Abstract
Background
Gouty arthritis is a common metabolic disease caused by long-term purine metabolic disorder and elevated serum uric acid. Jiang-Suan-Chu-Bi recipe (JSCBR), a traditional Chinese herbal formula prescribed according to utilization frequency and cluster analysis, has been clinically validated remedy for gouty arthritis. However, its therapeutic composition and mechanism remains unclear.
Methods
In the present study, a simple, rapid, and sensitive ultraperformance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UHPLC-QTOF-MS)-based chemical profiling was firstly established for comprehensively identifying the major constituents in JSCBR. A phytochemistry-based network pharmacology analysis was further performed to explore the potential therapeutic targets and pathways involved in JSCBR bioactivity. Finally, THP-1 cell model was used to verify the prediction results of network pharmacology by western blot analysis.
Results
A total of 139 compounds containing phenolic acids, flavonoids, triterpenoid saponins, alkaloids, amino acids, fatty acids, anthraquinones, terpenes, coumarins, and other miscellaneous compounds were identified, respectively. 175 disease genes, 51 potential target nodes, 80 compounds, and 11 related pathways based on network pharmacology analysis were achieved. Among these pathways and genes, NOD-like receptor signaling pathway may play an important role in the curative effect of JSCBR on gouty arthritis by regulation of NRLP3/ASC/CASP1/IL1B. The results of cellular and molecular experiments showed that JSCBR can effectively reduce the protein expression of ASC, caspase-1, IL-1β, and NRLP3 in monosodium urate-induced THP-1 cells, which indicated that JSCBR mediated inflammation in gouty arthritis by inhibiting the activation of NOD-like receptor signaling pathway.
Conclusion
Thus, the integrated approaches adopted in the present study could contribute to simplifying the complex system and providing directions for further research of JSCBR.
Keywords: Jiang-Suan-Chu-Bi recipe (JSCBR), gouty arthritis, chemical profile, network pharmacology, NOD-like receptor signaling pathway
Introduction
Gouty arthritis is a metabolic disease caused by the deposition of monosodium urate crystals in joints and soft tissues (Pascart et al., 2018) and closely associated with chronic hyperuricemia, which seriously affects quality of life of patients due to severe pain (Nielsen et al., 2018). Across the globe, the age of onset is getting younger and younger (Zhou et al., 2014). It is estimated that the number of gout patients in China will exceed 100 million in 2020. Now, gout has become the second largest metabolic disease in China (Wilson and Saseen, 2016).
At present, western medicines including colchicine, allopurinol, benzbromarone, and febuxostat have been used as the conventional therapy method for gouty arthritis. Although their short-term effect on inhibiting uric acid production, promoting uric acid excretion, or analgesic effect is optimistic, the side effects such as skin mucosal injury, kidney and liver injury, digestive tract injury, as well as some potential neurotoxicity and muscular toxicity caused by long-term use could not be ignored. Meanwhile, as the main form of traditional Chinese medicine (TCM) that based on the compatibility theory, traditional Chinese herbal formula has been applied for the treatment of gouty arthritis for thousands of years in view of remarkable therapeutic effect and little adverse reactions (Zhou et al., 2014).
Jiang-Suan-Chu-Bi recipe (JSCBR) is a formula prescribed according to utilization frequency and cluster analysis by retrieving gout-related data obtained from multiple databases (Xiao et al., 2019), which has been clinically validated remedy for gouty arthritis under the guidance of TCM doctors. It consists of 12 herbs, namely, Smilax glabra Roxb. (tu-fu-ling in Chinese, TFL, batch No. 20160925), Paeonia lactiflora Pall (chi-shao in Chinese, CS, batch No. 20160814), Reynoutria japonica Houtt. (hu-zhang in Chinese, HZ, batch No. 20160728), Cremastra appendiculata (D.Don) Makino (shan-ci-gu in Chinese, SCG, batch No. 20160723), Rheum officinale Baill. (da-huang in Chinese, DH, batch No. 20160719), Clematis chinensis Osbeck (wei-ling-xian in Chinese, WLX, batch No. 20160915), Angelica pubescens Maxim. (du-huo in Chinese, Dh, batch No. 20160822), Phellodendron chinense C.K.Schneid. (huang-bai in Chinese, HB, batch No. 20160918), Atractylodes lancea (Thunb.) DC. (cang-zhu in Chinese, CZ, batch No. 20160814), Glycyrrhiza glabra L. (gan-cao in Chinese, GC, batch No. 20160816), Codonopsis pilosula (Franch.) Nannf. (dang-shen in Chinese, DS, batch No. 20160723), and Angelica sinensis (Oliv.) Diels (dang-gui in Chinese, DG, batch No. 20160801). Although its prescription is scientific and the efficacy is definite. To date, no reports on the chemical composition and functional mechanisms of JSCBR contribute to its bioactivity has been published, which restrict its further application and development.
In the present study, a systematic dissection of JSCBR was employed by integrating chemical profile, network pharmacology, and experimental support using molecular cell biology. The schematic diagram of the present study was depicted in Figure 1 .
Figure 1.
The schematic diagram of the present study.
Materials and Methods
Chemicals, Reagents, and Materials
UHPLC-MS grade acetonitrile and methanol were supplied by Merck Company Inc., (Darmstadt, Germany). MS grade formic acid was purchased from Fisher Scientific Company (Inc., Fairlawn, NJ). Ultrapure water (18.2 MΩ) was prepared with a Milli-Q water purification system (Millipore, Milford, MA, USA). All other reagents were of analytical grade and purchased from Tianjin Concord Technology Co. Ltd. (Tianjin, China).
The reference compounds ferulic acid (56), polydatin (57), paeoniflorin (79), atractylodin (81), liquiritin (84), aloe-emodin (94), rhein (115), glycyrrhizic acid (119), osthole (126), columbianadin (127), and oleanolic acid (134) were purchased from Chengdu Pufei De Biotech (Chengdu, Sichuan, China). The reference compounds astilbin (60), chysophanol (93), physcion (100), and emodin (122) were purchased from Baoji Herbest Biotech (Baoji, Shanxi, China). The purity of each reference standard was determined to be over 98% by UHPLC analysis. All the 12 herbs of JSCBR were purchased from Beijing Tong-Ren-Tang Technologies Co., Ltd. (Taiyuan, Shanxi Province, China), and authenticated by Professor Yunpeng Diao (College of Pharmacy, Dalian Medical University). Voucher specimens were deposited at the authors’ laboratory.
THP-1 cell was purchased from Yipu Biological Technology Co., Ltd. (Wuhan, Hubei Province, China). RPMI 1640 cell culture mediums and pencillin-streptomycin were purchased from Gibco BRL, Invitrogen Corporation (Grand Island, NY). Fetal bovine serum was purchased from Zhejiang Tianhang Biotechnology Co., Ltd. (Hangzhou, Zhejiang Province, China). Cell Counting Kit -8 (CCK-8) was purchased from Yiyuan Biotechnology Co., Ltd. (Guangzhou, Guangdong Province, China). Phorbol-12-myristate-13-acetate (PMA) was purchased from Abcam Ltd., (Cambridge, UK). Monosodium urate (MSU) was purchased from Sigma-Aldrich (St Louis, MO, USA). Colchicine was supplied by Shengshi Kangpu Chemical Technology Research Institute (Beijing, China). Antibodies against caspase-1, β-actin, apoptosis-associated speck-like protein (ASC), IL-1β, and antirabbit IgG-HRP were purchased from Bioss (Beijing, China). The BCA Protein Quantification Kit was purchased from KeyGen Biotech Co., Ltd. (Nanjing, Jiangsu Province, China). RIPA lysis buffer and PMSF protease inhibitor were purchased from Beyotime Institute of Technology (Shanghai, China). ECL-plus chemiluminescence reagent was purchased from Bio-rad (Richmond, CA, USA).
Preparation of JSCBR Extract
The usage of each crude herb were accurately weighed as follows: Smilacis Glabrae Rhizome (30 g), Paeoniae Radix Rubra (10 g), Polygoni Cuspidati Rhizoma et Radix (15 g), Cremastrae Pseudobulbus (8 g), Rhei Radix et Rhizoma (10 g), Clematidis Radix et Rhizome (15 g), Angelicae Pubescentis Radix (10 g), Phellodendri Chinensis Cortex (10 g), Atractylodis Rhizome (10 g), Glycyrrhizae Radix et Rhizome (10 g), Codonopsis Radix (15 g), and Angelicae Sinensis Radix (10 g). They were mixed and prepared by the decocting method as described below: a total of 153 g mixture was immersed in sevenfold mass of water for 2 h, heated and refluxed for 2 h and then filtered with six-layer absorbent gauze. A fivefold mass of water was subsequently added to the residues and boiled for 2 h. After being filtered with six-layer absorbent gauze, the two filters were combined and concentrated to 150 ml (equal to 1 g crude herb/ml). The liquid was finally transformed into powder by vacuum freeze drying technology.
A 1.0 g of the freeze-dried powder was accurately weighted and extracted with 50 ml of methanol/water (1:1, v/v) for 30 min under ultrasound. The extract solution was centrifuged at 13,000 rpm for 10 min at 4°C and the supernatant was filtered through a 0.22 μm membrane filters. 1.0 μl of filtrate was injected to UHPLC-QTOF-MS for analysis.
UHPLC-QTOF-MS Analysis
The chromatography and MS conditions were almost the same as those reported in literature (Huang et al., 2019). The only difference is the change of elution gradient, which was listed as follows: 0–25 min, 5%–45% B; 25–35 min, 45%–99% B; 35–38 min, 99%–5% B; 38–40 min, 5% B.
Establishment of In-House Library for JSCBR and Generation of Empirical Molecular Formula
The in-house library that covers all previous reported compounds from the 12 formulated herbs was established in a Microsoft office excel table, which includes compound name, molecular formula, molecular weight, chemical structures, natural source, and related references (Chen et al., 2010; Liang et al., 2013; Ge et al., 2014; Ma et al., 2014; Bai et al., 2015; Guo et al., 2015; He et al., 2016; Sun et al., 2016; Liu et al., 2018; Wang et al., 2018; Xu et al., 2018). The empirical molecular formula that matched the criterion for ppm below 10 were deduced as potential candidates by the function “Find Compounds by Formula” of Agilent MassHunter qualitative analysis software. Only those compounds that had been compared with standards or characteristic fragment ions were finally selected as chemical composition of JSCBR.
Network Pharmacology Analysis
Identification of Candidate Targets of Gouty Arthritis
Gouty arthritis related targets were acquired from seven existing resources: 1. TTD database (http://bidd.nus.edu.sg/BIDD-Databases/TTD/TTD.asp); 2. OMIM database (http://omim.org/); 3. PharmGKB database (https://www.pharmgkb.org/); 4. DrugBank database (http://www.drugbank.ca/, version 4.3); 5. GAD database (https://geneticassociationdb.nih.gov/); 6. DisGeNET database (http://www.disgenet.org/web/DisGeNET/menu); 7. GeneCards database (https://www.genecards.org/). We searched these databases and acquired 175 genes totally after removing duplicates. The detailed information is provided in Supplementary Table S1 .
Identification of Compound Targets of JSCBR
After identifying the compounds contained in JSCBR by UHPLC-QTOF-MS, the CAS number and Canonical SMILES were collected by PubChem Compound (https://www.ncbi.nlm.nih.gov/pccompound/). In order to get as many targets as possible, two databases were employed to this study, including Swiss Target Prediction (http://www.swisstargetprediction.ch/) and Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://lsp.nwu.edu.cn/tcmsp.php). Then, the gene information including name, gene ID, and organism was confirmed using UniProt (http://www.uniprot.org/). The final selected genes are supplied in Supplementary Table S2 .
The Protein-Protein Interactions (PPIs) Network Analysis of Disease Targets
The protein-protein interactions (PPIs) network of gouty arthritis was constructed and analyzed by STRING database. Those PPIs with high confidence score (>0.95) should be selected for network construction and analysis to ensure the accuracy of the results.
Network Construction and Analysis
All the networks can be performed by employing the network visualization software Cytoscape 3.2.1. Three networks were constructed as follows: (1) PPIs of gouty arthritis targets; (2) Compound-potential targets-disease network analysis; (3) Pathways-targets network analysis. In these network plots, nodes represent herbs and chemicals, and edges indicate interactions between different chemicals, targets, and pathways. The “degree” of a node was defined as the number of its connected edges.
Enrichment Analysis
To clarify the signaling pathways and functions of potential target genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichments analysis was performed based on Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/home.jsp, ver. 6.8) and STRING.
Experimental Validation Using Molecular Cell Biology
Cell Culture
Human monocytic leukemia THP-1 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml) and streptomycin (100 μg/ml). Cells were maintained in an incubator at 37˚C with 5% CO2 and saturated humidity and the culture medium was replaced with complete culture medium every 2 or 3 days.
CCK-8 Assay for Cell Viability
THP-1 cells were harvested during the logarithmic growth phase and seeded into a 96-well plate at a density of 1x106 cells/well with a final volume of 100 μl. The cells were treated with PMA (100 nM) for 3 h to induce their differentiation into resting M0 macrophage. Fresh complete culture medium was added and cultured for 24 h after washing the cells with PBS solution. Differentiated THP-1 cells were then stimulated with MSU (100, 150, 200, 300, 400, and 500 μg/ml) with optimal concentration and treated with JSCBR extracts at final concentrations of 1, 2, 3, 4, and 5 mg/ml as well as colchicine (positive drug, 2 μg/ml) simultaneously at 37˚C for 24 h (Li et al., 2019). After treatment, 10 μl of CCK-8 was added to each well, and the plates were then incubated for additional 1.5 h. The absorbance at 450 nm was measured using 600 nm as a reference wavelength, which performed on a SpectraMax M2 reader (Molecular Devices, Silicon Valley, USA), and culture medium without cell was used as a blank. The experiments were performed in triplicate and repeated at least twice. The cell viability was calculated using the following formula:
where As, Ab, and Ac represent the absorbance difference of experimental, blank, and control groups, respectively.
Western Blot Analysis
Total protein was extracted by incubation of cell pellet with RIPA lysis buffer and PMSF protease inhibitor. The protein concentration was determined by the BCA Protein Quantification Kit according to the instructions of manufacturer. The cell lysate containing 68 μg of protein was fractionated by 10% SDS-PAGE and then transferred to a poly (vinylidene difluoride) (PVDF) membrane. After blocking with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween-20 at room temperature for 2 h, the membrane was incubated with primary antibodies at 4˚C overnight and horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature, respectively. Protein bands were probed with an ECL Plus chemiluminescence reagent and exposed to a Tanon-5200 Imaging System (Tanon, Shanghai, China).
Statistical Analysis
Data are presented as means ± SD for three independent experiments. Statistical differences between two groups were analyzed using a Student’s t test by GraphPad Prism 5.0 (San Diego, CA, USA). A significant difference was considered as P < 0.05.
Results
Chemical Profile of JSCBR by UHPLC-QTOF-MS
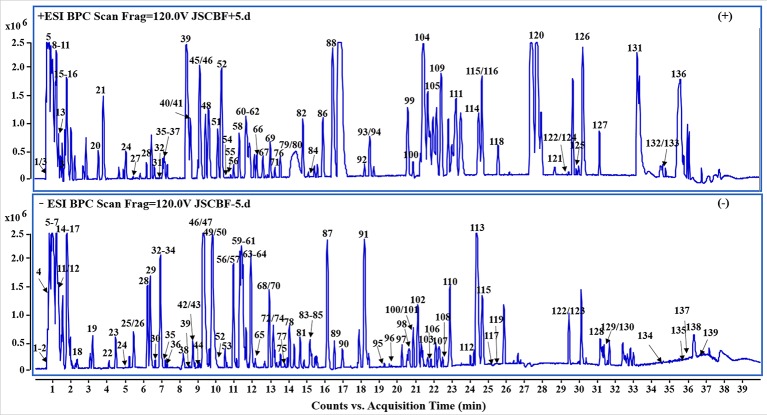
In the present study, a specific UHPLC-QTOF-MS protocol was performed to rapidly identify the compounds of JSCBR as many as possible after optimizing the LC and MS conditions systemically. As a result, a total of 139 compounds, including 39 phenolic acids, 19 flavonoids, 13 triterpenoid saponins, 11 alkaloids, 10 amino acids, 10 fatty acids, 8 anthraquinones, 8 terpenes, 6 coumarins, and 15 miscellaneous compounds were identified or tentatively characterized ( Figure 2 , Table 1 ).
Figure 2.
Representative base peak chromatogram (BPC) of Jiang-Suan-Chu-Bi recipe (JSCBR) in the positive and negative ions mode, respectively.
Table 1.
Characterization of the chemical constituents in Jiang-Suan-Chu-Bi recipe (JSCBR) by ultraperformance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UHPLC-QTOF-MS).
| Peak No | tR (min) | Identification | Formula ion | Negative ion | Positive ion | Source a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Quasi-molecular mass (Da) | Observed mass (Da) | Calculated molecular ion | ppm | Quasi- mass (Da) | Observed mass (Da) | Calculated | ppm | |||||
| 1 | 0.766 | Arginine | C6H14N4O2 | [M-H] – | 173.1044 | 173.1044 | -2.8 | [M+H] + | 175.1191 | 175.1190 | -0.6 | GC/DS/DG |
| 2 | 0.799 | Fructose | C6H12O6 | [M-H] – | 179.0566 | 179.0561 | -2.8 | — | — | — | — | DG |
| 3 | 0.804 | Pulegone | C10H16O | — | — | — | — | [M+Na] + | 175.1098 | 175.1093 | -2.9 | DS |
| 4 | 0.816 | Hypoxanthine | C5H4N4O | [M+CH3COO] – | 195.0517 | 195.0524 | 3.6 | — | — | — | — | DG |
| 5 | 0.853 | Proline | C5H9NO2 | [M-H] – | 114.0539 | 114.0561 | 19.3 | [M+H] + | 116.0694 | 116.0706 | 10.3 | GC/DG |
| 6 | 0.866 | Atractylenolide III sulfate | C15H20O6S | [M+CH3COO] – | 387.1152 | 387.1119 | -8.5 | DS | ||||
| 7 | 1.031 | Malic acid | C4H6O5 | [M-H] – | 133.0148 | 133.0142 | -4.5 | — | — | — | — | CZ |
| 8 | 1.268 | Pyroglutamic acid | C5H7NO3 | — | — | — | — | [M+H] + | 130.0499 | 130.0499 | 0 | GC/SCG |
| 9 | 1.350 | Tyrosine | C9H11NO3 | — | — | — | — | [M+H] + | 182.0794 | 182.0812 | 9.9 | GC/SCG/DG |
| 10 | 1.384 | Adenosine | C10H13N5O4 | — | — | — | — | [M+H] + | 268.1040 | 268.1040 | 0 | Dh/SCG/HB/DS/DG |
| 11 | 1.429 | Guanosine | C10H13N5O5 | [M-H] – | 282.0859 | 282.0844 | -5.3 | [M+H] + | 284.1001 | 284.0989 | -4.2 | DG |
| 12 | 1.479 | Succinic acid | C4H6O4 | [M-H] – | 117.0193 | 117.0193 | 0 | — | — | — | — | DS/SCG/DG |
| 13 | 1.483 | Guanine | C5H5N5O | — | — | — | — | [M+H] + | 152.0558 | 152.0567 | 5.9 | DG |
| 14 | 1.545 | Galloyl glucose | C13H16O10 | [M-H] – | 331.0684 | 331.0671 | -3.9 | — | — | — | — | DH |
| 15 | 1.715 | Leucine | C6H13NO2 | [M-H] – | 130.0874 | 130.0874 | 0 | [M+H] + | 132.1021 | 132.1019 | -1.5 | DG |
| 16 | 1.794 | Gallic acid | C7H6O5 | [M-H] – | 169.0146 | 169.0142 | -2.4 | [M+H] + | 171.0288 | 171.0288 | 0 | CS/DG |
| 17 | 1.976 | Gastrodine | C13H18O7 | [M+HCOO] – | 331.1037 | 331.1035 | -0.6 | — | — | — | — | SCG |
| 18 | 2.357 | Gallic acid -O-diglucoside | C19H26O15 | [M-H] – | 493.1211 | 493.1199 | -2.4 | — | — | — | — | DH |
| 19 | 3.335 | Woodorien | C14H18O9 | [M-H] – | 329.0890 | 329.0878 | -3.6 | — | — | — | — | DS |
| 20 | 3.621 | 9,10-dihydro-2-methyl-anthracen | C15H14 | — | — | — | — | [M+H] + | 195.1169 | 195.1168 | -0.5 | HZ |
| 21 | 3.870 | Codonopsine | C14H21NO4 | — | — | — | — | [M+H] + | 268.1503 | 268.1543 | 14.9 | DS |
| 22 | 4.130 | Catechin 7-O-β-D-glucopyranoside | C21H24O11 | [M-H] – | 451.1258 | 451.1246 | -2.7 | — | — | — | — | DH |
| 23 | 6.383 | Chlorogenic acid | C16H18O9 | [M-H] – | 353.0891 | 353.0878 | -3.7 | — | — | — | — | DG/HB |
| 24 | 5.080 | Tryptophan | C11H12N2O2 | [M-H] – | 203.0832 | 203.0826 | -3 | [M+H] + | 205.0952 | 205.0972 | 9.8 | DS/DG |
| 25 | 5.472 | p-Hydroxybenzyl malonic acid | C10H10O5 | [M-H] – | 209.0462 | 209.0455 | -3.3 | — | — | — | — | GC |
| 26 | 5.472 | Paeonol | C9H10O3 | [M-H] – | 165.0561 | 165.0557 | -2.4 | — | — | — | — | CS |
| 27 | 5.494 | Emodin-O-(O-acetyl)-glucopyranoside | C23H22O11 | — | — | — | — | [M+H] + | 475.1226 | 475.1235 | 1.9 | DH |
| 28 | 6.234 | (-)-epicatechin | C15H14O6 | [M-H] – | 289.0727 | 289.0718 | -3.1 | [M+H] + | 291.0850 | 291.0863 | 4.5 | DH/TFL |
| 29 | 4.478 | Neochlorogenic acid | C16H18O9 | [M-H] – | 353.0891 | 353.0878 | -3.7 | — | — | — | — | HB |
| 30 | 6.649 | digalloyl-glucoside | C27H24O18 | [M-H] – | 635.0905 | 635.0890 | -2.4 | — | — | — | — | DH |
| 31 | 6.654 | Umbelliferone | C9H6O3 | — | — | — | — | [M+H] + | 163.0381 | 163.0390 | 5.5 | Dh |
| 32 | 6.897 | Cryptochlorogenin acid | C16H18O9 | [M-H] – | 353.0891 | 353.0878 | -3.7 | [M+H] + | 355.1017 | 355.1024 | 2 | HB |
| 33 | 6.947 | 3-O-Feruloylquinic acid | C17H20O9 | [M-H] – | 367.1046 | 367.1035 | -3 | — | — | — | — | HB |
| 34 | 6.947 | Berberrubine | C19H16NO4 | [M+HCOO] – | 367.1052 | 367.1061 | 2.5 | — | — | — | — | HB |
| 35 | 7.046 | Caffeic acid | C9H8O4 | [M-H] – | 179.0357 | 179.0350 | -3.9 | [M+H] + | 181.0484 | 181.0495 | 6.1 | DG |
| 36 | 7.212 | Vanillic acid | C8H8O4 | [M-H] – | 167.0354 | 167.0350 | -2.4 | [M+H] + | 169.0490 | 169.0495 | 3 | DG/DS/SCG |
| 37 | 7.267 | O-(O-methoxyphenoxy) phenol | C13H12O3 | — | — | — | — | [M+H] + | 217.0871 | 217.0859 | -5.5 | DS |
| 38 | 8.223 | Isomer of liquiritin-O-glucoside | C32H40O18 | [M-H] – | 711.2143 | 711.2142 | -0.1 | — | — | — | — | GC |
| 39 | 8.510 | (+)-Catechin | C15H14O6 | [M-H] – | 289.0727 | 289.0718 | -3.1 | [M+H] + | 291.0850 | 291.0859 | 4.5 | DG/DH/TFL |
| 40 | 8.643 | (p-Hydroxybenzul)-6,7-dihydroxy- | C23H29NO8 | — | — | — | — | [M+H] + | 448.1957 | 448.1966 | 2 | HB |
| N-metylterahydroisoquinoline-7-O- | ||||||||||||
| β-D-glucopyranoside | ||||||||||||
| 41 | 8.659 | 7-O-Glucoptranoside of 1-(p-hydroxy | C23H30NO8 | — | — | — | — | [M+H]+ | 449.2013 | 449.2044 | 6.9 | HB |
| benzyl)-6,7-dihydroxy-N-methylisoquinoline | ||||||||||||
| 42 | 8.703 | Kaempferol-3-O-rutinose | C27H30O15 | [M-H] – | 593.1541 | 593.1512 | -4.9 | — | — | — | — | DG |
| 43 | 8.803 | Phellodendrine | C20H24NO4 | [M+CI] – | 376.1331 | 376.1321 | -2.7 | — | — | — | — | HB |
| 44 | 9.035 | Hexyl-β-D-glucoside sulfate | C12H24O9S | [M+CH3COO] – | 403.1250 | 403.1280 | 7.4 | — | — | — | — | DS |
| 45 | 9.057 | Apigenin-6,8-di-C-β-D-glucopyranoside | C27H30O15 | — | — | — | — | [M+H]+ | 595.1645 | 595.1657 | 2 | GC |
| 46 | 9.140 | Tetrehydropalmatrubine | C20H23NO4 | [M-H] – | 340.1497 | 340.1554 | 16.8 | [M+H]+ | 342.1690 | 342.1700 | 2.9 | HB |
| 47 | 9.233 | Amurenlaetone B | C17H20O9 | [M-H] – | 367.1045 | 367.1035 | -2.7 | — | — | — | — | HB |
| 48 | 9.538 | 1-O-galloyl-2-O-cinnamoyl-β-D-glucose | C22H22O11 | — | — | — | — | [M+H] + | 463.1225 | 463.1235 | 2.2 | DH |
| 49 | 9.797 | Albiflorin | C23H28O11 | [M+HCOO] – | 525.1634 | 525.1614 | -3.8 | — | — | — | — | CS/DG |
| 50 | 9.797 | Benzoylpaeoniflorin | C24H24O10 | [M+HCOO] – | 517.1324 | 517.1351 | 5.2 | — | — | — | — | DG |
| 51 | 10.151 | 4-Hydroxy-3-methyl-acetophenone | C9H10O2 | — | — | — | — | [M+H] + | 151.0740 | 151.0754 | 9.3 | CS/Dh |
| 52 | 10.350 | Isoliensinine | C19H23NO3 | [M-H] – | 312.1613 | 312.1605 | -2.6 | [M+H] + | 314.1743 | 314.1751 | 2.5 | HB |
| 53 | 10.592 | Galloylpaeoniflorin | C34H28O22 | [M-H] – | 787.1008 | 787.0999 | -1.1 | — | — | — | — | CS |
| 54 | 10.625 | Cyclohexanecarboxylic acid | C17H20O9 | — | — | — | — | [M+H] + | 369.1165 | 369.1180 | 4.1 | HB |
| 55 | 10.830 | Astragaloside V/VI/VII | C26H28O13 | — | — | — | — | [M+H] + | 549.1595 | 549.1603 | 1.5 | DG |
| 56 b | 10.973 | Ferulic acid | C10H10O4 | [M-H] – | 193.0510 | 193.0506 | -2.1 | [M+H] + | 195.0644 | 195.0652 | 4.1 | DG/Dh/HB/DS |
| 57 b | 10.975 | Polydatin | C20H22O8 | [M-H] – | 389.1258 | 389.1242 | -4.1 | — | — | — | — | HZ |
| 55 | 10.830 | Astragaloside V/VI/VII | C26H28O13 | — | — | — | — | [M+H] + | 549.1595 | 549.1603 | 1.5 | DG |
| 56 b | 10.973 | Ferulic acid | C10H10O4 | [M-H] – | 193.0510 | 193.0506 | -2.1 | [M+H] + | 195.0644 | 195.0652 | 4.1 | DG/Dh/HB/DS |
| 57 b | 10.975 | Polydatin | C20H22O8 | [M-H] – | 389.1258 | 389.1242 | -4.1 | — | — | — | — | HZ |
| 58 | 11.029 | Atractylenolide III | C15H20O3 | — | — | — | — | [M+H] + | 249.1469 | 249.1485 | 6.4 | CZ/DS |
| 59 | 11.404 | Raffinose | C18H32O16 | [M+HCOO] – | 549.1632 | 549.1672 | 7.3 | — | — | — | — | DS |
| 60 b | 11.470 | Astilbin | C21H22O11 | [M-H] – | 449.1109 | 449.1089 | -4.5 | [M+H] + | 451.1228 | 451.1235 | 1.6 | TFL |
| 61 | 11.570 | Cinnamic acid | C9H8O2 | [M-H] – | 147.0450 | 147.0452 | 1.4 | [M+H] + | 149.0588 | 149.0597 | 6 | SCG |
| 62 | 11.725 | Liquiritigenin | C15H12O4 | — | — | — | — | [M+H] + | 257.0800 | 257.0808 | 3.1 | GC |
| 63 | 11.818 | (+/-)-8-(4-Hydroxy-3-methoxyphenyl)- | C27H36O13 | [M-H] – | 567.2095 | 567.2083 | -2.1 | — | — | — | — | HB |
| 6,7-bis(hydroxymethyl)-3-methoxy-5,6, | ||||||||||||
| 7,8-tetrahydro-2-naphthalenyl-β-D- | ||||||||||||
| glucopyranoside | ||||||||||||
| 64 | 12.017 | Ferulic acid-1-O-glucoside | C16H20O9 | [M-H] – | 355.1047 | 355.1035 | -3.4 | — | — | — | — | DH |
| 65 | 12.166 | Galloylpaeoniflorinb | C30H32O15 | [M-H] – | 631.1678 | 631.1668 | -1.6 | — | — | — | — | DG |
| 66 | 12.289 | Taxifolin | C15H12O7 | — | — | — | — | [M+H] + | 305.0645 | 305.0656 | 3.6 | TFL |
| 67 | 12.769 | Galloyl cinnamoyl-glucose | C22H22O11 | — | — | — | — | [M+H] + | 463.1229 | 463.1235 | 1.3 | DH |
| 68 | 13.078 | Quercitrin | C21H20O11 | [M-H] – | 447.0944 | 447.0933 | -2.5 | — | — | — | — | TFL |
| 69 | 13.084 | Oxidized jatrorrhizine | C20H17NO5 | — | — | — | — | [M+H] + | 352.1169 | 352.1179 | 2.8 | HB |
| 70 | 13.160 | Laccaic acid D-O-glucose | C22H20O12 | [M-H] – | 475.0897 | 475.0882 | -3.2 | — | — | — | — | DH |
| 71 | 13.283 | Isomer of liquiritin apioside | C25H24O11 | — | — | — | — | [M+H] + | 501.1400 | 501.1391 | -1.8 | GC |
| 72 | 13.310 | Engeletin | C21H22O10 | [M-H] – | 433.1152 | 433.1140 | -2.8 | — | — | — | — | TFL |
| 73 | 13.343 | Procyanidine/isomer | C16H20O9 | [M-H] – | 355.1047 | 355.1035 | -3.4 | — | — | — | — | DH |
| 74 | 13.359 | Resveratrol-4-O-β-D-(2′-O-galloyl)- | C27H26O12 | [M-H] – | 541.1367 | 541.1351 | -3 | — | — | — | — | DH |
| glucopyranoside or resveratrol-4-O-β- | ||||||||||||
| D-(6′-O-galloyl)-glucopyranoside | ||||||||||||
| 75 | 13.558 | Dactylorhin A | C40H56O22 | [M-H] – | 887.3196 | 887.3190 | -0.7 | — | — | — | — | SCG |
| 76 | 13.581 | Columbianetin | C14H14O4 | — | — | — | — | [M+H] + | 247.0956 | 247.0965 | 3.6 | Dh |
| 77 | 13.790 | Azelaic acid | C9H16O4 | [M-H] – | 187.0982 | 187.0976 | -3.2 | — | — | — | — | DS |
| 78 | 13.989 | (6aR, 11aR)-3-hydroxy-9, 10-dimethoxy- | C25H24O12 | [M-H] – | 515.1209 | 515.1195 | -2.7 | — | — | — | — | DG |
| pterocarpan-3-O-β-D-glc-6′′-O-malonate | ||||||||||||
| 79 b | 14.394 | Paeoniflorin | C23H28O11 | — | — | — | — | [M+H] + | 481.1570 | 481.1704 | 27.8 | CS |
| 80 | 14.510 | 3-methoxy-4-hydroxyphenylethanol | C9H12O3 | — | — | — | — | [M+Na] + | 191.0692 | 191.0679 | -6.8 | SCG |
| 81 b | 14.718 | Atractylodin | C13H10O | [M+HCOO] – | 227.0721 | 227.0714 | -3.1 | — | — | — | — | CZ |
| 82 | 14.990 | Aloe-emodin-8-O-β-D-glucopyranoside | C21H20O10 | — | — | — | — | [M+Na] + | 455.0948 | 455.0949 | 0.2 | DH |
| 83 | 15.182 | 1-O-galloyl-2-O-cinnamoyl-β-D-glucose | C22H22O11 | [M-H] – | 461.1105 | 461.1089 | -3.5 | — | — | — | — | DH |
| 84 b | 15.265 | Liquiritin | C21H22O9 | [M-H] – | 417.1157 | 417.1191 | 8.2 | [M+H] + | 419.1332 | 419.1337 | 1.2 | GC |
| 85 | 15.281 | Acetyl-liquiritin/isoliquiritin | C23H24O10 | [M-H] – | 459.1306 | 459.1297 | -2 | — | — | — | — | GC |
| 86 | 15.935 | Ononin | C22H22O9 | — | — | — | — | [M+H] + | 431.1306 | 431.1337 | 7.2 | DG/GC |
| 87 | 16.160 | Licorice glycoside B | C35H36O15 | [M-H] – | 695.1989 | 695.1981 | -1.2 | — | GC | |||
| 88 | 16.482 | Hexyl-b-D-glucopyranosyl-(1/6)-β-D- | C18H34O11 | — | — | — | — | [M+H] + | 427.2146 | 427.2174 | 6.8 | DS |
| glucopyranoside | ||||||||||||
| 89 | 16.524 | Militarine | C34H46O17 | [M-H] – | 725.2670 | 725.26662 | -1.1 | — | — | — | — | SCG |
| 90 | 16.988 | Cinnamic-O-galloyl-glucoside | C22H22O11 | [M-H] – | 461.1104 | 461.1089 | -3.3 | — | — | — | — | DH |
| 91 | 18.181 | Isoengeletin | C21H21O10 | [M-H] – | 432.1029 | 432.1062 | 7.6 | — | — | — | — | TFL |
| 92 | 18.222 | Senkyunolide H | C12H16O4 | — | — | — | — | [M+Na] + | 247.0958 | 247.0941 | -6.9 | DG |
| 93 b | 18.413 | Chrysophanol | C15H10O4 | [M-H] – | 253.0509 | 253.0506 | -1.2 | [M+H] + | 255.0642 | 255.0652 | 3.9 | DH/SCG |
| 94 b | 18.520 | Aloe-emodin | C15H10O5 | — | — | — | — | [M+H] + | 271.0596 | 271.0601 | 1.8 | DH |
| 95 | 19.291 | 22-Acetoxyl licorice-saponin G2 | C44H64O19 | [M-H] – | 895.3978 | 895.3969 | -1 | — | — | — | — | GC |
| 96 | 19.556 | 22-Hydroxy-licorice-saponin G2 | C42H62O18 | [M-H] – | 853.3871 | 853.3863 | -0.9 | — | — | — | — | GC |
| 97 | 20.269 | Licorice-saponin A3 (or isomer) | C48H72O21 | [M-H] – | 983.4492 | 983.4493 | 0.1 | — | — | — | — | GC |
| 98 | 20.551 | Calycosin-7-O-β-D-glucoside | C22H22O10 | [M-H] – | 445.1148 | 445.1140 | -1.8 | — | — | — | — | DG/GC |
| 99 | 20.608 | N-Methylcanadine | C21H24NO4 | — | — | — | — | [M+Na] + | 377.1591 | 377.1598 | 1.9 | HB |
| 100 b | 20.857 | Physcion | C16H12O5 | [M-H] – | 283.0619 | 283.0612 | -2.5 | [M+H] + | 285.0751 | 285.0757 | 2.1 | DH/SCG |
| 101 | 20.865 | 22β-Acetoxyl-glycyrrhizin | C44H64O18 | [M-H] – | 879.4027 | 879.4020 | -0.8 | — | — | — | — | GC |
| 102 | 21.130 | Uralsaponin E | C42H60O17 | [M-H] – | 835.3799 | 835.3758 | -4.9 | — | — | — | — | GC |
| 103 | 21.313 | Uralsaponin U/Uralsaponin N | C42H62O17 | [M-H] – | 837.3922 | 837.3914 | -1 | — | — | — | — | GC |
| 104 | 21.470 | (6aR, 11aR)-3-Hydroxy-9, 10- | C17H16O5 | — | — | — | — | [M+H] + | 301.1070 | 301.1071 | 0.3 | DG |
| dimethoxypterocarpan | ||||||||||||
| 105 | 21.719 | Phellodenol D | C17H18O5 | — | — | — | — | [M+H] + | 303.1230 | 303.1227 | -1 | HB |
| 106 | 21.876 | Shancigusin H | C49H62O23 | [M-H] – | 1017.3613 | 1017.3609 | -0.4 | — | — | — | — | SCG |
| 107 | 22.307 | Laccaic acid D-8-O-(6′-O-cinnamoyl)- | C31H26O13 | [M-H] – | 605.1307 | 605.1301 | -1 | — | — | — | — | DH |
| glucopyranoside | ||||||||||||
| 108 | 22.539 | (6R, 9S)-3-Oxo-α-ionol-β-D- | C19H30O10S | [M-H] – | 449.1469 | 449.1487 | 4 | — | — | — | — | DS |
| glucopyranoside sulfate | ||||||||||||
| 109 | 22.597 | Bicyclo [3.1.0] hexan-3-one | C12H18O2 | — | — | — | — | [M+H] + | 195.1366 | 195.1380 | 7.2 | Dh |
| 110 | 22.920 | Licorice-saponin G2 | C42H62O17 | [M-H] – | 837.3932 | 837.3914 | −2.1 | — | — | — | — | GC |
| 111 | 23.244 | (+) N-methylcorydine | C21H26NO4 | — | — | — | — | [M+Na] + | 379.1740 | 379.1754 | 3.7 | HB |
| 112 | 24.014 | 22β-Acetoxy-licorice-saponin B2 | C44H66O17 | [M-H] – | 865.4229 | 865.4227 | −0.2 | — | — | — | — | GC |
| 113 | 24.279 | Macedonoside C | C42H62O16 | [M-H] – | 821.3972 | 821.3965 | −0.9 | — | — | — | — | GC |
| 114 | 24.487 | Obaculactone | C26H30O8 | — | — | — | — | [M+H] + | 471.2009 | 471.2013 | 0.8 | HB |
| 115 b | 24.676 | Rhein | C15H8O6 | [M-H] – | 283.0258 | 283.0248 | −3.5 | [M+H] + | 285.0390 | 285.0394 | 1.4 | DH |
| 116 | 24.702 | Glycyrrhetic acid | C30H46O4 | — | — | — | — | [M+H] + | 471.3467 | 471.3469 | 0.4 | GC |
| 117 | 25.588 | Glycyrflavoside C | C42H64O15 | [M-H] – | 807.4180 | 807.4172 | −1 | — | — | — | — | GC |
| 118 | 25.614 | trans-Resveratrol | C14H12O3 | — | — | — | — | [M+H] + | 229.0857 | 229.0859 | 0.9 | TFL/HZ |
| 119 b | 25.869 | Glycyrrhizic acid | C42H62O16 | [M-H] – | 821.3975 | 821.3965 | −1.2 | — | — | — | — | GC |
| 120 | 27.652 | Phytosphingosine | C18H39NO3 | — | — | — | — | [M+H] + | 318.2991 | 318.3003 | 3.8 | DG |
| 121 | 28.829 | Octanal | C8H16O | — | — | — | — | [M+H] + | 129.1266 | 129.1274 | 6.2 | DS/Dh |
| 122 b | 29.448 | Emodin | C15H10O5 | [M-H] – | 269.0465 | 269.0455 | −3.7 | [M+H] + | 271.0596 | 271.0601 | 1.8 | DS/DH/SCG/HZ |
| 123 | 29.448 | Z-11-hexadecenoic acid | C16H30O2 | [M+CH3COO] – | 313.2393 | 313.2384 | −2.9 | — | — | — | — | Dh |
| 124 | 29.458 | 2, 6-dimethoxy-4-(2’-propenyl) phenol | C11H14O3 | — | — | — | — | [M+H] + | 195.1016 | 195.1016 | 0 | DS |
| 125 | 29.889 | Ligustilide | C12H14O2 | — | — | — | — | [M+H] + | 191.1063 | 191.1067 | 2.1 | DG |
| 126 b | 30.254 | Osthole | C15H16O3 | — | — | — | — | [M+H] + | 245.1173 | 245.1172 | −0.4 | DG/Dh |
| 127 b | 31.116 | Columbianadin | C19H20O5 | — | — | — | — | [M+H] + | 329.1387 | 329.1384 | −0.9 | Dh |
| 128 | 31.271 | 9-Tetradecenoic acid | C14H26O2 | [M+CH3COO] – | 285.2081 | 285.2071 | −3.5 | — | — | — | — | Dh |
| 129 | 31.652 | (E)-2-Hexenyl-β-D-glucopyranosyl- | C18H32O11 | [M-H] – | 423.1860 | 423.1872 | 2.8 | — | — | — | — | DS |
| (1/2)-β-D-glucopyranoside | ||||||||||||
| 130 | 31.917 | α-Selinene | C14H20 | [M+HCOO] – | 233.1554 | 233.1547 | −3 | — | — | — | — | CZ |
| 131 | 33.353 | Dibutylphthalat | C16H22O4 | — | — | — | — | [M+H] + | 279.1598 | 279.1591 | −2.5 | HZ |
| 132 | 34.563 | Citronellyl acetate | C12H22O2 | — | — | — | — | [M+H] + | 199.1698 | 199.1693 | −2.5 | CZ |
| 133 | 34.646 | Litcubine | C19H22NO4 | — | — | — | — | [M+H] + | 329.1605 | 329.1622 | 5.2 | HB |
| 134 b | 34.834 | Oleanolic acid | C30H48O3 | [M-H] – | 455.3532 | 455.3531 | −0.2 | — | — | — | — | WLX |
| 135 | 35.546 | Tetradecanoic acid | C14H28O2 | [M-H] – | 227.2022 | 227.2017 | −2.2 | — | — | — | — | DS |
| 136 | 35.624 | 1,2-Benzenedicarboxylic acid | C24H38O4 | — | — | — | — | [M+H] + | 391.2852 | 391.2843 | −2.3 | DS |
| diisooctyl ester | ||||||||||||
| 137 | 36.192 | Linoleate | C18H32O2 | [M-H] – | 279.2340 | 279.2330 | −3.6 | — | — | — | — | DG |
| 138 | 36.325 | Octadecanoic acid | C18H36O2 | [M-H] – | 283.2650 | 283.2643 | −2.5 | — | — | — | — | DG |
| 139 | 37.170 | Hexadecanoic acid | C16H32O2 | [M-H] – | 255.2340 | 255.2330 | −3.9 | — | — | — | — | DS/DG/Dh/SCG |
a TFL, Smilacis Glabrae Rhizome; CS, Paeoniae Radix Rubra; HZ, Polygoni Cuspidati Rhizoma et Radix; SCG, Cremastrae Pseudobulbus; DH, Rhei Radix et Rhizoma; WLX, Clematidis Radix et Rhizome; Dh, Angelicae Pubescentis Radix; HB, Phellodendri Chinensis Cortex;CZ, Atractylodis Rhizome; GC, Glycyrrhizae Radix et Rhizome; DS, Codonopsis Radix; DG, Angelicae Sinensis Radix.
b Components identified with reference compounds comparison.
Among them, 15 compounds (compounds 56, 57, 60, 79, 81, 84, 93, 94, 100, 115, 119, 122, 126, 127, and 134) were identified as ferulic acid, polydatin, astilbin, paeoniflorin, atractylodin, liquiritin, chysophanol, aloe-emodin, physcion, rhein, glycyrrhizic acid, emodin, osthole, columbianadin, and oleanolic acid by comparing the retention time, quasi-molecular ions with authentic standards, respectively. While the others were tentatively deduced based on their high-accurate quasi-molecular ion such as [M−H]−, [M+HCOO]−, [M+CH3COO]−, [M]+, [M+H]+, and [M+Na]+ with those of the known published compounds recorded in the in-house library. Information regarding the 139 constituents, such as tR (min), identification, formula, negative ion (m/z), positive ion (m/z), and botanical source, is offered in Table 1 .
Network Pharmacology Analysis
Gouty Arthritis-Related Targets Network Analysis
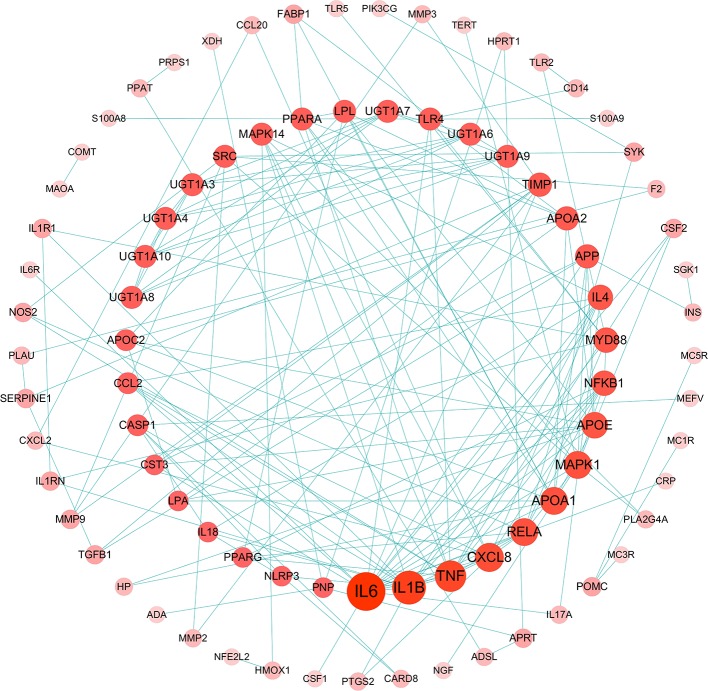
The relationship among 175 disease genes from PPI were subjected to STRING. And a gene-gene interaction network was accordingly achieved. Then, disease targets of PPI with high confidence score (>0.95) were screened for network construction and analysis ( Figure 3 ), 84 nodes and 171 edges were involved in this network. Among them, the central part notes connected by more than 4 edges, such as 19 in IL-6, 15 in IL-1β, 5 in CASP1, and 4 in NLRP3. It implies that these genes might be key nodes in this network.
Figure 3.
Gouty arthritis-related targets of protein-protein interaction (PPI) network (Confidence score >0.95, Node size represent the degree).
Potential Target Genes and Network Construction
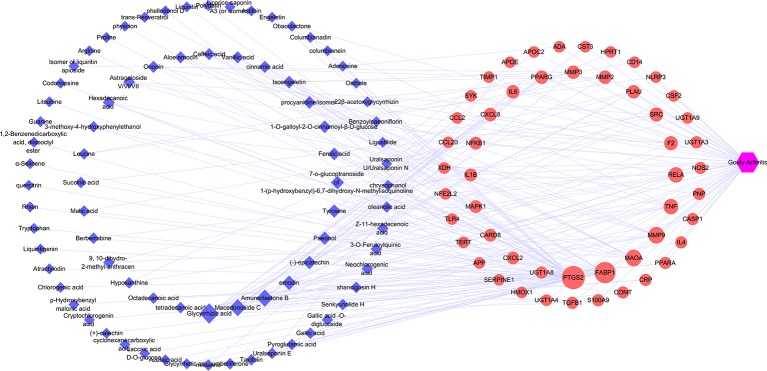
The target genes related to gouty arthritis and compounds were got from associated databases, and 51 overlapping genes were identified by matching the aforementioned genes. Then compound-potential targets-disease network constructed by Cytoscape was shown in Figure 4 , which comprise of 132 nodes (1 disease node, 51 potential target nodes, and 80 compound nodes) and 236 edges. From this network, we can conclude that glycyrrhizic acid, amurenlaetone B, and macedonoside C may be the main components of JSCBR in treating gouty arthritis due to higher degree.
Figure 4.
Compound-potential targets-disease network (Blue diamond represents compound target; red circle represents pathway; purple hexagon represents disease).
Potential Target Gene-Related Pathway Analysis
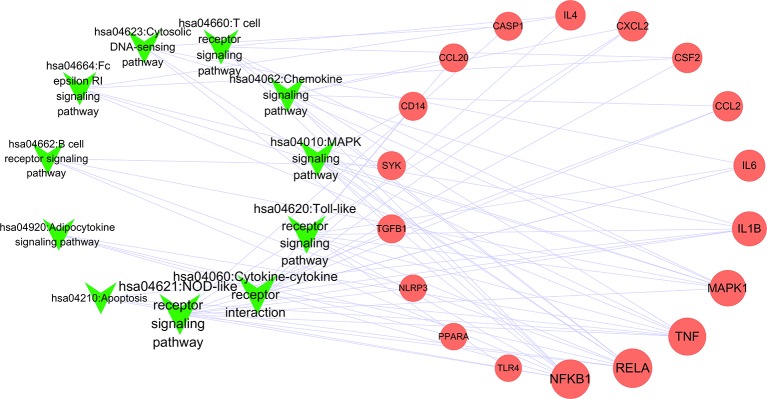
To better understand the signaling pathways and functions of these potential target genes, we performed functional enrichment analysis using KEGG pathways. The identified pathways and genes in which they are involved are shown in Figure 5 and Table 2 (p < 0.05). NOD-like receptor signaling pathway (hsa04621) is ranked first, which has 10 genes involved. In our present research, NRLP3/ASC/CASP1/IL1B was selected for experimental verification.
Figure 5.
Network of pathways and related genes (Green “V” represents pathways; red circle represents pathway; node size represents the degree).
Table 2.
Functions of potential target genes based on KEGG pathway analysis.
| No | Term | P Value | Genes |
|---|---|---|---|
| 1 | hsa04621: NOD-like receptor signaling pathway | 1.24E-11 | MAPK1, CARD8, IL6, TNF, CCL2, RELA, CXCL2, IL1B, NFKB1, CASP1, NLRP3 |
| 2 | hsa04620: Toll-like receptor signaling pathway | 7.54E-06 | MAPK1, IL6, TNF, RELA, IL1B, NFKB1, TLR4, CD14 |
| 3 | hsa04060: Cytokine-cytokine receptor interaction | 5.65E-04 | IL4, CSF2, IL6, TNF, CCL2, CCL20, CXCL2, IL1B, TGFB1 |
| 4 | hsa04623: Cytosolic DNA-sensing pathway | 6.88E-04 | IL6, RELA, IL1B, NFKB1, CASP1 |
| 5 | hsa04660: T cell receptor signaling pathway | 0.0011312 | IL4, CSF2, MAPK1, TNF, RELA, NFKB1 |
| 6 | hsa04664: Fc epsilon RI signaling pathway | 0.0025462 | IL4, CSF2, MAPK1, TNF, SYK |
| 7 | hsa04062: Chemokine signaling pathway | 0.011938 | MAPK1, CCL2, CCL20, RELA, CXCL2, NFKB1 |
| 8 | hsa04010: MAPK signaling pathway | 0.0132372 | MAPK1, TNF, RELA, IL1B, NFKB1, CD14, TGFB1 |
| 9 | hsa04920: Adipocytokine signaling pathway | 0.0132749 | PPARA, TNF, RELA, NFKB1 |
| 10 | hsa04662: B cell receptor signaling pathway | 0.0179629 | MAPK1, RELA, NFKB1, SYK |
| 11 | hsa04210: Apoptosis | 0.0265251 | TNF, RELA, IL1B, NFKB1 |
Experimental Validation Using Molecular Cell Biology
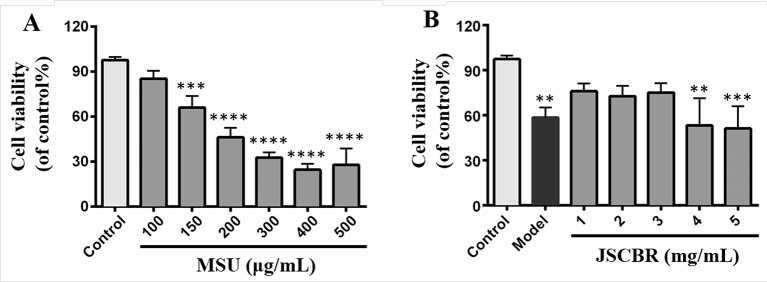
Effect of JSCBR Extracts on Cell Viability
The induction effect of MSU was evaluated in vitro with a CCK-8 assay and shown in Figure 6A . Notably, MSU acted as the strongest inducer decreased the viabilities of THP-1 cell in a concentration-dependent manner. Since the relatively low viability was observed in cells exposed to MSU with dosages greater than or equal to 200 μg/ml, 150 μg/ml was the optimum induction dosage in further experiments. For antiinflammatory activity, THP-1 cells treated with JSCBR extracts from 1 to 5 mg/ml exhibited viability of 67.8%~80.6% ( Figure 6B ). Since no further antiproliferation effect was observed in cells exposed to 4 and 5 mg/ml, the concentrations of extracs were defined to 1, 2, and 3 mg/ml for western Blot verification.
Figure 6.
Effects of Jiang-Suan-Chu-Bi recipe (JSCBR) extracts on monosodium urate (MSU)-induced THP-1 cell viability. (A) THP-1 cells were exposed to MSU at various concentrations for 24 h. (B) Protective effects of JSCBR extracts on the viabilities of MSU-induced THP-1 cells. Cell viability was assessed by CCK-8 assay and expressed relative to untreated control cells. **p < 0.01, ***p < 0.001, ****p < 0.0001 versus control group.
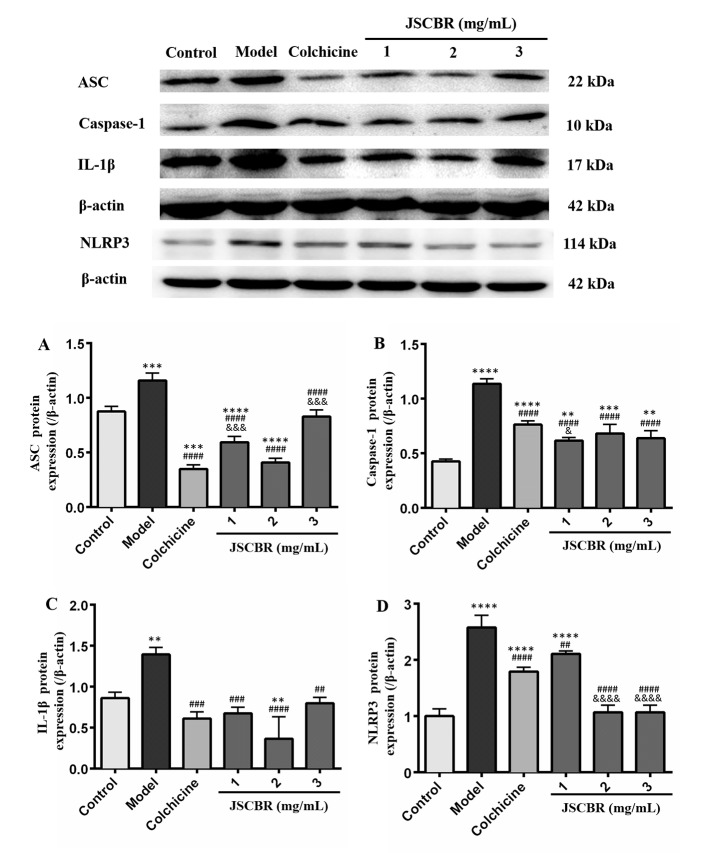
Western Blot Analysis
In order to validate the action mechanism of JSCBR screened out by phytochemistry-based network pharmacology, protein expression of ASC, caspase-1, IL-1β, and NLRP3 was examined by Western Blot Analysis. Compared with a control group, the expression of these three proteins in the model group was significantly increased ( Figure 7 , P < 0.01), while these protein expression changes were attenuated by treatment with colchicine and different concentrations of JSCBR extracts (1, 2, and 3 mg/ml) ( Figure 7 , P < 0.01). The results suggested that the antiinflammation of JSCBR on gouty arthriris was associated with inhibition of ASC, caspase-1, IL-1β, and NLRP3 protein expression, which belongs to NOD-like receptor signaling pathway.
Figure 7.
Jiang-Suan-Chu-Bi recipe (JSCBR) extracts protect THP-1 cells against monosodium urate (MSU)-induced inflammation by affecting the expression of proteins from the NOD-like receptor signaling pathway. (A) Effects of JSCBR extracts on ASC, caspase-1, IL-1β, and NLRP3 protein levels in MSU-induced THP-1 cells based on the western blotting assay; (B) Statistical analysis of the effects of JSCBR extracts on protein expressions levels. Data are presented as the mean ± SD (n = 3), **p < 0.01, ***p < 0.001, ****p < 0.0001 versus control group. ## p < 0.01, ### p < 0.001, #### p < 0.0001 versus model group. & p < 0.05, &&& p < 0.001, &&&& p < 0.0001 versus colchicine group.
Discussion
In recent years, prevalence of gouty arthritis annually increased with the continuous improvement of people’s living standards. Although some achievement has been made in reducing the mortality of the disease, it still imposed a huge economic burden on patients and society which also reduced the quality of life of patients. Colchicine, glucocorticoids, and nonsteroidal antiinflammatory drugs, acted as the current mainstay drugs for gouty arthritis, have been controversial due to their various side effects. It is very necessary to develop new drugs with remarkable curative effect and little side effect.
The pathogenesis of gouty arthritis is classified in the “damp-heat” category according to TCM theory, the therapeutic principles of “clearing heat, dispelling dampness, dissipating stasis, and relieving pain” should be used. JSCBR, a formula prescribed according to abovementioned principles and utilization frequency and made up of 12 kinds of herbs, which has been effective in treating gouty arthritis. However, its “multicomponents”, “multitargets” and “multipathways” features make it much difficult to decipher the molecular mechanisms of JSCBR in the treatment of gouty arthritis from a systematic perspective. In this study, we combined phytochemistry, network pharmacology and molecular cell biology to evaluate the effective components and possible molecular mechanisms of JSCBR in the treatment of gouty arthritis (Dong et al., 2019).
Firstly, chemical profile of JSCBR was characterized for the first time, which lays the material foundation for the follow-up network pharmacology research. Network pharmacological analysis of JSCBR identified 51 potential target nodes, 80 compounds, and 11 related pathways related to gouty arthritis. Among the target compounds linked to the network, two triterpenoid saponins glycyrrhizic acid (Compound 119) from Glycyrrhizae Radix et Rhizome and macedonoside C (Compound 113) from Rhei Radix et Rhizoma, as well as a phenolic acid amurenlaetone B (Compound 47) from Phellodendri Chinensis Cortex may be the effective components of JSCBR in treating gouty arthritis due to higher degree. The two triterpenoid saponins could act at least in part, as a glucocorticoid-like drug due to the similar chemical structure related to glucocorticoids. Additionally, reports have provided evidence that treatments with these two types of components can alleviate the symptoms of gouty arthritis by exerting antiinflammatory and antioxidant pharmacological effects (Ling and Bochu, 2014; Zhou et al, 2017; Jhang et al, 2018).
Among the 51 putative targets and 11 related pathways of JSCBR associate with gouty arthritis, it was discovered that IL-6, IL-1β, CASP1, and NLRP3 were relatively important targets evaluated by topological parameters, which are involved in NOD-like receptor, Toll-like receptor, cytokine-cytokine receptor interaction, and cytosolic DNA-sensing signaling pathways. Among which, NOD-like receptor signaling pathways was ranked first. Thus, a further confirmation was selected for further verification.
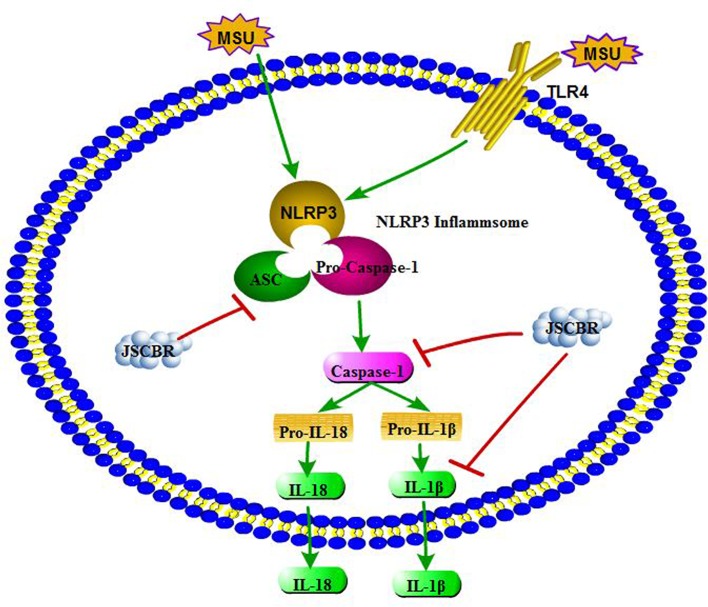
As illustrated in some research studies, NOD-like receptor protein-3 inflammasome (NLRP3) and toll-like receptor-4 (TLR4) plays an important role during gouty arthritis ( Figure 8 ). NLRP3 inflammasome, an assembly composed of NLRP3, apoptosis-associated speck-like protein (ASC) containing a C-terminal caspase recruitment domain (CARD) and the effector procaspase-1, which can be activated by a “danger signal” crystal MSU. Once activated, NLRP3 oligomerizes and recruits the ASC adaptor protein and procaspase-1 sequentially. Caspase-1 was then activated and proteolytic cleavaged by the complex, resulting in specialized maturation and secretion of IL-1β and IL-18. In addition, MSU could also be recognized by Toll-like receptors, Toll-like receptor pathway could active the NLRP3 inflammasome complex and further induce an activation of inflammatory cascade (Mccormack et al., 2009; Xiao et al, 2015).
Figure 8.
Overview of potential mechanisms underlying the protective effects of Jiang-Suan-Chu-Bi recipe (JSCBR) on monosodium urate (MSU)-induced gouty arthritis.
Our experiments in molecular cell biology had shown that the expressions of ASC, caspase-1, and IL-1β were significantly increased in the MSU-induced THP-1 cells, while they were significantly downregulated after treatment with JSCBR, which is consistent with network prediction. Therefore, it is speculated that NOD-like receptor signaling pathway is involved in the pathogenesis of gouty arthritis, and JSCBR could treat gouty arthritis by negative regulation of the inflammatory response. Besides, other potential mechanisms of JSCBR explored by above integrated strategy need for further investigation.
Conclusion
In the current study, an integrated approach based on chemical profile, network pharmacology and experimental support using molecular cell biology were carried out to reveal the therapeutic composition and mechanisms of JSCBR against gouty arthritis. Following identification of 139 chemical constituents in JSCBR by UHPLC-QTOF MS, 175 disease genes, 51 potential target nodes, 80 compounds, and 11 related pathways were achieved by network pharmacology analysis. Potential key targets NRLP3/ASC/CASP1/IL1B that associated with the negative regulation of in NOD-like receptor signaling pathway were further verified in monosodium urate-induced THP-1 cells, and a plausible mechanism for the multitarget effects of JSCBR on gouty arthritis was consequently proposed ( Figure 8 ). Taken together, our results provide a comprehensive insight into therapeutic composition and mechanistic of JSCBR, which make beneficial exploration for the research and development of traditional Chinese herbal formula.
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .
Author Contributions
NX and JQ wrote the manuscript and analyze data. NX, SH, PH, YQ conducted the research and developed the network pharmacology experiment. GL and TP revised the manuscript and edited the graphs. HS did the English proofreading of the manuscript. LZ designed the experiments and edited the manuscript. All the authors read and approved the final version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 81873195 and 81703675), Liaoning Natural Science Foundation (20180550980 and 20180550593), Liaoning Revitalization Talents Program (XLYC1907113), 2018 program for Liaoning Excellent Talents in University, the project of Dalian Young Star on Science and Technology in 2016 (2017RQ122) and Dalian Municipal Medical Research Foundation (no. 17Z2001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01626/full#supplementary-material
Therapeutic targets of gouty arthritis.
Compound targets for JSCBR.
References
- Bai Y. J., Xu J. D., Kong M., Gao Q., Liu L. F., Li S. L. (2015). Discovery of characteristic chemical markers for inspecting sulfur-fumigated Radix Angelicae Sinensis by ultra-high performance liquid chromatography-quadrupole/time-of-flight mass spectrometry based metabolomics and chemical profiling approach. Food Res. Int. 76 (3), 387–394. 10.1016/j.foodres.2015.05.055 [DOI] [PubMed] [Google Scholar]
- Chen L., Tang Y. P., Chen M. J., Deng H. S., Yan X. P., Wu D. K. (2010). Chemical correlation between Gegen Qinlian dispensing granule and its four raw herbs by LC fingerprint. Phytomedicine 17 (2), 100–107. 10.1016/j.phymed.2009.10.012 [DOI] [PubMed] [Google Scholar]
- Dong Y., Qiu P., Zhu R., Zhao L. S., Zhang P. H., Wang Y. Q., et al. (2019). A combined phytochemistry and network pharmacology approach to reveal the potential antitumor effective substances and mechanism of Phellinus igniarius. Front. Pharmacol. 10, 266. 10.3389/fphar.2019.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge A. H., Ma W. F., Wang C. P., Li J., He J., Liu E. W., et al. (2014). Ultra high performance liquid chromatography with photodiode array detector and quadrupole time-of-flight tandem mass spectrometry coupled with discriminant analysis to evaluate Angelicae pubescentis radix from different regions. J. Sep. Sci. 37 (18), 2523–2534. 10.1002/jssc.201400289 [DOI] [PubMed] [Google Scholar]
- Guo L. X., Li R., Liu K., Yang J., Li H. J., Li S. L., et al. (2015). Structural characterization and discrimination of Chinese medicinal materials with multiple botanical origins based on metabolite profiling and chemometrics analysis: Clematidis Radix et Rhizoma as a case study. J. Chromatogr. A. 1425, 129–140. 10.1016/j.chroma.2015.11.013 [DOI] [PubMed] [Google Scholar]
- He X., Yi T., Tang Y., Xu J., Zhang J., Zhang Y., et al. (2016). Assessing the quality of Smilacis Glabrae Rhizoma (Tufuling) by colormetrics and UPLC-Q-TOF-MS. Chin. Med. 11, 33. 10.1186/s13020-016-0104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Ke H. W., Qiu Y., Cai M. C., Qu. J. L., Leng A. J. (2019). Systematically characterizing chemical profile and potential mechanisms of Qingre Lidan Decoction acting on cholelithiasis by integrating UHPLC-QTOF-MS and network target analysis. Evid. Based. Complement. Alternat. Med. 2019, 2675287. 10.1155/2019/2675287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhang J. J., Lin J. H., Yen G. C. (2018). Beneficial properties of phytochemicals on NLRP3 Inflammasome-Mediated Gout and Complication. J. Agric. Food Chem. 66 (4), 765–772. 10.1021/acs.jafc.7b05113 [DOI] [PubMed] [Google Scholar]
- Li H. M., Ou G. C., He Y. L., Ren L., Yang X. H., Zeng M. (2019). Resveratrol attenuates the MSU crystal-induced inflammatory response through the inhibition of TAK1 activity. Int. Immunopharmacol. 67, 62–68. 10.1016/j.intimp.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Liang J., Xu F., Zhang Y. Z., Huang S., Zang X. Y., Zhao X., et al. (2013). The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MS (n) technique: a novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. Anal. 83, 108–121. 10.1016/j.jpba.2013.04.029 [DOI] [PubMed] [Google Scholar]
- Ling X., Bochu W. (2014). A review of phytotherapy of gout: perspective of new pharmacological treatments. Pharmazie 69 (4), 243–256. 10.1691/ph.2014.3642 [DOI] [PubMed] [Google Scholar]
- Liu Y., Li L., Xiao Y. Q., Yao J. Q., Li P. Y., Yu D. R., et al. (2018). Global metabolite profiling and diagnostic ion filtering strategy by LC–QTOF MS for rapid identification of raw and processed pieces of Rheum palmatum L. Food Chem. 192, 531–540. 10.1016/j.foodchem.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Ma X. Q., Leung A. K., Chan C. L., Su T., Li W. D., Li S. M., et al. (2014). UHPLC UHD Q-TOF MS/MS analysis of the impact of sulfur fumigation on the chemical profile ofCodonopsis Radix (Dangshen). Analyst 139 (2), 505–516. 10.1039/c3an01561k [DOI] [PubMed] [Google Scholar]
- Mccormack W. J., Parker A. E., ONeill L. A. (2009). Toll-like receptors and NOD-like receptors in rheumatic diseases. Arthritis Res. Ther. 11 (5), 243. 10.1186/ar2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S. M., Zobbe K., Kristensen L. E., Christensen R. (2018). Nutritional recommendations for gout: an update from clinical epidemiology. Autoimmun. Rev. 17, 1090–1096. 10.1016/j.autrev.2018.05.008 [DOI] [PubMed] [Google Scholar]
- Pascart T., Grandjean A., Capon B., Legrand J., Namane N., Ducoulombier V., et al. (2018). Monosodium urate burden assessed with dual-energy computed tomography predicts the risk of flares in gout: a 12-month observational study: MSU burden and risk of gout flare. Arthritis Res. Ther. 20 (1), 210. 10.1186/s13075-018-1714-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Wang H. Y., Zhang A. H., Yan G. L., Han Y., Li Y., et al. (2016). Chemical discrimination of Cortex Phellodendri amurensis and Cortex Phellodendri chinensis by multivariate analysis approach. Pharmacogn. Mag. 12 (45), 41–49. 10.4103/0973-1296.176023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Qin Y., Li G. Q., Chen S., Ma J. Q., Guo Y. L., et al. (2018). Study on chemical constituents in polygoni Cuspidati Folium and its Preparation by UPLC-ESI-Q-TOF MS/MS. J. Chromatogr. Sci. 56 (5), 425–435. 10.1093/chromsci/bmy017 [DOI] [PubMed] [Google Scholar]
- Wilson L., Saseen J. J. (2016). Gouty Arthritis: a review of acute management and prevention. Pharmacotherapy 36 (8), 906–922. 10.1002/phar.1788 [DOI] [PubMed] [Google Scholar]
- Xiao J., Fu C. S., Zhang X. L., Zhu D. Y., Chen W. J., Lu Y. J., et al. (2015). Soluble monosodium urate, but not its crystal, induces toll like receptor 4-dependent immune activation in renal mesangial cells. Mol. Immunol. 66 (2), 310–318. 10.1016/j.molimm.2015.03.250 [DOI] [PubMed] [Google Scholar]
- Xiao N., Dong Y. X., Liu J. W., Li M. Y., Niu X., Tang S. F. et al. (2019). Using the literature to study the medication feature of Chinese medicine treatment for gout. J. Shenyang Pharm. Univ. 36 (5), 427–432. 10.14066/j.cnki.cn21-1349/r.2019.05.009 [DOI] [Google Scholar]
- Xu S. Z., Qi X. J., Liu Y. Q., Liu Y. H., Lv X., Sun J. Z., et al. (2018). UPLC-MS/MS of Atractylenolide I, Atractylenolide II, Atractylenolide III, and Atractyloside A in Rat plasma after oral administration of raw and wheat Bran-Processed Atractylodis Rhizoma. Molecules 23 (12), E3234. 10.3390/molecules23123234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Liu L., Liu X., Chen P., Liu L., Zhang Y., et al. (2014). Systematic review and meta-analysis of the clinical efficacy and adverse effects of Chinese herbal decoction for the treatment of gout. PloS One 9 (1), e85008. 10.1371/journal.pone.0085008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Lin F. F., Liu S. M., Sui X. F. (2017). Influence of the total saponin fraction from Dioscorea nipponica Makino on TLR2/4-IL1R receptor singnal pathway in rats of gouty arthritis. J. Ethnopharmacol. 206, 274–282. 10.1016/j.jep.2017.04.024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Therapeutic targets of gouty arthritis.
Compound targets for JSCBR.
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .