The genome sequence of the acidophile Rhodovastum atsumiense was determined for comparison with that of Rhodopila globiformis. Both genomes are unusually large for purple bacteria (7.10 Mb and 7.25 Mb, respectively), and they have an average nucleotide identity of 72%. This value is remarkably similar to the average nucleotide identity values for Acidisphaera, Elioraea, and Paracraurococcus, all aerobic anoxygenic phototrophs.
ABSTRACT
The genome sequence of the acidophile Rhodovastum atsumiense was determined for comparison with that of Rhodopila globiformis. Both genomes are unusually large for purple bacteria (7.10 Mb and 7.25 Mb, respectively), and they have an average nucleotide identity of 72%. This value is remarkably similar to the average nucleotide identity values for Acidisphaera, Elioraea, and Paracraurococcus, all aerobic anoxygenic phototrophs.
ANNOUNCEMENT
Acidophilic purple photosynthetic bacteria are relatively unusual. There are two groups, which are only distantly related, i.e., those in the Bradyrhizobiaceae family, including Rhodoblastus acidophilus (1) and Rhodoblastus sphagnicola (2), and those in the Acetobacteraceae family, including Rhodopila globiformis (3) and Rhodovastum atsumiense (4). One of the most important characteristics of these bacteria is the optimal growth pH, which is 4.8 to 5.0 for R. globiformis and 6.0 to 6.5 for R. atsumiense (3, 4). The redox potentials of cytochrome c2 and high-potential iron-sulfur protein (HiPIP) are surprisingly high, in the region of 400 mV (5–7), but the reason for this observation remains unknown. If nothing else, it suggests that R. globiformis lives in a highly aerobic environment. Another observation is that cells die immediately if they are overgrown, which is generally not the case with other purple bacteria (8, 9). Acidisphaera rubrifaciens is an aerobic anoxygenic phototroph (AAP), whose rRNA is relatively closely related to that of R. globiformis (94.4% identity) and R. atsumiense (95.5% identity) (10). The genome sequence of R. globiformis was determined previously (11), and we now report the genome sequence of R. atsumiense.
Rhodovastum atsumiense was originally isolated from submerged paddy soil from the Atsumi Peninsula in Japan (4). Genomic DNA of R. atsumiense (strain DSM 21279) was obtained from DSMZ. DNA analysis using Qubit and NanoDrop instruments showed an A260/A280 ratio of 1.79. The sequencing library was prepared using the Illumina Nextera DNA Flex library preparation kit. The genome was sequenced with an Illumina MiniSeq system using 500 μl of a 1.8 pM library. Paired-end (2 × 150-bp) sequencing generated 2,592,590 reads and 202.3 Mbp (35× coverage). Quality control of the reads was performed using FastQC within BaseSpace (version 1.0.0; Illumina), using a k-mer size of 5 and contamination filtering. We assembled the genome de novo using SPAdes (version 3.10.0) (12) through PATRIC (13). This assembly yielded 226 contigs (>300 bp), with the largest being 264,348 bp; the N50 was 104,226 bp. The genome had a GC content of 68.7% and was 7,097,890 bp long, which is larger than the average size of purple bacterial genomes (2.5 to 5.5 Mbp) (14) but similar to the size of the Rhodopila globiformis genome (7.25 Mb) (11). The genome was annotated using the RAST tool kit (15) within PATRIC (13), and this showed our strain to have 6,942 coding sequences and 50 tRNAs.
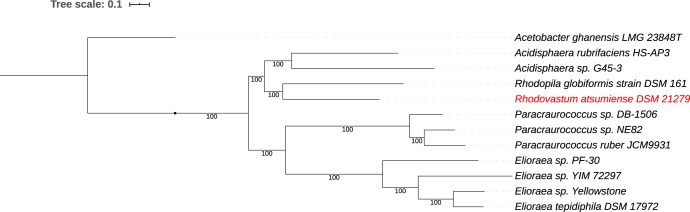
A JSpeciesWS comparison (16) of the average nucleotide identity (ANI) of the Rhodovastum atsumiense genome indicated 72.0% identity to the R. globiformis genome and 72.5% to Acidisphaera rubrifaciens DSM 16009. Other AAPs, namely, Elioraea tepidiphila DSM 17972 and Paracraurococcus ruber, both demonstrated ANI values of 70.5%. The ANI values for Rhodovastum atsumiense are clearly below the proposed 95% cutoff value for genome definition of a species (16). ANI analysis also showed that R. atsumiense and the AAPs mentioned above are not very closely related (<72% identity). Phylogenetic analysis of the R. atsumiense genome using RAxML within PATRIC (17, 18) showed R. globiformis as the closest relative, followed by Acidisphaera and, more distantly, Paracraurococcus and Elioraea (Fig. 1).
FIG 1.
Phylogenetic tree for the whole genome of Rhodovastum atsumiense, in comparison to its closest relatives. The phylogenetic tree was generated using the codon tree method within PATRIC (13), which used cross-genus families (PGFams) as homology groups; 385 PGFams were found among these selected genomes using Codon Tree analysis, and the aligned proteins and coding DNA from single-copy genes were used for RAxML analysis (17, 18). Acetobacter ghanensis was used as an outgroup. iTOL was used for the tree visualization.
R. atsumiense has both cytochrome c2 and HiPIP, and the HiPIP gene is located in the same place as in R. globiformis, downstream of PuhA and the cytochromes c2 that donate electrons to the photosynthetic reaction center in other species of purple bacteria. This finding suggests that HiPIP, rather than cytochrome c2, is the electron donor in these two species, although the proteins could react interchangeably.
Data availability.
This whole-genome shotgun project has been deposited in DDBJ/ENA/GenBank under accession number VWPK00000000; the version described in this paper is version VWPK01000000. The raw sequencing reads have been submitted to SRA under accession number SRR10679485.
ACKNOWLEDGMENT
This work was sponsored by the Wilson Enhancement Fund for Applied Research in Science at Bellevue University.
REFERENCES
- 1.Pfennig N. 1969. Rhodopseudomonas acidophila, sp. n., a new species of the budding purple nonsulfur bacteria. J Bacteriol 99:597–602. doi: 10.1128/JB.99.2.597-602.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulichevskaya IS, Guzev VS, Gorlenko VM, Liesack W, Dedysh SN. 2006. Rhodoblastus sphagnicola sp. nov., a novel acidophilic purple non-sulfur bacterium from a sphagnum peat bog. Int J Syst Evol Microbiol 56:1397–1402. doi: 10.1099/ijs.0.63962-0. [DOI] [PubMed] [Google Scholar]
- 3.Pfennig N. 1974. Rhodopseudomonas globiformis sp. n., a new species of the Rhodospirillaceae. Arch Microbiol 100:197–206. doi: 10.1007/BF00446317. [DOI] [Google Scholar]
- 4.Okamura K, Hisada T, Kanbe T, Hiraishi A. 2009. Rhodovastum atsumiense gen. nov., sp. nov., a phototrophic alphaproteobacterium isolated from paddy soil. J Gen Appl Microbiol 55:43–50. doi: 10.2323/jgam.55.43. [DOI] [PubMed] [Google Scholar]
- 5.Ambler RP, Meyer TE, Cusanovich MA, Kamen MD. 1987. The amino acid sequence of the cytochrome c2 from the phototrophic bacterium Rhodopseudomonas globiformis. Biochem J 246:115–120. doi: 10.1042/bj2460115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambler RP, Meyer TE, Kamen MD. 1993. Amino acid sequence of a high redox potential ferredoxin (HiPIP) from the purple phototrophic bacterium Rhodopila globiformis, which has the highest known redox potential of its class. Arch Biochem Biophys 306:215–222. doi: 10.1006/abbi.1993.1503. [DOI] [PubMed] [Google Scholar]
- 7.Benning MM, Meyer TE, Holden HM. 1996. Molecular structure of a high potential cytochrome c2 isolated from Rhodopila globiformis. Arch Biochem Biophys 333:338–348. doi: 10.1006/abbi.1996.0400. [DOI] [PubMed] [Google Scholar]
- 8.Pfennig N. 1993. Reflections of a microbiologist, or how to learn from the microbes. Annu Rev Microbiol 47:1–31. doi: 10.1146/annurev.mi.47.100193.000245. [DOI] [PubMed] [Google Scholar]
- 9.Girija KR, Vinay B, Sasikala C, Ramana CV. 2010. Novel heliobacteria of a few semi-arid tropical soils. Indian J Microbiol 50:17–20. doi: 10.1007/s12088-010-0069-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiraishi A, Matsuzawa Y, Kanbe T, Wakao N. 2000. Acidisphaera rubrifaciens gen. nov., sp. nov., an aerobic bacteriochlorophyll-containing bacterium isolated from acidic environments. Int J Syst Evol Microbiol 50:1539–1546. doi: 10.1099/00207713-50-4-1539. [DOI] [PubMed] [Google Scholar]
- 11.Imhoff JF, Rahn T, Künzel S, Neulinger SC. 2018. New insights into the metabolic potential of the phototrophic purple bacterium Rhodopila globiformis DSM161T from its draft genome sequence and evidence for a vanadium-dependent nitrogenase. Arch Microbiol 200:847–857. doi: 10.1007/s00203-018-1489-z. [DOI] [PubMed] [Google Scholar]
- 12.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu B, Zhang X, Zhao C, Chen S, Yang S. 2018. Comparative genome analysis of marine purple sulfur bacterium Marichromatium gracile YL28 reveals the diverse nitrogen cycle mechanisms and habitat-specific traits. Sci Rep 8:17803. doi: 10.1038/s41598-018-36160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: Rapid Annotations using Subsystems Technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter M, Rosselló-Móra R, Glöckner FO, Peplies J. 2016. JSpeciesWS: a Web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 18.Stamatakis AJB. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome shotgun project has been deposited in DDBJ/ENA/GenBank under accession number VWPK00000000; the version described in this paper is version VWPK01000000. The raw sequencing reads have been submitted to SRA under accession number SRR10679485.