results from several groups indicated that ABEs containing the ecTadA‐ecTadA*7.10‐nSpCas9 (D10A) fusion are quite inefficient at many target sites in plants, especially in rice (Hua et al., 2018; Li et al., 2018; Yan et al., 2018). It is not known whether the ABE architecture used for E.coli genome editing may work more efficiently in plants.
In this study, we used the ecTadA*7.10‐nSpCas9 (D10A) fusion to perform adenine base editing in rice and found that it has much higher editing efficiency compared to the widely used ABEs consisting of the ecTadA‐ecTadA*7.10‐nSpCas9 (D10A) fusions. Moreover, we demonstrate that this strategy can also be used to improve the editing efficiency of other ABEs containing SaCas9 or the engineered SaKKH‐Cas9 variant.
Results and discussion
To test whether the ABE architecture used for E. coli genome editing may work in plants, we fused ecTadA*7.10 to the N‐terminus of nSpCas9 (D10A), generating a simplified adenine base editor, ABE‐P1S (Figure 1a). In order to test the editing activity of ABE‐P1S in rice, we first targeted the OsSPL14 and SLR1 genes in the rice variety Nipponbare. In our previous study, both genes could be modestly edited by ABE‐P1, which contained the widely used ecTadA‐ecTadA*7.10‐nSpCas9 (D10A) fusion (Figure 1a) (Hua et al., 2018). At the sgRNA1 target site in OsSPL14 (Figure 1b), we surprisingly found that the base editing efficiency of ABE‐P1S (70.6%, 12/17) was 1.9‐fold higher than that of ABE‐P1 (37.5%, 18/48) (Table 1). We also found that the base editing window of ABE‐P1S at the OsSPL14 target site was expanded (Figure 1c). Adenines at positions 1, 3, 5, 7, 10 and 12 in the protospacer region of OsSPL14 could be efficiently edited by ABE‐P1S (Figure 1c), whereas ABE‐P1 only edited adenines at positions 5, 7 and 10 (Table S1). The base editing efficiency of ABE‐P1S at the sgRNA2 target site SLR1 (35%) was twice of that of ABE‐P1 (17.5%) (Table 1). Then, we designed an sgRNA (sgRNA3) that targets OsSERK2 (Figure 1d), which was previously reported to be edited by the pUbi:rBE14 editor containing the ecTadA‐ecTadA*7.10‐nSpCas9 (D10A) fusion (editing efficiency 32.1%; Yan et al., 2018). Our base editor ABE‐P1 with a similar architecture as pUbi:rBE14 achieved a much higher editing rate at this site (66.6%, 26/39) (Table 1), a 1.1‐fold increase compared to pUbi:rBE14. Interestingly, ABE‐P1S further improved the editing efficiency to 77.8% (21/27) (Table 1), a 20% increase compared to ABE‐P1. Importantly, we found that thymines at positions 6 and 8 in the protospacer of OsSERK2 could be efficiently edited by both ABE‐P1 and ABE‐P1S (Figure 1e and Table S1), whereas the previous study showed that only the thymine at position 6 in the protospacer could be edited (Yan et al., 2018). The above results suggest that the simplified base editor ABE‐P1S has much higher editing efficiency in rice compared to the widely used ABEs containing the ecTadA‐ecTadA*7.10‐nSpCas9 (D10A) fusion.
Figure 1.
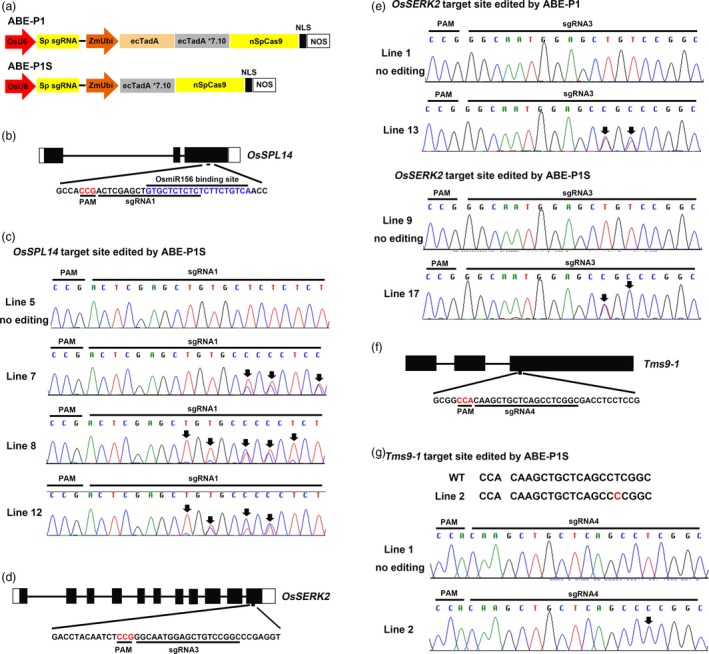
Adenine base editing at different target sites by ABE‐P1S in rice. a, Diagram of the ABE‐P1 and ABE‐P1S base editors used in this study. NLS, nuclear localization signal from Argobacterium VirD2 protein. b, Schematic view of the sgRNA1 target site in OsSPL14. c, Sequence chromatograms of Line 7, Line 8 and Line 12 at the OsSPL14 target site edited by ABE‐P1S. d, Schematic view of the sgRNA3 target site in OsSERK2. e, Sequence chromatograms of Lines 13 and 17 at the OsSERK2 target site edited by ABE‐P1 and ABE‐P1S, respectively. f, Schematic view of the sgRNA4 target site in Tms9‐1. g, One representative line, Line 2, edited by ABE‐P1S is a homozygous mutant. Sequence chromatogram of Line 2 at the target site in Tms9‐1 is shown here. The PAM sequence and miRNA binding site are highlighted in red and blue, respectively (b, d, f). Sequence chromatograms of lines with no mutations at the corresponding target sites are used as controls (c, e, g). Arrows point to the positions with an edited base (c, e, g).
Table 1.
Summary of base editing efficiencies at different sites for ABE‐P1 and ABE‐P1S
| sgRNA | Target gene | Base editor | Rice variety | Number of lines genotyped | Number of base edited lines | Editing efficiency (%) |
|---|---|---|---|---|---|---|
| sgRNA1 | OsSPL14 | ABE‐P1 | Nipponbare | 48 | 18 | 37.5 |
| ABE‐P1S | Nipponbare | 17 | 12 | 70.6 | ||
| sgRNA2 | SLR1 | ABE‐P1 | Nipponbare | 40 | 7 | 17.5 |
| ABE‐P1S | Nipponbare | 20 | 7 | 35 | ||
| sgRNA3 | OsSERK2 | ABE‐P1 | Nipponbare | 39 | 26 | 66.7 |
| ABE‐P1S | Nipponbare | 27 | 21 | 77.8 | ||
| sgRNA4 | Tms9‐1 | ABE‐P1 | Kitaake | 22 | 1 | 4.5 |
| ABE‐P1S | Kitaake | 36 | 4 | 11.1 | ||
| sgRNA5 | OsNRT1.1B | ABE‐P1 | Kitaake | 37 | 27 | 72.9 |
| ABE‐P1S | Kitaake | 54 | 52 | 96.3 | ||
| sgRNA6 | OsACC1 | ABE‐P1 | Kitaake | 36 | 25 | 69.4 |
| ABE‐P1S | Kitaake | 42 | 31 | 73.8 | ||
| sgRNA7 | OsDEP1 | ABE‐P1 | Kitaake | 50 | 40 | 80 |
| ABE‐P1S | Kitaake | 35 | 25 | 71.4 |
To test whether ABE‐P1S has a higher editing efficiency in other rice genetic backgrounds, we also chose the rice variety Kitaake for transformation. We designed an sgRNA (sgRNA4) that targets Tms9‐1 (Figure 1f), which failed to be edited by the pUbi:rBE14 editor in the Kitaake background in a previous report (Yan et al., 2018). However, using our ABE‐P1 base editor, we identified one editing event from 22 transgenic lines (Table 1). Thus, the adenine base editor ABE‐P1 used in our study was more efficient than the previously reported pUbi:rBE14 editor at the Tms9‐1 target site, although they have a similar architecture. We note that the nuclear localization sequence and the codon usage of SpCas9 and ecTadA in ABE‐P1 and pUbi‐rBE14 are different, which may affect their nuclear targeting efficiencies and expression levels. The different base editing rates between ABE‐P1 and pUbi‐rBE14 at the same target site may be a reflection of these differences. Using the ABE‐P1S base editor, 4 out of 36 lines were found to harbour a T‐C conversion at the 5th position in the protospacer of Tms9‐1, with the Line 2 having a homozygous substitution (Figure 1g and Table S1). Next, we selected three target sites with base editing efficiencies ranging between 15% and 40% from the study of Li et al. (2018). Interestingly, both ABE‐P1 and ABE‐P1S gave very high editing rates at these three target sites (Table 1). At the sgRNA5 target site OsNRT 1.1B, the base editing efficiency of ABE‐P1S was up to 96.3% (52/54), 1.3‐fold higher than that of ABE‐P1 (72.9%, 27/37; Table 1). We observed comparable base editing efficiencies at the sgRNA6 target site OsACC1 for ABE‐P1 (69.4%) and ABE‐P1S (73.8%) (Table 1). However, the editing efficiency of ABE‐P1S at the sgRNA7 target site OsDEP1 (71.4%) was slightly lower than that of ABE‐P1 (80%) (Table 1). Since ABE‐P1 already has high editing rates at the three target sites in the Kitaake background (averaging 74%), it seems difficult to improve the editing efficiencies further using ABE‐P1S.
To evaluate the specificity of ABE‐P1 and ABE‐P1S in rice, we sequenced the potential off‐target sites of sgRNA1 and sgRNA6. ABE‐P1 and ABE‐P1S did not induce any mutation at the potential off‐target sites of sgRNA6 in the Kitaake background (Table S4). Consistent with our previous study (Hua et al., 2018), ABE‐P1 was highly specific when guided by sgRNA1 (Table S3). However, we found that ABE‐P1S could induce base conversion at off‐target site 1 of sgRNA1, which has one mismatch to sgRNA1 at the fifth position upstream of the PAM sequence (Figure S1a, b and Table S3). As ABE‐P1 and ABE‐P1S both rely on SpCas9 to recognize target sites, we speculate that the base conversion at off‐target site 1 by ABE‐P1S may be due to its higher editing activity.
Since simplifying the adenine base editor ABE‐P1 containing SpCas9 can significantly improve base editing efficiency in rice, we hypothesized that this effect might extend to other adenine base editors containing different Cas9 proteins. So we sought to simplify the adenine base editor ABE‐P2 (Figure 2a), which relies on SaCas9 to recognize target sites. We have previously demonstrated that ABE‐P2 can edit endogenous genes in rice (Hua et al., 2018). We fused ecTadA*7.10 to the N‐terminus of nSaCas9 (D10A) to obtain the adenine base editor ABE‐P2S (Figure 3a). We first designed sgRNA8 to target the OsmiR827 binding site in SPX‐MFS2 (Figure 3b) to compare the editing efficiencies between ABE‐P2 and ABE‐P2S in rice. For the base editor ABE‐P2, 4 out of 41 transgenic lines were found to have a single T‐C substitution at the target site (Table 2). The thymine at position 1, 9 or 15 in the protospacer region could be edited (Table S2), but no transgenic line showed a homozygous substitution. The base editing efficiency of ABE‐P2S at the SPX‐MFS2 target site was 14.9% (7/47) (Table 2), which was 1.5‐fold higher than that of ABE‐P2 (9.8%). Base conversions were found at position (s) 3, 6, 9 or 15 in the protospacer region of SPX‐MFS2 (Table S2), with one transgenic line, Line 38, showing a homozygous T‐C substitution at position 6 in the protospacer of SPX‐MFS2 (Figure 2c). In addition, we designed sgRNA9 and sgRNA10 to test whether ABE‐P2S may outperform ABE‐P2 when multiple sites in the rice genome are simultaneously targeted. The sgRNA9 was designed to simultaneously target the OsmiR156 binding sites in OsSPL14 and OsSPL17 (Hua et al., 2018; Figure 2d). For ABE‐P2, we obtained 28 transgenic lines and 11 of them harboured base conversions at both the OsSPL14 and OsSPL17 target sites. The other two lines had T‐C substitutions at the OsSPL17 target site only. From 36 transgenic lines that were genotyped for ABE‐P2S, 20 and 22 lines had T‐C substitutions in the protospacer of OsSPL14 and OsSPL17, respectively (Table 2). Among the edited plants, 15 lines were simultaneously modified at both sites (Table 2). The base editing efficiency of ABE‐P2S at the OsSPL17 target site was 30% higher than that achieved by ABE‐P2 (61.1% vs 46.4%) (Table 2). At the OsSPL14 target site, the base editing efficiency of ABE‐P2S was 1.4‐fold as high as that of ABE‐P2 (55.6% vs 39.3%; Table 2). The sgRNA10 targets the OsmiR156 binding sites in OsSPL16 and OsSPL18 (Figure 3e). The base editing efficiencies of ABE‐P2S at these two target sites were 30.3% (10/33) and 39.4% (13/33) (Table 2), respectively, which were 78% and 68% higher compared to that achieved by ABE‐P2 in our previous work (17% at OsSPL14 and 23.4% at OsSPL18; Hua et al., 2018). Moreover, 5 lines were simultaneously edited at both sites (Table 2). Interestingly, we found that compared to ABE‐P2, the base editing window of ABE‐P2S at some target sites was also expanded, as even the thymine at position 17 in the protospacer region of OsSPL14 or OsSPL18 could be efficiently edited (Figure 2d, e and Table S2). Taken together, these results show that ABE‐P2S has a higher editing efficiency compared to ABE‐P2 in rice and the deamination window of ABE‐P2S is also expanded at some target sites. To evaluate the specificity of ABE‐P2S, we amplified and sequenced the potential off‐target sites of sgRNA9 and did not detect any off‐target editing at these sites, suggesting that ABE‐P2S is highly specific at the sgRNA9 target site (Table S5).
Figure 2.
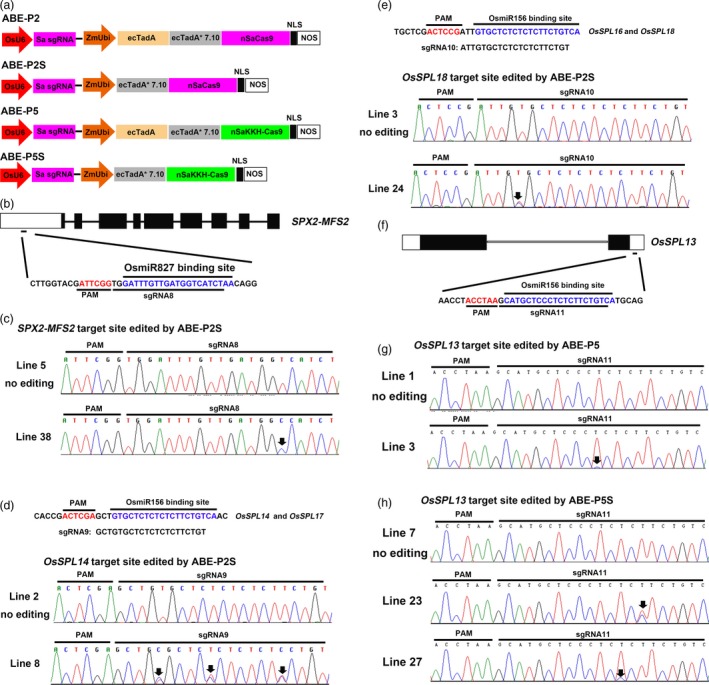
Adenine base editing in rice by ABE‐P2S and ABE‐P5S. a, Diagram of the ABE‐P2, ABE‐P2S, ABE‐P5 and ABE‐P5S base editors. b, Schematic view of the sgRNA8 target site in SPX‐MFS2. c, Sequence chromatogram of one homozygous line, Line 38, at the SPX‐MFS2 target site edited by ABE‐P2S. d, Sequence chromatogram of Line 8 at the OsSPL14 target site edited by ABE‐P2S. The sgRNA9 target site in OsSPL14 and OsSPL17 is shown. e, Sequence chromatogram of Line 24 at the OsSPL18 target site edited by ABE‐P2S. The sgRNA10 target site in OsSPL16 and OsSPL18 is shown. f, Schematic view of the sgRNA11 target site in OsSPL13. g, Sequence chromatogram of Line 3 at the OsSPL13 target site edited by ABE‐P5. h, Sequence chromatograms of Line 23 and Line 27 at the OsSPL13 target site edited by ABE‐P5S. The PAM sequence and miRNA binding site are highlighted in red and blue, respectively (b, d, e, f). Sequence chromatograms of lines with no mutation at the corresponding target sites are used as controls (c, d, e, g, h). Arrows point to the positions with an edited base (c, d, e, g, h).
Figure 3.
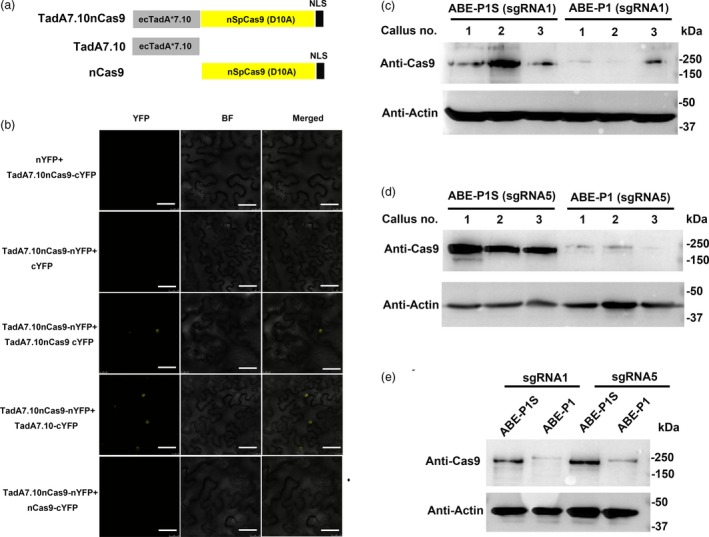
Detection of the self‐dimerization of ecTadA*7.10‐nSpCas9 (D10A) in tobacco leaf and protein levels of ABE‐P1 and ABE‐P1S in rice. a, Schematic diagrams of different coding sequences used in the BiFC assay. NLS, nuclear localization signal from the Agrobacterium VirD2 protein. b, BiFC assay testing interactions between TadA7.10‐nSpCas9 and TadA7.10nCas9, and between TadA7.10 and nCas9 in tobacco leaves. BF, bright field; nYFP, N‐terminal portion of enhanced yellow fluorescent protein; cYFP, C‐terminal portion of enhanced yellow fluorescent protein. Scale bars, 50 μm. c, Detection of ABE‐P1 and ABE‐P1S proteins in transgenic calli from Nipponbare. ABE‐P1 and ABE‐P1S vectors contain sgRNA1 that targets OsSPL14. Upper panel, ABE‐P1 and ABE‐P1S were immunoblotted with an anti‐SpCas9 antibody. Lower panel, actin was immunoblotted with an anti‐actin antibody and was used as a loading control. d, Detection of ABE‐P1 and ABE‐P1S proteins in transgenic calli from Kitaake. ABE‐P1 and ABE‐P1S vectors contain sgRNA5 that targets OsNRT1.1B. e, Protein levels of ABE‐P1 and ABE‐P1S in rice protoplasts. ABE‐P1 and ABE‐P1S vectors containing the sgRNA1 or sgRNA5 were transformed into the protoplasts isolated from the leaf sheaths of Nipponbare seedlings grown in the dark.
Table 2.
Summary of base editing efficiencies at different sites for ABE‐P2, ABE‐P2S, ABE‐P5 and ABE‐P5S
| sgRNA | Target gene | Base editor | Rice variety | Number of lines genotyped | Number of base edited lines* | Editing efficiency (%) |
|---|---|---|---|---|---|---|
| sgRNA8 | SPX‐MSF2 | ABE‐P2 | Nipponbare | 41 | 4 | 9.8 |
| ABE‐P2S | Nipponbare | 47 | 7 | 15.9 | ||
| sgRNA9 | OsSPL14 | ABE‐P2 | Nipponbare | 28 | 11 (11) | 39.3 |
| OsSPL17 | ABE‐P2 | Nipponbare | 28 | 13 (11) | 46.4 | |
| OsSPL14 | ABE‐P2S | Nipponbare | 36 | 20 (15) | 55.6 | |
| OsSPL17 | ABE‐P2S | Nipponbare | 36 | 22 (15) | 61.1 | |
| sgRNA10 | OsSPL16 | ABE‐P2S | Nipponbare | 33 | 10 (5) | 30.3 |
| OsSPL18 | ABE‐P2S | Nipponbare | 33 | 13 (5) | 39.4 | |
| sgRNA11 | OsSPL13 | ABE‐P5 | Nipponbare | 46 | 1 | 2.2 |
| ABE‐P5S | Nipponbare | 33 | 2 | 6.1 | ||
| sgRNA12 | SNB | ABE‐P5 | Nipponbare | 22 | 2 | 9.1 |
| ABE‐P5S | Nipponbare | 62 | 21 | 33.9 |
*The number in () indicates the number of lines that were edited at both target sites simultaneously.
Previously, we showed that the base editing efficiencies of ABE‐P4 (containing the VRER‐Cas9 variant) and ABE‐P5 (containing the SaKKH‐Cas9 variant) at all selected target sites were lower than 10% (Hua et al., 2019). We wondered whether the simplified ecTadA*7.10‐nCas9 fusion can also improve the base editing efficiency of these adenine base editors with engineered Cas9 variants. We chose ABE‐P5 (Figure 2a) to test this hypothesis in rice. We constructed the simplified base editor ABE‐P5S by fusing ecTadA*7.10 to the N‐terminus of nSaKKH‐Cas9 (D10A) (Figure 2a). In order to directly compare the base editing efficiencies of ABE‐P5 and ABE‐P5S in rice, we first selected OsSPL13 as a target. The sgRNA11 was designed to target the OsmiR156 binding site in OsSPL13 followed by an TTAGGT PAM sequence (Figure 2f). This site was totally resistant to editing by ABE‐P5 in our previous work (Hua et al., 2019). Here, we obtained 46 transgenic lines for ABE‐P5 and only one line was identified to have a slight fraction with T‐C substitution at position 11 in the protospacer of OsSPL13 (Figure 2g). For the base editor ABE‐P5S, 2 out of 33 lines were found to harbour an T‐C substitution at position 7 or 9 in the protospacer of OsSPL13 (Figure 2h and Table S2). Thus, the base editing efficiency of ABE‐P5S at OsSPL13 target site is 2.8‐fold as high as that of ABE‐P5 (6.1% for ABE‐P5S vs 2.2% for ABE‐P5; Table 2). To further compare the efficacy between ABE‐P5 and ABE‐P5S, we designed sgRNA12 that targets the OsmiR172 binding site in SNB. The ABE‐P5 achieved a base conversion rate of 9.1% (2/22) at this site, and only the adenine at position 4 in the protospacer region could be modestly edited (Table 2 and Table S2). In contrast, the base editing efficiency of ABE‐P5S was increased to 33.9% (21/62), a 2.7‐fold increase compared to ABE‐P5 (Table 2), and the adenine at position 4, 8, or 9 in the protospacer of SNB could be efficiently edited (Table S2). Collectively, these results suggest that simplifying ABE‐P5 to ABE‐P5S can also improve the adenine base editing efficiency in rice.
Our findings here appear to contradict previous adenine base editing results in mammalian and plant systems because it has been widely accepted that eukaryotic cells need an additional ecTadA monomer to assist ecTadA*7.10‐nSpCas9 (D10A) for efficient base editing (Gaudelli et al., 2017). However, it should be noted that this inference was based on an early‐stage evolved ecTadA* variant (ecTadA*2.1), which showed relatively low adenine base editing activity even when provided with an additional ecTadA monomer (Gaudelli et al., 2017). Whether the later‐stage evolved ecTadA* monomers, especially the highly active ecTadA*7.10 variant, alone can support efficient base editing when fused with nSpCas9 (D10A), had not been tested.
The native TadA deaminase functions as a homodimer (Losey et al., 2006). Therefore, it is possible that the ecTadA*2.1‐nSpCas9 (D10A) relies on intermolecular dimerization to support a low efficiency adenine base editing in mammalian cells (Gaudelli et al., 2017). This dimerization may occur between two ecTadA*2.1 monomers. We speculated that ecTadA*7.10‐nSpCas9 (D10A) may intermolecularly dimerize in the nuclei of plant cells. Bimolecular fluorescence complementation (BiFC) assays suggested that ecTadA*7.10‐nSpCas9 (D10A) indeed can dimerize in the nuclei of tobacco leaf cells and the intermolecular dimerization is mediated by ecTadA*7.10 (Figure 3a and b).
Although the ecTadA*7.10‐nSpCas9 (D10A) can self‐dimerize to support adenine base editing, the underlying reasons for its higher base editing efficiency was not known. Recent studies in mammalian cells and plants suggested that the protein levels and nuclear targeting efficiencies of base editors are highly correlated with their editing efficiencies (Koblan et al., 2018; Wang et al., 2019; Zafra et al., 2018). The ABE‐P1S and ABE‐P1 used the same nuclear localization signal (NLS) from the Agrobacterium VirD2 protein (Data S1). BiFC assays indicated that the NLS from VirD2 is highly efficient for nuclear targeting as ecTadA*7.10‐nCas9 (D10A) dimerizes only in the nucleus, as evidenced by the yellow fluorescence signal exclusively in the nucleus (Figure 3b). Therefore, we believe that ABE‐P1S and ABE‐P1 both have a high nuclear targeting efficiency, although we cannot exclude that an additional wild‐type ecTadA at the N‐terminus of ABE‐P1 may compromise its nuclear targeting. Since the nucleotide sequence of wild‐type ecTadA is nearly the same as the ecTadA*7.10 variant (97.2% identity, see Data S1), we wondered whether the tandem repeat of ecTadA sequences at the N‐terminus of ABE‐P1 may reduce its expression levels in rice due to potential gene silencing. Therefore, we examined the protein levels of ABE‐P1 and ABE‐P1S in rice calli after a 1‐month selection. In the Nipponbare background, we found that the protein level of ABE‐P1S was much higher than that of ABE‐P1 (Figure 3c). In the Kitaake background, ABE‐P1S also accumulated to a much higher level than ABE‐P1 (Figure 3d). To exclude the position effect of T‐DNA random insertion on the expression level of the different base editors, we transiently expressed ABE‐P1S and ABE‐P1 in rice protoplasts and compared their protein levels. We found that the protein level of ABE‐P1S was also substantially higher than that of ABE‐P1 in rice protoplasts (Figure 3e). Collectively, our results suggest that removing wild‐type ecTadA from ABE‐P1 helps to improve the protein level of the base editor, which may account for the higher editing efficiency of the simplified base editor in rice.
In summary, we have demonstrated that the ecTadA*7.10‐nCas9 fusion is a general strategy that can be used to improve the base editing efficiency of ABEs containing different Cas9 proteins or variants in rice. The newly developed ABEs can achieve base conversion rates over 50% at many target sites, with the highest ones up to 96.3% (Tables 1 and 2). Moreover, our improved ABEs showed expanded base editing window at some target sites and did not induce any unintended base changes or indels at any of the tested target sites. The improved efficiencies of the simplified adenine base editors may be explained at least in part by their improved protein levels (Figure 3c–e). It would be worth to test whether the simplified ABEs may also have higher protein levels and base editing efficiencies in Arabidopsis and other plants. Our findings will boost the application of adenine base editing in crop trait improvements. Nevertheless, we note that some target sites in rice were still inefficiently edited (Tables 1 and 2), implying a need for further optimization of these ABEs. In mammalian and plant systems, it has been reported that the base editing efficiency is highly correlated with the expression levels of adenine and cytosine base editors (Koblan et al., 2018; Wang et al., 2019; Zafra et al., 2018). Through changing the codon usage of base editors, modifying the number and position of the nuclear localization signals (NLS) and using ancestral deaminase component, the expression level and nuclear targeting of base editors can be significantly improved to increase the base editing efficiency (Koblan et al., 2018; Wang et al., 2019; Zafra et al., 2018). The TadA deaminase and different Cas9 proteins used in this study were human‐codon optimized (Data S1), indicating that there are opportunities for further codon optimization to improve the editing efficiency of adenine base editors in plants.
Methods
Plasmid construction
The nucleotide sequences of key components in the new adenine base editors developed in this study have been published (Hua et al., 2019) and are provided in the Data S1. The simplified base editors ABE‐P1S and ABE‐P2S were constructed based on our previous reported vectors pRSp‐OsU6 and pRSa‐OsU6 (Hua et al., 2018). The ecTadA*7.10 coding sequence with a 96 bp linker was inserted between two AarI sites of pRSp‐OsU6 and pRSa‐OsU6 by the Golden Gate method, leading to the pRSABESp‐OsU6 (ABE‐P1S) and pRSABESa‐OsU6 vectors. A PCR fragment comprised of the OsU6 promoter, two BsaI sites and the sgRNA scaffold for SaCas9 was used to replace the OsU6‐SpsgRNA cassette in the pRSABESp‐OsU6 vector, leading to the vector pRSABESa‐OsU6Sa (ABE‐P2S). Then, the SaCas9 (D10A) nickase in the pRSABESa‐OsU6sa vector was replaced by the SaKKH‐Cas9 (D10A) nickase by Gibson Assembly method, resulting in the vector pRSABESa‐SaKKH (ABE‐P5S).
All the sgRNAs used in this study were selected from previous reports (Hua et al., 2018, 2019; Li et al., 2018; Yan et al., 2018), except the sgRNA8 that targets to SPX‐MFX2. The sgRNAs inserted into the binary vectors through Golden Gate method. Before rice transformation, the accuracy of sgRNA sequences was confirmed by Sanger sequencing. Primers for sgRNAs used in this study are shown in Table S6.
Rice transformation
Two rice varieties Nipponbare (Oryza sativa L. japonica. cv. Nipponbare) and Kitaake (Oryza sativa L. japonica. cv. Kitaake) were used for Agrobacterium‐mediated rice transformation. The embryogenic calli were induced from mature seeds of these two varieties and infected by Agrobacterium tumefaciens strain EH105 at 22°C in the dark for 3 days. After repeat washing the infected calli with water containing 250 mg/L carbenicillin, rice calli were selected on the selection medium (containing 40 mg/L hygromycin and 250 mg/L carbenicillin) for 2 weeks at 30°C in the dark. Then, the hygromycin‐resistant calli were transferred to the regeneration medium (also containing 40 mg/L hygromycin and 250 mg/L carbenicillin) for shoot induction at 30°C under cold fluorescent light. To examine the protein level of ABE‐P1s and ABE‐P1 in rice calli, these hygromycin‐resistant calli were selected for another 2 weeks on the fresh selection medium. All resistant calli derived from one infected callus were pooled together for protein extraction. The young plantlets that grew vigorously on the regeneration medium were transferred to rooting medium for root induction. Two weeks later, the plantlets were ready for transplanting to soil and grew in glasshouseunder the standard conditions for rice (12‐h light 28°C and 12‐h darkness at 22°C). The tissue culture medium that we used for rice transformation was previously reported (Nishimura et al., 2007).
BiFC assay
The coding sequences of ecTadA*7.10‐nCas9 (D10A)‐NLS, ecTadA*7.10 and nCas9 (D10A)‐NLS were PCR amplified from the pRSABESp‐OsU6 vector and in‐frame cloned into p2YN and/or p2YC vectors by Gibson assembly. The accuracy of all recombinant vectors was confirmed by Sanger sequencing. The empty and recombinant vectors were then transformed into A. tumefaciens GV3101 by the freeze‐thaw method. Single clone of Agribacterium was first inoculated into 5 mL LB medium with antibiotics and grown overnight at 28°C in the dark, and then, 100 μL of the culture was transferred to fresh 5 mL LB medium containing antibiotics. The fresh culture was kept growing at 28°C until the OD600 reaches 3.0. The Agrobacteria cells were harvested by centrifugation, and the pellets were resuspended in the infiltration buffer (10 mm MgCl2, 10 mm MES, pH5.6, 100 μm acetosyringone) to an OD600 of 0.8. All p2YN/p2YC combinations were mixed in equal volume and incubated at room temperature for 2 h. The prepared suspensions were infiltrated into N. benthamiana leaves with a needleless syringe. Three days after infiltration, the yellow fluorescence signals were monitored using a confocal scanning laser microscopy (Leica SMD FLCS Confocal System).
Rice protoplast isolation and transformation
The de‐husked Nipponbare seeds were surface‐sterilized in 2% sodium hypochlorite for 30 min followed by rinsing with sterile water. The healthy seeds were selected for germination on 1/2 MS medium and then grown at 28°C in the dark for 10 days. Fifty rice seedlings were used for protoplast isolation. Rice protoplast isolation was performed as previously described (He et al., 2016). For protoplast transformation, 10 μL (1 μg/μL) plasmid DNA was mixed with 200 μL protoplasts and 210 μL PEG solution (40% w/v PEG4000, 0.2 m mannitol and 0.1 m CaCl2). The mixture was incubated at room temperature for 20 min, and the transformation reaction was quenched by adding 1 mL W5 solution. Protoplasts were harvested by centrifugation at 183 g for 5 min and resuspended in 1 mL W5 solution. The protoplasts were incubated at 28°C in the dark for 24 h before harvesting for protein extraction.
Protein extraction and Western blotting
Total proteins of rice calli or protoplasts were extracted using the protein extraction buffer (50 mm Tris‐HCl pH 8.0, 150 mm NaCl, 10 mm MgCl2, 1 mm EDTA pH 8.0, 10% glycerol) with Protease inhibitor cocktail (sigma). Equal amount of total proteins from different samples were resolved on an 8% SDS‐PAGE gel and then transferred to a PVDF membrane. ABE‐P1S and ABE‐P1 proteins were immunoblotted with an anti‐Cas9 antibody (1 : 5000) (Active motif), whereas the actin was immunodetected with an anti‐actin antibody (1 : 5000) (Sangon Biotech, Shanghai). The membranes were rinsed three times with TBST buffer and then blotted with secondary antibodies. The signals were detected using High‐signal ECL Western Blotting Substrate from Tanon (Shanghai).
Genotyping at on‐target and off‐target sites
Rice genome DNA was extracted from the young leaves of all T0 transgenic lines. The on‐target sites for sgRNA1‐sgRNA12 were amplified by PCR, and the PCR products were purified for Sanger sequencing by a nest primer. Primers for on‐target sites amplification and sequencing are listed in Table S7. The potential off‐target sites for sgRNA1, sgRNA6 and sgRNA9 were predicted by the online tool CRISPR‐GE (Xie et al., 2017). We chose sequences with up to 4 or 5 bp mismatches to target sites in the rice genome as potential off‐target sites. All the potential off‐target sites were each amplified from eight randomly selected base edited lines and sequenced by a nest primer. Primers for potential off‐target sites amplification and sequencing are listed in Table S8.
Author contributions
K.H. designed and performed most of the experiments, analysed the data and wrote the manuscript. X.T. conducted rice transformation. W.L, Z.Z. and R.G. performed some genotyping experiments. J.‐K. Z. supervised the project and edited the manuscript.
Conflict of interest statement
The authors declare no conflict of interests.
Supporting information
Figure S1 Off‐target editing at OsSPL17 by ABE‐P1S.
Table S1 The base editing windows of ABE‐P1 and ABE‐P1S at different target sites.
Table S2 The base editing windows of ABE‐P2, ABE‐P2S, ABE‐P5 and ABE‐P5S at different target sites.
Table S3 The base editing frequencies of ABE‐P1 and ABE‐P1S at potential off‐target sites of sgRNA1.
Table S4 The base editing frequencies of ABE‐P1 and ABE‐P1S at potential off‐target sites of sgRNA6.
Table S5 The base editing frequencies of ABE‐P2S at potential off‐target sites of sgRNA9.
Table S6 The target sites and primers for all sgRNAs used in this study.
Table S7 Primers for on‐target site amplification and sequencing.
Table S8 Primers for potential off‐target site amplification and sequencing.
Data S1 The DNA sequences of key components in the different adenine base editors.
Acknowledgements
This work was supported by the Chinese Academy of Sciences.
References
- Carroll, D. (2014) Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. and Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli, N.M. , Komor, A.C. , Rees, H.A. , Packer, M.S. , Badran, A.H. , Bryson, D.I. and Liu, D.R. (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 551, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. , Chen, S. , Ning, Y. and Wang, G.L. (2016) Rice (Oryza sativa) protoplast isolation and its application for transient expression analysis. Curr. Protoc. Plant Biol. 1, 373–383. [DOI] [PubMed] [Google Scholar]
- Hille, F. , Richter, H. , Wong, S.P. , Bratovic, M. , Ressel, S. and Charpentier, E. (2018) The biology of CRISPR‐Cas: backward and forward. Cell, 172, 1239–1259. [DOI] [PubMed] [Google Scholar]
- Hua, K. , Tao, X. , Yuan, F. , Wang, D. and Zhu, J.K. (2018) Precise A•T to G•C base editing in the rice genome. Mol. Plant, 11, 627–630. [DOI] [PubMed] [Google Scholar]
- Hua, K. , Tao, X. and Zhu, J.‐K. (2019) Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 17, 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.C. , Yun, J.Y. , Kim, S.T. , Shin, Y. , Ryu, J. , Choi, M. , Woo, J.W. et al (2018) Precision genome engineering through adenine base editing in plants. Nat. Plants, 4, 427–431. [DOI] [PubMed] [Google Scholar]
- Koblan, L.W. , Doman, J.L. , Wilson, C. , Levy, J.M. , Tay, T. , Newby, G.A. , Maianti, J.P. et al (2018) Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Packer, M.S. , Zuris, J.A. and Liu, D.R. (2016) Programmable editing of a target base in genomic DNA without double‐stranded DNA cleavage. Nature, 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A.C. , Badran, A.H. and Liu, D.R. (2017) CRISPR‐based technologies for the manipulation of eukaryotic genomes. Cell, 168, 20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Sun, Y. , Du, J. , Zhao, Y. and Xia, L. (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant, 10, 526–529. [DOI] [PubMed] [Google Scholar]
- Li, C. , Zong, Y. , Wang, Y. , Jin, S. , Zhang, D. , Song, Q. , Zhang, R. et al (2018) Expanded base editing in rice and wheat using a Cas9‐adenosine deaminase fusion. Genome Biol. 19, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, P. , Sun, H. , Zhang, X. , Xie, X. , Zhang, J. , Bai, Y. , Ouyang, X. et al (2018) Effective and precise adenine base editing in mouse zygotes. Protein Cell, 9, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Chen, M. , Chen, S. , Deng, J. , Song, Y. , Lai, L. and Li, Z. (2018a) Highly efficient RNA‐guided base editing in rabbit. Nat. Commun. 9, 2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Lu, Z. , Yang, G. , Huang, S. , Li, G. , Feng, S. , Liu, Y. et al (2018b) Efficient generation of mouse models of human diseases via ABE‐ and BE‐mediated base editing. Nat. Commun. 9, 2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losey, H.C. , Ruthenburg, A.J. and Verdine, G.L. (2006) Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159. [DOI] [PubMed] [Google Scholar]
- Lu, Y. and Zhu, J.K. (2017) Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant, 10, 523–525. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Yu, L. , Zhang, X. , Xin, C. , Huang, S. , Bai, L. , Chen, W. et al (2018) Highly efficient and precise base editing by engineered dCas9‐guide tRNA adenosine deaminase in rats. Cell Discov. 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz, M.M. , Piatek, A. and Stewart, C.N. Jr . (2014) Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives. Plant Biotechnol. J. 12, 1006–1014. [DOI] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , Norville, J.E. et al (2013) RNA‐guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y. , Botella, J.R. and Zhu, J.‐K. (2017) Heritability of targeted gene modifications induced by plant‐optimized CRISPR systems. Cell. Mol. Life Sci. 74, 1075–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi, K. , Kaya, H. , Abe, K. , Hara, N. , Saika, H. and Toki, S. (2019) An adenine base editor with expanded targeting scope using SpCas9‐NGv1 in rice. Plant Biotechnol. J. 17, 1476–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A. , Aichi, I. and Matsuoka, M. (2007) A protocol for Agrobacterium‐mediated transformation in rice. Nat. Protoc. 1, 2796–2802. [DOI] [PubMed] [Google Scholar]
- Qin, L. , Li, J. , Wang, Q. , Xu, Z. , Sun, L. , Alariqi, M. , Manghwar, H. et al (2019a) High‐efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 10.1111/pbi.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, R. , Li, J. , Li, H. , Zhang, Y. , Liu, X. , Miao, Y. , Zhang, X. et al (2019b) Developing a highly efficient and wildly adaptive CRISPR‐SaCas9 toolset for plant genome editing. Plant Biotechnol. J. 17, 706–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, H.A. and Liu, D.R. (2018) Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, B. , Yan, F. , Kuang, Y. , Li, N. , Zhang, D. , Zhou, X. , Lin, H. et al (2018) Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9‐guided hyperactive hAID mutant. Mol. Plant, 11, 623–626. [DOI] [PubMed] [Google Scholar]
- Ryu, S.M. , Koo, T. , Kim, K. , Lim, K. , Baek, G. , Kim, S.T. , Kim, H.S. et al (2018) Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 36, 536–539. [DOI] [PubMed] [Google Scholar]
- Shimatani, Z. , Kashojiya, S. , Takayama, M. , Terada, R. , Arazoe, T. , Ishii, H. , Teramura, H. et al (2017) Targeted base editing in rice and tomato using a CRISPR‐Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. [DOI] [PubMed] [Google Scholar]
- Teng, F. , Cui, T. , Feng, G. , Guo, L. , Xu, K. , Gao, Q. , Li, T. et al (2018) Repurposing CRISPR‐Cas12b for mammalian genome engineering. Cell Discov. 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Wang, Z. , Mao, Y. , Lu, Y. , Yang, R. , Tao, X. and Zhu, J.‐K. (2019) Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol. J. 17, 1697–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. , Ma, X. , Zhu, Q. , Zeng, D. , Li, G. and Liu, Y.‐G. (2017) CRISPR‐GE: a convenient software toolkit for CRISPR‐based genome editing. Mol. Plant, 10, 1246–1249. [DOI] [PubMed] [Google Scholar]
- Yan, F. , Kuang, Y. , Ren, B. , Wang, J. , Zhang, D. , Lin, H. , Yang, B. et al (2018) High‐efficient A.T to G.C base editing by Cas9n‐guided tRNA adenosine deaminase in rice. Mol. Plant, 11, 631–634. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Zhang, X. , Wang, L. , Yin, S. , Zhu, B. , Xie, L. , Duan, Q. et al (2018) Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell, 9, 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra, M.P. , Schatoff, E.M. , Katti, A. , Foronda, M. , Breinig, M. , Schweitzer, A.Y. , Simon, A. et al (2018) Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 36, 888–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B. , Gootenberg, J.S. , Abudayyeh, O.O. , Slaymaker, I.M. , Makarova, K.S. , Essletzbichler, P. , Volz, S.E. et al (2015) Cpf1 is a single RNA‐guided endonuclease of a class 2 CRISPR‐Cas system. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Massel, K. , Godwin, I.D. and Gao, C. (2018) Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Y. , Wang, Y. , Li, C. , Zhang, R. , Chen, K. , Ran, Y. , Qiu, J.L. et al (2017) Precise base editing in rice, wheat and maize with a Cas9‐cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440. [DOI] [PubMed] [Google Scholar]
- Zong, Y. , Song, Q. , Li, C. , Jin, S. , Zhang, D. , Wang, Y. , Qiu, J.L. et al (2018). Efficient C‐to‐T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol., 36, 950–953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Off‐target editing at OsSPL17 by ABE‐P1S.
Table S1 The base editing windows of ABE‐P1 and ABE‐P1S at different target sites.
Table S2 The base editing windows of ABE‐P2, ABE‐P2S, ABE‐P5 and ABE‐P5S at different target sites.
Table S3 The base editing frequencies of ABE‐P1 and ABE‐P1S at potential off‐target sites of sgRNA1.
Table S4 The base editing frequencies of ABE‐P1 and ABE‐P1S at potential off‐target sites of sgRNA6.
Table S5 The base editing frequencies of ABE‐P2S at potential off‐target sites of sgRNA9.
Table S6 The target sites and primers for all sgRNAs used in this study.
Table S7 Primers for on‐target site amplification and sequencing.
Table S8 Primers for potential off‐target site amplification and sequencing.
Data S1 The DNA sequences of key components in the different adenine base editors.


