Abstract
Primary lateral sclerosis and amyotrophic lateral sclerosis are primarily associated with motor cortex and corticospinal tract pathology. A standardised, prospective, single-centre neuroimaging protocol was used to characterise thalamic, hippocampal and basal ganglia involvement in 33 patients with primary lateral sclerosis (PLS), 100 patients with amyotrophic lateral sclerosis (ALS), and 117 healthy controls. “Widespread subcortical grey matter degeneration in primary lateral sclerosis: a multimodal imaging study with genetic profiling” [1] Imaging data were acquired on a 3 T MRI system using a 3D Inversion Recovery prepared Spoiled Gradient Recalled echo sequence. Model based segmentation was used to estimate the volumes of the thalamus, hippocampus, amygdala, caudate, pallidum, putamen and accumbens nucleus in each hemisphere. The hippocampus was further parcellated into cytologically-defined subfields. Total intracranial volume (TIV) was estimated for each participant to aid the interpretation of subcortical volume alterations. Group comparisons were corrected for age, gender, TIV, education and symptom duration. Considerable thalamic, hippocampal and accumbens nucleus atrophy was detected in PLS compared to healthy controls and selective dentate, molecular layer, CA1, CA3, and CA4 hippocampal pathology was also identified. In ALS, additional volume reductions were noted in the amygdala, left caudate and the hippocampal-amygdala transition area of the hippocampus. Our imaging data provide evidence of extensive and phenotype-specific patterns of subcortical degeneration in PLS.
Keywords: Primary lateral sclerosis, Amyotrophic lateral sclerosis, Neuroimaging, MRI, Thalamus, Hippocampus, Basal ganglia, Biomarkers
Specifications Table
| Subject | Primary Lateral Sclerosis, Radiology, Neuroimaging |
| Specific subject area | MRI, Grey matter volumetry, Hippocampus |
| Type of data | Volumetric neuroimaging data with standardised acquisition |
| How data were acquired | Imaging data were acquired on a Philips Achieva 3T MRI scanner (Philips Medical Systems, Best, The Netherlands) with an 8-channel head coil. |
| Data format | Estimated marginal means and standard error for subcortical grey matter structures and hippocampal subfields adjusted for total-intracranial volume, age, gender, and education. |
| Parameters for data collection | 3D–T1-weighted sequence: spatial resolution: 1 × 1 × 1 mm, Field of view: 256 × 256 × 160 mm, TR/TE = 8.5/3.9 ms, TI = 1060 ms, flip angle = 8°, SENSE factor = 1.5, sagittal acquisition; 256 slices. |
| Description of data collection | The protocol, consent forms, recruitment procedures, and data management were approved by the institutional ethics committee. All participants provided informed consent prior to inclusion. Participating ALS patients were diagnosed according to the El Escorial research criteria and PLS patients were diagnosed according to the Gordon criteria. Patients underwent standardised neurological assessments and MRI data were acquired with uniform pulse sequence settings and anonymised. |
| Data source location | Institution: Computational neuroimaging group, Trinity Biomedical Sciences Institute, Trinity College Dublin City/Town/Region: Dublin Country: Ireland |
| Data accessibility | The subcortical grey matter profile and hippocampal subfield features of the three groups are presented as raw data in box plots and contrasted in statistical tables using the relevant covariates. |
| Related research article | Authors: Eoin Finegan, Stacey Li Hi Shing, Rangariroyashe H. Chipika, Mark A. Doherty, Jennifer C. Hengeveld, Alice Vajda, Colette Donaghy, Niall Pender, Russell L. McLaughlin, Orla Hardiman, Peter Bede Title: Widespread subcortical grey matter degeneration in primary lateral sclerosis: a multimodal imaging study with genetic profiling Journal: Neuroimage Clinical DOI: https://doi.org/10.1016/j.nicl.2019.102089 |
Value of the Data
|
1. Data
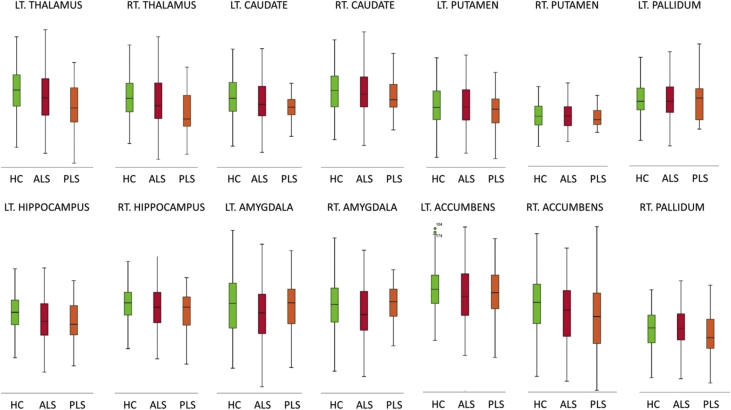
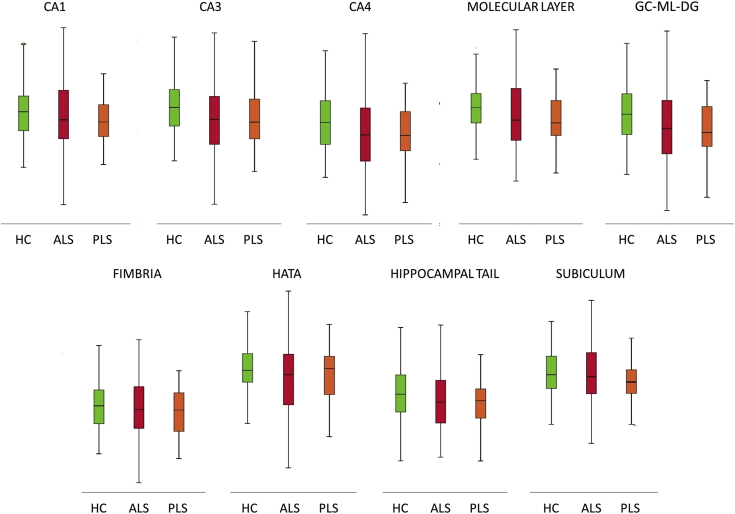
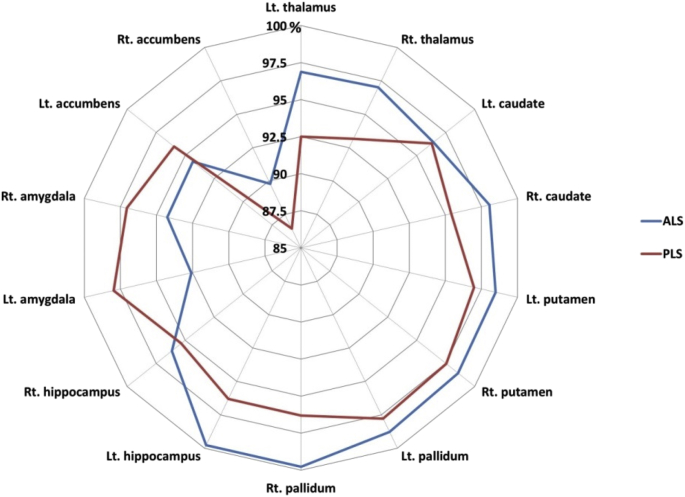
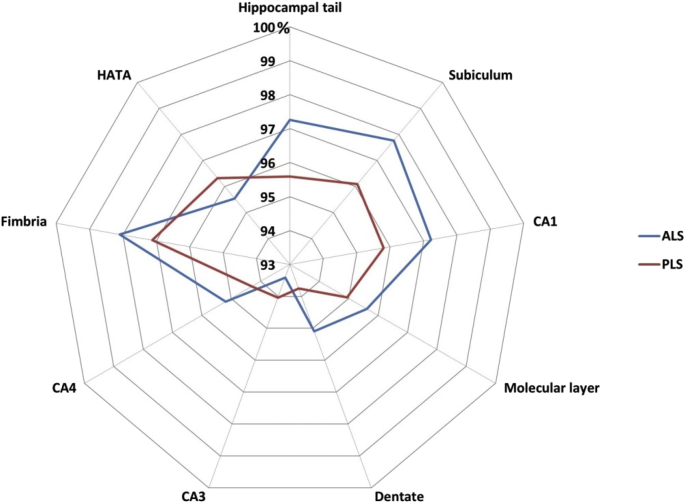
The majority of primary lateral sclerosis and amyotrophic lateral sclerosis studies focus on the motor cortex and corticospinal tracts [[2], [3], [4], [5]]. In this dataset (Table 1) we present total intracranial volume, thalamus, hippocampus, amygdala, caudate, pallidum, putamen and accumbens nucleus volumes for 33 patients with primary lateral sclerosis, 100 patients with amyotrophic lateral sclerosis and 117 age and gender-matched healthy controls. Data for bilateral structures are presented separately. Additionally, we provide hippocampal subfield volume estimates for the CA1, CA2/3, CA4, fimbria, subiculum, hippocampal tail, molecular layer, dentate gyrus, and hippocampal-amygdala transition area (HATA). Accompanying clinical and demographic characteristics are presented to highlight that the patient groups were matched for age, gender and ALSFRS-r and differed in years of education and symptom duration. Raw subcortical volumetric data (Fig. 1) and raw hippocampal subfield characteristics (Fig. 2) are presented in box plots for each cohort. The estimated marginal means and standard error of subcortical structures adjusted for the relevant clinical, radiological and demographic variables (age, gender, education, total intracranial volume, symptom duration) are summarised in Table 2, and hippocampal subfield profiles are presented in Table 3 using the same covariates. Based on estimated marginal means corrected for age, gender, total intracranial volumes and education, the comparative profile of the two patient groups are also illustrated in radar plots with reference to healthy controls. Contrary to our original paper [1] where percentage change was presented as absolute values, here, ‘100%’ represents the estimated marginal means of healthy controls as normative values and the concentric circles depict the selective atrophy profiles of the ALS and PLS cohorts (Fig. 3, Fig. 4).
Table 1.
Data categories and measures ALS = amyotrophic lateral sclerosis; ALSFRS-R = amyotrophic lateral sclerosis functional rating scale-revised; PLS = Primary lateral sclerosis; CA = Cornu Ammonis; GC-DG = granule cell layer of the dentate gyrus; HATA = hippocampus-amygdala transition area, Lt = Left, Rt = Right.
| Data categories | Specific measures |
|---|---|
| Demographic variables | Age (year) |
| Gender (Male/Female) | |
| Years of education (years) | |
| Handedness (Rt/Lt) | |
| Clinical data for ALS and PLS | Symptom duration (months) |
| ALSFRS-R (max 48) | |
| Subcortical grey matter structure volumes | hippocampus (mm3) |
| amygdala (mm3) | |
| thalamus (mm3) | |
| nucleus accumbens (mm3) | |
| caudate nucleus (mm3) | |
| putamen (mm3) | |
| pallidum (mm3) | |
| Hippocampal subfield volumes | CA1 (mm3) |
| CA2/CA3 (mm3) | |
| CA4 (mm3) | |
| Fimbria (mm3) | |
| Subiculum (mm3) | |
| Molecular layer (mm3) | |
| GC-ML-DG (mm3) | |
| HATA (mm3) |
Fig. 1.
Subcortical grey matter volumes in primary lateral sclerosis (PLS), amyotrophic lateral sclerosis (ALS) and healthy controls (HC).
Fig. 2.
Hippocampal subfield volumes in primary lateral sclerosis (PLS), amyotrophic lateral sclerosis (ALS) and healthy controls (HC). CA = Cornu Ammonis; GC-ML-DG = Granule Cell and Molecular Layer of the Dentate Gyrus; HATA = hippocampus-amygdala transition area.
Table 2.
The demographic, clinical and subcortical grey matter profile of the three groups. ALSFRS-r = the revised ALS functional rating scale, EMM = estimated marginal mean, M = Mean, N/a = Not applicable, SE = standard error, SD = standard deviation. Estimate marginal means are adjusted with the following values age = 58.77, gender = 1.45, education = 13.78, and total intracranial volume = 1429699.38.
| PLS n = 33 |
ALS n = 100 |
Healthy Controls n = 117 |
p value | |
|---|---|---|---|---|
| Age M/SD | 60.5 (10.5) | 59.8 (11.2) | 57.4 (11.9) | 0.19 |
| Gender (M/F) | 19/14 | 62/38 | 56/61 | 0.11 |
| Education (years) M/SD | 12.9 (3.4) | 13.5 (3.2) | 14.3 (3.3) | 0.04 |
| Handedness (R/L) | 29/4 | 90/10 | 109/8 | 0.55 |
| ALSFRS-r (max 48) M/SD | 34.4 (5.3) | 36.6 (7.5) | N/a | 0.11 |
| Symptom Duration (months) M/SD | 121.76 (68.7) | 20.6 (14.3) | N/a | 7.1E-16 |
| Total intracranial volume (mm3) M/SD | 1439675.5 (145614.1) | 1440623.0 (149085.7) |
1417549.2 (128172.3) | 0.432 |
| Hippocampus (Left) EMM/SE | 3566.6 (84.4) | 3624.1 (48.5) | 3805.1 (45.1) | 0.007 |
| Hippocampus (Right) EMM/SE | 3707.4 (85.4) | 3739.1 (49.1) | 3888.5 (45.7) | 0.045 |
| Amygdala (Left) EMM/SE | 1189.4 (42.3) | 1124.0 (24.4) | 1213.9 (22.7) | 0.03 |
| Amygdala (Right) EMM/SE | 1144.2 (44.0) | 1111.2 (25.3) | 1178.8 (23.5) | 0.16 |
| Thalamus (Left) EMM/SE | 7044.0 (97.9) | 7376.1 (56.3) | 7614.0 (52.4) | 2E-6 |
| Thalamus (Right) EMM/SE | 6896.5 (81.5) | 7181.9 (52.6) | 7402.8 (48.9) | 5E-6 |
| Nucleus accumbens (Left) EMM/SE | 473.8 (19.6) | 465.8 (11.3) | 493.8 (10.5) | 0.19 |
| Nucleus accumbens (Right) EMM/SE | 326.9 (19.0) | 339.6 (10.9) | 378.2 (10.2) | 0.01 |
| Caudate nucleus (Left) EMM/SE | 3283.3 (60.0) | 3287.5 (34.5) | 3409.6 (32.1) | 0.02 |
| Caudate nucleus (Right) EMM/SE | 3412.4 (66.3) | 3506.8 (38.1) | 3576.7 (35.5) | 0.08 |
| Putamen (Left) EMM/SE | 4550.8 (82.3) | 4620.7 (47.3) | 4692.2 (44.0) | 0.27 |
| Putamen (Right) EMM/SE | 4625.6 (86.7) | 4674.1 (50.0) | 4741.9 (46.3) | 0.41 |
| Pallidum (Left) EMM/SE | 1733.7 (36.3) | 1751.6 (20.8) | 1773.1 (19.4) | 0.58 |
| Pallidum (Right) EMM/SE | 1713.7 (40.5) | 1775.4 (23.3) | 1779.5 (21.7) | 0.34 |
Table 3.
The hippocampal profile of the three groups. ALSFRS-r = the revised ALS functional rating scale, EMM = estimated marginal mean, M = Mean, N/a = Not applicable, SE = standard error, SD = standard deviation. Estimate marginal means are adjusted with the following values age = 58.77, gender = 1.45, education = 13.78, and total intracranial volume = 1429699.38.
| PLS n = 33 |
ALS n = 100 |
Healthy Controls n = 117 |
p value | |
|---|---|---|---|---|
| CA1 (mm3) EMM/SE | 632.8 (10.8) | 642.2 (6.2) | 660.5 (5.8) | 0.03 |
| CA2/CA3 (mm3) EMM/SE | 214.1 (5.0) | 212.6 (2.9) | 227.6 (2.7) | 5.2E-4 |
| CA4 (mm3) EMM/SE | 254.6 (4.6) | 257.0 (2.6) | 270.0 (2.5) | 4.2E-4 |
| Fimbria (mm3) EMM/SE | 74.9 (2.9) | 75.6 (1.6) | 77.1 (1.5) | 0.72 |
| Subiculum (mm3) EMM/SE | 421.6 (7.4) | 428.9 (4.3) | 438.7 (4.0) | 0.08 |
| Molecular layer (mm3) EMM/SE | 559.5 (9.3) | 563.5 (5.4) | 589.3 (5.0) | 6.5E-4 |
| GC-ML-DG (mm3) EMM/SE | 292.3 (5.4) | 296.5 (3.1) | 311.8 (2.9) | 2.9E-4 |
| HATA (mm3) EMM/SE | 62.7 (1.5) | 62.2 (0.9) | 65.1 (0.8) | 0.04 |
Fig. 3.
The subcortical volumetric profile of PLS and ALS with reference to healthy controls. Estimated marginal means of volumes were calculated for each structure with the following values age = 58.77, gender = 1.45, education = 13.78, and total intracranial volume = 1429699.38. The estimated marginal means of healthy controls represent 100%.
Fig. 4.
The hippocampal volumetric profile of PLS and ALS with reference to healthy controls. Estimated marginal means of volumes were calculated for each structure with the following values age = 58.77, gender = 1.45, education = 13.78, and total intracranial volume = 1429699.38. The estimated marginal means of healthy controls represent 100%.
2. Experimental design, materials, and methods
Following institutional ethics approval, patients were recruited from a national motor neuron disease clinic and data were acquired with a standardised protocol [6]. Participating ALS patients were diagnosed according to the El Escorial research criteria and PLS patients were diagnosed according to the Gordon criteria. The protocol was specifically designed to characterise subcortical grey matter degeneration in PLS based on evidence of extra-motor involvement in other motor neuron diseases [[7], [8], [9], [10], [11]]. T1-weighted images were acquired with a spatial resolution of 1 × 1 × 1mm and field of view of 256 × 256 × 160 mm using a 3D Inversion Recovery prepared Spoiled Gradient Recalled echo (IR-SPGR) sequence. Pulse sequence settings are as follows: repetition time (TR) = 8.5 ms, echo time (TE) = 3.9 ms, Inversion time (TI) = 1060 ms, flip angle = 8°, SENSE factor = 1.5 [12]. Raw MRI data underwent thorough quality control before pre-processing. Following ‘deskulling’ and spatial registration, model based segmentation was used the estimate subcortical volumes using FSL-FIRST of the FMRIB's Software Library (FSL) [13,14]. Raw volumetric data were recorded for the hippocampus, amygdala, thalamus, nucleus accumbens, caudate nucleus, putamen, and pallidum in each hemisphere. Subsequently, the hippocampus of each participant was segmented into cytologically-defined subfields using version 6.0 of the FreeSurfer image analysis suite to estimate volumes of the CA1, CA2/3, CA4, fimbria, subiculum, hippocampal tail, molecular layer, dentate gyrus, and the hippocampal-amygdala transition area [15]. Analyses of covariance (ANCOVA) were used to explore intergroup volumetric differences using age, education, gender and TIV as covariates [16,17]. PLS versus ALS contrasts were also corrected for symptom duration. To illustrate disease-specific volumetric traits in PLS and ALS, the estimated marginal means of each structure were plotted on radar charts with reference to healthy controls.
Acknowledgments
We thank all the patients, their families and the healthy controls for participating in this research project. Without their support, this project would not have been possible. Peter Bede and the computational neuroimaging group is supported by the Health Research Board (HRB – Ireland; HRB EIA-2017-019), the Andrew Lydon scholarship, the Iris O'Brien Foundation, and the Research Motor Neuron (RMN-Ireland) Foundation. Russell L McLaughlin is supported by the Motor Neurone Disease Association (957-799) and Science Foundation Ireland (17/CDA/4737). Mark A Doherty is supported by Science Foundation Ireland (15/SPP/3244).
Conflict of Interest
Peter Bede is the associate editor of Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, member of the UK Motor Neuron Disease Association (MNDA) Research Advisory Panel, the steering committee of Neuroimaging Society in ALS (NiSALS) and the medical patron of the Irish Motor Neuron Disease Association (IMNDA). These affiliations had no impact on the analyses or interpretation of the presented data.
References
- 1.Finegan E., Li Hi Shing S., Chipika R.H., Doherty M.A., Hengeveld J.C., Vajda A., Donaghy C., McLaughlin R.L., Pender N., Hardiman O., Bede P. Widespread subcortical grey matter degeneration in primary lateral sclerosis: a multimodal imaging study with genetic profiling. NeuroImage Clin. 2019;24:1–11. doi: 10.1016/j.nicl.2019.102089. Epub 12 November 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuster C., Hardiman O., Bede P. Development of an automated MRI-based diagnostic protocol for amyotrophic lateral sclerosis using disease-specific pathognomonic features: a quantitative disease-state classification study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167331. Epub 2016/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bede P., Hardiman O. Longitudinal structural changes in ALS: a three time-point imaging study of white and gray matter degeneration. Amyotroph Lateral Scler. Frontotemporal Degeneration. 2018;19:232–241. doi: 10.1080/21678421.2017.1407795. Epub 2017/12/08. [DOI] [PubMed] [Google Scholar]
- 4.Finegan E., Chipika R.H., Shing S.L.H., Hardiman O., Bede P. Primary lateral sclerosis: a distinct entity or part of the ALS spectrum? Amyotroph Lateral Scler. Frontotemporal Degeneration. 2019:1–13. doi: 10.1080/21678421.2018.1550518. Epub 2019/01/19. [DOI] [PubMed] [Google Scholar]
- 5.Bede P., Querin G., Pradat P.F. The changing landscape of motor neuron disease imaging: the transition from descriptive studies to precision clinical tools. Curr. Opin. Neurol. 2018;31:431–438. doi: 10.1097/WCO.0000000000000569. Epub 2018/05/12. [DOI] [PubMed] [Google Scholar]
- 6.Schuster C., Elamin M., Hardiman O., Bede P. The segmental diffusivity profile of amyotrophic lateral sclerosis associated white matter degeneration. Eur. J. Neurol. 2016;23:1361–1371. doi: 10.1111/ene.13038. Epub 2016/05/22. [DOI] [PubMed] [Google Scholar]
- 7.Christidi F., Karavasilis E., Rentzos M., Kelekis N., Evdokimidis I., Bede P. Clinical and radiological markers of extra-motor deficits in amyotrophic lateral sclerosis. Front. Neurol. 2018;9:1005. doi: 10.3389/fneur.2018.01005. Epub 2018/12/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christidi F., Karavasilis E., Velonakis G., Ferentinos P., Rentzos M., Kelekis N., Evdokimidis I., Bede P. The clinical and radiological spectrum of hippocampal pathology in amyotrophic lateral sclerosis. Front. Neurol. 2018;9:523. doi: 10.3389/fneur.2018.00523. Epub 2018/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finegan E., Chipika R.H., Li Hi Shing S., Hardiman O., Bede P. Pathological crying and laughing in motor neuron disease: pathobiology, screening, intervention. Front. Neurol. 2019;10:260. doi: 10.3389/fneur.2019.00260. Epub 2019/04/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omer T., Finegan E., Hutchinson S., Doherty M., Vajda A., McLaughlin R.L., Pender N., Hardiman O., Bede P. Neuroimaging patterns along the ALS-FTD spectrum: a multiparametric imaging study. Amyotroph Lateral Scler. Frontotemporal Degeneration. 2017:1–13. doi: 10.1080/21678421.2017.1332077. Epub 2017/06/01. [DOI] [PubMed] [Google Scholar]
- 11.Elamin M., Pinto-Grau M., Burke T., Bede P., Rooney J., O'Sullivan M., Lonergan K., Kirby E., Quinlan E., Breen N., Vajda A., Heverin M., Pender N., Hardiman O. Identifying behavioural changes in ALS: validation of the Beaumont Behavioural Inventory (BBI) Amyotroph Lateral Scler. Frontotemporal Degeneration. 2017;18:68–73. doi: 10.1080/21678421.2016.1248976. Epub 2016/11/30. [DOI] [PubMed] [Google Scholar]
- 12.Finegan E., Chipika R.H., Li Hi Shing S., Doherty M.A., Hengeveld J.C., Vajda A., Donaghy C., McLaughlin R.L., Pender N., Hardiman O., Bede P. The clinical and radiological profile of primary lateral sclerosis: a population-based study. J. Neurol. 2019;266:2718–2733. doi: 10.1007/s00415-019-09473-z. Epub 2019/07/22. [DOI] [PubMed] [Google Scholar]
- 13.Bede P., Omer T., Finegan E., Chipika R.H., Iyer P.M., Doherty M.A., Vajda A., Pender N., McLaughlin R.L., Hutchinson S., Hardiman O. Connectivity-based characterisation of subcortical grey matter pathology in frontotemporal dementia and ALS: a multimodal neuroimaging study. Brain Imag. Behav. 2018;12:1696–1707. doi: 10.1007/s11682-018-9837-9. Epub 2018/02/10. [DOI] [PubMed] [Google Scholar]
- 14.Bede P., Iyer P.M., Schuster C., Elamin M., McLaughlin R.L., Kenna K., Hardiman O. The selective anatomical vulnerability of ALS: 'disease-defining' and 'disease-defying' brain regions. Amyotroph Lateral Scler. Frontotemporal Degeneration. 2016;17:561–570. doi: 10.3109/21678421.2016.1173702. Epub 2016/04/19. [DOI] [PubMed] [Google Scholar]
- 15.Christidi F., Karavasilis E., Rentzos M., Velonakis G., Zouvelou V., Xirou S., Argyropoulos G., Papatriantafyllou I., Pantolewn V., Ferentinos P., Kelekis N., Seimenis I., Evdokimidis I., Bede P. Hippocampal pathology in amyotrophic lateral sclerosis: selective vulnerability of subfields and their associated projections. Neurobiol. Aging. 2019;84:178–188. doi: 10.1016/j.neurobiolaging.2019.07.019. Epub 2019/10/20. [DOI] [PubMed] [Google Scholar]
- 16.Schuster C., Hardiman O., Bede P. Survival prediction in Amyotrophic lateral sclerosis based on MRI measures and clinical characteristics. BMC Neurol. 2017;17:73. doi: 10.1186/s12883-017-0854-x. Epub 2017/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bede P., Elamin M., Byrne S., Hardiman O. Sexual dimorphism in ALS: exploring gender-specific neuroimaging signatures. Amyotroph Lateral Scler. Frontotemporal Degeneration. 2014;15:235–243. doi: 10.3109/21678421.2013.865749. Epub 2013/12/19. [DOI] [PubMed] [Google Scholar]