Abstract
MicroRNAs are small non-coding RNAs that range in length from 18 to 24 nucleotides. As one of the most extensively studied microRNAs, microRNA-21 (miR-21) is highly expressed in many mammalian cell types. It regulates multiple biological functions such as proliferation, differentiation, migration, and apoptosis. In this review, we summarized the mechanism of miR-21 in the pathogenesis of various liver diseases. While it is clear that miR-21 plays an important role in different types of liver diseases, its use as a diagnostic marker for specific liver disease or its therapeutic implication are not ready for prime time due to significant variability and heterogeneity in the expression of miR-21 in different types of liver diseases depending on the studies. Additional studies to further define miR-21 functions and its mechanism in association with each type of chronic liver diseases are needed before we can translate the bedside observations into clinical settings.
Keywords: miRNA-21, viral hepatitis, non-alcoholic fatty liver disease, alcohol liver disease (ALD), hepatocellular carcinoma
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs with 18–24 nucleotides in length. MiRNAs can bind to target mRNAs and negatively regulate gene expression (1). MiRNAs are transcribed by RNA polymerase II as part of capped and polyadenylated primary transcripts (pri-miRNAs) that can be either protein-coding or non-coding (2). The biogenesis of miRNAs can be regulated either at the transcriptional level by specific transcription factors or at the post-transcriptional level by changes in processing (3, 4). MiRNAs target and regulate essentially all biological processes and cell types, and influence complex programs of gene expression in several cellular processes. Particular miRNAs emerge as principal regulators that control major cell functions in various physiological and pathophysiological settings.
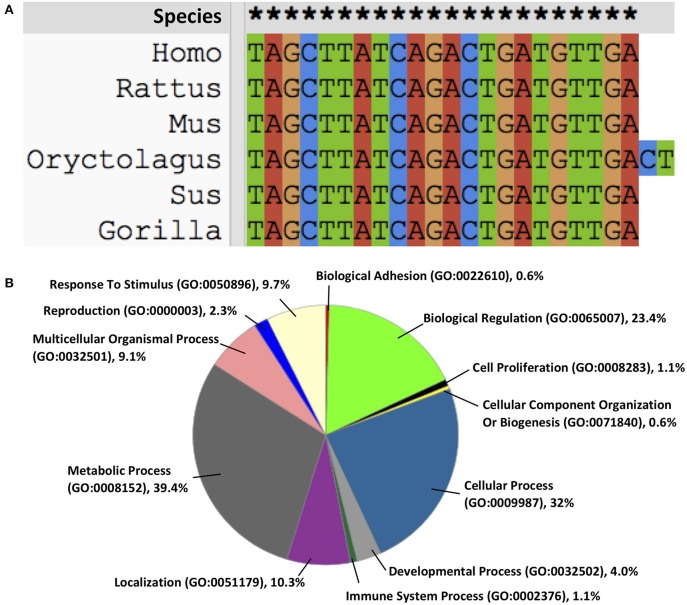
MicroRNA-21 (miR-21) gene is located on chromosome 17 of Homo sapiens and highly conserved (Figure 1A). Its promoter described by Fujita et al. has several conserved enhancer elements including binding sites for activation protein 1 (AP-1; composed of Fos and Jun family proteins), E26 transformation-specific family transcription factor PU1 (Ets/PU1), CCAAT/enhancer binding proteins α (C/EBPα), nuclear factor I (NFI), serum response factor (SRF), p53 and signal transducer and activator of transcription 3 (STAT3) (5, 6). At the cellular level, miR-21 is located in the cytosol (7), extracellular exosome (8), and at the organ level, miR-21 is found in peripheral blood, bone marrow, liver, lung, kidney, Intestine, colon, and thyroid (9). Functionally, miR-21 regulates its targets via interaction with the 3′ untranslated region (UTR) binding involving in post-transcriptional gene silencing (10). It is predicted using computational algorithms that 175 genes involving in biological regulation, cellular and metabolic processes are under regulation of miR-21 [Figure 1B; (11)], however, relatively few have been experimentally validated (Table 1).
Figure 1.
(A) miR-21 is highly conserved across the species, (B) the functions of miR-21 on intracellular biological processes.
Table 1.
Reported gene targets known to be regulated by miR-21.
| Targets | Gene name | Mainly function | Disease | References |
|---|---|---|---|---|
| FASLG | Fas ligand | Regulation of the immune system and the progression of cancer | ALD, HCC | (12, 13) |
| PTEN | Phosphatase and tensin homolog | Regulation of the cell cycle | lung squamous carcinoma, HCC | (14, 15) |
| TFDP3 | Transcription Factor Dp Family Member 3 | Regulation of the cell cycle | Lung cancer | (16) |
| HBP1 | HMG-Box Transcription Factor 1 | Transcriptional repressor, regulation of the cell cycle | NAFLD and HCC | (17) |
| HMGCR | 3-Hydroxy-3-Methylglutaryl-CoA Reductase | A key enzyme of mevalonate pathway, which produce cholesterol and isoprenoids. | NAFLD | (18) |
| FABP7 | Fatty Acid Binding Protein 7 | Fatty acid uptake, transport, and metabolism | NAFLD | (19) |
| HIF-1a | Hypoxia-inducible factor 1-alpha | A transcriptional regulator of cell response to hypoxia, involving cell survival, tumor invasion, and angiogenesis | ovarian cancer | (20) |
| PDCD4 | Programmed Cell Death 4 | Plays a role in apoptosis | breast cancer | (21, 22) |
| PPARα | Peroxisome proliferator-activated receptor alpha | Regulation of lipid metabolism in liver | NAFLD | (23) |
| TGF-β | Transforming growth factor, beta | Multifunctional cytokine, regulation of immune cells, cell growth. | spinal cord injury, colon cancer | (24, 25) |
| SMAD7 | SMAD Family Member 7 | Inhibitor of the TGF-β signaling | NASH | (26) |
| IL-12 | Interleukin 12 | A T cell-stimulating factor, activation of immune response | HCC | (27) |
| RECK | Reversion-inducing cysteine-rich protein with Kazal motifs precursor | Metalloendopeptidase inhibitor, wnt-protein binding | HCC | (28) |
| TIMP-3 | Tissue inhibitors of metalloproteinases 3 | Inhibitor of the matrix metalloproteinases | Liver fibrosis, HCC | (28, 29) |
MiR-21 is upregulated in many biological processes, including inflammation, fibrosis, and cancer (5). Increasing evidence has demonstrated the important role of miR-21 in several types of liver diseases. In this current review, we summarized the mechanism of miR-21 in common liver diseases, such as viral hepatitis, non-alcoholic fatty liver disease (NAFLD), alcohol-associated liver disease (ALD), and hepatocellular carcinoma (HCC).
MiR-21 in Viral Hepatitis
Host miRNAs may target viral genomes or cellular factors, positively or negatively regulating viral infection (30). Viral infections can affect cellular miRNA expression levels and create a favorable environment for their survival and pathogenic effects (30). Serum levels of miR-21 were increased in patients infected with Hepatitis B virus (HBV) (31, 32). Although there was no direct evidence to prove that miR-21 was responsible for HBV infection or replication, some studies showed that miR-21 was essential in the HBV x protein (HBx) induced non-tumor to tumor transformation (27, 31, 33), mechanically through phosphatase and tensin homolog/phosphoinositide 3-kinase/protein kinase B (PTEN/PI3k/Akt) signaling pathway (34).
Hepatitis C virus (HCV) increases the expression of miR-21 in hepatocyte cell lines and primary human hepatocytes (35). Clinical data showed that miR-21 expression in liver tissues was associated with viral load and the level of fibrosis in liver biopsies of patients with HCV infection (36). Chen et al., showed that during HCV infection miR-21 negatively regulated IFN-α signaling by inhibiting myeloid differentiation primary response 88 (MyD88) and Interleukin 1 Receptor Associated Kinase 1 (IRAK1) (37).
MiR-21 in Non-Alcoholic Fatty Liver Disease (NAFLD)
NAFLD is one of the most common chronic liver diseases which is associated with metabolic syndrome. It represents a broad spectrum of histopathological changes ranging from simple steatosis, steatohepatitis (NASH), and cirrhosis (38, 39). Hepatic miR-21 expression is increased in animal models and patients with NAFLD/NASH (23, 40, 41); however, serum miR-21 levels in NAFLD patients when compared to controls were varied depending on the studies. One study showed that serum miR-21 level was lower in 25 NAFLD patients than those in 12 healthy controls (18), the other study claimed that serum level of miR-21 was higher in patients with NAFLD (42). Several studies showed that miR-21 relies on a complex transcription network to regulate glucose and lipid metabolism in hepatocytes. MiR-21, in part, promotes hepatic lipid accumulation by interacting with several factors, such as sterol regulatory element binding protein (SREBP1) (17, 43), 3-hydroxy-3-methylglutaryl-co-enzyme A reductase (HMGCR) (18), fatty acid binding protein 7 (FABP7) (19). In addition, Calo et al. (44) revealed a new role for miR-21 in hepatocytes in promoting hepatic insulin resistance and steatosis in diet-induced obese mice through regulation of forkhead box protein O1 (Foxo1), insulin induced gene 2 (Insig2), STAT3 and Hepatocyte nuclear factor 4 alpha (HNF4-α). Lack of hepatic miR-21 was sufficient to improve glucose tolerance, insulin sensitivity as well as to prevent hepatic steatosis and fatty acid uptake. MiR-21 also contributes to cell injury, inflammation and fibrosis, through its inhibition of peroxisome proliferator-activated receptor alpha (PPARα) signal pathway (23). Taken together, miR-21 may therefore be implicated at different steps of the NAFLD progression in a cell-specific manner: (1) early steps of lipid accumulation and steatosis onset in hepatocytes and/or (2) inflammation and fibrosis at later stages of the disease (45).
MiR-21 in Alcohol Associated Liver Disease (ALD)
ALD comprises of histopathological changes similar to those of NAFLD in patients with excessive alcohol use. Several miRNAs are aberrantly expressed after alcohol-induced liver injury. In animal models of mice fed with ethanol via intragastric ethanol feeding (12) or 5 weeks Lieber Decarli ethanol feeding (12), the levels of hepatic miR-21 were found to be differentially overexpressed in mice fed with ethanol compared to pair-fed controls. The induction of hepatic miR-21 is believed to exert its protective effect against liver injury secondary to alcohol. First, overexpression of miR-21 increases cell survival during alcohol-induced liver injury (12). Second, alcoholic hepatitis and alcoholic cirrhosis lead to alterations of tissue repair; a process involving a series of death receptor signaling pathways (46, 47). MiR-21 is a putative mediator of hepatic damage and crucial in tissue repair during alcohol exposure (12). Third, miR-21 may serve as a key regulator of liver regeneration in response to liver injury secondary to alcohol consumption (48). In addition to the findings in animal model, there are 2 lines of evidence supporting the important role of miR-21 in ALD. Integrative miRNA profiling of human liver tissues revealed an important dysregulation of miRNA expression among patients with AH compared to controls (49). Among miRNAs which were differentially expressed from miRNA profiling, hepatic miR-21 was confirmed and validated to be significantly upregulated in patients with AH (49). Despite the evidence suggesting the protective role of miR-21 in ALD, an in-depth analysis to further study the molecular mechanism on the role of miR-21 on the 3 key histological pathologies commonly observed in alcoholic hepatitis; steatosis, inflammation, and fibrosis, are lacking. The processes involving in the spectrum of alcohol-induced liver injury are complex and involved the cross talk between the hepatocytes, kupffer cells (KCs), and stellate cells. Apoptotic hepatocytes secondary to alcohol-induced liver injury promote secretion of inflammatory and pro-fibrogenic cytokines from KCs (47). The role of KCs in the pathogenesis of liver fibrosis has been shown indispensable since macrophage depletion blunts the development of fibrosis (50). As miR-21 is present in the hepatocyte (12) and inflammatory cells/macrophage (51), and stellate cells (52), the specific role of miR-21 from different cell types contributing to ALD pathogenesis should be further studied.
MiR-21 in Liver Fibrosis and Hepatocellular Carcinoma (HCC)
MiR-21 has been shown to promote fibrogenesis in muscles and various organs including heart, kidneys, lungs, and liver (53). Clinical data also showed that miR-21 expression was up-regulated in liver of patients with biliary atresia-induced liver fibrosis (54). In liver, miR-21 induces fibrosis by activating hepatic stellate cells (HSCs) and collagen synthesis (52, 55, 56). Mechanically, the over expression of miR-21 promotes oxidation, increases in collagen production and activates angiotensin via sprouty RTK Signaling Antagonist 1 (Spry1)/ERK/NF-κB, PTEN/Akt, programmed cell death 4 (PDCD4)/AP-1, Smad7/Smad2/3/NADPH oxidase 4 (NOX4) pathways (52, 57, 58). Recently, research showed that in an methionine choline deficient diet model of NASH-associated liver damage, miR-21 knockout results in decrease of steatosis, inflammation, and lipoapoptosis, with impairment of fibrosis (59). Similarly, in a different study, the loss of miR-21 expression resulted in decreased collagen deposition and expression of fibrotic markers transforming growth factor-β1 and α-smooth muscle actin in bile duct ligation mice model (60). Despite the evidence on the role of miR-21 and fibrosis, a recent study found that antisense inhibition or genetic deletion of miR-21 does not alter HSC activation or liver fibrosis in CCL4 induced liver fibrosis mice models (29).
MiR-21 is an “onco-miR,” and miR-21 is frequently up-regulated in human solid malignancies, such as tumors of breast, colon, lung, pancreas, prostate, liver, and stomach (61). MiR-21 is an established survival factor during liver injury and hepatocellular carcinoma development. Clinical data showed that miR-21 was significantly upregulated in both HCC tissues and serum (62–64). Although miR-21 expression in HCC tissues did not predict overall survival (64), studies showed that increased expression of miR-21 was significantly correlated with tumor progression and could be a novel potential biomarker for HCC prognosis (63–65). Mechanically, miR-21 promotes migration and invasion in HCC through the miR-21-PDCD4-AP-1 feedback loop (66). Upregulation of miR-21 can activate phosphatase and tensin homolog (PTEN), which activates phosphatidylinositol 3-kinase signaling to AKT and contributes to progression of HCC (67). Moreover, miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting Kruppel Like Factor 5 (KLF5) (68). In addition, HCC cells secreted exosomal miRNA-21 that directly targeted PTEN, leading to activation of pyruvate dehydrogenase kinase 1 (PDK1)/AKT signaling in HSCs; then promoted cancer progression by secreting angiogenic cytokines, including vascular endothelial growth factor (VEGF), matrix metallopeptidase 2 (MMP2), MMP9, basic fibroblast growth factor (bFGF), and transforming growth factor-β (TGF-β) (69). In two separate HCC tumor xenograft models, treatment with specific single-stranded oligonucleotide inhibitors of miR-21 (anti-miRNAs) suppresses HCC growth (70).
Potential Roles of miR-21 as Diagnostic and Therapeutic Targets for Liver Diseases
While it is clear that miR-21 plays an important role in different types of liver diseases, its use as a diagnostic marker for specific liver disease or its therapeutic implication are not ready for prime time. Circulating miR-21 as a diagnostic marker for disease staging such as in patients with NAFLD yielded contradicting results (18, 42). More importantly, the lack of standard operating procedures and the uniform method to normalize the level of miR-21 with gatekeeping genes are also problematic to adopt to use of miR-21 as the diagnostic tool. Targeting miRNA has previously been conducted for the treatment of hepatitis C infection (36), however, more studies are needed to further explore specific mechanisms of miR-21 in the pathogenesis of various types of liver diseases before its use as a therapeutic intervention.
Summary
Dysregulation of MiR-21 is common in several types of chronic liver diseases (Table 2). However, in each type of liver disease, there is a variability and heterogeneity in the expression of miR-21 depending on the studies. The underlying explanation may be due to the use of different animal models and lack of standardized procedures and methods to normalize its level. There are several pitfalls in using miR-21 as the therapeutic target or as potential biomarkers for specific types of liver diseases. Additional studies to further define miR-21 functions and its mechanism in association with each type of chronic liver diseases are needed before we can translate the bedside observations into clinical settings.
Table 2.
Summary of miRNA-21 dysregulation in various liver diseases.
| MiR-21 dysregulation | Sample type | Detect methods | Liver diseases | References |
|---|---|---|---|---|
| Up-regulation | Human serum | RT-qPCR | HBV | (31, 32) |
| Up-regulation, correlated with fibrotic stage, viral load | Human liver | RT-qPCR | HCV | (36) |
| Up-regulation | Cell | RT-qPCR | HCV | (36) |
| Up-regulation | Mice liver | RT-qPCR | High fat diet model | (23) |
| Up-regulation | Human liver | microarray | NASH | (23) |
| Down-regulation | Human serum | RT-qPCR | NAFLD | (18) |
| Up-regulation | Human serum | RT-qPCR | NAFLD | (42) |
| Up-regulation | Mice liver | microarray | ALD | (12) |
| Up-regulation | Human liver | microarray | AH | (49) |
| Up-regulation | Human liver | RT-qPCR | Liver fibrosis with biliary atresia | (54) |
| Up-regulation | Human liver, serum | RT-qPCR | HCC | (62, 63) |
Author Contributions
TZ, ZY, and SL contribute article design. TZ, ZY, PK, and SH contribute data collection. TZ contributes first drafts and final submission. SL contributes revision articles.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- ALD
alcohol-associated liver disease
- AP-1
activation protein 1
- C/EBPα
CCAAT/enhancer binding proteins α
- Ets/PU1
E26 transformation-specific family transcription factor PU1
- FABP7
fatty acid binding protein 7
- FASLG
Fas ligand
- Foxo1
forkhead box O1
- HBP1
HMG-Box transcription factor 1
- HCC
hepatocellular carcinoma
- HMGCR
3-hydroxy-3-methylglutaryl-coA reductase
- HNF4-α
hepatocyte nuclear factor 4 alpha
- IL-12
interleukin 12
- Insig2
insulin induced gene 2
- IRAK1
interleukin 1 Receptor Associated Kinase 1
- KC
Kupffer cells
- KLF5
Kruppel Like Factor 5
- MMP
matrix metallopeptidase
- MyD88
myeloid differentiation primary response 88
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- NFI
nuclear factor I
- NOX4
NADPH oxidase 4
- PDCD4
programmed cell death 4
- PPARα
peroxisome proliferator-activated receptor alpha
- PTEN
phosphatase and tensin homolog
- RECK
reversion inducing cysteine rich protein with kazal motifs
- SMAD
a family of proteins similar to the gene products of the Drosophila gene 'mothers against decapentaplegic’ (Mad) and the C. elegans gene Sma.
- Spry1
sprouty RTK signaling antagonist 1
- SREBP1
sterol regulatory element binding protein
- SRF
serum response factor
- STAT3
signal transducer and activator of transcription 3
- TFDP3
transcription factor Dp family member 3 TGF-β transforming growth factor-β
- TIMP-3
tissue inhibitors of metalloproteinases 3
- VEGF
vascular endothelial growth factor.
Footnotes
Funding. This work was partly supported by grants R01DK107682, R01AA025208, U01 AA026917, UH2 AA026903, and I01CX000361 (to SL) and K01AA026385 (to ZY).
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Ann Rev Biochem. (2010) 79:351–79. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 3.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 4.O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. (2018) 9:402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. (2011) 8:706–13. 10.4161/rna.8.5.16154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. (2008) 378:492–504. 10.1016/j.jmb.2008.03.015 [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai J, et al. MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. J Clin Immunol. (2013) 33:1100–9. 10.1007/s10875-013-9896-z [DOI] [PubMed] [Google Scholar]
- 8.Beltrami C, Besnier M, Shantikumar S, Shearn AI, Rajakaruna C, Laftah A, et al. Human pericardial fluid contains exosomes enriched with cardiovascular-expressed MicroRNAs and promotes therapeutic angiogenesis. Mol Ther. (2017) 25:679–93. 10.1016/j.ymthe.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. (2016) 44:3865–77. 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. (2008) 27:4373–9. 10.1038/onc.2008.72 [DOI] [PubMed] [Google Scholar]
- 11.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. (2007) 39:1278. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- 12.Francis H, McDaniel K, Han Y, Liu X, Kennedy L, Yang F, et al. Regulation of the extrinsic apoptotic pathway by microRNA-21 in alcoholic liver injury. J Biol Chem. (2014) 289:27526–39. 10.1074/jbc.M114.602383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Ren L, Liu W, Xiao Z. MiR-21 regulates the apoptosis of keloid fibroblasts by caspase-8 and the mitochondria-mediated apoptotic signaling pathway via targeting FasL. Biochem Cell Biol. (2018) 96:548–55. 10.1139/bcb-2017-0306 [DOI] [PubMed] [Google Scholar]
- 14.Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS ONE. (2014) 9:e103698. 10.1371/journal.pone.0103698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. (2010) 103:1617–26. 10.1038/sj.bjc.6605958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Zhang L. Possible molecular mechanisms for the roles of MicroRNA-21 played in lung cancer. Technol Cancer Res Treat. (2019) 18:1533033819875130. 10.1177/1533033819875130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Ng R, Chen X, Steer CJ, Song GS. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut. (2016) 65:1850–60. 10.1136/gutjnl-2014-308430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, Huang F, Liu X, Xiao X, Yang M, Hu G, et al. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int J Mol Med. (2015) 35:847–53. 10.3892/ijmm.2015.2076 [DOI] [PubMed] [Google Scholar]
- 19.Ahn J, Lee H, Jung CH, Ha T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res. (2012) 56:1665–74. 10.1002/mnfr.201200182 [DOI] [PubMed] [Google Scholar]
- 20.Xie Z, Cao L, Zhang J. miR-21 modulates paclitaxel sensitivity and hypoxia-inducible factor-1alpha expression in human ovarian cancer cells. Oncol Lett. (2013) 6:795–800. 10.3892/ol.2013.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Xu J, Choi HH, Han C, Fang Y, Li Y, et al. Targeting 17q23 amplicon to overcome the resistance to anti-HER2 therapy in HER2+ breast cancer. Nat Commun. (2018) 9:4718. 10.1038/s41467-018-07264-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Yuan YC, Wang Y, Liu Z, Chan HJ, Chen S. Down-regulation of programmed cell death 4 (PDCD4) is associated with aromatase inhibitor resistance and a poor prognosis in estrogen receptor-positive breast cancer. Breast Cancer Res Treat. (2015) 152:29–39. 10.1007/s10549-015-3446-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loyer X, Paradis V, Henique C, Vion AC, Colnot N, Guerin CL, et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPAR alpha expression. Gut. (2016) 65:1882–94. 10.1136/gutjnl-2014-308883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, et al. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis. (2012) 33:68–76. 10.1093/carcin/bgr246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Wang W, Wang S, Xie W, Li H, Ning B. microRNA-21 regulates astrocytic reaction post-acute phase of spinal cord injury through modulating TGF-beta signaling. Aging. (2018) 10:1474–88. 10.18632/aging.101484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dattaroy D, Pourhoseini S, Das S, Alhasson F, Seth RK, Nagarkatti M, et al. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. (2015) 308:G298–312. 10.1152/ajpgi.00346.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin D, Wang Y, Sai W, Zhang L, Miao Y, Cao L, et al. HBx-induced miR-21 suppresses cell apoptosis in hepatocellular carcinoma by targeting interleukin-12. Oncol Rep. (2016) 36:2305–12. 10.3892/or.2016.5026 [DOI] [PubMed] [Google Scholar]
- 28.Chen M, Liu Y, Varley P, Chang Y, He XX, Huang H, et al. High-mobility group box 1 promotes hepatocellular carcinoma progression through miR-21-mediated matrix metalloproteinase activity. Cancer Res. (2015) 75:1645–56. 10.1158/0008-5472.CAN-14-2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caviglia JM, Yan J, Jang MK, Gwak GY, Affo S, Yu L, et al. MicroRNA-21 and Dicer are dispensable for hepatic stellate cell activation and the development of liver fibrosis. Hepatology. (2018) 67:2414–29. 10.1002/hep.29627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. (2006) 4:651–9. 10.1038/nrmicro1473 [DOI] [PubMed] [Google Scholar]
- 31.Damania P, Sen B, Dar SB, Kumar S, Kumari A, Gupta E, et al. Hepatitis B virus induces cell proliferation via HBx-induced microRNA-21 in hepatocellular carcinoma by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN). PLoS ONE. (2014) 9:e91745. 10.1371/journal.pone.0091745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei YF, Cui GY, Ye P, Chen JN, Diao HY. MicroRNAs may solve the mystery of chronic hepatitis B virus infection. World J Gastroenterol. (2013) 19:4867–76. 10.3748/wjg.v19.i30.4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li CH, Xu F, Chow S, Feng L, Yin D, Ng TB, et al. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur J Cancer. (2014) 50:2560–9. 10.1016/j.ejca.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 34.Hou Z, Quan J. Hepatitis B virus X protein increases microRNA21 expression and accelerates the development of hepatoma via the phosphatase and tensin homolog/phosphoinositide 3kinase/protein kinase B signaling pathway. Mol Med Rep. (2017) 15:3285–91. 10.3892/mmr.2017.6363 [DOI] [PubMed] [Google Scholar]
- 35.Bandiera S, Pernot S, El Saghire H, Durand SC, Thumann C, Crouchet E, et al. Hepatitis C virus-induced upregulation of MicroRNA miR-146a-5p in hepatocytes promotes viral infection and deregulates metabolic pathways associated with liver disease pathogenesis. J Virol. (2016) 90:6387–400. 10.1128/JVI.00619-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Investig. (2010) 90:1727–36. 10.1038/labinvest.2010.126 [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, et al. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. (2013) 9:e1003248. 10.1371/journal.ppat.1003248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. (2018) 67:328–57. 10.1002/hep.29367 [DOI] [PubMed] [Google Scholar]
- 39.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. (2012) 55:2005–23. 10.1002/hep.25762 [DOI] [PubMed] [Google Scholar]
- 40.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. (2008) 48:1810–20. 10.1002/hep.22569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Xiao Y, Wu XK, Jiang LC, Yang SR, Ding ZM, et al. A circulating microRNA signature as noninvasive diagnostic and prognostic biomarkers for nonalcoholic steatohepatitis. Bmc Genomics. (2018) 19:188. 10.1186/s12864-018-4575-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. (2013) 424:99–103. 10.1016/j.cca.2013.05.021 [DOI] [PubMed] [Google Scholar]
- 43.Kennedy L, Venter J, Glaser SS, Standeford HA, Karstens A, Zhou TH, et al. Knockout of microRNA-21 increases high fat diet-induced lipid accumulation, biliary damage, and liver fibrosis via modulation of the SREBP1/HMGCR pathway. Hepatology. (2016) 64:801a 10.1002/hep.28798 [DOI] [Google Scholar]
- 44.Calo N, Ramadori P, Sobolewski C, Romero Y, Maeder C, Fournier M, et al. Stress-activated miR-21/miR-21* in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut. (2016) 65:1871–81. 10.1136/gutjnl-2015-310822 [DOI] [PubMed] [Google Scholar]
- 45.Benhamouche-Trouillet S, Postic C. Emerging role of miR-21 in non-alcoholic fatty liver disease. Gut. (2016) 65:1781–3. 10.1136/gutjnl-2015-310044 [DOI] [PubMed] [Google Scholar]
- 46.Molina PE, Gardner JD, Souza-Smith FM, Whitaker AM. Alcohol abuse: critical pathophysiological processes and contribution to disease burden. Physiology. (2014) 29:203–15. 10.1152/physiol.00055.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guicciardi ME, Gores GJ. Apoptosis: a mechanism of acute and chronic liver injury. Gut. (2005) 54:1024–33. 10.1136/gut.2004.053850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dippold RP, Vadigepalli R, Gonye GE, Hoek JB. Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am J Physiol Gastrointest Liver Physiol. (2012) 303:G733–43. 10.1152/ajpgi.00019.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaya D, Coll M, Rodrigo-Torres D, Vila-Casadesus M, Altamirano J, Llopis M, et al. Integrative microRNA profiling in alcoholic hepatitis reveals a role for microRNA-182 in liver injury and inflammation. Gut. (2016) 65:1535–45. 10.1136/gutjnl-2015-311314 [DOI] [PubMed] [Google Scholar]
- 50.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. (2016) 44:450–62. 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thulasingam S, Massilamany C, Gangaplara A, Dai H, Yarbaeva S, Subramaniam S, et al. miR-27b*, an oxidative stress-responsive microRNA modulates nuclear factor-kB pathway in RAW 264.7 cells. Mol Cell Biochem. (2011) 352:181–8. 10.1007/s11010-011-0752-2 [DOI] [PubMed] [Google Scholar]
- 52.Wei J, Feng L, Li Z, Xu G, Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother. (2013) 67:387–92. 10.1016/j.biopha.2013.03.014 [DOI] [PubMed] [Google Scholar]
- 53.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. (2009) 10:399–416. 10.2217/14622416.10.3.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makhmudi A, Kalim AS, Gunadi. microRNA-21 expressions impact on liver fibrosis in biliary atresia patients. BMC Res Notes. (2019) 12:189. 10.1186/s13104-019-4227-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao J, Tang N, Wu K, Dai W, Ye C, Shi J, et al. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLoS ONE. (2014) 9:e108005. 10.1371/journal.pone.0108005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitano M, Bloomston PM. Hepatic stellate cells and microRNAs in pathogenesis of liver fibrosis. J Clin Med. (2016) 5:E38. 10.3390/jcm5030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ning ZW, Luo XY, Wang GZ, Li Y, Pan MX, Yang RQ, et al. MicroRNA-21 mediates angiotensin II-induced liver fibrosis by activating NLRP3 inflammasome/IL-1β axis via targeting Smad7 and Spry1. Antioxidants & Redox Signaling. (2017) 27:1–20. 10.1089/ars.2016.6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z, Zha Y, Hu W, Huang Z, Gao Z, Zang Y, et al. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J Biol Chem. (2013) 288:37082–93. 10.1074/jbc.M113.517953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodrigues PM, Afonso MB, Simao AL, Carvalho CC, Trindade A, Duarte A, et al. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. (2017) 8:e2748 10.1038/cddis.2017.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kennedy LL, Meng FY, Venter JK, Zhou TH, Karstens WA, Hargrove LA, et al. Knockout of microRNA-21 reduces biliary hyperplasia and liver fibrosis in cholestatic bile duct ligated mice. Lab Investig. (2016) 96:1256–67. 10.1038/labinvest.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang XW, Heegaard NHH, Orum H. MicroRNAs in liver disease. Gastroenterology. (2012) 142:1431–43. 10.1053/j.gastro.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. (2013) 52:297–303. 10.1002/mc.21864 [DOI] [PubMed] [Google Scholar]
- 63.Guo X, Lv X, Lv X, Ma Y, Chen L, Chen Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget. (2017) 8:44050–8. 10.18632/oncotarget.17211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon JS, Kim G, Lee YR, Park SY, Tak WY, Kweon YO, et al. Clinical significance of microRNA-21 expression in disease progression of patients with hepatocellular carcinoma. Biomark Med. (2018) 12:1105–14. 10.2217/bmm-2018-0096 [DOI] [PubMed] [Google Scholar]
- 65.Huang CS, Yu W, Cui H, Wang YJ, Zhang L, Han F, et al. Increased expression of miR-21 predicts poor prognosis in patients with hepatocellular carcinoma. Int J Clin Exp Pathol. (2015) 8:7234–8. [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Q, Wang Z, Hu Y, Li J, Li X, Zhou L, et al. miR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncol Rep. (2012) 27:1660–8. 10.3892/or.2012.168 [DOI] [PubMed] [Google Scholar]
- 67.Meng FY, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. (2007) 133:647–58. 10.1053/j.gastro.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Chu YF, Xu M, Zhang XM, Zhou YM. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol Lett. (2019) 17:2221–7. 10.3892/ol.2018.9843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. (2018) 37:324. 10.1186/s13046-018-0965-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, et al. Anti-miR-21 Suppresses hepatocellular carcinoma growth via broad transcriptional network deregulation. Mol Cancer Res. (2015) 13:1009–21. 10.1158/1541-7786.MCR-14-0703 [DOI] [PubMed] [Google Scholar]