Abstract
Large-scale chromosomal translocations are frequent oncogenic drivers in acute myeloid leukemia (AML). These translocations often occur in critical transcriptional/epigenetic regulators and contribute to malignant cell growth through alteration of normal gene expression. Despite this knowledge, the specific gene expression alterations that contribute to the development of leukemia remain incompletely understood. Here, through characterization of transcriptional regulation by the RUNX1-ETO fusion protein, we have identified Ras-association domain family member 2 (RASSF2) as a critical gene that is aberrantly transcriptionally repressed in t(8;21)-associated AML. Re-expression of RASSF2 specifically inhibits t(8;21) AML development in multiple models. Through biochemical and functional studies, we demonstrate RASSF2-mediated functions to be dependent on interaction with Hippo kinases, MST1 and MST2, but independent of canonical Hippo pathway signaling. Using proximity-based biotin labeling we define the RASSF2-proximal proteome in leukemia cells and reveal association with Rac GTPase-related proteins, including an interaction with the guanine nucleotide exchange factor, DOCK2. Importantly, RASSF2 knockdown impairs Rac GTPase activation, and RASSF2 expression is broadly correlated with Rac-mediated signal transduction in AML patients. Together, these data reveal a previously unappreciated mechanistic link between RASSF2, Hippo kinases, and Rac activity with potentially broad functional consequences in leukemia.
Subject terms: Acute myeloid leukaemia, Oncogenes
Introduction
Hematologic cancer is dominated by mutations and large-scale chromosome abnormalities affecting transcription factors, splicing factors, and epigenetic regulators, which promote malignancy through alteration of gene expression1–4. These altered gene expression events effectively “re-program” signaling pathways in cancer cells to promote transformation; but also cause the acquisition of dependencies that can be therapeutically exploited to inhibit malignant cell growth and survival5. Despite knowledge of the recurring somatic mutations affecting transcriptional regulators across cancer, the specific gene expression changes that contribute to development of individual malignancies, such as acute myeloid leukemia (AML), remain incompletely understood.
One such example that is frequent in AML is t(8;21), which occurs between the RUNX1 (AML1) and RUNX1T1 (ETO) genes on chromosomes 21 and 8, respectively6. This recurring chromosomal translocation results in generation of the aberrant oncofusion protein, RUNX1-ETO, which is an established driver of leukemia development. RUNX1-ETO primarily functions to promote leukemogenesis through assembly of large transcriptional regulatory complexes that globally alter gene expression patterns and mediate aberrant transcriptional repression of critical hematopoietic genes7–12.
By performing gene expression analysis using a murine AML model driven by the t(8;21)-associated oncofusion protein, we previously identified Ras-association domain family member 2 (Rassf2) as a putative target gene that is downregulated in RUNX1-ETO-expressing leukemic blasts13. The “classical” RASSFs (RASSF1-6) are among the most frequent transcriptionally altered genes in human cancer14. RASSF proteins are non-enzymatic adaptors defined by the presence of both a Ras-association (RA) domain and a C-terminal Salvador-Rassf-Hippo (SARAH) domain15. Family member RASSF1A, in particular, functions as a ubiquitous tumor suppressor that experiences transcriptional silencing through promoter hypermethylation at high frequencies in cancer16,17, which contributes to tumorigenesis via several mechanisms18,19. Other individual RASSFs display more nuanced cell-type-specific expression patterns and functions20,21. RASSF2, similar to RASSF1A, is transcriptionally silenced in several cancer types and knockdown has previously been demonstrated to promote cell proliferation and more aggressive phenotypes in lung cancer22. Although linked with both oncogenic Ras signaling23 and the Hippo tumor suppressor pathway24,25, the molecular functions of RASSF2 remain poorly understood and have not been investigated in leukemia. Here, we define RASSF2 as a transcriptionally repressed target gene of the RUNX1-ETO fusion protein and demonstrate the unique functional importance of this event to the pathogenesis of t(8;21) AML. Through biochemical and functional characterization of RASSF2 in leukemia models, we establish that a SARAH domain-dependent interaction with Hippo kinases MST1 and MST2 is absolutely essential for stabilization of RASSF2 protein and for its tumor-suppressive function against RUNX1-ETO-mediated leukemogenesis. A proximity-based biotin labeling approach demonstrates that RASSF2 forms a discrete complex with MST1 and MST2 that is independent of the canonical Hippo pathway in leukemia cells. Furthermore, we reveal an association with the Rac GTPase-specific guanine nucleotide exchange factor, DOCK2, and demonstrate a functional role for RASSF2 in contributing to Rac GTPase activation in AML. These findings link RASSF2 with an emerging function of Hippo kinases in regulating Rac GTPase activity through DOCK family proteins in the context of normal and malignant hematopoietic cells.
Materials and methods
Please see Supplementary Methods for additional experimental details.
siRNA nucleofection
RUNX1-ETO and control-targeting siRNAs, as previously described26, were synthesized and purchased from GE Dharmacon (Lafayette, CO). Nucleofections were performed by addition of 2 μl siRNA to 100 μl cells (1.5 × 107/ml) in Amaxa buffer V using program P-019 in an Amaxa nucleofector (Lonza, Cologne, Germany).
Serial replating/colony formation assay
Serial replating/colony formation assays were performed essentially as described previously27, but with use of MSCV-PGK-NeoR or MSCV-RUNX1-ETO-PGK-NeoR retroviral expression vectors, and MSCV-IRES-PuroR or MSCV-Rassf2-IRES-PuroR (and variants), together.
RUNX1-ETO9a/Rassf2 primary leukemia model
Transfections of HEK293T cells were conducted by combining 2.5 μg of MSCV-IRES-GFP or MSCV-RUNX1-ETO9a-IRES-GFP retroviral expression vector, 2.5 μg of MSCV-IRES-tdTomato or MSCV-Rassf2-IRES-tdTomato, along with packaging components. For transduction, primary E16.5–E18.5 murine fetal liver cells were resuspended in supplemented retroviral supernatant at densities of 2–3 × 106 cells/ml and centrifuged (2000 × g) in six-well plates at 32 °C for 3 h (Allegra X-12R centrifuge, Beckman Coulter), followed by overnight culture at 37 °C. Two consecutive retroviral transductions were performed in this manner on subsequent days. Transduction efficiencies were measured (GFP+ and tdTomato+ frequencies) by flow cytometry. Transduced cell populations were flow sorted and then pooled with untransduced “helper” fetal liver cells at a ratio of 1:6. For the primary leukemia model, 150,000 total cells per mouse were then transplanted into lethally irradiated (9.5 Gy) recipients and leukemia was monitored. Recipients were randomly allocated to experimental groups with equal distribution based on sex. Recipient mice found to develop GFP-negative/non-myeloid malignancies were excluded from analysis. Investigators were not blinded during group allocation or assessment of experimental outcomes.
Peripheral blood collection and analysis
Peripheral blood samples (approximately 100 μl per mouse) were collected and analyzed exactly as described previously28.
Plasmids and molecular cloning
Murine Rassf2 cDNA was purchased from Open Biosystems (#MMM1013-9202192). Human RASSF2 cDNA vector was kindly provided by Dr. Geoffrey J. Clark. CMV-MCS-BioID2-HA-NeoR (Addgene Plasmid #74224), CMV-MCS-13xLinker-BioID2-HA-NeoR (Addgene Plasmid #80899), and CMV-myc-BioID2-MCS-NeoR (Addgene Plasmid #74223) were kindly provided by Kyle Roux. pJ3H-MST1 (Addgene Plasmid #12203) and pJ3H-MST1-K59R (Addgene Plasmid #12204) expression plasmids were kindly provided by Jonathan Chernoff. Relevant cDNA inserts were inserted into MSCV-based retroviral expression vector backbones by PCR cloning. RASSF2 and MST1 truncation mutant cDNA fragments were generated by overlap PCR.
Cycloheximide chase assay
Forty-eight hours following transfection with indicated constructs, HEK293T cells were washed and seeded in six-well plates containing 25 μM cycloheximide (Sigma, St. Louis, MO) and cultured for indicated lengths of time before harvesting lysates for protein analysis. Where applicable for proteasome experiments, 10 μM lactacystin (Sigma, St. Louis, MO) was also added to culture media.
PAK1-based pull-down assay for active Rac-GTP measurement
Rac1 GTPase Activation Assay (Cell Biolabs # STA-401-1, San Diego, CA) was performed essentially as described by manufacturer’s instructions.
Proximity-based biotin labeling and protein identification
AML cell lines stably expressing RASSF2-BioID2 or Control-BioID2 cDNA were established via retroviral transduction followed by 5 days of selection (1.5 µg/ml) with puromycin. Stable lines were then cultured in media (RPMI, 10% FBS, 1% Pen/Strep) supplemented with 50 µM biotin for 48 h (approximately one population doubling). Approximately 20 million cells per sample were washed, lysed, and subjected to streptavidin-based co-immunoprecipitation for identification of proximal proteins using the BioID2 method as described in Roux et al.29. Mass spectrometry was performed by the Sanford Burnham Prebys Proteomics Shared Resource.
Differential expression analyses
Cell lysis and RNA isolation were performed using Trizol reagent (ThermoFisher Scientific, #15596026) according to the manufacturer’s instructions; isolated RNA was aliquoted and stored at −80 °C. Library preparation and sequencing were performed by Novogene (Sacramento, CA). For differential gene expression, six independent shRASSF2 replicates (three each, two shRNAs) are compared against three independent shCTRL replicates. Differentially expressed genes (DESeq2) are defined by:log2(fold-change) > 1.0 or <−1.0, and –log10(adjusted p value) > 3.0. Gene ontology term enrichment within the differentially expressed genes from TCGA AML patient cohorts was performed using ClusterProfiler30.
Bone marrow mononuclear cell collection
Bone marrow mononuclear cell isolation and enrichment have been described previously28.
Cell lines
Human AML cell lines Kasumi-1, SKNO-1, THP-1, MV4-11, U937, NB-4, K562, and HEK293T cells were purchased from ATCC and maintained as low passage stocks in the lab. OCI-AML3 AML cell line was kindly provided by Suming Huang (University of Florida, Gainesville, FL).
Mice
All animal protocols were approved by the UCSD Institutional Animal Care and Use Committee (IACUC). Mice were housed in standard conditions in accordance with IACUC guidelines. C57BL/6J (stock #000664), Vav1-Cre (stock #008610)31 transgenic, and Stk4- and Stk3-floxed (stock #017635)32 mice were obtained from The Jackson Lab. Genotyping for floxed alleles and Cre transgenes was conducted as described on The Jackson Lab website.
Results
RASSF2 is a transcriptionally repressed target of the RUNX1-ETO fusion protein in t(8;21) AML
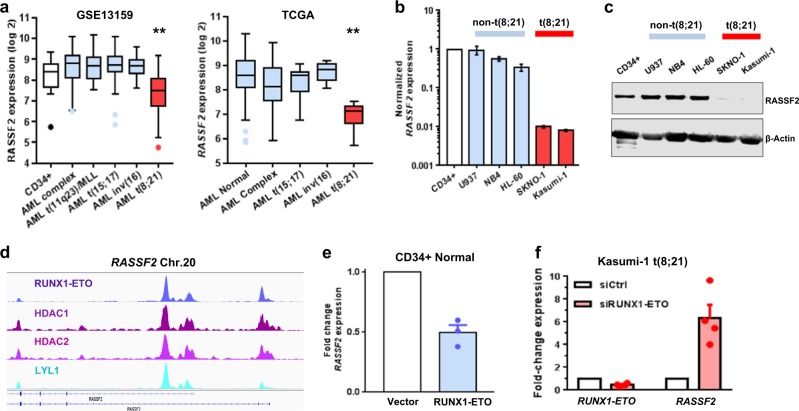
Using a mouse model of t(8;21) AML, we previously identified Rassf2 as a gene that is transcriptionally downregulated greater than 20-fold in RUNX1-ETO expressing leukemic blasts compared to normal hematopoietic progenitors13. We therefore set out to study RASSF2 in the context of myeloid leukemia development. Consistent with an initial report23, we found RASSF2 transcript expression to be most abundant in normal human peripheral blood tissue (Supplementary Fig. 1a), supportive of important normal functions in hematopoietic cell populations. To follow-up the murine leukemia study we assessed RASSF2 transcript expression in two cohorts of AML patients (GSE13159 (ref. 33) and TCGA1, Fig. 1a). Consistent with the mouse model, RASSF2 is uniquely downregulated in t(8;21) AML patients relative to other AML subtypes and healthy-donor CD34+ hematopoietic cells. We also found both RASSF2 mRNA (Fig. 1b) and protein (Fig. 1c) expression to be drastically downregulated (~10–50 fold) specifically in t(8;21) AML cell lines relative to non-t(8;21) AML cell lines and healthy human hematopoietic CD34+ cells. We next sought to determine whether this transcriptional repression was directly mediated by the RUNX1-ETO oncofusion protein in t(8;21) AML6. Supporting this hypothesis, both the RASSF2 promoter CpG island34 and a downstream alternative transcription start site demonstrated chromatin co-occupancy of RUNX1-ETO, repressive histone deacetylases (HDAC1/2), and RUNX1-ETO transcription factor complex component LYL1 (ref. 7), in t(8;21) AML cells (Fig. 1d). These genomic regions are bound by the major hematopoietic transcription factors RUNX1, ERG, and FLI1 in healthy human CD34+ cells35, demonstrating these as major regulatory elements for controlling RASSF2 expression during hematopoiesis (Supplementary Fig. 1b). Retroviral transduction of cord-blood-derived human CD34+ cells with a RUNX1-ETO expression vector was sufficient to reduce RASSF2 transcript (Fig. 1e). Similarly, RUNX1-ETO knockdown using siRNA targeted to the fusion site26 was sufficient to increase RASSF2 mRNA expression in a t(8;21) AML cell line (Fig. 1f). These findings are consistent with published studies in which RASSF2 is reproducibly one of the most dynamically upregulated genes following RUNX1-ETO knockdown7,8,36. Together, these results demonstrate that RASSF2 is differentially expressed across AML subtypes and is a direct transcriptional target of the RUNX1-ETO oncofusion protein in t(8;21) AML.
Fig. 1. RASSF2 is a transcriptionally repressed target of RUNX1-ETO in t(8;21) AML.
a Normalized RASSF2 expression from two independent cohorts of AML patients (GSE13159 (ref. 33), left and TCGA1, right). For GSE13159, CD34+ cells from healthy donors (n = 18) are indicated in white, non-t(8;21) AML patients (n = 192) are indicated in blue, and t(8;21) AML patients (n = 60) are indicated in red. For TCGA, non-t(8;21) AML patients (n = 164) are indicated in blue and t(8;21) AML patients (n = 7) are indicated in red. Data are presented as Tukey boxplots with outliers indicated by individual points. **p < 0.01, ANOVA followed by post hoc Tukey test. b RT-qPCR data showing RASSF2 expression in non-t(8;21) (blue) and t(8;21) (red) AML cell lines. Data are normalized relative to healthy CD34+ cord blood cells and presented as mean ± s.e.m. of three experiments. c Western blot showing protein from whole-cell lysates as indicated, data are representative of three experiments. d ChIP-seq tracks showing binding of indicated transcription/epigenetic factors within the RASSF2 genomic locus in the Kasumi-1 t(8;21) AML cell line. e Normalized RT-qPCR data showing fold-change RASSF2 expression in cord-blood CD34+ cells 72 h post-transduction with empty or RUNX1-ETO expression vectors. Data are mean ± s.e.m. of three experiments (indicated by points). f Normalized RT-qPCR data showing fold-change RASSF2 expression in Kasumi-1 cells 72 h post-nucleofection with control or RUNX1-ETO targeting siRNA. Data are mean ± s.e.m. of four experiments (indicated by points).
Transcriptional repression of RASSF2 contributes to the t(8;21)-associated gene expression signature in AML
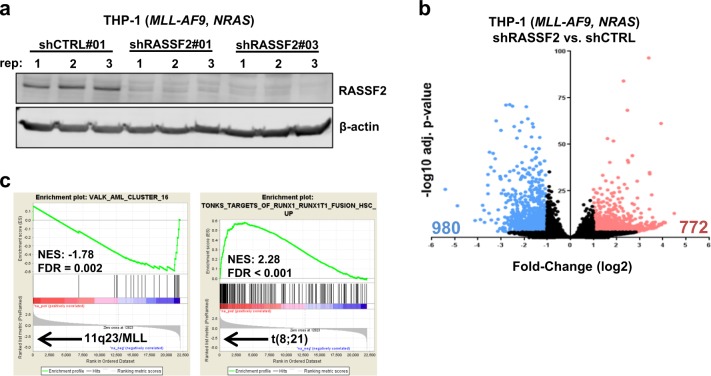
To further understand the impact of differential transcriptional regulation of RASSF2 in AML, we performed gene expression profiling in a non-t(8;21) AML cell context following shRNA-mediated depletion of RASSF2 transcript (Fig. 2a). RASSF2 knockdown in MLL-rearranged THP-1 AML cells profoundly altered the global transcriptional signature (Fig. 2b, Supplementary Table 1). Remarkably, gene-set enrichment analysis (GSEA)37 revealed RASSF2 knockdown to cause loss of a gene expression signature specifically associated with AML patients harboring MLL rearrangements38, and gain of a gene expression signature defined by expression of the t(8;21) oncofusion protein RUNX1-ETO39 (Fig. 2c). This observation highlights the contribution of a single-gene transcriptional repression event to overall AML subtype-specific downstream transcriptional output, and supports a model in which at least part of the RUNX1-ETO upregulated gene expression signature is mediated through indirect signaling consequences caused by transcriptional repression of key target genes. Furthermore, this overlap in gene expression signatures with a defined RUNX1-ETO expression signature in primary cells highlights the relevance of continued study of RASSF2 in the context of t(8;21) AML.
Fig. 2. Transcriptional repression of RASSF2 contributes to the t(8;21)-associated gene expression signature in AML.
a Western blot showing RASSF2 protein in THP-1 cells following lentiviral transduction with indicated shRNAs. Data for nine independent replicates (three shCTRL, six shRASSF2) that were submitted for RNA-sequencing analysis are included. b Volcano plot of RNA-sequencing analysis depicting fold-change expression and significance for all detected genes in human THP-1 cells with stable transduction of two independent RASSF2-targeting shRNAs (shRASSF2, three replicates each) compared to control-targeting shRNA (shCTRL, three replicates). Significantly changed genes are highlighted (downregulated, blue; upregulated, red). c Gene-set enrichment plots as indicated derived from gene expression analysis in b. NES normalized enrichment score, FDR false discovery rate.
Re-expression of RASSF2 is tumor-suppressive specifically in t(8;21) AML
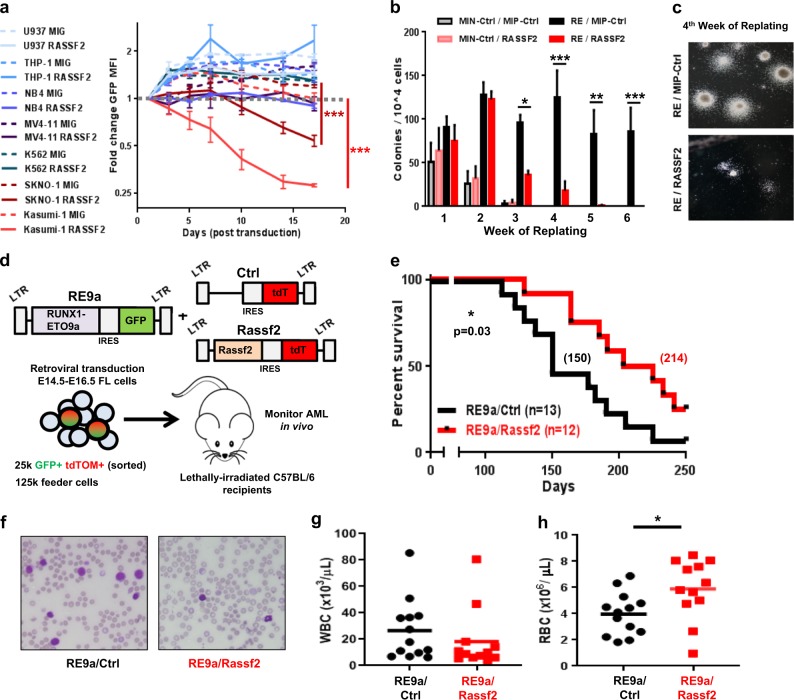
We next explored the functional consequences of transcriptional repression of RASSF2 to leukemogenesis. Using retroviral GFP-based reporter vectors, we screened the effect of RASSF2 expression across a panel of AML cell lines, using change in GFP fluorescence intensity over time as a functional readout for cell proliferation. Only upon expression in the t(8;21) AML cell lines, Kasumi-1 and SKNO-1, did RASSF2 impart a competitive growth disadvantage relative to control-transduced (GFP only) populations (Fig. 3a). Importantly, assessment of protein expression in RASSF2-transduced t(8;21) cell populations showed that re-expression occurred in a physiologically relevant manner and was not overexpressed compared to endogenous amounts observed in non-t(8;21) AML cell lines (Supplementary Fig. 2a). These results suggest that RASSF2 is a context-specific, rather than general, tumor suppressor protein in myeloid leukemia. We next assessed the ability of RASSF2 to suppress RUNX1-ETO-mediated leukemic transformation of primary murine hematopoietic progenitors using a serial replating/colony formation assay (Supplementary Fig. 2b). Re-expression of RASSF2 was sufficient to block RUNX1-ETO-mediated long-term clonogenic self-renewal after 3–4 weeks (Fig. 3b, c). We also assessed RASSF2-mediated tumor suppression in vivo using a murine retroviral transduction and transplantation model of t(8;21) AML with the alternatively spliced leukemogenic protein variant, RUNX1-ETO9a (RE9a, Fig. 3d)40. Co-expression of Rassf2 significantly delayed RE9a leukemia onset (median survival 214 vs 150 days) compared to an empty-vector control (Fig. 3e). The delay in leukemia onset was associated with reduced leukemic burden in the peripheral blood (Fig. 3f, g, Supplementary Figs. 2c, d) and less severe anemia (Fig. 3h, Supplementary Fig. 2e). Terminal assessment of moribund mice revealed AML development in both populations with no significant morphologic differences between cohorts (Supplementary Fig. 2f). These data reveal RASSF2 to be a physiologically relevant tumor suppressor capable of inhibiting t(8;21) AML development both in vitro and in vivo.
Fig. 3. Re-expression of RASSF2 is tumor suppressive specifically in t(8;21) AML.
a Fold-change in population mean GFP fluorescence intensity (MFI) over time was measured by flow cytometry in the indicated cell lines following transduction (efficiencies ~50–60%) with retroviral MSCV-IRES-GFP (MIG), or MSCV-RASSF2-IRES-GFP (RASSF2) vectors. Data are mean ± s.e.m. of four experiments. ***p < 0.001, two-tailed Student’s t-test performed at day 17 for cell lines in which GFP MFI of one vector-transduced population dropped below initial measurement value (day 1 post-transduction), which is indicated by dashed gray line. b Number of colonies (per 10,000 plated cells) for each week of serial replating of primary murine bone marrow cells transduced with vectors as indicated. Data are mean ± s.e.m. of five experiments. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t-test. See also Supplementary Fig. 2b. c Representative images of colonies from fourth week of replating (b) for indicated populations. d Schematic for primary RUNX1-ETO9a retroviral transduction/transplantation murine model of t(8;21) AML with re-expression of Rassf2 (RE9a/Rassf2) or vector control (RE9a/Ctrl). e Survival analysis for experiment described in d; significance determined by log-rank (Mantel-Cox) test. f Representative peripheral blood smears of indicated mice from experiment described in d at 125 days post-transplantation. g, h Peripheral (g) white blood cell (WBC) and (h) red blood cell (RBC) counts of indicated mice from experiment described in d at 125 days post-transplantation, solid lines indicate population mean. *p < 0.05, two-tailed Student’s t-test.
RASSF2 function is independent of nucleo-cytoplasmic shuttling and oncogenic Ras signaling in hematologic cancer
We next sought to explore the mechanism of RASSF2 function in myeloid leukemogenesis. RASSF proteins possess no enzymatic activity and are primarily thought to function as scaffolds through their protein–protein interaction domains15. RASSF2 is reported to exert tumor-suppressive functions as a nucleo-cytoplasmic shuttling protein41. We could not, however, detect any evidence for nuclear localization of endogenous or exogenous RASSF2 in myeloid leukemia cell lines, while still replicating nucleo-cytoplasmic shuttling upon expression in HEK293T cells (Supplementary Figs. 3a–c). These results are consistent with data from the Human Protein Atlas42, in which RASSF2 is exclusively localized to the cytoplasmic/membranous compartments in hematopoietic cells (www.proteinatlas.org). Several RASSFs have been implicated in negative regulation of oncogenic Ras function through the Ras-association (RA) domain14,15,23. Activating point mutations in NRAS and KRAS are frequent in AML and often co-occur with t(8;21)43. Despite this, re-expression of RASSF2 in t(8;21) AML cells did not affect canonical Ras signaling through the MAPK/ERK or AKT pathways following stimulation with cytokines (Supplementary Fig. 3d). Furthermore, we found no correlation between RASSF2 expression and the presence of oncogenic RAS mutations more broadly in AML patients (Supplementary Fig. 3e). These data warranted investigation of RASSF2 function through additional mechanisms.
RASSF2-mediated tumor suppression is dependent on interaction with Hippo kinases MST1/2
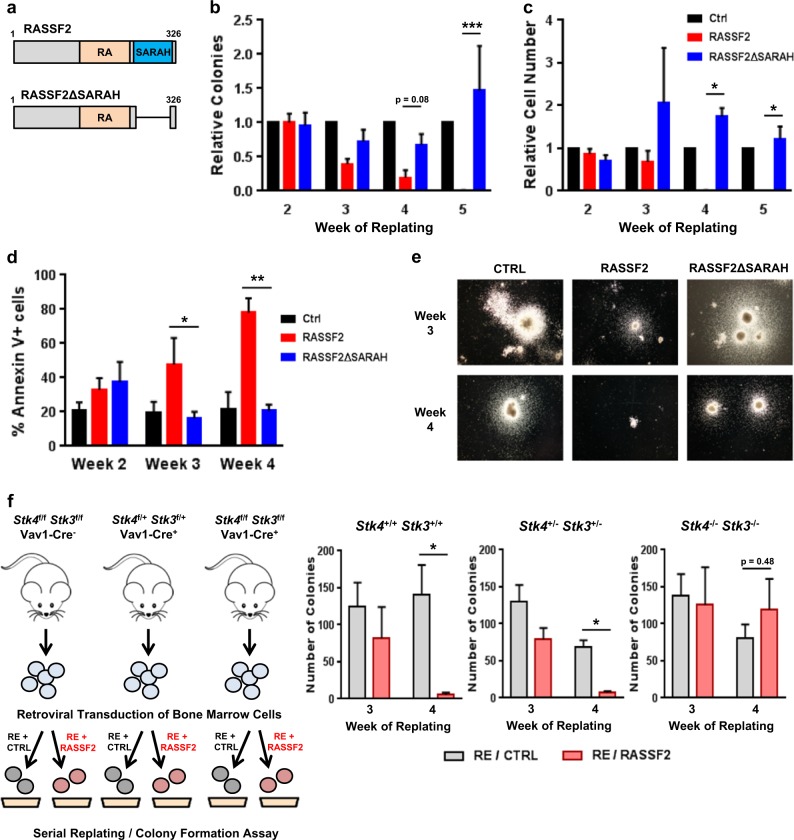
All six classical RASSFs contain a highly-conserved C-terminal Salvador-Rassf-Hippo (SARAH) domain that is unique to protein components of the Hippo signaling pathway and mediates homo- or hetero-dimerization between SARAH domain-containing proteins15. Structural and biochemical analyses suggest that the mammalian Hippo kinases, MST1 and MST2, primarily exist as SARAH-mediated heterodimers with RASSF proteins under basal cellular conditions44. To determine whether interaction with MST1/2 was required for RASSF2 function, we generated a RASSF2 deletion mutant lacking the SARAH domain (RASSF2ΔSARAH) for use in the serial replating/colony formation assay (Fig. 4a, Supplementary Fig. 2b). RASSF2ΔSARAH completely lost the ability to suppress RUNX1-ETO leukemic transformation and induce apoptosis in RUNX1-ETO-expressing cells (Fig. 4b–e). To further confirm the requirement of Hippo kinases for RASSF2-mediated tumor suppression we generated mice with conditional gene inactivation of both MST1 (Stk4) and MST2 (Stk3) in hematopoietic cells (Vav1-Cre). By co-expression of RUNX1-ETO and RASSF2 in the background of control (Vav1-Cre−), heterozygous (Stk4f/+Stk3f/+;Vav1-Cre+), and homozygous (Stk4f/fStk3f/f;Vav1-Cre+) knockout mice, we found that the presence of MST1/2 was essential for the ability of RASSF2 to inhibit RUNX1-ETO leukemic transformation (Fig. 4f). These data revealed the importance of the Hippo kinase-RASSF2 interaction and we set out to further characterize this.
Fig. 4. RASSF2-mediated tumor suppression is dependent on interaction with Hippo kinases MST1/2 in t(8;21) AML.
a Schematic of RASSF2 proteins expressed in experiments b–e, RA, Ras-association domain, SARAH, Salvador-Rassf-Hippo domain. b Colony number (per 10,000 cells plated), c total viable cell number (per 10,000 cells plated), d frequency of Annexin-V+ apoptotic cells, and e representative images of colonies for RUNX1-ETO serial replating/colony formation assay with co-transduction of indicated vectors. Data are normalized relative to empty-vector control-transduced populations and presented as mean ± s.e.m. of four independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t-test. f RUNX1-ETO primary murine serial replating/colony formation assay was performed in mice of three indicated genotypes, with expression of RASSF2 or empty-vector control, as shown in schematic (left). Colony numbers (per 10,000 cells plated) are shown for third and fourth week of replating for each genotype (right). Data are mean ± s.e.m. of three experiments. *p < 0.05, two-tailed Student’s t-test.
RASSF2 is completely dependent on Hippo kinases for stabilization in vitro and in vivo
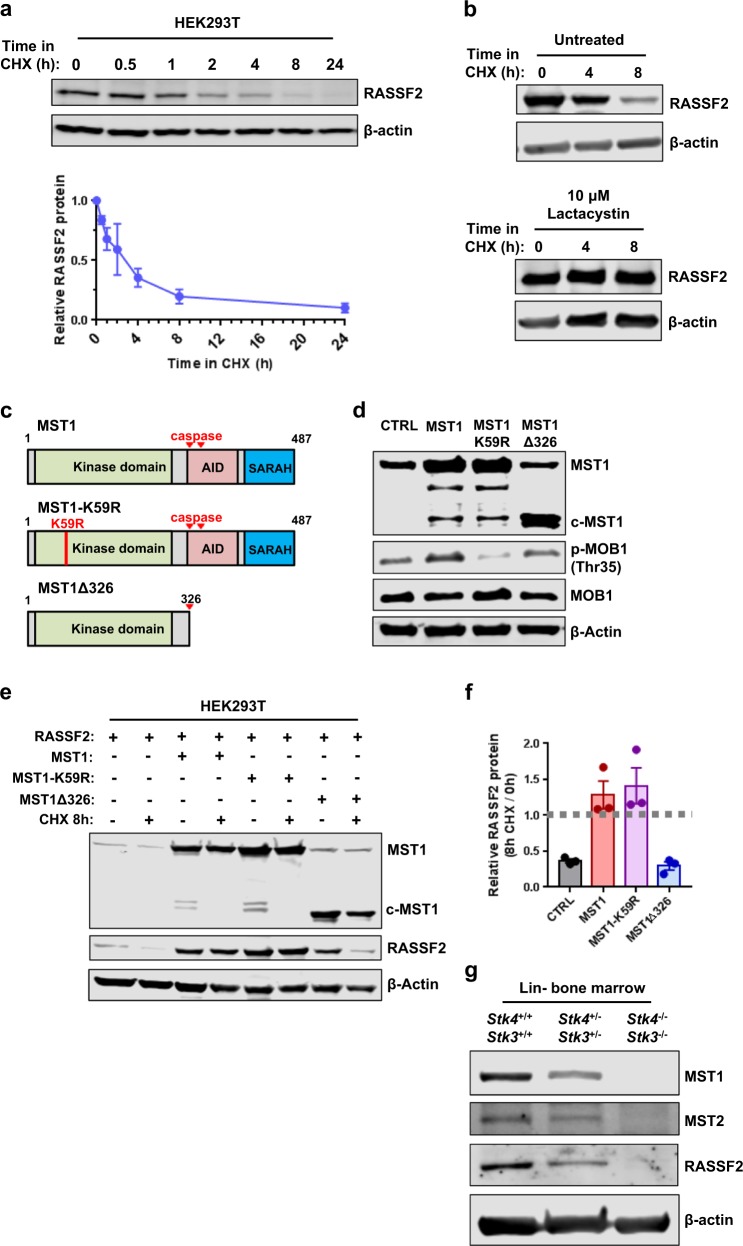
Despite a modest stabilizing effect on MST1 and MST2, RASSF2 expression did not alter signaling through canonical Hippo pathway targets MOB1 and LATS1 (ref. 25) in t(8;21) AML cells (Supplementary Figs. 4a, b). These findings are consistent with another report in which RASSF2 expression did not affect canonical MST1/2 phosphorylation targets24. Therefore, we hypothesized that RASSF2-mediated tumor suppression against RUNX1-ETO-expressing cells may be dependent on SARAH domain-dependent stabilization by the Hippo kinases. Consistent with this, we found RASSF2 protein to have a relatively short half-life of 2–3 h in HEK293T cells in cycloheximide chase assays (Fig. 5a). This degradation was proteasome-dependent (Fig. 5b). Based on this, we further explored the kinetics of RASSF2 protein stability and its relationship with Hippo kinase MST1. To determine whether kinase activity was required for RASSF2 function we utilized a kinase-dead point-mutant protein, MST1-K59R (Fig. 5c). We also included a biologically relevant C-terminal truncation mutant of MST1 that mimics caspase-cleavage at amino acid 326 (MST1Δ326), which is widely accepted to exert increased proapoptotic kinase activity through removal of an auto-inhibitory domain (AID), while also losing its ability to dimerize with RASSF proteins through the SARAH domain (Fig. 5c)45. We confirmed MST1-K59R to act as a dominant negative kinase towards MOB1 phosphorylation (Fig. 5d). We then assessed the ability of MST1 and its protein variants to protect RASSF2 from proteasomal degradation. Consistent with being independent of kinase activity, MST1 and MST1-K59R, but not MST1Δ326, effectively protected RASSF2 from degradation following cycloheximide treatment (Fig. 5e, f). This revealed that the ability of MST1 to interact with RASSF2 through the SARAH domain, rather than its kinase activity towards canonical effectors, contributes to its function in myeloid leukemia cells. To confirm the relevance of this non-canonical Hippo kinase function in vivo we assessed protein expression in murine hematopoietic progenitor cells from conditional knockout mice described above, and found endogenous RASSF2 protein to be completely dependent on the presence of MST1/2 for stabilization (Fig. 5g). These data demonstrate that a previously unappreciated kinase-independent function of the Hippo kinases MST1/2 can contribute to regulation of cancer cell growth through SARAH domain-dependent stabilization of RASSFs.
Fig. 5. RASSF2 is completely dependent on stabilization by Hippo kinases in vitro and in vivo.
a Western blot showing time-course of protein expression following addition of 25 μM cycloheximide (CHX) from HEK293T whole cell lysates beginning 48 h post-transfection with RASSF2 expression vector. Quantification of three experiments is shown below. b Western blot showing time-course of protein expression following addition of 25 μM cycloheximide (CHX) from HEK293T whole-cell lysates beginning 48 h post-transfection with RASSF2 expression vector, with (bottom) or without (top) the addition of 10 μM Lactacystin. Data are representative of three experiments. c Schematic of MST1 protein variants expressed in experiments d–f. K59R, substitution of arginine for lysine at amino acid residue 59 results in kinase-dead protein that functions as dominant negative; AID, auto-inhibitory domain; SARAH, Salvador-Rassf-Hippo domain. Red arrows represent endogenous caspase-cleavage sites. d Western blot showing protein from whole-cell lysates in the SKNO-1 t(8;21) cell line transduced with MST1-variant retroviral expression vectors as indicated. c-MST1 caspase-cleaved MST1. e Western blot showing protein from HEK293T whole-cell lysates 72 h post-transfection with indicated expression vectors, with or without the addition of 25 μM cycloheximide (CHX) for the final 8 h as indicated. Data are representative of three experiments. f Quantification of relative change in RASSF2 protein abundance with or without cycloheximide treatment from three experiments described in e. g Western blot showing protein from whole-cell lysates of Lineage-marker negative (Lin−) primary murine hematopoietic cells derived from bone marrow of mice of indicated genotypes. Cells from three mice per genotype are pooled for analysis.
Proximity-dependent biotin labeling defines the RASSF2-proximal proteome and reveals a role in regulation of Rac GTPase activity in AML
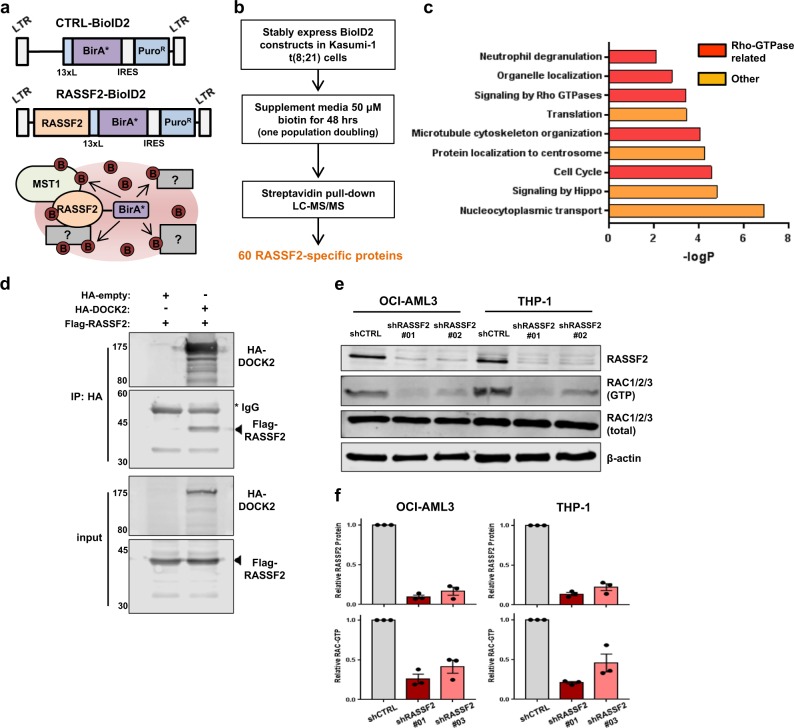
RASSF proteins lack enzymatic activity and instead mediate their functions through various protein–protein interactions. These potential interactions have not previously been characterized in the context of hematologic cancer. Our data revealed RASSF2 to suppress cancer growth in the context of t(8;21) AML so we set out to identify proteins/complexes associated with RASSF2 in leukemia cells. To do this, we employed a proximity-dependent biotin labeling (BioID) approach29 to identify physiologically relevant proximal proteins upon re-expression of RASSF2 in t(8;21) AML cells. We tested three different orientations for expression of the RASSF2-BioID246 fusion cassette, and ultimately chose a C-terminal fusion separated with a linker peptide based on its retained ability to interact with and biotinylate MST1 as a positive control (Fig. 6a, Supplementary Figs. 5a–c). Affinity purification of biotinylated proteins in Kasumi-1 t(8;21) AML cells was performed using steptavidin-conjugated beads and samples were submitted for mass spectrometry analysis following stringent washing conditions (Fig. 6b, Supplementary Fig. 5d). After strict filtering based on the CRAPome47 we identified 60 unique RASSF2-specific proximal proteins across three independent replicates in Kasumi-1 cells (Supplementary Table 2). Hippo kinases MST1 and MST2 were the first- and sixth-ranked hits, respectively, highlighting the accuracy of this approach. Gene ontology (GO) analysis of the full list of proteins revealed RASSF2 to be associated with several pathways and cellular functions related to signaling by Rho-family GTPases (Fig. 6c). This proximal proteome includes general downstream Rho-effectors related to regulation of microtubule polymerization, including PPP1CC, NUDC, SEH1L, STMN1, STMN2, FKBP4, and MAP4, as well as proteins involved in specific-regulation of the localization and activation of Rac-family GTPases, DVL2, ANXA2, and DOCK2. No additional canonical Hippo pathway components or SARAH domain-containing proteins were identified, demonstrating the discrete nature of a Hippo kinase-RASSF2 non-canonical signaling complex in leukemia cells.
Fig. 6. Proximity-dependent biotin labeling (BioID) identifies a role for RASSF2 in regulation of Rac GTPase activation in AML.
a Schematic showing retroviral expression vectors and general assay principle for performing proximity-based biotin labeling and identification using an improved biotin ligase (BioID2, see Methods). b Strategy for performing proximity-based biotin labeling in Kasumi-1 t(8;21) cell line. Experiment was performed in three independent replicates that were submitted for mass-spectrometric analysis, and 60 unique high-confidence RASSF2-proximal proteins were identified across three replicates. c Bar graph showing significantly enriched Gene Ontology (GO) terms associated with the RASSF2-specific protein hits identified by proximity-based biotin labeling. d, Western blot showing interaction between HA-DOCK2 and Flag-RASSF2 as assessed in HEK293T cell lysates by immunoblotting following HA-immunoprecipitation (IP) ~48 h post-transfection with indicated constructs. Inputs are shown below. Molecular weight markers (kDa) are indicated at left. Data are representative of three experiments. e Immunoblotting for active (GTP-bound) Rac by PAK1 pull-down assay performed in two representative AML cell lines 4 days post-transduction with lentiviral shRNAs targeting control sequence (shCTRL) or RASSF2 (shR2#1, shR2#2) as indicated. Data are representative of three experiments. f Quantification of experiments performed in e. Data are normalized relative to β-actin protein abundance and presented as mean ± s.e.m. of three experiments.
One specific identified protein of interest was the Rac1/2-specific atypical guanine nucleotide exchange factor (GEF), DOCK2. DOCK2 functions as an essential positive regulator of Rac1/2 activation in hematopoietic cells by catalyzing nucleotide exchange via a DHR2 domain48. We confirmed the interaction between RASSF2 and DOCK2 via co-immunoprecipitation experiments (Fig. 6d). Based on an association with DOCK2, we examined whether RASSF2 perturbation may have an effect on endogenous Rac GTPase activity in AML. Strikingly, lentiviral shRNA-mediated knockdown of RASSF2 in myeloid leukemia cells with two independent shRNAs was sufficient to profoundly reduce endogenous Rac-GTP to ~ 30% of basal amounts (Fig. 6e, f). These data demonstrate a previously unappreciated link between Hippo kinases, RASSF2, and Rac GTPases in the context of AML.
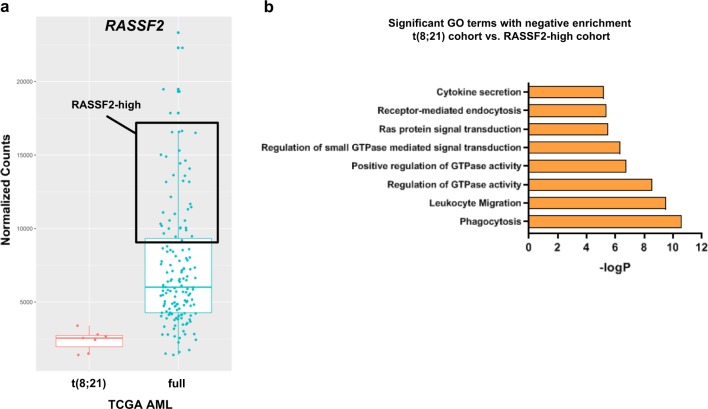
To determine whether this mechanistic finding could be applied more broadly in AML, we utilized published RNA-sequencing data (TCGA cohort)1. Based on the marked RUNX1-ETO-dependent transcriptional repression of RASSF2 (Fig. 1), we hypothesized that comparing gene expression between t(8;21) AML patients and patients with high RASSF2 transcript expression (RASSF2-high) would reveal underlying differences in cellular biology and signal transduction pathways. Indeed, differential expression analysis and subsequent GO enrichment revealed the top-two most significantly e cellular processes in t(8;21) AML patients (relative to the RASSF2-high cohort) to be signatures of phagocytosis and leukocyte migration (Fig. 7a, b), which are both well-established to be dependent on Rac GTPases49,50. Furthermore, several additional significant GO terms directly indicated reduced Rho-family GTPase activation in t(8;21) AML patients compared to the RASSF2-high cohort (Fig. 7b). Together these data demonstrate a meaningful biologic role for RASSF2 expression in regulating Rac GTPase signal transduction and cellular processes in AML.
Fig. 7. RASSF2 expression is broadly predictive of Rac GTPase signal transduction in AML.
a Normalized RASSF2 expression (counts) in t(8;21) AML patient cohort (left) and full TCGA AML cohort (right) showing stratification of patients into two sub-cohorts for purpose of differential gene expression analysis: t(8;21) and RASSF2-high (defined as upper quartile by gene expression). b Bar graph showing top-nine significantly enriched Gene Ontology (GO) terms, with negative enrichment (downregulation) in t(8;21) cohort, as determined by differential gene expression analysis of t(8;21) AML patient cohort vs. RASSF2-high AML patient cohort as defined in a.
Discussion
In summary, we identify RASSF2 as a critical transcriptionally repressed target gene of the RUNX1-ETO fusion protein in t(8;21) AML. RASSF2 is abundantly expressed in normal hematopoietic cell populations and the majority of AML subtypes, but is specifically downregulated in the presence of RUNX1-ETO. RUNX1-ETO directly binds at multiple sites within the RASSF2 genomic locus, and depletion of RUNX1-ETO reproducibly results in upregulation of RASSF2 transcript. This transcriptional repression has downstream functional consequences to t(8;21)-associated leukemogenesis, as RASSF2 knockdown in a non-t(8;21) AML context is sufficient to reproduce an established RUNX1-ETO gene expression signature. Importantly, re-expression of RASSF2 in a physiologically relevant manner specifically inhibits t(8;21) leukemia cell growth, in both in vitro and in vivo assays. These findings not only demonstrate an important role for this specific transcriptional repression event, but also highlight a context-specific nature of RASSF2 in suppression of AML.
RASSF2 and other RASSFs have previously been linked with oncogenic Ras signaling, through the presence of their Ras-association (RA) domain, in a variety of cancer contexts; however, we found no evidence of this in AML. Similarly, we did not find evidence of Ras pathway-associated proteins or GTPases in our proximal biotin labeling experiments. These findings are consistent with Ras-centric proteomic studies that have not identified RASSF proteins as endogenous interactors of either wild type or mutant Ras proteins51. Instead, we here define the RASSF2-proximal proteome in leukemia cells and demonstrate the critical importance of its endogenous interaction with Hippo kinases, MST1 and MST2, through the C-terminal SARAH domain. This interaction is required to protect RASSF2 from proteasomal degradation, which we find evidence for both in vitro and in vivo. Expression of full-length MST1 or a kinase-dead variant (MST1-K59R), but not a C-terminally truncated protein that mimics physiologic caspase-cleavage and lacks the SARAH domain (MST1Δ326), is sufficient for stabilization of RASSF2. These findings suggest the potential for an underappreciated non-canonical signaling mechanism of MST1 and MST2 that functions independently of kinase activity, and instead relies on SARAH domain-dependent stabilization of RASSF2 or other RASSF proteins. Future studies may wish to further characterize this relationship in human cancer.
Through proximity-based biotin labeling (BioID2) and co-immunoprecipitation experiments, we find evidence that RASSF2 interacts with DOCK2 and positively contributes to regulation of Rac GTPase activity mediated by DOCK2 in AML cells. Importantly, several recent studies have revealed a mechanistic link between the Hippo kinases and regulation of Rac GTPase activity in hematopoietic cell populations. For example, MST1 and MST2 have been linked with positive regulation of Rac during the generation of reactive oxygen species and bactericidal activity in macrophages52. MST1 and MST2 were also recently identified in complex with the DOCK family Rac-GEF, DOCK8, in regulatory T cell populations, where they were found to be essential for Rac activation in response to IL-2 stimulation53. Recently, rare families harboring homozygous germline inactivating mutations in STK4 (MST1) have been identified with clinical presentation of a lethal combined immunodeficiency syndrome and autoimmune reactions54,55. Remarkably, there is striking phenotypic overlap between individuals with STK4 deficiency, and individuals with immunodeficiencies caused by homozygous inactivation of the atypical Rac-GEFs, DOCK2 and DOCK8 (refs. 56,57). This is compelling functional evidence in humans supporting a mechanistic link between Hippo kinases and sustained Rac GTPase activation in immune cell populations. Our data suggest an important contribution by RASSF2 in cooperation with DOCK2 to this process in myeloid leukemia cells, which we reveal to have important consequences in AML.
We find that knockdown of RASSF2 in a non-t(8;21) cell context potently reduces GTP-bound Rac, and conversely, Rac GTPase-deficient gene expression signatures are readily apparent in t(8;21) AML patients as compared to a RASSF2 highly expressing cohort. Although the precise mechanisms through which RASSF2 is capable of suppressing t(8;21) AML cell growth remain incompletely understood, our data suggest a role for Rac GTPase signal transduction in this process. The downstream outputs of activated Rac signaling vary depending on the cell type or cancer context and include (1) cell cycle regulation; (2) signaling through JNK/SAPKs, p38-MAPKs, PAKs, the NF-kB pathway, WASP/WAVE complexes, and the BCL-2 family; (3) regulation of actin dynamics during leukocyte migration and phagocytosis; and (4) NADPH oxidase activation for reactive oxygen species (ROS) generation58,59. Interestingly, Rac GTPases have previously been shown to be essential for sustained leukemia cell growth in AML driven by different oncogenes, such as in MLL-rearranged leukemias60, further demonstrating the context-specific nature of RASSF2 function in AML. We propose that one or more of these Rac GTPase signal transduction pathways are uniquely tumor suppressive in the context of t(8;21) AML, and therefore contribute to the importance of transcriptional repression of RASSF2 for this specific leukemia subtype. For example, RUNX1-ETO has previously been shown to independently induce ROS and senescence-like growth arrest61, hinting that additional ROS generation through Rac/NADPH oxidase activity may have detrimental effects on t(8;21) AML cell growth. Future studies may seek to delineate which of these pathways downstream of Rac activation are critical for suppression of t(8;21) AML cell growth. Exploration of potential synthetic lethal genetic interactions in Rac-deficient cells may also be worth consideration for treatment of t(8;21) AML. The findings herein represent a significant step towards a more detailed understanding of mechanisms that uniquely oppose leukemic transformation by the RUNX1-ETO fusion protein in t(8;21) AML. Furthermore we identify a previously unappreciated function of RASSF2 in regulating Rac GTPase signal transduction that is more broadly applicable in AML.
Supplementary information
Acknowledgements
This work is supported by research funding from the National Cancer Institute (NCI) of the National Institutes of Health (NIH) Award Numbers R01CA104509 (to D.E.Z.) and F31CA189452 (to S.A.S.). The authors additionally acknowledge an NIH National Library of Medicine Training Grant T15LM011271 (awarded to M.D.). The authors thank Geoffrey J. Clark (University of Louisville) for providing human RASSF2 cDNA expression plasmids and additional reagents. The authors thank Alex R. Campos and the Sanford Burnham Prebys Proteomics Shared Resource for assistance with proximity-based biotin labeling experiments and mass spectrometry analysis. The authors thank J. Silvio Gutkind and Kun-Liang Guan (UC San Diego Moores Cancer Center) for helpful discussions related to the manuscript.
Author contributions
S.A.S., R.C.D. and D.-E.Z. conceived of the initial study design and experimental plan. S.A.S., K.T.H.L., E.T.A., K.-I.A., A.G.D., S.W. and R.C.D. performed in vitro biological experimentation and interpretation. S.A.S. and M.Y. performed in vivo mouse leukemia modeling and interpretation. S.A.S., M.L., M.D., S.X. and H.C. performed RNA-sequencing and other bioinformatics data analyses. S.A.S. and D.E.Z. prepared the manuscript. All authors approved of final manuscript version.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41408-020-0282-9).
References
- 1.Ley TJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyner JW, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 4.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front. Biosci. 2012;17:1120–1139. doi: 10.2741/3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun XJ, et al. A stable transcription factor complex nucleated by oligomeric AML1-ETO controls leukaemogenesis. Nature. 2013;500:93–97. doi: 10.1038/nature12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ptasinska A, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 2014;8:1974–1988. doi: 10.1016/j.celrep.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Soria N, et al. The oncogenic transcription factor RUNX1/ETO corrupts cell cycle regulation to drive leukemic transformation. Cancer Cell. 2018;34:626–642. doi: 10.1016/j.ccell.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pabst T, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat. Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 11.Vangala RK, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101:270–277. doi: 10.1182/blood-2002-04-1288. [DOI] [PubMed] [Google Scholar]
- 12.Amann JM, et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 2001;21:6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo MC, et al. Combined gene expression and DNA occupancy profiling identifies potential therapeutic targets of t(8;21) AML. Blood. 2012;120:1473–1484. doi: 10.1182/blood-2011-12-395335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim. Biophys. Acta. 2009;2:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Iwasa H, Hossain S, Hata Y. Tumor suppressor C-RASSF proteins. Cell. Mol. Life Sci. 2018;75:1773–1787. doi: 10.1007/s00018-018-2756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dammann R, et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 17.Dammann R, Yang G, Pfeifer GP. Hypermethylation of the cpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 2001;61:3105–3109. [PubMed] [Google Scholar]
- 18.Pefani DE, et al. RASSF1A-LATS1 signalling stabilizes replication forks by restricting CDK2-mediated phosphorylation of BRCA2. Nat. Cell. Biol. 2014;16:962–971. doi: 10.1038/ncb3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt ML, Hobbing KR, Donninger H, Clark GJ. RASSF1A deficiency enhances RAS-driven lung tumorigenesis. Cancer Res. 2018;78:2614–2623. doi: 10.1158/0008-5472.CAN-17-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katagiri K, Imamura M, Kinashi T. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat. Immunol. 2006;7:919–928. doi: 10.1038/ni1374. [DOI] [PubMed] [Google Scholar]
- 21.Song H, et al. Ablation of Rassf2 induces bone defects and subsequent haematopoietic anomalies in mice. EMBO J. 2012;31:1147–1159. doi: 10.1038/emboj.2011.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark J, Freeman J, Donninger H. Loss of RASSF2 enhances tumorigencity of lung cancer cells and confers resistance to chemotherapy. Mol. Biol. Int. 2012;705948:24. doi: 10.1155/2012/705948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vos MD, et al. RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J. Biol. Chem. 2003;278:28045–28051. doi: 10.1074/jbc.M300554200. [DOI] [PubMed] [Google Scholar]
- 24.Song H, Oh S, Oh HJ, Lim DS. Role of the tumor suppressor RASSF2 in regulation of MST1 kinase activity. Biochem. Biophys. Res. Commun. 2010;391:969–973. doi: 10.1016/j.bbrc.2009.11.175. [DOI] [PubMed] [Google Scholar]
- 25.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidenreich O, et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101:3157–3163. doi: 10.1182/blood-2002-05-1589. [DOI] [PubMed] [Google Scholar]
- 27.DeKelver RC, et al. Cooperation between RUNX1-ETO9a and novel transcriptional partner KLF6 in upregulation of Alox5 in acute myeloid leukemia. PLoS Genet. 2013;9:e1003765–e1003765. doi: 10.1371/journal.pgen.1003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoner SA, et al. Hippo kinase loss contributes to del(20q) hematologic malignancies through chronic innate immune activation. Blood. 2019;134:1730–1744. doi: 10.1182/blood.2019000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux KJ, Kim DI, Burke B, May DG. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 2018;91:1–19. doi: 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogilvy S, et al. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94:1855–1863. doi: 10.1182/blood.V94.6.1855. [DOI] [PubMed] [Google Scholar]
- 32.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haferlach T, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hesson LB, et al. CpG island promoter hypermethylation of a novel Ras-effector gene RASSF2A is an early event in colon carcinogenesis and correlates inversely with K-ras mutations. Oncogene. 2005;24:3987–3994. doi: 10.1038/sj.onc.1208566. [DOI] [PubMed] [Google Scholar]
- 35.Beck D, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:2013–2003. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Ami O, et al. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 2013;4:1131–1143. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valk PJ, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 39.Tonks A, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007;21:2495–2505. doi: 10.1038/sj.leu.2404961. [DOI] [PubMed] [Google Scholar]
- 40.Yan M, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 41.Donninger H, et al. The Ras effector RASSF2 controls the PAR-4 tumor suppressor. Mol. Cell. Biol. 2010;30:2608–2620. doi: 10.1128/MCB.00208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 43.Faber ZJ, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat. Genet. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang E, et al. Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway. Acta Crystallogr. D Biol. Crystallogr. 2014;70:1944–1953. doi: 10.1107/S139900471400947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avruch J, et al. Protein kinases of the Hippo pathway: regulation and substrates. Semin Cell Dev. Biol. 2012;23:770–784. doi: 10.1016/j.semcdb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DI, et al. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukui Y, et al. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412:826–831. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- 49.Cox D, et al. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J. Exp. Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cancelas JA, Jansen M, Williams DA. The role of chemokine activation of Rac GTPases in hematopoietic stem cell marrow homing, retention, and peripheral mobilization. Exp. Hematol. 2006;34:976–985. doi: 10.1016/j.exphem.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 51.Kovalski JR, et al. The functional proximal proteome of oncogenic Ras includes mTORC2. Mol Cell. 2019;73:830–844. doi: 10.1016/j.molcel.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geng J, et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat. Immunol. 2015;16:1142–1152. doi: 10.1038/ni.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi H, et al. Hippo kinases Mst1 and Mst2 sense and amplify IL-2R-STAT5 signaling in regulatory T cells to establish stable regulatory activity. Immunity. 2018;49:899–914. doi: 10.1016/j.immuni.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdollahpour H, et al. The phenotype of human STK4 deficiency. Blood. 2012;119:3450–3457. doi: 10.1182/blood-2011-09-378158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nehme NT, et al. MST1 mutations in autosomal recessive primary immunodeficiency characterized by defective naive T-cell survival. Blood. 2012;119:3458–3468. doi: 10.1182/blood-2011-09-378364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q, et al. Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 2009;361:2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobbs K, et al. Inherited DOCK2 Deficiency in Patients with Early-Onset Invasive Infections. N. Engl. J. Med. 2015;372:2409–2422. doi: 10.1056/NEJMoa1413462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kazanietz MG, Caloca MJ. The Rac GTPase in cancer: from old concepts to new paradigms. Cancer Res. 2017;77:5445–5451. doi: 10.1158/0008-5472.CAN-17-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 60.Mizukawa B, et al. Inhibition of Rac GTPase signaling and downstream prosurvival Bcl-2 proteins as combination targeted therapy in MLL-AF9 leukemia. Blood. 2011;118:5235–5245. doi: 10.1182/blood-2011-04-351817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolyniec K, et al. RUNX1 and its fusion oncoprotein derivative, RUNX1-ETO, induce senescence-like growth arrest independently of replicative stress. Oncogene. 2009;28:2502–2512. doi: 10.1038/onc.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.