Abstract
Molecular dynamics (MD) simulations were used to investigate the binding of four ligands to the Val122Ile mutant of the protein transthyretin. Dissociation, misfolding, and subsequent aggregation of mutated transthyretin proteins are associated with the disease Familial Amyloidal Cardiomyopathy. The ligands investigated were the drug candidate AG10 and its decarboxy and N-methyl derivatives along with the drug tafamidis. These ligands bound to the receptor in two halogen binding pockets (HBP) designated AB and A’B’. Inter-ligand distances, solvent accessible surface areas, root mean squared deviation measurements, and extracted structures showed very little change in the AG10 ligands' conformations or locations within the HBP during the MD simulation. In addition, the AG10 ligands experienced stable, two-point interactions with the protein by forming hydrogen bonds with Ser-117 residues in both the AB and A’B’ binding pockets and Lysine-15 residues found near the surface of the receptor. Distance measurements showed these H-bonds formed simultaneously during the MD simulation. Removal of the AG10 carboxylate functional group to form decarboxy-AG10 disrupted this two-point interaction causing the ligand in the AB pocket to undergo a conformational change during the MD simulation. Likewise, addition of a methyl group to the AG10 hydrazone functional group also disrupted the two-point interaction by decreasing hydrogen bonding interactions with the receptor. Finally, MD simulations showed that the tafamidis ligands experienced fewer hydrogen bonding interactions than AG10 with the protein receptor. The tafamidis ligand in pocket A’B’ was also found to move deeper into the HBP during the MD simulation.
Keywords: Molecular dynamics simulation, Transthyretin, Tafamidis, AG10, Val122Ile mutant
Abbreviations: MD, Molecular Dynamics; TTR, Transthyretin; HBP, Halogen Binding Pocket; V122I, Val122Ile mutant of transthyretin; FAC, Familial Amyloidal Cardiomyopathy; SASA, Solvent Accessible Surface Area; RMSD, Root Mean Squared Deviation; DO-AG10, Decarboxy-AG10
Graphical abstract
Highlights
-
•
The ligand AG10 forms a two-point interaction with the Val122Ile mutant of transthyretin.
-
•
The two point interaction is comprised of hydrogen bonds to Ser-117 and Lys-15 residues.
-
•
Two point interactions are interrupted when structural modifications are made to AG10.
1. Introduction
Familial Amyloidal Cardiomyopathy (FAC) is a rare genetic disorder caused by the deposition of amyloid plaques in the heart [1]. These plaques infiltrate the cardiac muscle and lead to diastolic dysfunction, the malfunctioning of the ventricles, and eventually to heart failure. They can also damage the nerves, lungs, and other organs. Most cases of FAC onset after the age of sixty and have a prior onset of carpal tunnel syndrome [2]. Symptom relief treatments include pacemakers and both drug and gene therapies [1].
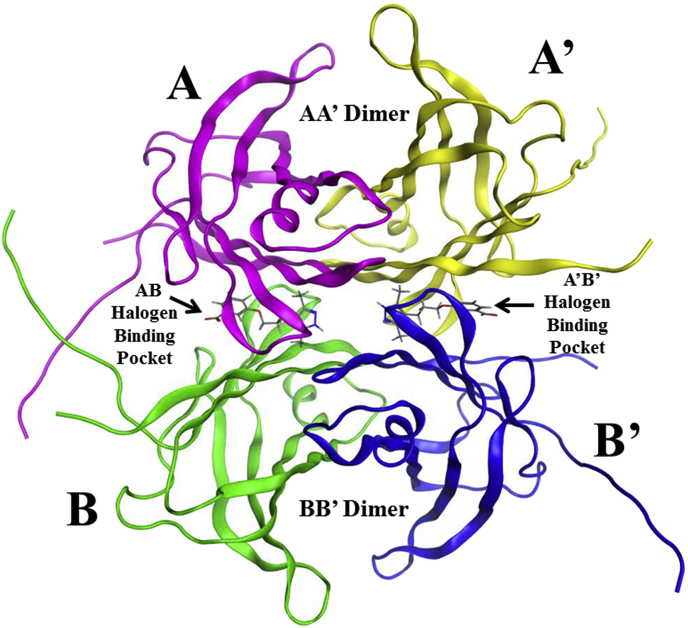
The amyloid plaques associated with FAC result from the misfolding and subsequent aggregation of the protein transthyretin (TTR). TTR is produced in the liver and the brain's choroid plexus. Its main function is the transport of the hormone thyroxine in both the blood and the cerebrospinal fluid. TTR is a tetramer composed of four identical 127 amino acid monomers. Each monomer has eight β-sheets and a shorter α-helix [3]. As shown in Fig. 1, two TTR monomers associate via hydrogen bonds to form stable dimers designated AA′ and BB’. The two dimers associate further to form the TTR tetramer. Thyroxine binds to the tetramer in two identical T4-binding sites or Halogen Binding Pockets (HBP) found at the interface of the two dimers. T4 and HBP are different designations or labels given to the same binding sites. The later designation, i.e. halogen binding pocket, is used here. Furthermore, in order to distinguish between the two ligands, we will also refer to the binding pockets as AB and A’B’ (see Fig. 1) for the pockets formed at the interface of monomers A and B and A′ and B′, respectively. [3]. While the pockets are identical, over a short time period during an MD simulation the two ligands may show different behavior. Designating the HBP as AB and A’B’ will allow these differences to be described.
Fig. 1.
Transthyretin (TTR) Structure. Halogen binding pockets (HBP) and AA′ and BB′ dimers are shown.
As stated above, the disease FAC is associated with the stability, dissociation, and misfolding of TTR [[1], [2], [3], [4]]. In addition, many point mutations of the TTR protein have been reported [3,5]. Some of these mutations destabilize the tetramer, while others have a stabilizing effect. Tetramer stability is also affected by external factors such as pH and temperature [3]. Point mutations that destabilize the TTR protein have been shown to facilitate its disassociation into monomers [[3], [4], [5], [6], [7]]. These monomers can then undergo a conformational change which initiates a cascade of events leading to the misfolded monomers aggregating to form amyloid plaques [[8], [9], [10]]. Plaque formation by mutated forms of TTR is associated with the hereditary form of FAC. While over 100 point mutations of TTR have been reported, the TTR mutant investigated here is V122I, or a substitution of Val-122 with an Isoleucine. Approximately 3% of the African American community has been shown to carry this mutation [11].
Liver transplantation has been used as a treatment for FAC because it suppresses production of the mutant TTR protein [12]. In addition, the drug tafamidis (Fig. 2(c)) was shown in 2012 to bind to the V122I mutant in its HBP and inhibit V122I tetramer disassociation [13]. Subsequent work investigated the safety and clinical efficacy of the drug [14]. The long-term efficacy and safety of tafamidis for periods up to 5–8 years were investigated in 2016. While at that time the risk of adverse events were difficult to evaluate based on the small number of patients who received the drug, the overall results overall appeared to be favorable [14]. Pfizer in fact recently won FDA approval for the use of two formulations of tafamidis to treat transthyretin amyloids [15,16].
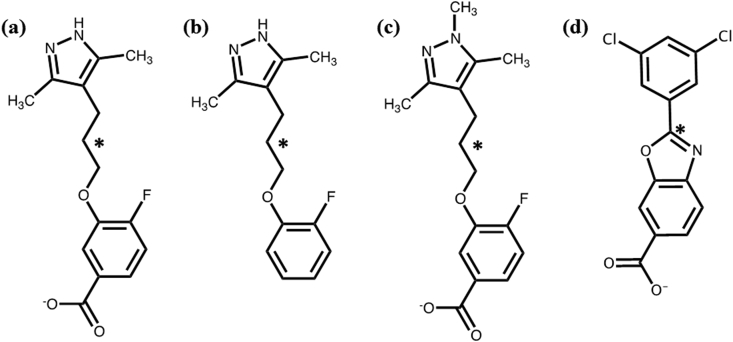
Fig. 2.
Structures of (a) AG10, (b) decarboxy-AG10 (DO-AG10), (c) N-methyl AG10, and (d) tafamidis.
However, since there are still relatively few drug treatments for FAC, the development of additional FAC drug therapies is an active area of research [[17], [18], [19], [20], [21], [22], [23], [24], [25]]. In this context, the drug candidate AG10 was investigated by Penshala et al., in 2013 [18]. The structure of this compound is shown in 2(a). The structure in Fig. 2(b) was also investigated by Penshala et al. This compound differs from AG10 only by the absence of a carboxylate functional group. Therefore, we refer to this compound as decarboxy-AG10 or DO-AG10. It has been reported that both AG10 and DO-AG10, like tafamidis, bind to V122I in the HBP, stabilize the V122I tetramer, and prevent transthyretin amyloids [18]. Since 2013, additional studies have also investigated AG10 binding to TTR [[17], [18], [19]] and Phase 1 clinical trials of the drug's safety and tolerability are currently underway [22].
As stated above, DO-AG10 and AG10 differ only by the presence of a carboxylate (-CO2-) functional group para to the fluorine atom on the drugs’ aromatic rings. It was hypothesized by Penshala et al. that the presence of this –CO2- in AG10 would allow additional favorable interactions to occur between the ligand and the V122I receptor [18]. Tafamidis also has a –CO2- functional group in a similar location [13]. In this project, molecular dynamics (MD) simulations were used to investigate this hypothesis and to characterize how both DO-AG10 and AG10 interact with the V122I mutant. MD simulations of the tafamidis: V122I complex are presented as well.
The work reported here used the AG10:V122I crystal structure as a starting point for the MD simulations. This crystal structure was also published Penshala et al., in 2013 [18]. It showed that AG10 inserted into the V122I HBP and experienced a two-point interaction with the amino acids present within, respectively the inner and outer HBP [18]. The primary inner pocket interaction was a hydrogen-bond formed between V122I Ser-117 and the NH atom of AG10's 5-membered ring. The primary outer-pocket interactions were water mediated electrostatic attractions and hydrogen bonds between Lys-15 of V122I and the AG10 carboxylate functional group [18].
Finally, the work presented here builds on previous MD simulation research with TTR mutants and TTR receptor: ligand complexes. For example, MD simulation studies of wild type (WT) and mutant TTR proteins have investigated the protein's conformational flexibility [8] and conformational intermediates that may lead to amyloid fibril formation [10]. MD simulations of V30 M, L55P, and Y116S variants of TTR have been reported as well [[6], [7], [8]] along with thermodynamic investigations of thyroxine binding to the wt protein [26]. Finally, a more recent study by Yee et al. combined MD simulation, neutron crystallography, and mass spectrometry to show that the T119 M mutation destabilizes the protein's dimer-dimer interface and thus overall tetramer stability [27]. Finally, studies by Yee et al. with receptor: ligand complexes showed that ligands like tafamidis and protective TTR mutations stabilize the TTR tetramer in similar ways [27].
2. Materials and methods
Structures of the V122I: AG10 and V122I: tafamidis complexes were taken from the Protein Data Bank. The PDB codes were 4HIQ and 4HIS, respectively [28]. DO-AG10 and N–CH3-AG10 structures were generated by modifying the ligand's structure with the software package MOE (Molecular Operating Environment, Chemical Computing Group. Inc.) [29]. MOE was also used to add missing residues to the proteins' N and C termini and to prepare the structures for MD simulation runs. AMBER 14 and the ff14SB force field [30,31] were then used to carry out a 90–100 ns MD simulation on each system containing the V122I: ligand complex, sodium counter-ions, and approximately 18000 TIP3P water molecules. All MD simulation analyses included an energy minimization step followed by a 20 ps MD simulation to bring the system to 300 K. A one ns MD simulation was used to equilibrate to a pressure of 1 atm and finally the 90–100 ns MD simulation production run was carried out. The production run employed cubic periodic boundary conditions, the MD simulation time step was two fs, and structures were stored every 0.2 ps. The system was at physiological pH in each MD simulation performed. The mm-PBSA method was used to calculate the binding free energies for AG10, DO-AG10, N–CH3-AG10, and tafamidis [32]. These free energy values represent the difference between the mm-PBSA free energy of the V122I:ligand complex and the sum of the individual free energies of the ligand and the receptor [32]. The cpptraj utility in AMBER 14 was used to perform all trajectory analyses. In the hydrogen bond analyses, the Amber software used both the distance and angle between the H-bond donor and acceptor atoms to determine if a given hydrogen bond was present in each frame of the MD simulation. The H-bond percent occupancies reported in Table 1, Table 2, Table 3 are the ratio of the frames in which a given H-bond was present over the total frames in the MD simulation [30].
Table 1.
Hydrogen bonds detected in the AG10: V122I MD simulation. The H-bonds reported for Ser and Lys residues form with the amino acids’ side chain –OH and –NH3+ functional groups.
| AG10 in AB Pocket | ||
|---|---|---|
| Acceptor | Donor | Percent Occupancy |
| Ser-117 (B) –OH | AG10 NH | 59.4 |
| AG10 N | Ser-117 (A) –OH | 55.8 |
| AG10 CO2− |
Lys-15 (A) |
18.1, 16.0, 14.8, 13.7, 13.5, 13.4 |
|
AG10 in A’B’ Pocket | ||
| AG10 N | Ser-117(B′) –OH | 64.4 |
| Ser-117(A′) –OH | AG10 NH | 35.9 |
| AG10 CO2− | Lys-15(A′) –NH3+ | 15.5, 15.3, 13.0 |
| AG10 CO2− | Lys-15(B′) –NH3+ | 12.2, 10.0, 8.3 |
Table 2.
Intermolecular hydrogen bonds detected in the DO-AG10: V122I MD simulation. The H-bonds reported for Ser and Thr residues form with the amino acid's side chain –OH.
| DO-AG10 in AB Pocket | ||
|---|---|---|
| Acceptor | Donor | Percent Occupancy |
| Thr-118 (B) | DO-AG10 NH | 43.8 |
| DO-AG10 –O- | Thr-119 (A) | 27.1 |
| Ser-117(B) |
DO-AG10 NH |
10.1 |
|
DO-AG10 in A’B’ Pocket | ||
| Ser-117(A′) | DO-AG10 NH | 55.7 |
| DO-AG-10 N | Ser-117(B′) | 47.5 |
Table 3.
Hydrogen bonds detected in the Tafamidis: V122I MD simulation. The H-bonds reported for Thr and Lys residues form with the amino acid's side chain –OH and –NH3+ functional groups.
| Tafamidis in AB Pocket | ||
|---|---|---|
| Acceptor | Donor | Percent Occupancy |
| Tafamidis N | Thr-119(A) | 63.7 |
| Tafamidis –CO2- | Lys-15 (A) | 8.7, 8.2, 8.1 |
| Tafamidis –CO2- |
Lys-15 (A) |
5.9, 5.4, 5.3 |
|
Tafamidis in A’B’ Pocket | ||
| Tafamidis N | Thr-106(A′) | 10.0, 7.1 |
| Tafamidis –CO2- | Lys-15(B′) | 5.4 |
3. Results and discussion
To validate the results obtained from the MD simulations, the protein's secondary structure in the AG10, DO-AG10, and tafamidis V122I protein: ligand complexes were compared to the protein secondary structure from an MD simulation containing only V122I. We would expect the V122I secondary structures to be largely the same in each of these cases. The protein's secondary structure in the V122I:AG10 complex was found to contain 45.7% of residues in β-sheets, 5.3% in α-helices, and 15.5% in turns. The secondary structure of the V122I: DOAG10 complex was found to contain 44.5% of residues in β-sheets, 4.7% in α helices, and 15.5% in turns and the secondary structure of the tafamidis: V122I complex contained 46.7% β-sheets, 3.9% α-helices, and 16.5% turns. Finally, in the MD simulation containing only the V122I receptor, the protein had 45.7% of its residues in β-sheets, 6.3% in α-helices, and 16.9% in turns. We can, therefore conclude that the V122I receptor and the protein in the receptor: ligand complexes had very similar secondary structures. In other words, the protein's secondary structure did not change significantly during the MD simulations when either AG10, DO-AG10, or tafamidis were bound to the HBP. Further validation can be carried out by comparing the averageV122I radius of gyration in the V122I (23.1 ± 0.1 Å), AG10:V122I (22.6 ± 0.1 Å), DOAG10:V122I (22.6 ± 0.1 Å), and tafamidis: V122I (22.7 ± 0.1 Å) MD simulations. Again we see that the radii are nearly identical, further indicating that presence of the AG10, DO-AG10, or tafamidis ligands in the MD simulations had very little effect on the V122I structure.
The final validation experiment was done by comparing the free energy of ligand binding calculated for AG10, DO-AG10, and tafamidis to corresponding literature values for thyroxine. Since all these ligands bind in the HBP, we would expect the free energies of binding to be similar for all three ligands. Sorensen et al. used MD simulations to calculate a binding free energy of −14.0 kcal mol−1 for thyroxine binding to wild type TTR [26]. Recall the TTR receptor has two identical HBP. The ΔG reported above corresponds to the free energy change associated with a thyroxine ligand binding to a protein receptor which already contained a ligand in the other HBP. When a corresponding value was calculated for AG10, a ΔG of −10.3 kcal mol−1 was obtained. The ΔG of binding for DO-AG10 was found to be −10.0 kcal mol−1 and the ΔG of binding for tafamidis was −8.3 kcal mol−1. As expected, the literature thyroxine and the calculated AG10, DO-AG10, and tafamidis ΔG values were found to be similar to one another.
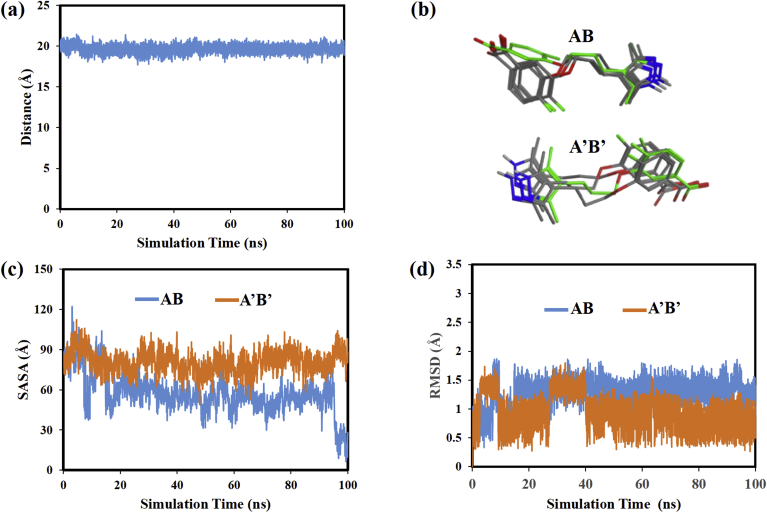
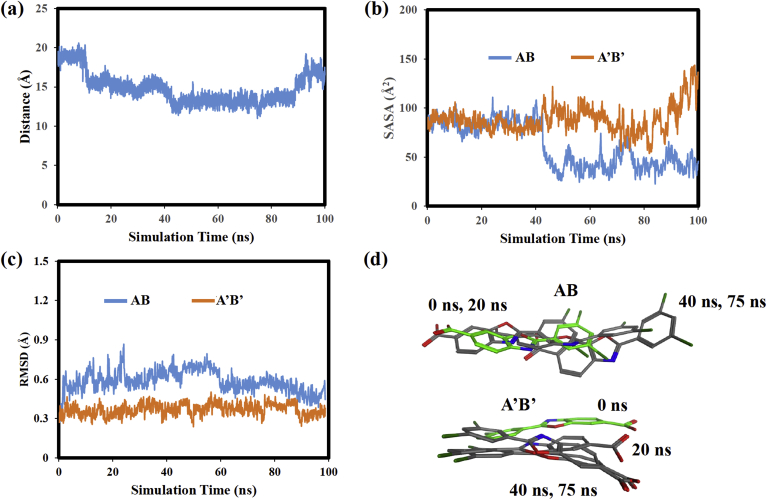
MD simulation analyses were also done to assess whether the two AG10 ligands remained at a fixed location within the HBP throughout the MD simulation or whether they showed more mobile or dynamic behavior within the HBP. In order to quantify the AG10 ligand's movement, the distance between the C atoms annotated with a star in Fig. 2(a) for the AG10 ligands in the two HBP was monitored as a function of stimulation time. This distance plot is shown in Fig. 3(a). Note that the distance monitored remained at approximately 20 Ȧ from 0 to 100 ns. This result suggests that the AG10 ligands do not move closer to one another during the MD simulation, but rather remain at a relatively fixed location within the binding pocket.
Fig. 3.
(a) distance between AG10 ligands versus simulation time, (b) superimposed AG10 structures from the V122I:AG10 MD simulation, (c) AG10 solvent accessible surface area versus simulation time, (d) AG10 RMSD values versus simulation time.
AG10 structures were then extracted and superimposed to compare the location of the ligands within the HBP at different time steps of the MD simulation. Fig. 3(b) shows superimposed structures at 0, 20, 80, and 90 ns. The structure at 0 ns is colored green. The Fig. 3(b) structures show that the N-containing 5-membered ring of the AG10 ligand in the AB HBP, remains at a relatively fixed location at all time steps. This ligand's fluorine-containing 6-membered ring, though, rotates downward toward V122I monomer B early in the MD simulation and then remains in that position until 100 ns. This movement likely places the ligand's CO2− functional group in a position that enables it to more favorably interact with V122I Lys-15 residues. The AG10 ligand in the A’B’ HBP, in contrast showed less mobility and no obvious conformational changes during the MD simulation. Overall, the results from these superimposed structures are consistent with the distance plot in Fig. 3(a) which also showed that the ligands did not move further into or out of the HBP during the MD simulation.
The solvent accessible surface area (SASA) of the AG10 ligand was then calculated and plotted vs simulation time (Fig. 3(c)). SASA is a measurement of the ligand's surface area that is accessible to solvent molecules when it is in the HBP. SASA versus MD simulation time plots were used throughout the study to investigate changes in the position and conformation of the ligands within the HBP. For example, if during the MD simulation a ligand moved deeper into the HBP or changed its conformation in such a way as to more effectively shield its atoms from solvent exposure, we would expect the SASA to decrease. Increases in SASA would be expected if the ligand moved into a more solvent exposed environment. Finally, when comparing SASA values for different ligands it must be recognized that the ligands contained different numbers of atoms and, therefore, different free solution SASA values. For example, the free solution surface areas of AG10, DO-AG10, N-methyl-AG10, and tafamidis were found to be 303 Å2, 272 Å2, 326 Å2, and 280 Å2, respectively. These values were calculated by performing a short MD simulation containing each respective ligand and solvent molecules, but no TTR receptor.
Fig. 3(c) shows that the SASA of the AG10 ligand in the AB pocket decreased from approximately 80 Ȧ2 to approximately 60 Ȧ2 during the first 20 ns of the MD simulation. From 20 to 90 ns the SASA then remained relatively constant. The SASA decrease observed early in the MD simulation is likely associated with movement of the ligand's fluorine-containing aromatic ring into an environment that is more shielded from solvent. This movement is discussed above and is shown in the Fig. 3(b) superimposed structures. The SASA of the AG10 ligand in the A’B’ pocket in contrast remains at ∼80 Ȧ2 from 0 to 100 ns. This result suggests that the ligand remains in largely the same environment throughout the MD simulation. The SASA analyses are also consistent with the superimposed structures in Fig. 3(b) which show that the AG10 ligand moves very little in the A’B’ pocket.
The RMSD of both AG10 ligands was then calculated to assess the conformational flexibilities of the ligands within the respective HBP. The RMSD plot for AG10 in pocket AB changes early in the MD simulation, likely due to the reorientation of the ligand's aromatic ring shown in Fig. 3(b). The RMSD value for this ligand then remains constant for the remainder of the MD simulation. The RMSD values for AG10 in the A’B’ pocket are also relatively constant and less than 1.5 Å during most of the MD simulation time. Therefore, the RMSD plots suggest that the bound AG10 ligands experience very little conformational flexibility during the MD simulation.
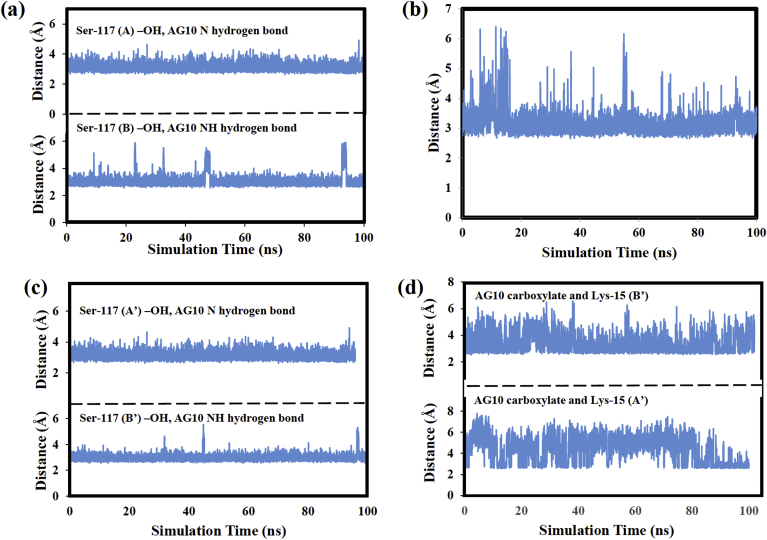
The intermolecular receptor: ligand hydrogen bonds responsible for holding the AG10 ligands in the halogen binding pockets will now be analyzed. Table I presents the donor and acceptor atoms making up each intermolecular hydrogen-bond along with each H-bond's percent occupancy. The latter quantity is the percentage of the total MD simulation time that the H-bond was present. Only hydrogen bonds with occupancies greater than five percent are shown. The H-bond analysis for the AG10 ligand in the AB pocket shows that the highest occupancy intermolecular hydrogen bond (59.4%) formed between the hydroxyl group of Ser-117 on V122I monomer chain B and the NH atom of AG10's hydrazone functional group. A similar H-bond with an occupancy of 55.8% also formed between the nitrogen atom in the same AG10 functional group and the hydrogen atom of V122I monomer chain A's Ser-117 –OH. The Ser-117 residues that are involved in both H-bonds are in the inner V122I HBP.
Distance measurements were then made between the heavy atoms making up these two high occupancy H-bonds (Fig. 4(a)) to determine if they formed at the same or at different times during the MD simulation. If they formed at different times, we would expect the atoms in the two hydrogen bonds to be close to one another at different times as well. Instead, the distance plots in Fig. 4(a) show that the heavy atoms making up these hydrogen bonds remained close to one another throughout the MD simulation, with average distances of 3.1 ± 0.4 Å and 3.1 ± 0.2 Å for the 59.4% and 55.8% hydrogen bonds, respectively. Therefore, it is likely that the atoms in the AG10 hydrozone functional group form simultaneous hydrogen bonds with V122I residues Ser-117(B) and Ser-117(A). The AG10 ligand's ability to hydrogen bond to residues on both the AA′ and BB′ V122I dimers may contribute to its ability to stabilize the V122I tetramer dissociation and inhibit tetramer dissociation [18,19].
Fig. 4.
(a) Distance between the heavy atoms in the H-bond between both Ser-117 (B) –OH and Ser-117 (A) –OH and AG10, (b) Distance between the heavy atoms in the H-bond between AG10 CO2− and Lys-15 (A), (c) Distances between the heavy atoms in H-bonds between both Ser-117(B′) and Ser-117(A′) and AG10, (d) Distances between the AG10 carboxylate oxygen and Lys-15(A′) and Lys-15(B′).
The AG10 ligand in V122I pocket AB also formed six H-bonds between the oxygen atoms of its carboxylate functional group and the NH3+ hydrogen atoms of Lys-15(A). The percent occupancies of these hydrogen bonds ranged from 18.1% to 13.4%. The structures in Fig. 3(b) show that the AG10 carboxylate is oriented upward toward V122I monomer chain A. Therefore, it seems reasonable that hydrogen bonds were detected between the AG10 carboxylate and Lys-15 of the same monomer chain. Fig. 4(b) plots the average distance between the Lys-15(A) side chain nitrogen atom and the two AG10 carboxylate oxygens. The plot shows some variability from 0 to 20 ns, but overall these atoms remain close to one another, with an average separation of 3.2 ± 0.4 Å. Therefore, these distance measurements suggest that the ligand carboxylate spends a considerable amount of the MD simulation either hydrogen bound to or interacting electrostatically with the residue Lys-15(A). In other words, this interaction which was present in the V122I: AG10 crystal-structure, persisted throughout the MD simulation. In addition, the H-bonds formed by the AG10 hydrozone and carboxylate functional groups which are at opposite ends of the molecule create a two-point ligand: receptor interaction within the HBP. This two-point interaction is likely responsible for the ligand remaining at a relatively fixed location throughout the MD simulation.
Table 1 also reports the intermolecular hydrogen bonds formed by the AG10 ligand in the A’B’ pocket. Overall the hydrogen bonds detected and their percent occupancies are very similar to those discussed above for AG10 in the AB pocket. The A’B’ ligand formed two H-bonds with occupancies of 64.4% and 35.9% between the N and NH atoms of its hydrozone functional group and, respectively V122I residues Ser-117(B′) and Ser-117(A′). Plots of the distances between the heavy atoms forming these H-bonds shown in Fig. 4(c) suggest that those atoms remain close to one another throughout the MD simulation, with average distances of 3.0 ± 0.2 Å (64.4% H-bond) and 3.1 ± 0.2 Å (35.9% H-bond). Therefore, like the ligand in the AB pocket, the A’B’ ligand likely forms simultaneous hydrogen bonds between its hydrozone functional group and Serine residues on both the V122I AA′ and BB′ dimers.
Finally, the AG10 ligand in the A’B’ pocket also forms five hydrogen bonds with Lys-15 residues of V122I. However, unlike the ligand in the AB pocket, all of the H-bonds do not form with the same V122I residue. For example, three H-bonds with occupancies of 15.5%, 15.3%, and 13.0% form between the AG10 carboxylate and Lys-15 of monomer A′ and two additional H-bonds with occupancies of 12.2% and 10.0% form between the same AG10 functional group and Lys-15 of monomer B’. Again these interactions with two different V122I dimers, like the H-bonds formed by the hydrozone functional group, may stabilize the V122I tetramer. The average distances between the AG10 A’B’ carboxylate oxygen and Lys-15(A′) and Lys-15(B′) nitrogen atoms were 5.6 ± 1 Å and 5.0 ± 1 Å, respectively. These average distances are somewhat larger than corresponding values calculated for the AG10 ligand in the AB pocket. Fig. 4(d) plots the distances between the heavy atoms forming these H-bond versus simulation time. The plot shows that there are relatively few time steps when the AG10 carboxylate is close to both Lys-15 nitrogen atoms. Therefore, it seems unlikely that this AG10 ligand forms simultaneous H-bonds to both Lys-15 residues.
In order to further investigate the importance of the two-point ligand: receptor interaction discussed above, an MD simulation was carried out with a ligand we designate N-methyl-AG10. This compound's structure is shown in Fig. 2(c). Note that a methyl group has been added to the ligand's hydrazone functional group. This change would be expected to disrupt the ligand's ability to hydrogen bond to the receptor. A hydrogen bond analysis showed that the N-methyl-AG10 ligand in the V122I AB pocket formed a single H-bond (occupancy 54.2%) between its N atom and the Thr-119 –OH on V122I monomer A. Likewise the A’B’ ligand formed a 50.1% occupancy H-bond with Ser-117 on V122I monomer B’. Both ligands also experienced numerous low percent occupancy H-bonding interactions with Lys-15 residues in the outer HBP. The complete N-methyl-AG10 hydrogen-bond analysis is given in the Supplemental Information.
Recall however, that the MD simulation with AG10 showed two high occupancy H-bonds formed between each ligand and Ser-117 residues in the inner halogen binding pockets. These H-bonds had percent occupancies of 59.4% and 55.8% and 64.4 and 35.9% for the AB and A’B’ pockets, respectively. Therefore, as expected methylating the AG10 ligand caused it to experience fewer hydrogen bonding interactions with the V122I receptor. This change also resulted in a less favorable ligand binding free energy. Recall from above that the AG10 binding free energy was −10.3 kcal mol−1 compared to −8.8 kcal mol−1 for N-methyl-AG10. Therefore, disrupting the ligand:receptor hydrogen bonding interactions and the two-point interaction discussed above lead to overall less energetically favored ligand:receptor interactions.
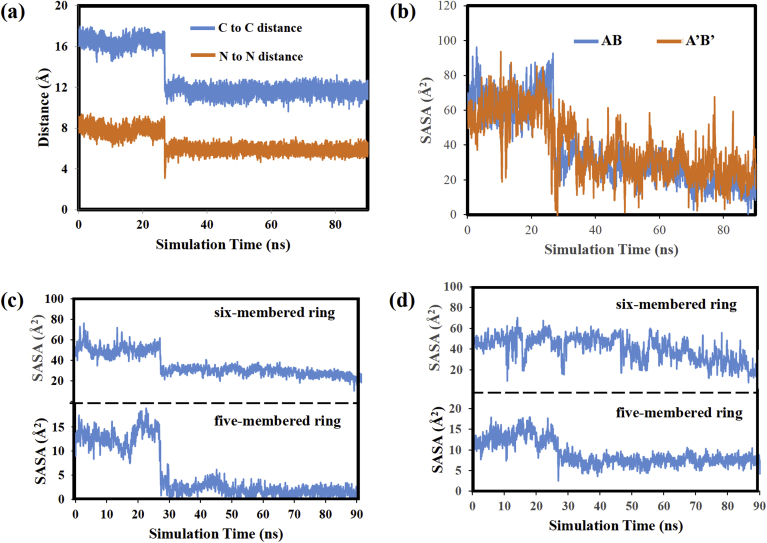
The results from the DO-AG10: V122I MD simulation will now be discussed. Fig. 5(a) plots the distance between the two DO-AG10 ligands versus simulation time. The carbon atom chosen for these distance measurements is starred in Fig. 2(b). The distance between the nitrogen atoms in the ligands' five-membered rings is also plotted. Recall that in the AG10: V122I analysis the ligands in the two HBP remained ∼20 Å from one another throughout the MD simulation. In contrast, the DO-AG10 carbon atoms start 16.5 Å apart, but at 27 ns the separation changed to 11.6 Å. This result suggests that one or both DO-AG10 ligands move deeper into the HBP at this time step of the MD simulation. The distance between the nitrogen atoms in each ligand's five-membered ring decreased as well from an average value of 7.8 Å early in the MD simulation to 5.9 Å after 27 ns.
Fig. 5.
(a) Plots of the distances between selected atoms of DO-AG10 ligands in the AB and A’B’ pockets, (b) SASA versus simulation time plots for DO-AG10 ligands, (c) Plots of SASA versus simulation time for atoms in the five and six-membered rings of DO-AG10 in the AB pocket, (d) Plots of SASA versus simulation time for atoms in the five and six-membered rings of DO-AG10 in the A’B’ pocket.
Fig. 5(b) plots the SASA for the DO-AG10 ligands in the AB and A’B’ binding pockets. Both plots show a decrease in the SASA values from 60 Å2 (AB pocket) and 55 Å2 (A’B’ pocket) early in the MD simulation to 10 Å2 (AB pocket) and 29 Å2 (A’B’ pocket) near the end. These results suggest that during the MD simulation both DO-AG10 ligands move in such a way as to more effectively shield their atoms from solvent exposure. Finally, Fig. 5(c) and (d) plot separately the SASA of the atoms making up the DO-AG10 five-membered and six membered rings for, respectively the ligands in the AB and A’B’ HBP. Here we see a difference in the behavior of the two ligands. The DO-AG10 ligand in the AB pocket shows a sudden decrease in both ring's SASA at 27 ns. Recall this is the same time step when the separation between the ligands decreased from 16.5 to 11.6 Å. The DO-AG10 ligand in the A’B’ pocket, however, shows a more gradual decrease in the SASA of the atoms in both rings.
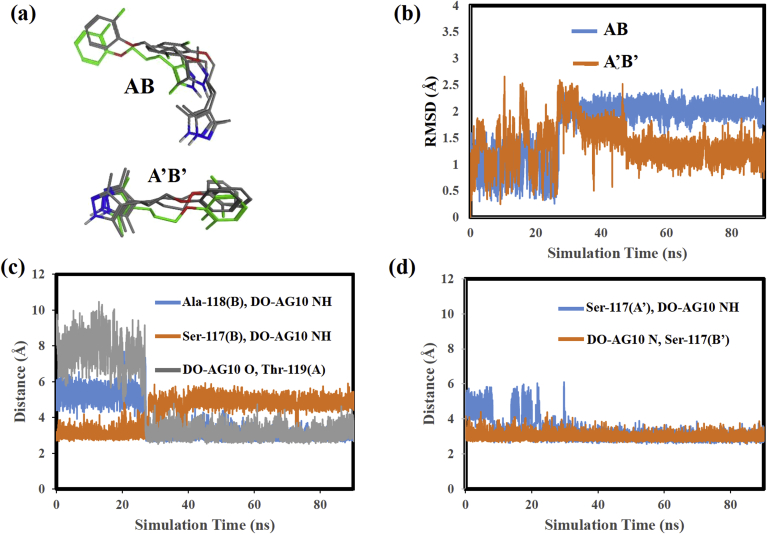
Fig. 6(a) shows superimposed structures for the DO-AG10 ligands at 0, 20, 30, and 80 ns of the MD simulation. In each superposition, the structure at 0 ns is colored green. We will begin by discussing the behavior of the ligand in the A’B’ pocket. The Fig. 6(a) structures show that, as in the AG10 analyses described above, the DO-AG10 ligand in the A’B’ HBP remained in a similar location throughout the MD simulation. The ligand's five-membered ring moves very little, while the six-membered ring and methylene chain show greater mobility. Overall, though, the center of mass of the ligand remains relatively stationary. The superimposed structures in Fig. 6(a) are also consistent with the ligand RMSD values plotted in Fig. 6(b). This plot shows that the A’B’ ligand's RMSD values change from 1.0 to 2.5 Å at 27 ns and then gradually decreases back to slightly greater than 1.0 Å during the rest of the MD simulation.
Fig. 6.
(a) Superimposed structures for DO-AG10 in the V122I binding pockets, (b) RMSD versus simulation time for DO-AG10 in the V122I binding pockets, (c) Distances between the heavy atoms in intermolecular hydrogen bonds formed by DO-AG10 in V122I pocket AB, (d) Distances between the heavy atoms in intermolecular hydrogen bonds formed by DO-AG10 in V122I pocket A’B’.
In addition, the Fig. 6(a) structures extracted at 0 and 20 ns show that at these time steps the DO-AG10 ligand in the AB pocket adopts an extended conformation similar to that of the AG10 ligand in the same HBP. By 30 ns, however, the location of the ligand within the pocket and its conformation have changed considerably. For example, at 30 ns the ligand's five-membered ring points downward toward V122I monomer B and the ligand has adopted a folded conformation. This folding of the ligand's methylene chain in turn allows its six-membered ring to move both downward toward V122I monomer B and further into the HBP.
The Fig. 6(a) DO-AG10 AB pocket structures are consistent with the distance and SASA measurements in Fig. 5(a) and (c), respectively. Fig. 5(a) shows that during the MD simulation the distance between nitrogen atoms in the ligands' hydrozone functional groups and the distance between the starred carbon atoms in Fig. 2(b) each change at 27 ns. These changes can be attributed to the ligand's five-membered ring moving downward toward V122I monomer B at the same time as the conformational change described above occurs. The plots in Fig. 5(c) show that the SASA of the atoms in the DO-AG10 five-membered and six-membered rings both decrease at 27 ns. These decreases can be attributed to the ligand simultaneously moving deeper into the A’B’ pocket and changing its conformation as shown in Fig. 6(b). Both changes move the ring atoms into locations that decrease their solvent exposure. Finally, the RMSD plot in Fig. 6(b) shows that the DO-AG10 A’B’ ligand's RMSD values change sharply at 27 ns from 1.0 to 2.5 Å and remain at 2.5 A for the rest of the MD simulation.
The intermolecular hydrogen bonds formed during the DO-AG10: V122I MD simulation are shown in Table 2. The DO-AG10 ligand in the A’B’ pocket forms two intermolecular hydrogen bonds with the V122I receptor. One of these H-bonds formed between Serine-117 on V122I monomer chain A′ and the NH atom of the ligand's hydrozone functional group, while the other formed between the N atom of the DO-AG10 hydrozone functional group and Serine-117 of chain B’. These same H-bonds formed in the AG10: V122I MD simulation. Fig. 6(d) plots the distances between the heavy atoms involved in these hydrogen bonds. The average distances between these atoms were 3.3 ± 0.7 and 3.0 ± 0.2 Å for the 55.7% and 47.5% hydrogen bonds, respectively. Therefore, as in the AG10: V122I analyses it is likely that these H-bonds are present simultaneously for a significant portion of the MD simulation.
The DO-AG10 ligand in the AB pocket formed three intermolecular hydrogen bonds. The highest percent occupancy H-bond (43.8%) formed between Thr-118 of V122I monomer chain A and the DO-AG10 NH atom. Additional H-bonds with occupancies of 27.1% and 10.1% formed between, respectively, the DO-AG10 ether oxygen and Thr-119 of monomer chain A and between Ser-117 on monomer chain B and the DO-AG10 NH atom. Fig. 6(c) plots the distances between the heavy atoms involved in each of these hydrogen bonds. The atoms forming the Ser-117(A) to DO-AG10 H-bond begin close to one another with an average separation of 3.1 Å during the first 27 ns of the MD simulation. The atoms forming the 43.8% and 27.1% H-bonds are farther from one another during this time with, respective average separations of 5.3 and 7.8 Å, respectively. Then at 27 ns, the atoms forming the 43.8% and 27.1% H-bonds move closer to one another and the atoms involved in the 10.1% H-bonds move farther away. For the remainder of the MD simulation the average separation between the atoms forming the 43.8% and 27.1% H-bonds are, respectively 3.0 and 3.1 Å.
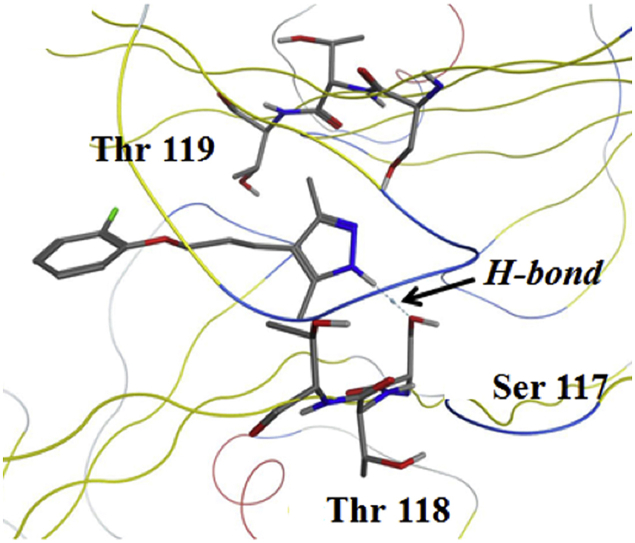
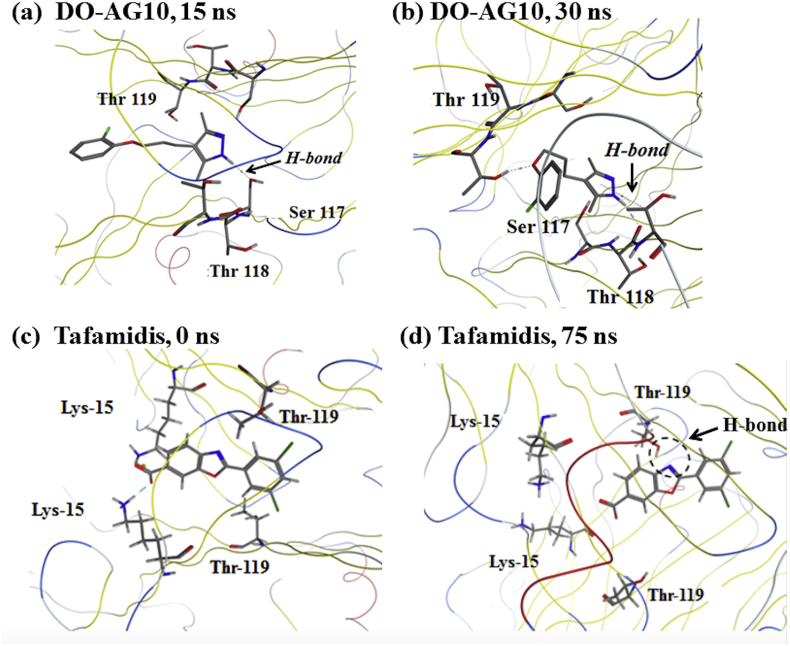
The distance plot in Fig. 6(c) suggests that early in the MD simulation the DO-AG10 ligand in pocket AB is held in place by a hydrogen bond between its hydrozone functional group and the Ser-117(A) –OH. However, since DO-AG10 does not have a carboxylate on its six-membered ring, no two-point ligand: receptor interaction, like that observed in the AG10 MD simulations, was present with DO-AG10. At 27 ns, the 10.1% H-bond breaks and the DO-AG10 ligand moves further into the receptor's AB HBP. Recall this was also the time step when the distance between the two DO-AG10 ligands and the SASA of the AB ligand's ring atoms decreased. After moving into the V122I AB HBP, the DO-AG10 ligand then formed two new H- bonds between, respectively its ether oxygen and Thr-119(A), and the NH atom of its hydrozone functional group and Thr-118(B). Since the heavy atoms involved in these hydrogen bonds remained close to one another for the remainder of the MD simulation, it is likely that there are times when these H-bonds formed simultaneously. Therefore, the DO-AG10 ligand was likely able during the last 73 ns of the MD simulation to form a two-point hydrogen bonding interaction with V122I residues Thr-118(B) and Thr-119(A). These interactions in turn likely hold the ligand in place during the remainder of the MD simulation. Finally, Fig. 7 shows structures extracted from the DO-AG10 pocket AB MD simulation at 15 and 30 ns. Note the H-bond between Ser-117(B) and the DO-AG10 NH atom in the Fig. 7(a) structure. The Fig. 7(b) structure shows simultaneous H-bonds formed between both Thr-118(B) and the DO-AG10 NH atom and between the DO-AG10 ether oxygen and Thr-119(A).
Fig. 7.
Structures extracted from the DO-AG10 MD simulation at (a) 15 ns and (b) 30 ns and from the tafamidis MD simulation at (c) 0 ns and (d) 75 ns showing intermolecular H-bonds between the ligand and V122I receptor.
To complete the AG10 analyses, MD simulations were also carried out with complexes containing either AG10 or DO-AG10 bound to the wild type (WT) TTR protein. In these MD simulations, the AG10 ligands formed a two-point hydrogen bonding interaction with the wt TTR receptor by interacting with Ser-117 residues in the inner HBP and Lys-15 residues nearer the receptor surface. The AG10 ligands also showed no significant movement within the HBP or conformational flexibility during the MD simulation. These results were analogous to those obtained with the V122I variant. Therefore, since the V122I mutation is outside of the binding pocket and since Valine and Isoleucine have very similar structures, MD simulations showed the V122I mutation had relatively little effect on AG10 binding to the receptor. Likewise, MD simulations with the DO-AG10: wt TTR complex showed that the DO-AG10 ligand in the A’B’ pocket experienced a conformational change analogous to that seen in the V122I experiments. Therefore, as with the AG10 MD simulations, DO-AG10 interactions with the wt and V122I variant TTR proteins were very similar. Results from MD simulations with wt TTR complexes are presented in the Supplemental Information.
Results from a tafamidis: V122I MD simulation will now be presented and compared to the behavior of the AG10 and DO-AG10 ligands described above. We first note that the Fig. 2 structures show both similarities and differences between the AG10 and tafamidis ligands. For example, the ligands both contain a carboxylate functional group and would, therefore, be expected to interact similarly with Lys-15 residues in the outer HBP. AG10, however, also contains a hydrozone functional group which, as described above, formed hydrogen bonds with Ser-117 residues in the inner HBP. Tafamidis in contrast has no H-bond donor or acceptor atoms in a comparable position. We would, therefore, expect tafamidis to not experience a two-point hydrogen bonding interaction with the receptor, as was seen in the AG10 MD simulation, but instead experience hydrophobic interactions within the inner HBP.
Fig. 8(a) plots the distance between the tafamidis ligands located in the two V122I HBP. The atom chosen for the distance measurements is starred in Fig. 2(d). This plot shows that the ligands begin the MD simulation 18 Å apart. The separation decreases to 15 Å at approximately 10 ns and decreases again to 12 Å at 43 ns. The SASA plots in Fig. 8(b) also show an abrupt drop at 43 ns in the solvent exposure of the tafamidis ligand in HBP AB. The SASA of the tafamidis ligand in HBP A’B’, in contrast, remained relatively constant throughout the MD simulation. Finally, the RMSD plots in Fig. 8(c) demonstrate that both ligands have very little conformational flexibility throughout the MD simulation because their RMSD values are small and relatively constant from 0 to 100 ns. This result is expected given tafamidis' rigid, fused ring structure.
Fig. 8.
(a) Distance versus simulation plot for tafamidis ligands in V122I AB and A’B’ pockets, (b) SASA versus simulation plot for tafamidis ligands in V122I AB and A’B’ pockets, (c) RMSD versus simulation plot for tafamidis ligands in V122I AB and A’B’ pockets, (d) Superimposed structures from 0, 20, 40, and 75 ns for tafamidis ligands in V122I AB and A’B’ pockets.
The superimposed structures in Fig. 8(d) give further insight into the behavior of the two tafamidis ligands. The structures at 0 and 20 ns for the tafamidis ligand in the AB HBP show that the ligand's position changes very little during the first 20 ns of the MD simulation. By 40 ns, however, the AB ligand has begun to move deeper into the HBP while remaining in an extended conformation. The structure at 75 ns shows the ligand in a conformation and orientation similar that of the 0 ns structure, however, at 75 ns the tafamidis ligand is located deeper within the HBP. This horizontal movement of the AB tafamidis ligand within the HBP likely accounts for the simultaneous decrease in inter-ligand distance and SASA observed around 40 ns in Fig. 8(a) and (b), respectively.
This movement of the tafamidis ligand within the HBP is likely unrelated to the V122I mutation in that the mutation lies outside the binding pocket. Instead, the movement likely results from the different hydrogen-bonding interactions present in tafamidis and AG10. As described above, tafamidis contains a carboxylate functional group that interacts with receptor Lys-15 residues, but there are no H-bond donor/acceptor atoms on the tafamidis six-membered ring that is located in the inner HBP. Therefore, if the tafamidis carboxylate momentarily moves away from a receptor Lys-15 residue, there is no additional H-bonding interaction in the inner HBP holding the ligand in place. At this point, the ligand is free to move deeper into the HBP.
Compared to the tafamidis ligand in the AB HBP, the position of the ligand in the A’B’ pocket changed relatively little during the MD simulation. The superimposed structures in Fig. 8(d) show that during the first 20 ns of the MD simulation the ligand in the A’B’ pocket moves downward toward V122I monomer B′ and slightly deeper into the HBP. This movement may account for the decrease in inter-ligand separation observed at 10 ns. The ligand in the A’B’ pocket then remained in largely the same position, orientation, and conformation for the remainder of the MD simulation. This behavior is consistent with the relatively constant SASA and RMSD values shown in Fig. 8(c) and (d), respectively.
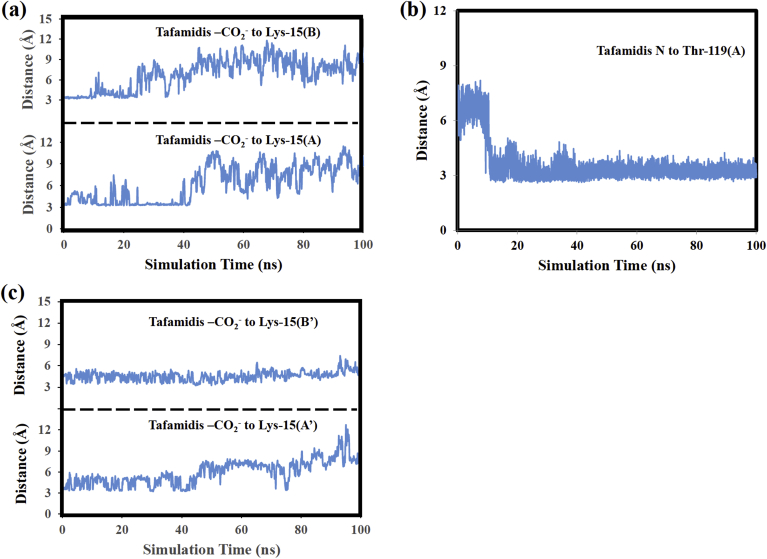
Table 3 lists the intermolecular hydrogen bonds observed in the tafamidis: V122I MD simulation. The highest occupancy intermolecular H-bond for tafamidis in the AB pocket formed between its nitrogen atom and Thr-119 of V122I monomer A. Three H-bonds with percent occupancies of 8.7%, 8.2% and 8.1% formed between the tafamidis carboxylate and Lys-15 of the same V122I monomer. Three additional H-bonds (5.9%, 5.4%, and 5.3%) formed between the same tafamidis functional group and Lys-15(B). Fig. 9(a) plots the distance between the heavy atoms involved in the H-bonds between the pocket AB tafamidis ligand's carboxylate and V122I Lys-15 residues. The plot shows that these atoms are only close to one another early in the MD simulation. After 40 ns they have moved too far from one another to form hydrogen bonds. This result suggests that as expected the ligand's hydrogen-bonding interactions with the Lys-15 residues occur early in the MD simulation and before the ligand moves deeper into the HBP.
Fig. 9.
(a) Distance between the heavy atoms for the H-bonds formed between the tafamidis (AB pocket) carboxylate and V122I Lys-15 residues, (b) Distance between the heavy atoms forming the tafamidis (AB pocket) N and Thr-119(A) hydrogen bonds, (c) Distance between the tafamidis carboxylate oxygens (A’B’ pocket) and V122I Lys-15 residues.
Fig. 9(b) plots the distance between the heavy atoms forming the 63.7% H-bond between the tafamidis nitrogen atom and Thr-119 of V122I monomer A. The plot shows that these atoms are on average 6.7 Å apart during the first 10 ns of the MD simulation. After 10 ns, these atoms move closer to one another to a separation where hydrogen bonding could occur. The average separation of these atoms is in fact 3.2 Å during the last 80 ns of the MD simulation. Fig. 7(c) and (d) show the V122I: tafamidis complex at 0 and 75 ns. At zero ns, the ligand is located in outer part of HBP and its N atom is relatively far from the hydroxyl group of the Thr-119(A) side chain. However, at 75 ns the Thr-119 side chain has reoriented and the tafamidis ligand has moved deeper into the HBP. Both changes facilitate the formation of the H-bond shown in Fig. 7(d). The Fig. 7(c) and (d) structures and the distance plot in Fig. 9(b) suggest that the Thr-119(A) side chain reorients, moving its hydroxyl group closer to the tafamidis nitrogen atom around 10 ns of the MD simulation. These atoms then remain relatively close to one another for the remainder of the MD simulation, both before and after the tafamidis ligand moves deeper into the HBP at 43 ns.
The tafamidis ligand in the A’B’ pocket formed relatively few intermolecular hydrogen bonds during the MD simulation. Two H-bonds with occupancies of 10.0% and 7.1% formed between Thr-106 of V122I monomer A′ and the tafamidis nitrogen atom and a 5.4% occupancy H-bond formed between the tafamidis carboxylate the Lys-15 of V122I monomer B’. While only a single H-bond was detected between the tafamidis A’B’ pocket carboxylate and Lys-15, Fig. 9(c) shows that the tafamidis carboxylate oxygens and the Lys-15 nitrogens of monomers A′ and B′ remain relatively close throughout the MD simulation. For example, the average distance between the tafamidis –CO2- and Lys-15(A′) nitrogen is 3.8 Å and 5.9 Å during, respectively the first 40 ns and last 60 ns of the MD simulation. The average separation between the tafamidis carboxylate and Lys-15(B′) nitrogen is 4.5 Å. These results suggest that even if relatively few H-bonds form between these functional groups, they do remain close to one another throughout the MD simulation and likely experience favorable electrostatic interactions.
4. Conclusions
Molecular dynamics simulations were used to investigate the binding of the drug candidate AG10, its N-methyl and decarboxy derivatives, and the drug tafamidis to the V122I mutant of transthyretin. Distance measurements, along with solvent accessible surface area, RMSD, and hydrogen bond analyses showed that AG10 formed a stable two-point interaction with the protein by hydrogen bonding to Ser-117 residues in the receptor's inner HBP and interacting electrostatically and through H-bonds with Lys-15 residues nearer the protein's surface. Methylating the AG10 hydrazone nitrogen or removing the compound's carboxylate functional group were both found to disrupt this two point interaction. The former reduced hydrogen bonding interactions with inner pocket residues, while the latter disrupted interactions with outer pocket Lys-15 side chains. Finally, tafamidis: V122I MD simulations showed that tafamidis formed fewer hydrogen bonding interactions with the receptor than AG10. Also, no stable two-point receptor interaction was observed because the tafamidis ligand, unlike AG10, does not have hydrogen-bond donor/acceptor atoms at opposite ends of the molecule. Finally, the tafamidis ligand in the V122I AB binding pocket also moved deeper into the HBP during the MD simulation.
CRediT authorship contribution statement
Kevin F. Morris: Conceptualization, Methodology, Investigation, Validation, Visualization, Writing - original draft, Writing - review & editing, Supervision. Riley M. Geoghegan: Formal analysis, Writing - original draft. Emily E. Palmer: Formal analysis, Writing - original draft. Matthew George: Conceptualization, Writing - review & editing. Yayin Fang: Conceptualization, Methodology, Investigation, Validation, Writing - original draft, Writing - review & editing, Supervision, Funding acquisition.
Acknowledgements
This work was partially supported by a grant from the National Institutes of Health (No.2U54MD007597) to RCMI program at the Howard University. This work was also supported by grants from the HUMAA and Office of Naval Research (N00014-18-1-2145 and N00014-19-1-2602) and HUMAA Endowed Founder's Chair in Basic Science award to Dr. Yayin Fang. We also acknowledge the generosity of the Ralph E. Klingenmeyer family.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100721.
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100721
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Transparency document
References
- 1.Ruberg F.L., Berk J.L. Transthyretin (TTR) cardiac amyloidosis. Circulation. 2012;126:1286–1300. doi: 10.1161/CIRCULATIONAHA.111.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koike H., Morozumi S., Kawagashira Y., Lijima M., Yamamoto M., Hattori N., Tanka F., Nakamura T., Hirayama M., Ando Y., Ikeda S., Sobue G. The significance of carpal tunnel syndrome in transthyretin Val30Met familial amyloid polyneuropathy. Amyloid. 2009;16:142–148. doi: 10.1080/13506120903094074. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton J.A., Benson M.D. Transthyretin: a review from a structural perspective. Cell. Mol. Life Sci. 2001;58:1491–1521. doi: 10.1007/PL00000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekijima Y., Wiseman R.L., Matteson J., Hammarström P., Miller S.R., Sawkar A.R., Balch W.E., Kelly J.W. The biological and chemical basis for tissue-selective amyloid sisease. Cell. 2005;121(1):73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Lei M., Yang M., Huo S. Intrinsic versus mutation dependent instability/flexibility: a comparative analysis of the structure and dynamics of wild-type transthyretin and its pathogenic variants. J. Struct. Biol. 2004;148:153–168. doi: 10.1016/j.jsb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee A., Bairagya H.R., Mukhopadhyay B.P., Nandi T.K., Bera A.K. Structural insight to mutated Y116S transthyretin by molecular dynamics simulation. Indian J. Biochem. Biophys. 2010;47:197–202. http://nopr.niscair.res.in/handle/123456789/10118 [PubMed] [Google Scholar]
- 7.Yang M., Lei M., Huo S. Why is Leu55➝Pro55 transthyretin variant the most amyloidogenic: insights from molecular dynamics simulations of transthyretin monomers. Protein Sci. 2003;12:1222–1231. doi: 10.1110/ps.0239703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das J.K., Mall S.S., Bej A., Mukherjee S. Conformational flexibility tunes the propensity of transthyretin to form fibrils through non-native intermediate states. Angew. Chem. Int. Ed. 2014;53:12781–12784. doi: 10.1002/anie.201407323. [DOI] [PubMed] [Google Scholar]
- 9.Sebastiao M.P., Saraiva M.J., Damas A.M. The crystal structure of amyloidogenic Leu➝Pro transthyretin variant reveals a possible pathway for transthyretin polymerization into amyloid fibrils. Int. J. Biol. Chem. Int. 1998;273:24715–24722. doi: 10.1074/jbc.273.38.24715. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues J.R., Simoes C.J.V., Silva C.G., Brito R.M.M. Potentially amyloidogenic conformational intermediates populate the unfolding landscape of transthyretin: insights from molecular dynamics simulations. Protein Sci. 2009;19:202–219. doi: 10.1002/pro.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damas A.M., Riberiro S., Lamzin V.S., Palha J.A., Saraiva M.J. Structure of Val122Ile variant transthyretin- a cardiomyopathic mutant. IUCr. 1996;52:966–972. doi: 10.1107/S0907444996003307. [DOI] [PubMed] [Google Scholar]
- 12.Plante-Bordeneuve V., Said G. Familial amyloid polyneuropathy. Lacent Neurol. 2011;10:1086–1097. doi: 10.1016/S1474-4422(11)70246-0. [DOI] [PubMed] [Google Scholar]
- 13.Bulawa C.E., Connelly S., DeVit M., Wang L., Weigel C., Fleming J.A., Packman J., Powers E.T., Wiseman R.L., Foss T.R., Wilson I.A., Kelly J.W., Labaudiniere R. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl. Acad. Sci. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coelho T., Merlini G., Bulawa C.E., Fleming J.A., Judge D.P., Kelly J.W., Maurer M.S., Plantè-Bordeneuve V., Labaudinière R., Mundayat R., Riley S., Lombardo I., Huertas P. Mechanism of action and clinical application of tafamidis in hereditary transthyretin amyloidosis. Neurol. Ther. 2016;5:1–25. doi: 10.1007/s40120-016-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keown A. Pfizer's tafamidis approved in first for rare heart disease. 2019. https://www.biospace.com/article/pfzer-s-tafamidis-approved-for-rare-heart-disease/
- 16.Erman M. Pfizer gets U.S. Approval for $225,000 a year heart drug. 2019. https://www.reuters.com/article/us-pfizer-approval/pfizer-gets-u-s-approval-for-225000-a-year-heart-drug-idUSKCN1SC0VX
- 17.Sant'Anna R., Gallego P., Robinson L.Z., Pereira-Henriques A., Ferreira N., Pinheiro F., Esperante S., Pallares I., Huertas O., Almeida M.R., Reixach N., Insa R., Velazquez-Campoy A., Reverter D., Reig N., Ventura S. Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat. Commun. 2016;7:1–13. doi: 10.1038/ncomms10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penchala S.C., Connelly S., Wang Y., Park M.S., Zhao L., Baranczak A., Rappley I., Vogel H., Liedtke M., Witteles R.M., Powers E.T., Reixach N., Chan W.K., Wilson I.A., Kelly J.W., Graef I.A., Alhamadsheh M.M. AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin. Proc. Natl. Acad. Sci. 2013;110:9992–9997. doi: 10.1073/pnas.1300761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller M., Pal A., Albusairi W., Joo H., Pappas B., Haque T., Liang D., Jampala R., Liu F., Kahn J., Faaij M., Park M., Chan W., Graef I., Zamboni R., Kumar N., Fox J., Sinha U., Alhamadsheh M. Enthalpy-driven stabilization of transthyretin by AG10 mimics a naturally occurring genetic variant that protects from transthyretin amyloidosis. J. Med. Chem. 2018;61:7862–7876. doi: 10.1021/acs.jmedchem.8b00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Judge D.P., Heitner S.B., Falk R.H., Maurer M.S., Shah S.J., Witteles R.M., Grogan M., Seiby V.N., Jacoby D., Hanna M., Nativi-Nicolau J., Patel J., Rao S., Sinha U., Turtle C.W., Fox J.C. Transthyretin stabilization by AG10 in symptomatic transthyretin amyloid Cardiomyopathy. J. Am. Coll. Cardiol. 2019;74:285–295. doi: 10.1016/j.jacc.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Hellawell J.L., Rao S., O'Reilly T., Lumpkin R., Jernelius J., Gretler D., Sinha U., Fox J.C. AG10, A novel, potent, and selective transthyretin stabilizer, is well tolerated at doses resulting in target therapeutic blood levels, and demonstrates clinical proof-of-concept in healthy volunteers. J. Card. Fail. 2018;24:S31–S32. [Google Scholar]
- 22.Fox J.C., Hellawell J.L., Rao S., Lumpkin R., Jernelius J., Gretler D., Sinha U., O'Reilly T. First-in-Human study of AG10, a novel, oral, specific, selective, and potent transthyretin stabilizer for the treatment of transthyretin amyloidosis: a Phase 1 safety, tolerability, pharmacokinetic, and pharmacodynamic study in healthy adult volunteers. Clin. Pharmacol. Drug Dev. 2019 doi: 10.1002/cpdd.700. https://accp1.onlinelibrary.wiley.com/doi/epdf/10.1002/cpdd.700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoyama T., Kosaka Y., Mizuguchi M. Crystal structures of human transthyretin complexed with glabridin. J. Med. Chem. 2014;57:1090–1096. doi: 10.1021/jm401832j. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama T., Ueda M., Ando Y., Mizuguchi M. Discovery of γ-mangostin as an amyloidogenesis inhibitor. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palaninathan S.K., Mohamedmohaideen N.N., Orlandini E., Ortore G., Nencetti S., Lapucci A., Rossello A., Freundlich J.S., Sacchettini J.C. Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis. PLoS One. 2009;4:1–13. doi: 10.1371/journal.pone.0006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorensen J., Hamelberg D., Schiott B., McCammon J.A. Comparative MD analysis of the stability of transthyretin providing insight into the fibrillation mechanism. Biopolymers. 2007;86:73–82. doi: 10.1002/bip.20705. [DOI] [PubMed] [Google Scholar]
- 27.Yee A.W., Aldeghi M., Blakeley M.P., Ostermann A., Mas P.J., Moulin M., de Sanctis D., Bowler M.W., Mueller-Dieckmann C., Mitchell E.P., Haertlein M., de Groot B.L., Erba E.B., Forsyth V.T. A molecular mechanism for transthyretin amyloidogenesis. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-08609-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.RCSB protein data bank. https://www.rcsb.org/
- 29.Molecular Operating Environment (MOE) Chemical computing group. 2018. https://www.chemcomp.com/
- 30.Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Gohlke H., Goetz A.W., Gusarov S., Homeyer N., Janowski P., Kaus J., Kolossvary I., Kovalenko A., Lee T.S., LeGrand S., Luchko T., Luo R., Madej B., Merz K.M., Paesani F., Roe D.R., Roitberg A., Sagui C., Salomon-Ferrer R., Seabra G., Simmerling C.L., Smith W., Swails J., Walker R.C., Wang J., Wolf R.M., Wu X., Kollman P.A. University of California; San Francisco: 2014. AMBER 14. [Google Scholar]
- 31.Maier J.A., Martinez C., Kasavajhala K., Hauser K.E., Wickstrom L., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3693–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D.A., Cheatman T.E. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.