Abstract
Acute lymphoblastic leukemia is marked by aberrant transcriptional features that alter cell differentiation, self-renewal, and proliferative features. We sought to identify the transcription factors exhibiting altered and subtype-specific expression patterns in B-ALL and report here that SOX11, a developmental and neuronal transcription factor, is aberrantly expressed in the ETV6-RUNX1 and TCF3-PBX1 subtypes of acute B-cell leukemias. We show that a high expression of SOX11 leads to alterations of gene expression that are typically associated with cell adhesion, migration, and differentiation. A high expression is associated with DNA hypomethylation at the SOX11 locus and a favorable outcome. The results indicate that SOX11 expression marks a group of patients with good outcomes and thereby prompts further study of its use as a biomarker.
Subject terms: Haematological cancer, Acute lymphocytic leukaemia
Introduction
SOX11 is a transcription factor (TF) encoded by the SOX11 gene located in chromosome 2p251. It is a member of the SoxC (sex-determining region Y-related HMG box) group of genes and consists of two functional domains—the N-terminal DNA-binding and the C-terminal transactivation domains2,3. Other SoxC family members include SOX4 and SOX12. Of these, SOX4 is a crucial TF in B lymphopoiesis and is expressed in the B- and T-cell lineages2,4,5. SOX11 is normally expressed in the developing central nervous system of the embryo, in keratinocytes, and in some other epithelial tissues1,6–8. It is also expressed in ovarian and breast cancer, in which both tumor suppressor and oncogenic functions have been suggested9,10. A knockout mouse model revealed the vital role of SOX11 during embryonic development, as SOX11-deficient mice died after birth likely from multiple heart defects, asplenia, and organ hypoplasia in the lungs, stomach, and pancreas11. SOX11 deletions and mutations are associated with neurodevelopmental disorders12.
Previous studies have shown that SOX11 mRNA and nuclear protein expression is a specific marker for conventional but not indolent mantle cell lymphoma (MCL)8,13–15. In MCL, SOX11 has been associated with either increased or reduced cell proliferation16–23 and either good or bad prognosis24–27. In a cohort of 50 adult acute myeloid leukemia (AML) patients, a high SOX11 expression was associated with FLT/ITD and NPM1 mutations and a shortened disease-free survival28.
There is other evidence linking SOX11 with B-lineage malignancies. Dictor et al. (2009) reported that the nuclear expression of SOX11 was found in eight cases of B-lymphoblastic neoplasias29. Another study reported the strong nuclear expression of SOX11 in a single B-cell and five T-cell lymphoblastic lymphoma/leukemias13. Vegliante et al. (2011) demonstrated the increased expression of SOX11 mRNA in ETV6-RUNX1 (E/R) and TCF3-rearranged B-cell precursor acute lymphoblastic leukemia (BCP-ALL)30, whereas Nordlund et al. (2012) and Busche et al. (2013) observed the prominent expression of SOX11 in the E/R subtype of ALL31,32.
We investigated the expression of SOX11 across lymphoid malignancies, focusing on B-lymphoblastic leukemias. The function of SOX11 in leukemias and its clinical significance as a biomarker were further explored.
Materials and Methods
Microarray datasets
We used three independent datasets to study SOX11 expression—a combined microarray dataset (“Hemap”) retrieved from Gene Expression Omnibus (GEO)33,34, the GEO series GSE4705135, and the publicly available BCP-ALL data from the recent PanALL study36. The sample sizes for each dataset are shown in Supplementary Table 1.
Cell lines, cell culture, and drug treatments
NALM-6, REH, 697, RCH-ACV, KOPN-8, KASUMI-2, JURKAT, MOLT-16, P12-ICHIKAWA, HPB-ALL, and CCRF-CEM were cultured in RPMI Medium 1640 (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) with 2 mM L-glut, 100 U penicillin, 100 µg/ml streptomycin with 10% FBS (Gibco), and MOLT-4, PEER, and MHH-CALL3 with 20% FBS (Gibco) at 37 °C in 5% CO2. An inducible E/R fusion in the NALM-6 cell line and a knockdown of E/R by a short hairpin RNA (shRNA) in the REH cell line have been previously described37. E/R expression was induced with 500 ng/ml doxycycline (Clontech). E/R expression changes were confirmed with RT-qPCR, with fusion gene-specific primers (Table S2). Mycoplasma tests were done regularly for the cell lines, and Eurofins Genomics (Ebersberg, Germany) services were used to authenticate the cell lines by STR genotyping. All cell lines used in this study were purchased from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany).
For the methyltransferase inhibition experiments, the cultured cells were treated for 72 h with decitabine, 5-Aza-2′-deoxycytidine solved in DMSO (A3656, Sigma-Aldrich, St. Louis, MO, USA) at 0, 0.1, and 1 µM concentrations. The media were changed at a 24 h interval to compensate for decitabine instability under cell culture conditions. After the treatment cells were collected, RNA was extracted for RT-qPCR analyses. Corticosteroid and chemotherapy treatments were conducted with the indicated concentration ranges, and cell viabilities were measured after either 72 (697 and RCH-ACV) or 96 h (REH). The corticosteroids included prednisolone (P6004, Sigma-Aldrich) and dexamethasone (D8893, Sigma-Aldrich), and the chemotherapy agents used were asparaginase (A3809, Sigma-Aldrich) and vincristine (V8879, Sigma-Aldrich). The applied concentrations for each cell line are indicated in Table S3.
Quantitative real-time PCR
Total RNA was extracted using the PureLink™ RNA Mini Kit, and the On-Column PureLink® DNase Treatment Protocol was used for DNA removal (Ambion® by Life Technologies and Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA); 100–500 ng of the extracted RNA was used as a starting material for cDNA synthesis, which was performed with iScript (Bio-Rad, Hercules, CA, USA). RT-qPCR reactions were conducted according to the manufacturer’s instructions with SsoFast EvaGreen® Supermix (Bio-Rad). The following program was performed with the Bio-Rad CFX96TM Real-Time System (Bio-Rad): initial denaturation at 96 °C for 30 s, 39 cycles of denaturation at 96 °C for 2 s, annealing at 60 °C for 5 s, and plate read. Independent experiments performed in triplicate were used as the starting material for the RT-qPCR measurements, and the relative 2−ΔΔCT method was used for quantification38. The primer sequences used in RT-qPCR are listed in Table S2.
Western blot
Protein extraction was performed using M-PER reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions to lyse the cells, and protein concentrations were measured with DC Protein Assay (Bio-Rad); 15–20 µg of a protein sample was loaded into the precast 10% Mini-PROTEAN® TGX Stain-FreeTM Gels (Bio-Rad). After the electrophoresis run, Trans-Blot® TurboTM Pack (Bio-Rad) was used to transfer the proteins from the gel to the nitrocellulose membrane. Transfer was done with the Trans-Blot® TurboTM Transfer system according to the manufacturer’s instructions (Bio-Rad). A prestained protein ladder, PageRuler Plus (#26619, Thermo Fischer Scientific), was used as a protein size marker. We utilized antibodies against SOX11 (1:1,000) (HPA000536, Sigma-Aldrich, Lot # BB107024) and Histone H3 (1:75,000) (ab4729, Abcam, Cambridge, UK, Lot # GR167613-1). Horseradish peroxidase conjugated anti-rabbit (1:2,000) (P0217, Lot # 00069121) was used as a secondary antibody (Agilent Technologies, Santa Clara, CA, USA). Chemiluminescence reaction by Amersham ECL reagent was detected with ChemiDocTM XRS+using Image LabTM Software (Bio-Rad).
Gene silencing with nucleofection
Knockdowns for suspension cells were performed using 4D-Nucleofector™ (Lonza, Basel, Switzerland) for transfections. SOX11 knockdown was done using gene sequence-specific small interfering RNA (siRNA) (Sigma-Aldrich), and a non-specific siRNA was used as a control (Table S4). Before nucleofection, 20 µM stock solutions were diluted in Resuspension Buffer (SR30005, OriGene, Rockville, MD, USA) so that the final concentration per reaction was 300 nM. One million cells were used for each nucleofection reaction. Nucleofection reactions were conducted in proper solutions and programs according to the manufacturer’s instructions in single nucleocuvettes (Table S5). Then, the cells were transferred to 12-well plates with prewarmed fresh media. The transfected cells were used in cell viability assays and RNA sequencing (RNA-seq). Western blot and RT-qPCR were used to assess knockdown levels.
RNA sequencing of cell lines
SOX11-specific and control siRNAs were used in the nucleofection for the 697, RCH-ACV, and REH cell lines to create SOX11 knockdown and control samples. After 48 h of nucleofection, three million cells per sample were collected for the total RNA extraction. RNA extraction was performed with the PureLink™ RNA Mini Kit, and the On-Column PureLink® DNase Treatment Protocol was used to avoid contamination by genomic DNA (Ambion® by Life Technologies and Invitrogen). Protein samples were also collected, and the SOX11 knockdown level was verified by both RT-qPCR and Western blotting. Three independent biological replicates were collected for each transfected cell line, and each sample had 25 ng of RNA in 40 µl. Library preparation and RNA-seq (GSE123943) were performed in the Finnish Functional Genomics Centre (Turku, Finland). See more details in Supplementary Information.
The quality of the raw sequencing reads was ensured with FastQC (v0.10.1). Based on the FastQC results, reads were trimmed and their quality was filtered using the FASTX Toolkit (0.0.14). The reads were mapped to the human reference genome version hg19 using STAR aligner software (2.5.3a modified); reads aligning to more than two locations were discarded39. The alignment file was turned into tag directories, and read counts were calculated using the HOMER tool kit (v4.8).
Differential gene expression was analyzed using the quasi-likelihood F-test from edgeR, an R package40. Differentially expressed genes from all cell lines were filtered using the adjusted p-value < 0.05 (Benjamini–Hochberg method) as a cut-off, and the resulting gene lists from three different cell lines were compared by drawing a Venn diagram with an interactive Venn online tool (http://www.interactivenn.net/). Heatmaps, presenting all biological replicates, were drawn by using the z-scores of reads per kilobase of transcript, the per million mapped reads (RPKM) normalized count matrix, and the ComplexHeatmap R-package41. Gene set enrichment was analyzed by Gene Set Enrichment Analysis (GSEA) 3.0 software using logFC ranked lists of genes from differential gene expression analysis, and the results presented had an adjusted p-value < 0.02 (Benjamini–Hochberg method)42,43. Gene ontology (GO) term enrichment was studied with two approaches—with GSEA software using f-statistics ranked lists of genes and with the GOrilla online tool using unranked target and background lists44,45. The target lists were created with a threshold of adjusted p-value < 0.05 and logFC >0.5 or <−0.5.
RNA sequencing and methylation analysis of the patient samples
Previous transcriptome sequencing of pediatric ALL cohort by Marincevic-Zuniga et al.46 included 116 BCP-ALL cases, of which 115 cases were used to assess the SOX11 mRNA expression level in this study. DNA methylation data (GSE49031) were available for 112 of the 115 BCP-ALL cases35. See more details in Supplementary Information.
Cell viability and proliferation assays
Fresh media were replaced on the transfected cells after 24 h of transfection. For the cell viability assay, the cells were counted, and 10,000 cells per well were used in a 96-well plate. The cells were allowed to grow up to 72 (697 and RCH-ACV) and 120 h (REH), and cell viability was measured every 24 h with 10 µl of Alamar Blue reagent per well with a 2 h incubation before fluorescence measurement with excitation of 560 nm and emission of 590 nm using the Tecan fluorometer Infinite 200 (Tecan, Männedorf, Switzerland). For each time point, we used four technical replicates per sample to calculate the mean.
Cell proliferation assays were performed by counting the cells every 24 h after the transfection up to 96 (697 and RCH-ACV) and 120 h (REH).
Clinical data and the patient samples
The data on pediatric ALL cases below 18 years of age and treated in the Tampere University Hospital were retrieved from the clinical registry from years 1990 to 2017. Essential clinical information, such as age, leukocyte count, gender, relapse, death, central nervous system leukemia, immunophenotype based on flow cytometry, and clinical genetic information, was collected. The study obtained permission from the local ethical committee, and informed consent was sought, as needed (Pirkanmaa Hospital District Ethical Committee, R16054 and R13109). The use of old biopsy samples was approved by the National Supervisory Authority for Welfare and Heath (Valvira), and the samples were handled in accordance with relevant guidelines and regulations.
A total of 126 representative diagnostic formalin-fixed and paraffin-embedded (FFPE) decalcified bone marrow trephine biopsy samples were collected from the pathology department archives on the basis of the primary sample reports. For a proportion of cases, remission and relapse samples were also retrieved. Plastic-embedded and inadequate samples were excluded. The cases were classified based on the WHO 2017 Classification of Tumors and Haematopoietic and Lymphoid Tissues47.
Immunohistochemistry
Four micron-thick whole tissue sections were used for immunohistochemistry. All cases were stained with anti-SOX11 antibody (clone MRQ-58, Cell Marque, Sigma-Aldrich, Lot # 1331005 A and 1430213 C) using Ventana Benchmark Classic at a dilution of 1:50. FFPE human MCL was used as a positive control material, and remission bone marrow samples were used as negative controls.
Staining intensity was graded, and cases with less than 20% of positivity of leukemic blast cell nuclei were interpreted as negative. Cases with immunohistochemical positivity ranging from 20% to 50% in the blast cell nuclei were graded positive, and cases with immunohistochemical nuclear positivity of over 50% were considered strongly positive. SOX11 expression was independently analyzed with a light microscope by two experienced pathologists without knowledge of the clinical data. Cases with discrepant scores were further analyzed by a third pathologist.
Fluorescence in situ hybridization
For cases lacking the genetic subtype information, fluorescence in situ hybridization analysis was performed on either the bone marrow aspiration samples or the FFPE samples. The following probes were used: Metasystems E2A Break Apart Probe 19p13 (Lot # 18216), Metasystems XL MLL plus Break Apart Probe 11q23 (Lot # 18451), Metasystems XL BCR/ABL1 plus (Lot # 19082), and Metasystems XL t(12;21) (Lot # 19133).
Flow cytometry analysis
For flow cytometry analysis, the cells were permeabilized using Fix&Perm reagent according to the manufacturer’s instructions (GAS003, Invitrogen). The cells were then stained with Alexa Fluor 647® conjugated rabbit anti-human SOX11 antibody [EPR8191 (2)] (ab198540, Abcam), while CD3-APC antibody (345767) (Becton Dickinson, Franklin Lakes, NJ, USA) served as a negative control. The data were acquired with the Beckman Coulter Navios cytometer (Beckman Coulter, Brea, CA, USA) using Red laser (638 nm) and 660/20 bandpass filter. Data analysis was conducted using Kaluza software (Beckman Coulter).
Statistical analysis
IBM SPSS Statistics (v. 22) and R (v. 3.40) were used for the statistical analysis. Kaplan–Meier survival analysis was performed, and Log-rank test was used, with a p-value < 0.05 considered statistically significant. Event-free survival (EFS) was analyzed using death, relapse, resistant disease (blast count >25% at the end of induction), and secondary malignancy as events. Cox proportional hazards models were fitted to estimate the effects of potential risk factors on survival. Chi-squared test was performed on SOX11 expression and clinicopathological prognostic variables. Kruskal–Wallis H and Mann–Whitney U tests were used to evaluate the differential expression of SOX11 in distinct leukemia subgroups (Table S6). All performed statistical tests were two tailed, and no corrections for multiple testing were used.
Results
SOX11 is overexpressed in acute lymphoblastic leukemias
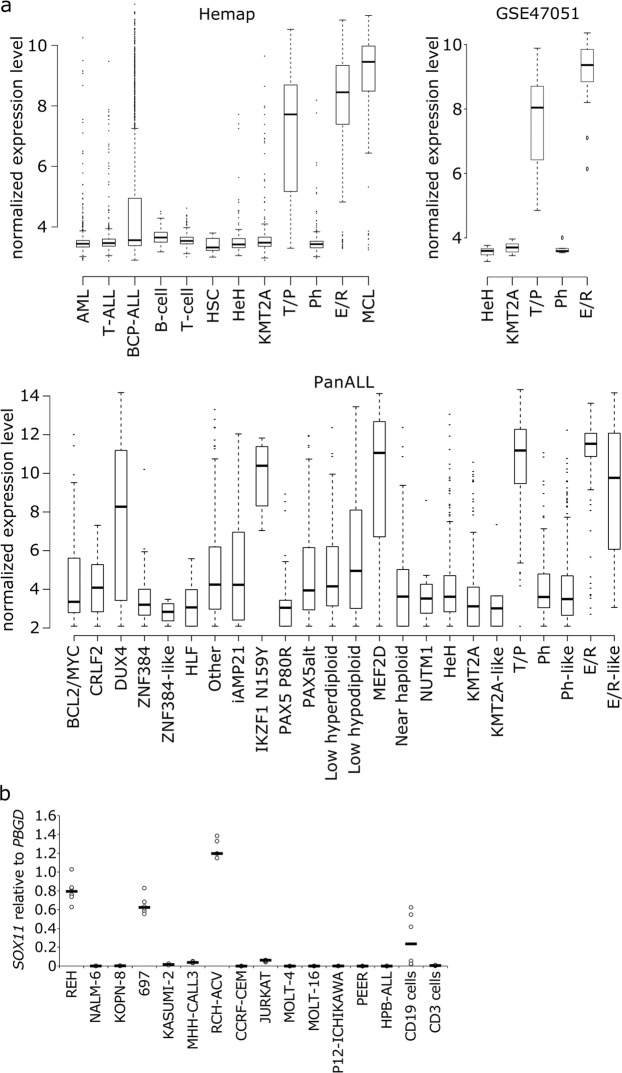
Derailed differentiation and abnormal proliferation of B-cells are thought to underlie the genesis of BCP-ALL. As TFs are key cell differentiation drivers, we sought to identify aberrantly expressed TFs in BCP-ALL. We utilized a large, curated dataset of microarray-based gene expression profiles retrieved from GEO34. This dataset comprises a total of 9,544 hematological gene expression profiles, including 4,418 leukemias, 428 healthy controls, and 862 cell lines. Among the genes with altered expression, TF SOX11 showed prominent expression in MCL and in the E/R and T/P subtypes of BCP-ALL (Fig. 1a). Compared with healthy hematopoietic cells, SOX11 had a 4.7- and 4-fold higher expression in the E/R and T/P subtypes, respectively, with an expression comparable to that in MCL (Fig. 1a). A similar subtype-specific expression of SOX11 was replicated in two additional ALL gene expression datasets (Fig. 1a). Interestingly, the PanALL study revealed that SOX11 expression is also elevated in novel subtypes, such as E/R-like, IKZF1 N159Y (IKZF1 missense alteration encoding p.Asn159Tyr), MEF2D rearrangement, and DUX4 rearrangement.
Figure 1.
SOX11 expression in hematological malignancies. (a) Expression boxplots of SOX11 in healthy cells, leukemias, and mantle cell lymphoma. Data sources: Hemap microarray dataset34, GSE4705135, and the PanALL study36. See Supplementary Information for more details. (b) SOX11 expression in ALL cell lines and healthy B- and T-cells, as measured by RT-qPCR (N = 2, black lines indicate the median). ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BCL2/MYC, BCL2/MYC rearranged; BCP-ALL, B-cell precursor ALL; CRLF2, CRLF2 (non-Ph-like); DUX4, DUX4 rearranged; E/R, ETV6-RUNX1; HeH, high hyperdiploid; HLF, TCF3/TCF4-HLF; HSC, hematopoietic stem cell; iAMP21, intrachromosomal amplification of chromosome 21; IKZF1 N159Y, IKZF1 missense alteration encoding p.Asn159Tyr; KMT2A, KMT2A rearranged; MCL, mantle cell lymphoma; MEF2D, MEF2D rearranged; NUTM1, NUTM1 rearranged; PAX5alt, PAX5 alterations; PAX5 P80R, PAX5 p.Pro80Arg (P80R) alteration; Ph, Philadelphia chromosome; T/P, TCF3-PBX1; ZNF384, ZNF384 rearranged. Reproduced with permission66.
As SOX11 belongs to the SoxC family of TFs, we investigated the expression of the two other members of this family. SOX4, which has been reported to affect survival, progression, and proliferation in BCP-ALL48, was highly expressed in BCP-ALL and T-ALL, but it showed no subtype-specific expression pattern. The expression of SOX12 did not vary markedly between the studied subtypes (Fig. S1a).
A subtype-specific expression pattern was also present in BCP-ALL cell lines, as REH cells, representing the E/R subtype, and RCH-ACV and 697 cells, representing the T/P subtype, showed an increased expression of SOX11 by RT-qPCR (Fig. 1b). Neither knockdown of E/R in REH cells nor their overexpression in NALM-6 cells had any effect on SOX11 expression levels (Fig. S2a).
Immunohistochemical analysis of SOX11 expression in ALL
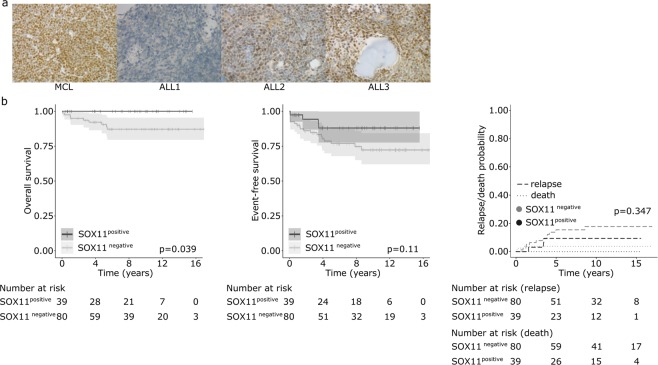
In order to confirm the SOX11 expression at the protein level and relate it to clinical features, we collected a retrospective cohort of 119 B-ALL cases with available bone marrow biopsies and associated clinical data (Table 1). We performed immunohistochemical staining of the biopsy samples by using a SOX11-specific antibody. Staining intensity was graded from 0 to 2, with 0 marking negative, 1 positive, and 2 strongly positive samples. Eighty out of 119 primary B-ALL bone marrow samples stained negative for SOX11, whereas 34 cases were positive and 5 were strongly positive (Fig. 2a and Table 2). A total of 29/39 (74.4%) of the positively staining B-ALL cases represented either the E/R or T/P subtype. A statistically significant association was observed with the E/R subtype (p-value < 0.001) but not with the T/P subtype (likely because of the low number of cases: N = 4, 3 positive cases). Cases with either KMT2A rearrangement or the Philadelphia chromosome (BCR-ABL1-translocation) did not express the SOX11 protein, and the majority of hyperdiploid cases were also SOX11 negative. Additionally, seven T-ALL cases included in the whole ALL patient cohort were all negative.
Table 1.
Summary of clinical characteristics of B-ALL patients.
| WHO Subtype | Total/Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Burkitt | NOS | Ph | KMT2A | ETV6-RUNX1 | Hyper-diploid | Hypo-diploid | TCF3-PBX1 | ||
| N of cases | 2 | 42 | 2 | 5 | 33 | 30 | 1 | 4 | 119 |
| Age, Md (min/max) | 12.7 | 8.1 | 10.1 | 1.3 | 4 | 3.7 | 4.1 | 7.4 | 4.3 (0.9/17.6) |
| WBC count (109/l), Md (min/max) | 18.9 | 6.7 | 156.3 | 109.7 | 7 | 7.1 | 1.9 | 43.4 | 7.1 (1/311) |
| Deceased | 2 | 6 | 1 | 0 | 1 | 1 | 0 | 0 | 9 |
| Relapse | 0 | 7 | 1 | 0 | 3 | 3 | 0 | 1 | 15 |
| CNS disease | 0 | 2 | 1 | 0 | 1 | 2 | 0 | 0 | 6 |
| Resistant disease | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 4 |
| MRD (%) at EOI, Md (min/max) | 0 | 0.05 | 1.75 | 0 | 0.06 | 0 | 0 | 0 | 0.02 (0/44) |
| Follow-up (years), Md (min/max) | 7.5 | 9.7 | 8.4 | 4 | 7.1 | 9.2 | 11.5 | 7.6 | 8.2 (0.1/17) |
CNS, central nervous system; EOI, end of induction therapy; KMT2A, KMT2A rearrangement; Max, maximum; Md, median; Min, minimum; MRD, minimal residual disease; NOS, not otherwise specified; Ph, Philadelphia chromosome; WBC, white blood cell.
Figure 2.
SOX11 protein expression and impact on the outcome in BCP-ALL. (a) Expression of SOX11 protein by immunohistochemistry. Mantle cell lymphoma (MCL), a strongly positive case for SOX11 (400×); ALL1, a negative B-ALL case (400×); ALL2, a positive B-ALL case (400×); ALL3, a strongly positive B-ALL case (400×). (b) Kaplan–Meier survival curves and Log-rank p-values for OS, EFS, and RFS in the SOX11-positive (high) and -negative (low) groups. Reproduced with permission66.
Table 2.
Summary of SOX11 protein expression by immunohistochemical staining in B-ALL.
| SOX11 IHC | WHO Subtype | All cases | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Burkitt | NOS | Ph | KMT2A | ETV6-RUNX1 | Hyper-diploid | Hypo-diploid | TCF3-PBX1 | ||
| Negative | 1 | 35 | 2 | 5 | 7 | 29 | 0 | 1 | 80 |
| Positive | 1 | 6 | 0 | 0 | 24 | 0 | 1 | 2 | 34 |
| Strong positive | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 5 |
| Total | 2 | 42 | 2 | 5 | 33 | 30 | 1 | 4 | 119 |
IHC, immunohistochemistry; KMT2A, KMT2A rearrangement; NOS, not otherwise specified; Ph, Philadelphia chromosome.
SOX11 expression is associated with a favorable outcome
We next sought to evaluate the clinical significance of SOX11 expression in B-ALL (N = 119). The overall survival (OS) was better in the SOX11-positive group, and no deaths occurred among SOX11-positive cases (p = 0.039) (Fig. 2b). The EFS and relapse-free survival (RFS) adjusted for the competing event (death) showed similar trends but did not reach statistical significance (Fig. 2b). SOX11 positivity was not associated with good early therapy response, as measured by a minimal residual disease below 0.1% at the end of induction therapy (OR = 0.54, 95% CI 0.22, 1.28, p = 0.17). In multivariate analysis of EFS with covariates (age, WBC, MRD and subtypes), a positive immunohistochemical staining for SOX11 protein showed a favorable trend (Table 3).
Table 3.
Multivariate and univariate analyses of the event-free survival based on expression level of SOX11 protein in pediatric B-ALL.
| N | MULTIVARIATE | UNIVARIATE | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | ||
| Age (years) | |||||||
| ≥1 and ≤10 | 93 | 1.00* | 1.00* | ||||
| <1 and >10 | 26 | 0.87 | 0.31–2.38 | 0.79 | 1.27 | 0.50–3.23 | 0.61 |
| WBC count (109/l) | |||||||
| <50 | 101 | 1.00* | 1.00* | ||||
| ≥50 | 18 | 0.55 | 0.16–1.92 | 0.35 | 0.90 | 0.27–3.04 | 0.87 |
| Subtype | |||||||
| Other B-ALL | 56 | 1.00* | 1.00* | ||||
| High hyperdiploidy | 30 | 0.27 | 0.07–1.00 | 0.05 | 0.45 | 0.15–1.36 | 0.16 |
| ETV6-RUNX1 | 33 | 0.72 | 0.18–2.83 | 0.64 | 0.50 | 0.17–1.51 | 0.22 |
| SOX11 expression | |||||||
| negative | 80 | 1.00* | 1.00* | ||||
| positive | 39 | 0.37 | 0.10–1.43 | 0.15 | 0.43 | 0.15–1.26 | 0.12 |
| MRD at EOI | |||||||
| <0.1% | 78 | 1.00* | 1.00* | ||||
| ≥0.1% | 37 | 1.70 | 0.68–4.24 | 0.26 | 2.18 | 0.94–5.06 | 0.07 |
Cox proportional hazards regression calculated for known risk factors. SOX11 expression was treated as a binary variable. 95% CI, 95% confidence interval; B-ALL, B-cell acute lymphoblastic leukemia; HR, hazard ratio; WBC, white blood cell. *Marks reference groups of each categorical variable.
When SOX11 immunostaining positivity was analyzed separately within the E/R subtype, SOX11-positive cases had a better OS (Log-rank test p = 0.004), but EFS did not show a statistically significant difference with a hazard ratio of 0.67 (95% CI 0.07, 6.43).
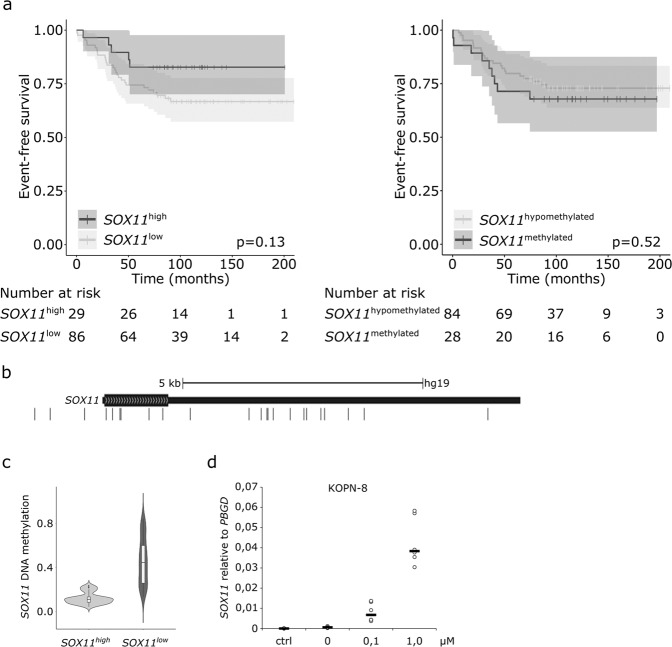
We replicated the survival findings in another dataset. Transcriptomic data from a patient cohort comprising 115 BCP-ALL cases were analyzed for SOX11 expression and patients’ survival status46. Figure 3a shows that a high SOX11 mRNA expression was associated with a favorable trend in EFS analysis.
Figure 3.
Survival analysis and methylation status of cases with either a high or low SOX11 mRNA expression. (a) Kaplan–Meier survival curves and p-values of Log-rank test for EFS in patients with a low or high expression of SOX11 and in patients with a low or high methylation of the SOX11 gene locus. (b) CpG sites at the SOX11 locus in chromosome 235. (c) DNA methylation at the SOX11 gene locus among patients with either a low or high expression of SOX11. (d) Effect of decitabine treatment on SOX11 mRNA expression in the KOPN-8 cell line. Reproduced with permission66.
SOX11 overexpression is associated with DNA hypomethylation
We next investigated the biology behind the increased SOX11 expression in leukemia. Neither primary transcription (GRO-seq, N = 8) nor whole-genome sequencing (WGS, N = 8) of the SOX11 gene in BCP-ALL cases revealed any aberrant enhancer activity or somatic mutations, respectively, in the SOX11 gene or nearby regions (data not shown), prompting us to look for other mechanisms. In MCL, hypomethylation (partly) drives the increased SOX11 expression30. We utilized the above-mentioned BCP-ALL patient cohort with readily available genome-wide CpG methylation data35,46. Altogether, 23 CpG sites were located within the SOX11 gene locus, and a strong pattern of DNA hypomethylation was seen in patients with a high SOX11 mRNA expression (Fig. 3b,c). Nevertheless, DNA hypomethylation of the CpG sites at the SOX11 locus was not associated with a better EFS (Fig. 3a, right panel). We also tested whether a methyltransferase inhibitor, decitabine, could reverse SOX11 expression in leukemia cell lines. After 72 h of decitabine treatment, a marked increase in SOX11 expression was observed in a concentration-dependent manner in KOPN-8 and REH cells (Figs. 3d and S2b).
Knockdown of SOX11 alters gene sets related to cell development, motility, and drug response pathways
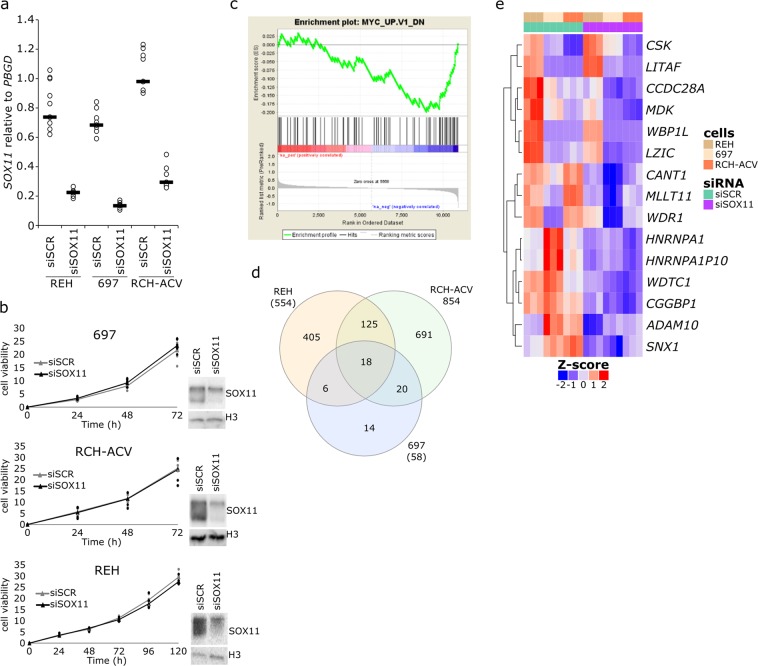
We next silenced SOX11 expression in three cell lines that overexpress SOX11 (REH, RCH-ACV, and 697) by using siRNA oligos. Figure 4a,b show that SOX11 expression was decreased to 20–40% at both the mRNA and protein levels compared with scrambled siRNA-transfected cells. Cell viability and proliferation assays did not demonstrate any significant changes (Figs. 4b and S4), and SOX11 knockdown did not have any impact on sensitivity to known leukemia drugs, such as dexamethasone, prednisolone, vincristine, and asparaginase (Fig. S5).
Figure 4.
Cell viability and transcriptomic changes after knockdown of SOX11. (a) Expression level of SOX11 after knockdown, as analyzed by RT-qPCR (N = 3, black lines indicate the median). (b) Knockdown of SOX11 caused no evident changes in cell viability. Curves are drawn from the biological replicates using the median value at given time points. 697 and RCH-ACV cells represent the T/P subtype, and REH cells represent the E/R subtype. SOX11 knockdown was confirmed by Western blotting, and cell viability assessments were conducted with the AlamarBlue assay. Measured absorbance intensities are reported as x103 (697 N = 6; RCH-ACV and REH N = 4). Western blot gel figures are cropped per cell line from the original blot figures shown in Fig. S3. (c) SOX11 knockdown in 697 cells caused downregulation of genes that are known to be altered after MYC upregulation. (d) Venn diagram of differentially expressed genes in the REH, 697, and RCH-ACV cell lines after knockdown of SOX11 (adjusted p-value < 0.05). (e) Heatmap of 15 concordantly differentially expressed genes in all three cell lines after knockdown of SOX11. Reproduced with permission66.
As SOX11 is a TF and it regulates gene expression, we measured changes in gene expression after SOX11 knockdown by using RNA-seq (GSE123943). Three biological replicates were used for each cell line (REH, RCH-ACV, and 697), and the data were analyzed using the R package EdgeR. In GO annotations, many of the altered terms were related to cell migration, adhesion, and differentiation (Fig. S6 and Supplementary Dataset). On the other hand, GSEA implied the altered expression of MYC and EF2 target genes (Figs. 4c and S6). SOX11 knockdown did not have a significant effect on the expression of other SoxC family members, such as SOX4 and SOX12 (Fig. S7).
The Venn diagram in Fig. 4d shows that 18 genes were differentially expressed in all of the three cell lines after SOX11 knockdown (with an adjusted p-value < 0.05). Of these genes, 15 were concordantly down-regulated in all three cell lines and are shown in the heatmap (Fig. 4e, Supplementary Dataset). SOX11 knockdown led to the downregulation of the WD Repeat Domain 1 (WDR1) gene, which is involved in the remodeling of the actin cytoskeleton, regulation of cell migration, motility of neutrophils, and maturation of megakaryocytes49,50. Another gene down-regulated by SOX11 knockdown is Midkine (MDK), a secreted growth factor that promotes cell migration and growth and is associated with an adverse prognosis in ALL possibly via increased drug resistance51,52. WW Domain Binding Protein 1 Like (WBP1L), also known as OPAL1 (outcome predictor for acute leukemia 1), which was recently identified as a direct target of ETV6 in ALL53, was down-regulated by SOX11 silencing. Previous reports on WBP1L’s association with a favorable prognosis have since been refuted54. However, WBP1L expression had a 2.8-fold increase in the E/R subtype55, which coincides with the overexpression of SOX11 in the same subtype and may suggest co-regulation. MLLT11 is involved in lymphoid regulation and is a known partner gene in rare leukemia translocations56,57. Similarly, Coiled-coil domain containing 28 A (CCDC28A) is a fusion partner to NUP98 in AML58. CANT1 is a calcium-dependent nucleotidase involved in pyrimidine metabolism whose regulation by SOX11 could be related with drug metabolism (cytarabine) and therapy response in ALL59. The lipopolysaccharide-induced TNF factor (LITAF) has been suggested to sensitize leukemia cells to chemotherapeutic drugs, especially in cells with a lower expression of LITAF60. Both LITAF and Sorting nexin 1 (SNX1) are involved in endosomal trafficking and regulation of cell-surface receptor signaling61,62. Taken together, SOX11 knockdown leads to alterations in genes and cellular processes related to leukemia cell motility, adhesion, differentiation, and drug response.
SOX11 protein can be detected by flow cytometry
Finally, we searched for flow cytometry markers that could serve as surrogates of SOX11 positivity. In MCL, the surface expression of CD5 is correlated with SOX11 positivity22, but in our immunohistochemical staining and microarray dataset34, no positive correlation was observed in leukemias (Fig. 5a). Alternative surrogate markers were searched for among the routinely studied cell surface proteins, but none were associated with SOX11 positivity (data not shown). Therefore, we explored the suitability of SOX11 antibodies to discriminate SOX11-positive cases by flow cytometry. Figure 5b shows that the intensity of SOX11 antibody staining could readily separate the high expressors (REH and RCH-ACV) from the low expressors (NALM-6 and KOPN-8) in leukemia cell lines, suggesting that SOX11 antibodies could possibly be used as a biomarker in the future.
Figure 5.
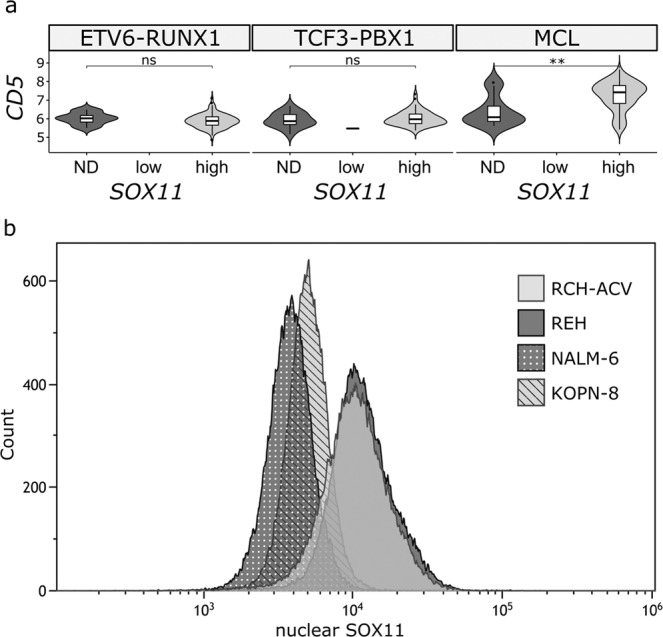
Detection of SOX11-positive cells by flow cytometry in BCP-ALL. (a) Correlation of CD5 expression with SOX11 mRNA expression in the E/R and T/P subtypes of B-ALL and MCL34. SOX11 expression was categorized into three groups, not detected (ND), low and not detected (ND). (b) Flow cytometry analysis of nuclear SOX11 expression in cell lines with either a low (NALM-6, KOPN-8) or high (REH, RCH-ACV) expression of SOX11. ND, not detected, MCL, mantle cell lymphoma. Reproduced with permission66.
Discussion
BCP-ALL is caused by a relatively small number of genetic mutations that impede normal B-cell differentiation, entail self-renewal capacity, and increase proliferative activity. This is evident in transcriptional programs that govern leukemic B-cells. We report here that SOX11, a developmental and neuronal TF6, is overexpressed in the E/R and T/P subtypes of BCP-ALL and also in novel E/R-like, IKZF1 N159Y, MEF2D rearrangement, and DUX4 rearrangement subtypes. A high expression is associated with DNA hypomethylation and a favorable clinical outcome. The results suggest that a SOX11-associated transcriptional program is related with a less-aggressive disease and indicates that SOX11 warrants further study as a biomarker for low-risk ALL patients.
We observed the high expression of SOX11 in the E/R and T/P subtypes of BCP-ALL in three separate gene expression datasets at the mRNA level34–36 and confirmed this association at the protein level by immunohistochemistry, confirming previously published data8,13,29,30 and adding further evidence at the protein level. We showed that the expression may be regulated epigenetically, that is, by hypomethylation of DNA at the SOX11 locus, similar to what was earlier reported for MCL and five cases of BCP-ALL by Vegliante et al.30. We did not have material available for epigenetics studies, such as histone modifications, as these could also have a role in SOX11 regulation30. Direct manipulation of the E/R fusion did not have any impact on SOX11 expression, suggesting that regulation is indirect.
An interesting finding relates to the clinical significance of SOX11 expression in BCP-ALL, as we observed a better OS in the SOX11-positive cases. A similar trend was also seen in EFS, RFS, and early therapy response. This finding was supported (similar trend) in another dataset with transcriptome expression profiles46. In the subgroup analysis of E/R cases, SOX11 positivity retained its prognostic significance, suggesting that (a high) SOX11 expression could possibly be utilized as a biomarker for cases with a very good prognosis. It is noteworthy that our immunohistochemical staining series spans almost two decades, and patients have been treated using several distinct NOPHO ALL chemotherapy protocols63,64. As the most recent protocols have conferred the best survival results [63, trying to replicate these findings in the most recent protocols is necessary in the future.
To aid in the screening of SOX11 positivity at diagnosis, we also successfully tested a flow cytometry-based assay in cell lines. SOX11 inclusion into the flow cytometry panel would be convenient compared with immunohistochemical staining of bone marrow biopsies, which is slow and not routinely done in all treatment centers.
SOX11 knockdown did not markedly influence cell viability or proliferation, nor did it affect chemotherapy sensitivity. In MCL, conflicting reports have been made about the effect of knockdown or SOX11 overexpression on cell proliferation and tumor growth16–23. As cell viability measurement is a relatively insensitive assay, we performed transcriptional profiling of SOX11 knockdown cells by using RNA-seq and noticed changes in the genes associated with cell migration, adhesion, oxidative phosphorylation, hypoxia, glycolysis, and differentiation, which could explain the association of SOX11 with favorable clinical outcomes. Notably, the changes observed were mostly mild to moderate. There were only few overlapping genes with previous profiling studies in both pro-B-cells18 and MCL cells16,17,20,21,65. For example, we did not see marked changes in the expression of either PAX5, as seen in MCL21, or Id1 and Tal1 in pro-B-cells18. Interestingly, MDK, which is involved in cell migration and growth, was downregulated by SOX11 knockdown in leukemia cell lines here and in an MCL cell line Z13816.
In conclusion, the association of SOX11 expression with a favorable prognosis invites further studies to confirm its prognostic value and applicability as a part of the diagnostic workup.
Supplementary information
Acknowledgements
We thank the Finnish Functional Genomics Centre, which is supported by the University of Turku, Åbo Akademi University, and Biocenter Finland. This work was supported by the Competitive State Research Financing of the Expert Responsibility area of Tampere University Hospital, as well as by grants from the Academy of Finland (project no. 276634 (M.H.), no. 277816 (O.L.)), the Finnish Cultural Foundation (Interdisciplinary Science Workshops (M.H.), Pirkanmaa Regional Fund, (T.G.)), the Sigrid Juselius Foundation (M.H. and O.L.), and the Cancer Society of Finland (MH, OL), the Paulo Foundation (O.L.), the Foundation for Pediatric Research (O.L.), the Jane and Aatos Erkko Foundation (O.L.), Tampere University (O.L.) and University of Eastern Finland (M.H.), Väre Foundation for Pediatric Cancer Research (T.G., A.M.), and the Finnish Hematology Association (T.G.). Grönroos Toni, Transcriptional regulation and cell signaling in acute lymphoblastic leukemia and hematopoiesis, Tampere University Dissertations: 74 (Tampere University, 2019)66.
Author contributions
T.G., A.M.: Study design, data collection, data interpretation, figure designing, literature search, and writing (equal contribution as first authors). S.L.: Data analysis, data interpretation, figure designing, literature search, and writing. J.M.: Data collection, data analysis, figure designing, and writing. A.N.: Data collection, data analysis, data interpretation, figure designing, and writing. L.O.: Data analysis, data interpretation, figure designing, and writing. S.R.: Data collection, data analysis, data interpretation, figure designing, and writing. Y.M-Z., J.N.: Data collection, data analysis, and data interpretation. V.P.: Data collection and data interpretation. T.P.: Data analysis. M.H.: Study design, data interpretation, and writing. O.L.: Study design, data interpretation, literature search, and writing. All authors reviewed the manuscript.
Data availability
The datasets generated and analyzed in the current study are available in the GEO repository, GSE47051 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47051), GSE123943 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123943), and GSE49031 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49031), or are included in the article or supplementary files. RNA-seq data of the patient samples46 are not publicly available, as the patient/parent consent does not cover depositing data into repositories; however, they are available from the authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Toni Grönroos and Artturi Mäkinen.
Supplementary information
is available for this paper at 10.1038/s41598-020-58970-z.
References
- 1.Jay P, et al. The human SOX11 gene: cloning, chromosomal assignment and tissue expression. Genomics. 1995;29:541–545. doi: 10.1006/geno.1995.9970. [DOI] [PubMed] [Google Scholar]
- 2.Dy P, et al. The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penzo-Méndez AI. Critical roles for SoxC transcription factors in development and cancer. Int. J. Biochem. Cell Biol. 2010;42:425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Yoshitomi H, et al. Human Sox4 facilitates the development of CXCL13-producing helper T cells in inflammatory environments. Nat. Commun. 2018;9:3762. doi: 10.1038/s41467-018-06187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattaram P, et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ek S, Dictor M, Jerkeman M, Jirström K, Borrebaeck CA. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood. 2008;111:800–805. doi: 10.1182/blood-2007-06-093401. [DOI] [PubMed] [Google Scholar]
- 9.Shepherd JH, et al. The SOX11 transcription factor is a critical regulator of basal-like breast cancer growth, invasion, and basal-like gene expression. Oncotarget. 2016;7:13106–13121. doi: 10.18632/oncotarget.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sernbo S, et al. The tumour suppressor SOX11 is associated with improved survival among high grade epithelial ovarian cancers and is regulated by reversible promoter methylation. BMC Cancer. 2011;11:405. doi: 10.1186/1471-2407-11-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sock E, et al. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hempel A, et al. Deletions and de novo mutations of SOX11 are associated with a neurodevelopmental disorder with features of Coffin-Siris syndrome. J. Med. Genet. 2016;53:152–162. doi: 10.1136/jmedgenet-2015-103393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mozos A, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica. 2009;94:1555–1562. doi: 10.3324/haematol.2009.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng W, et al. Cyclin D1-negative blastoid mantle cell lymphoma identified by SOX11 expression. Am. J. Surg. Pathol. 2012;36:214–219. doi: 10.1097/PAS.0b013e318241f050. [DOI] [PubMed] [Google Scholar]
- 15.Fernàndez V, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70:1408–1418. doi: 10.1158/0008-5472.CAN-09-3419. [DOI] [PubMed] [Google Scholar]
- 16.Conrotto P, Andréasson U, Kuci V, Borrebaeck CA, Ek S. Knock-down of SOX11 induces autotaxin-dependent increase in proliferation in vitro and more aggressive tumors in vivo. Mol. Oncol. 2011;5:527–537. doi: 10.1016/j.molonc.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo PY, et al. High-resolution chromatin immunoprecipitation (ChIP) sequencing reveals novel binding targets and prognostic role for SOX11 in mantle cell lymphoma. Oncogene. 2015;34:1231–1240. doi: 10.1038/onc.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lord M, et al. Impact of Sox11 overexpression in Ba/F3 cells. Haematologica. 2018;103:e594–e597. doi: 10.3324/haematol.2018.197467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustavsson E, et al. SOX11 expression correlates to promoter methylation and regulates tumor growth in hematopoietic malignancies. Mol. Cancer. 2010;9:187. doi: 10.1186/1476-4598-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuci V, Nordström L, Conrotto P, Ek S. SOX11 and HIG-2 are cross-regulated and affect growth in mantle cell lymphoma. Leuk. Lymphoma. 2016;57:1883–1892. doi: 10.3109/10428194.2015.1121257. [DOI] [PubMed] [Google Scholar]
- 21.Vegliante MC, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood. 2013;121:2175–2185. doi: 10.1182/blood-2012-06-438937. [DOI] [PubMed] [Google Scholar]
- 22.Kuo PY, et al. SOX11 augments BCR signaling to drive MCL-like tumor development. Blood. 2018;131:2247–2255. doi: 10.1182/blood-2018-02-832535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beekman R, Amador V, Campo E. SOX11, a key oncogenic factor in mantle cell lymphoma. Curr. Opin. Hematol. 2018;25:299–306. doi: 10.1097/MOH.0000000000000434. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, et al. The subcellular Sox11 distribution pattern identifies subsets of mantle cell lymphoma: correlation to overall survival. Br. J. Haematol. 2008;143:248–252. doi: 10.1111/j.1365-2141.2008.07329.x. [DOI] [PubMed] [Google Scholar]
- 25.Meggendorfer M, Kern W, Haferlach C, Haferlach T, Schnittger S. SOX11 overexpression is a specific marker for mantle cell lymphoma and correlates with t(11;14) translocation, CCND1 expression and an adverse prognosis. Leukemia. 2013;27:2388–2391. doi: 10.1038/leu.2013.141. [DOI] [PubMed] [Google Scholar]
- 26.Navarro A, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72:5307–5316. doi: 10.1158/0008-5472.CAN-12-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nygren L, et al. Prognostic role of SOX11 in a population-based cohort of mantle cell lymphoma. Blood. 2012;119:4215–4223. doi: 10.1182/blood-2011-12-400580. [DOI] [PubMed] [Google Scholar]
- 28.Tosic N, et al. Prognostic significance of SOX2, SOX3, SOX11, SOX14 and SOX18 gene expression in adult de novo acute myeloid leukemia. Leuk. Res. 2018;67:32–38. doi: 10.1016/j.leukres.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Dictor M, et al. Strong lymphoid nuclear expression of SOX11 transcription factor defines lymphoblastic neoplasms, mantle cell lymphoma and Burkitt’s lymphoma. Haematologica. 2009;94:1563–1568. doi: 10.3324/haematol.2009.008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vegliante MC, et al. Epigenetic activation of SOX11 in lymphoid neoplasms by histone modifications. PLoS One. 2011;6:e21382. doi: 10.1371/journal.pone.0021382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordlund J, et al. Digital gene expression profiling of primary acute lymphoblastic leukemia cells. Leukemia. 2012;26:1218–1227. doi: 10.1038/leu.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busche S, et al. Integration of high-resolution methylome and transcriptome analyses to dissect epigenomic changes in childhood acute lymphoblastic leukemia. Cancer Res. 2013;73:4323–4336. doi: 10.1158/0008-5472.CAN-12-4367. [DOI] [PubMed] [Google Scholar]
- 33.Heinäniemi M, et al. Transcription-coupled genetic instability marks acute lymphoblastic leukemia structural variation hotspots. Elife. 2016;5:e13087. doi: 10.7554/eLife.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pölönen P, et al. Hemap: An Interactive Online Resource for Characterizing Molecular Phenotypes across Hematologic Malignancies. Cancer Res. 2019;79:2466–2479. doi: 10.1158/0008-5472.CAN-18-2970. [DOI] [PubMed] [Google Scholar]
- 35.Nordlund J, et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14:r105. doi: 10.1186/gb-2013-14-9-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu Z, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teppo S, et al. Genome-wide repression of eRNA and target gene loci by the ETV6-RUNX1 fusion in acute leukemia. Genome Res. 2016;26:1468–1477. doi: 10.1101/gr.193649.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Dobin A, et al. Bioinformatics. 2013. STAR: ultrafast universal RNA-seq aligner; pp. 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 42.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 44.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering Motifs in Ranked Lists of DNA sequences. PLoS Computational Biology. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A Tool For Discovery And Visualization of Enriched GO Terms in Ranked Gene Lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marincevic-Zuniga Y, et al. Transcriptome sequencing in pediatric acute lymphoblastic leukemia identifies fusion genes associated with distinct DNA methylation profiles. J. Hematol. Oncol. 2017;10:148. doi: 10.1186/s13045-017-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swerdlow, S. H. et al. (Eds): WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (Revised 4th Edition) IARC: Lyon (2017).
- 48.Ramezani-Rad P, et al. SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia. Blood. 2013;121:148–155. doi: 10.1182/blood-2012-05-428938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhns DB, et al. Cytoskeletal abnormalities and neutrophil dysfunction in WDR1 deficiency. Blood. 2016;128:2135–2143. doi: 10.1182/blood-2016-03-706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Standing ASI, et al. Autoinflammatory periodic fever, immunodeficiency, and thrombocytopenia (PFIT) caused by mutation in actin-regulatory gene WDR1. J. Exp. Med. 2017;214:59–71. doi: 10.1084/jem.20161228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia M, et al. High expression of Midkine (MK) indicates poor prognosis in childhood acute lymphoblastic leukemia. Hematology. 2016;21:69–77. doi: 10.1179/1607845415Y.0000000050. [DOI] [PubMed] [Google Scholar]
- 52.Hu R, et al. Increased drug efflux along with midkine gene high expression in childhood B-lineage acute lymphoblastic leukemia cells. Int. J. Hematol. 2010;92:105–110. doi: 10.1007/s12185-010-0613-x. [DOI] [PubMed] [Google Scholar]
- 53.Neveu B, et al. CLIC5: a novel ETV6 target gene in childhood acute lymphoblastic leukemia. Haematologica. 2016;101:1534–1543. doi: 10.3324/haematol.2016.149740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holleman A, et al. Expression of the outcome predictor in acute leukemia 1 (OPAL1) gene is not an independent prognostic factor in patients treated according to COALL or St Jude protocols. Blood. 2006;108:1984–1990. doi: 10.1182/blood-2006-04-015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeoh EJ, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/S1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 56.Tse W, Zhu W, Chen HS, Cohen A. A novel gene, AF1q, fused to MLL in t(1;11) (q21;q23), is specifically expressed in leukemic and immature hematopoietic cells. Blood. 1995;85:650–656. doi: 10.1182/blood.V85.3.650.bloodjournal853650. [DOI] [PubMed] [Google Scholar]
- 57.Ney Garcia DR, et al. Molecular characterization of KMT2A fusion partner genes in 13 cases of pediatric leukemia with complex or cryptic karyotypes. Hematol. Oncol. 2017;35:760–768. doi: 10.1002/hon.2299. [DOI] [PubMed] [Google Scholar]
- 58.Petit A, et al. Functional analysis of the NUP98-CCDC28A fusion protein. Haematologica. 2012;97:379–387. doi: 10.3324/haematol.2011.047969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fridley BL, et al. Weinshilboum RM. Gene set analysis of purine and pyrimidine antimetabolites cancer therapies. Pharmacogenet. Genomics. 2011;21:701–712. doi: 10.1097/FPC.0b013e32834a48a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, et al. PIG7 promotes leukemia cell chemosensitivity via lysosomal membrane permeabilization. Oncotarget. 2016;7:4841–4859. doi: 10.18632/oncotarget.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chin LS, Raynor MC, Wei X, Chen HQ, Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J. Biol. Chem. 2001;276:7069–7078. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- 62.Chen YS, et al. Tiger frog virus ORF080L protein interacts with LITAF and impairs EGF-induced EGFR degradation. Virus Res. 2016;217:133–142. doi: 10.1016/j.virusres.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Toft N, et al. Results of NOPHO ALL2008 treatment for patients aged 1-45 years with acute lymphoblastic leukemia. Leukemia. 2018;32:606–615. doi: 10.1038/leu.2017.265. [DOI] [PubMed] [Google Scholar]
- 64.Schmiegelow K, et al. Nordic Society of Paediatric Haematology and Oncology. Long-term results of NOPHO ALL-92 and ALL-2000 studies of childhood acute lymphoblastic leukemia. Leukemia. 2010;24:345–354. doi: 10.1038/leu.2009.251. [DOI] [PubMed] [Google Scholar]
- 65.Wang X, et al. Gene expression profiling and chromatin immunoprecipitation identify DBN1, SETMAR and HIG2 as direct targets of SOX11 in mantle cell lymphoma. PLoS One. 2010;5:e14085. doi: 10.1371/journal.pone.0014085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grönroos, T. Transcriptional regulation and cell signaling in acute lymphoblastic leukemia and hematopoiesis. Tampere University Dissertations: 74 (Tampere University, (2019)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in the current study are available in the GEO repository, GSE47051 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47051), GSE123943 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123943), and GSE49031 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49031), or are included in the article or supplementary files. RNA-seq data of the patient samples46 are not publicly available, as the patient/parent consent does not cover depositing data into repositories; however, they are available from the authors upon reasonable request.