Abstract
Mesenchymal stem cells (MSCs) have been reported to hold promise to accelerate the wound-healing process in diabetic foot ulcer (DFU) due to the multilineage differentiation potential. Hence, this study intended to explore the wound healing role of MSC-derived exosomes containing long noncoding RNA (lncRNA) H19 in DFU. lncRNA H19 was predicated to bind to microRNA-152-3p (miR-152-3p), which targeted phosphatase and tensin homolog (PTEN) deleted on chromosome ten. Fibroblasts in DFU samples exhibited highly expressed miR-152-3p and poorly expressed lncRNA H19 and PTEN, along with an activated phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/protein kinase B (Akt1) signaling pathway. The fibroblasts were cocultured with lncRNA H19-transfected MSCs and MSC-derived exosomes to assess the effect of the lncRNA H19/miR-152-3p/PTEN axis on the biological activities and inflammation in fibroblasts. Mouse models of DFU were developed by streptozotocin, which was injected with MSC-derived exosomes overexpressing lncRNA H19. lncRNA H19 in MSCs was transferred through exosomes to fibroblasts, the mechanism of which improved wound healing in DFU, corresponded to promoted fibroblast proliferation and migration, as well as suppressed apoptosis and inflammation. Wound healing in mice with DFU was facilitated following the injection of MSC-derived exosomes overexpressing lncRNA H19. Taken together, MSC-derived exosomal lncRNA H19 prevented the apoptosis and inflammation of fibroblasts by impairing miR-152-3p-mediated PTEN inhibition, leading to the stimulated wound-healing process in DFU.
Keywords: diabetic foot ulcer, long noncoding RNA H19, microRNA-152-3p, PTEN, mesenchymal stem cells, exosome
Introduction
Diabetic foot ulcer (DFU) is a devastating medical problem that results from diabetes mellitus, which affects 15% of diabetic patients and leads to a risk of amputation.1 The 5-year mortality of patients with DFU is 2.5 times higher than those without DFU.2 As one of the major causes of DFU, peripheral arterial disease (PAD) deteriorates the body response to foot ulcerations and results in nonhealing ulcerations. The bone marrow-derived stem cells have been reported as a favorable treatment target for PAD therapy management due to the therapeutic potential, including angiogenesis and ulcer healing.3 Among adult stem cells, mesenchymal stem cells (MSCs) are an extensively investigated subgroup, considering their unique biological properties.4 MSCs should be plastic adherent when exposed to standard culture conditions; be positive for CD105, CD73, and CD90 and negative for CD45, CD34, CD14 or CD11b, CD79a or CD19, and human leukocyte antigen-G (HLA-DR); and most importantly, have the potency to differentiate into osteoblasts, adipocytes, and chondroblasts in vitro.5 The unique differentiation ability of MSCs may contribute therapeutic significance to the wound-repair process.6 The role of MSCs in tissue repair has been demonstrated to be dependent on exosomes secretion, which consists of bioactive molecules to modulate the cellular activities in recipient cells as a connection between cells.7 The transportation of nano-sized exosomes between cells and the capability of transport proteins, microRNAs (miRNAs), and mRNAs have been well documented in a previous study.8
Intriguingly, the existence of long noncoding RNAs (lncRNAs) in exosomes has been reported, suggesting that lncRNAs may also be loaded with exosomes during cell-to-cell communication and further regulate gene expression in host cells.9 The lncRNA H19 was located in chromosome 11 and has been indicated to play a significant role in osteogenic differentiation of ectomesenchymal stem cells, which resulted in tooth tissue regeneration in a rat model.10 Besides, the participation of lncRNA H19 in the activation of gluconeogenesis has been elucidated to be beneficial to the regulation of endogenous glucose production in diabetic hyperglycemia.11 In addition, lncRNA H19 has also been found to serve as a competing endogenous RNA (ceRNA) for miR-138 in bone marrow MSCs during the regulation of osteogenic differentiation.12 Such crosstalk involving miR-152-3p has also been suggested previously in that the binding to miR-152-3p by the long non-coding RNA HOX (lncRNA HOX) transcript antisense RNA is capable of affecting the malignant melanoma progression.13 A target gene of miR-152-3p, phosphatase and tensin homolog (PTEN) deleted on chromosome ten, has been implicated in rat myocardium-derived H9C2 cells.14 PTEN is a phosphatase that can be encoded by a tension-homolog gene on chromosome 10 and function as an antagonist of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) signaling pathway, whereas the absence of PTEN may cause abnormal phosphorylation of AKT.15 The PI3K/AKT signaling pathway has been discussed to play a part in the protective effects on high glucose-induced podocytes against apoptosis in the context of diabetic nephropathy.16 Therefore, the present study was performed to investigate the potential therapeutic effect of the lncRNA H19/miR-152-3p/PTEN axis in DFU to provide a feasible therapeutic target for DFU treatment.
Results
Characterization of Fibroblasts of DFU Samples
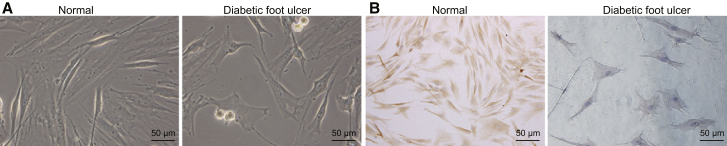
Initially, we resected foot tissue from patients with DFU and healthy participants, and then fibroblasts were isolated. During the culture of tissue blocks, there were tiny, elongated adhered cells crawling around the tissue block, 5–7 days later. According to inverted microscopic observation, normal human skin fibroblasts arranged tightly in a radial, interwoven, or swirling pattern. Some cells overlapped with each other. The cell body was elongated or an irregular polygon shape. Cell protrusion was short. Few or even no intracellular granules were observed in the cytoplasm with clear cell membrane without vacuoles. The nucleus was oval shaped (Figure 1A). Fibroblasts with DFU arranged irregularly with larger volume in a flat shape. The area of cytoplasm was larger. More and larger cell processes were observed with intracellular granules and vacuoles. The boundary was blurred, and shape was irregular (Figure 1A). Results of immunohistochemical staining for vimentin were positive, and a number of dark-brown particles were detected in the cytoplasm of fibroblasts (Figure 1B). Keratin antibody was negative, which was in accordance with the characteristics of mesoderm-derived fibroblasts (Figure 1B). The successful isolation of fibroblasts guaranteed the implementation of subsequent experiments in our study.
Figure 1.
Culture and Characterization of Fibroblasts
(A) Morphological observation of fibroblasts (200×). (B) Immunohistochemical analysis of fibroblasts (200×).
miR-152-3p Targets PTEN to Affect Fibroblast Proliferation, Migration, and Apoptosis
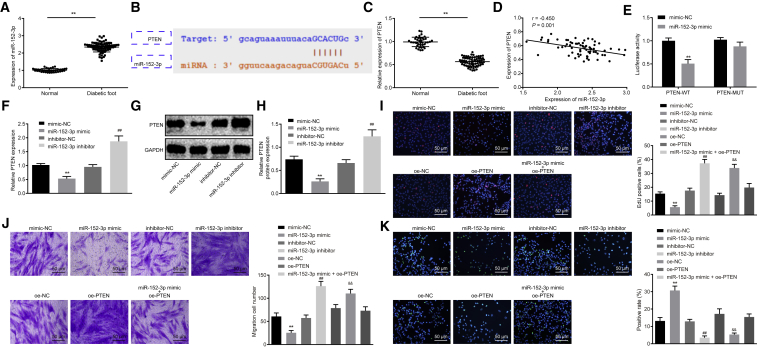
A functional study has indicated that multiple miRNAs are highly expressed in wound tissues of DFU.17 In our study, qRT-PCR was performed to determine miR-152-3p expression in ulcer tissues of patients with DFU and normal foot tissues, the results of which revealed significantly higher miR-152-3p expression in ulcer tissues of patients with DFU than that in normal foot tissues (p < 0.01; Figure 2A). The starBase database provided specific binding sites between PTEN and miR-152-3p (Figure 2B). PTEN has been reported as an anti-oncogene in association with an inflammatory response.18,19 qRT-PCR results showed that compared with normal foot tissues, PTEN was poorly expressed in foot tissues of DFU (p < 0.01; Figure 2C). The correlation analysis revealed a negative correlation between miR-152-3p and PTEN (Figure 2D). A dual-luciferase reporter gene assay was conducted to verify whether PTEN was a target gene of miR-152-3p. The results demonstrated that compared with the negative control (NC) group, luciferase activity of PTEN-wild-type (WT) was weakened in the miR-152-3p mimic group (p < 0.01), whereas no significant difference was detected with regard to luciferase activity of PTEN-mutant (Mut) (p > 0.05), suggesting that miR-152-3p could specifically bind to PTEN (Figure 2E). Furthermore, the expression of miR-152-3p in fibroblasts was altered, and the expression of PTEN was determined by qRT-PCR and western blot analysis. The results showed that compared with fibroblasts transfected with mimic-NC, fibroblasts in presence of miR-152-3p mimic had lower mRNA and protein expression of PTEN (p < 0.01; Figures 2F–2H). Opposite changing tendency was found in fibroblasts treated with the miR-152-3p inhibitor when compared with inhibitor-NC treatment (p < 0.01; Figures 2F–2H), indicating that PTEN is negatively regulated by miR-152-3p. Subsequently, effects of miR-152-3p and PTEN on proliferation, migration, and apoptosis of fibroblasts were investigated, and results indicated that treatments of the miR-152-3p inhibitor or oe-PTEN could both strengthen the proliferative and migration abilities as well as weaken apoptotic ability, whereas miR-152-3p mimic induced opposite changing tendency (p < 0.01; Figures 2I–2K). Taken together, these findings elucidate that miR-152-3p inhibits proliferation and migration and induces apoptosis of fibroblasts by regulating PTEN.
Figure 2.
miR-152-3p Exerts Effects on Proliferation, Migration, and Apoptosis of Fibroblasts by Targeting PTEN
(A) Expression of miR-152-3p in normal foot tissue (n = 40) and foot tissue from patients with DFU (n = 69), determined by qRT-PCR. (B) Predicted binding sites between miR-152-3p and PTEN by the starBase database. (C) mRNA expression of PTEN in normal foot tissue (n = 40) and foot tissue from patients with DFU (n = 69), determined by qRT-PCR. (D) Correlation between miR-152-3p and PTEN analyzed by Pearson correlation coefficient. (E) Luciferase activity of PTEN-WT and PTEN-Mut detected by dual-luciferase reporter gene assay. (F) mRNA expression of PTEN in fibroblasts after transfection, determined by qRT-PCR. (G) Protein bond diagram of PTEN in fibroblasts after transfection determined by western blot analysis. (H) Relative protein expression of PTEN normalized to GAPDH in fibroblasts after transfection determined by western blot analysis. (I) Fibroblast proliferation detected by EdU assay (200×). (J) Fibroblast migration detected by Transwell assay (200×). (K) Fibroblast apoptosis detected by TUNEL assay (200×). **p < 0.01 versus the normal group (normal foot tissue) or the mimic-NC group (fibroblasts treated with mimic-NC); ##p < 0.01 versus the inhibitor-NC group (fibroblasts treated with inhibitor-NC); &&p < 0.01 versus the oe-NC group (fibroblasts treated with oe-NC). Measurement data were expressed as mean ± SD. Comparison between two groups was conducted using independent sample t test. One-way ANOVA was used for data comparison among multiple groups, followed by Tukey’s post hoc test. The experiment was repeated independently three times.
lncRNA H19 Competitively Binds to miR-152-3p to Affect Fibroblast Proliferation, Migration, and Apoptosis
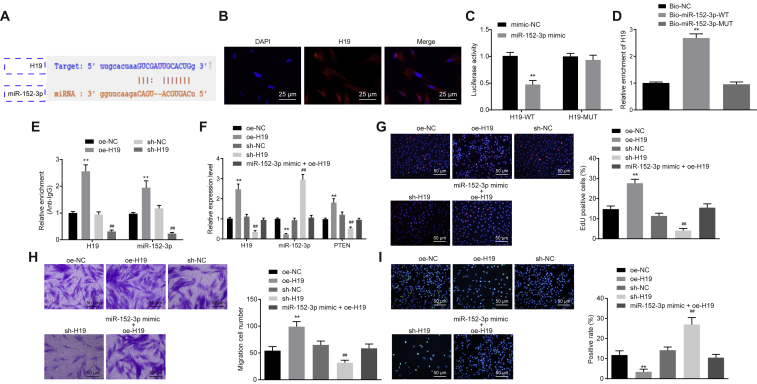
Available evidence has highlighted that lncRNAs can function as ceRNAs to regulate expression of miRNAs.20 With the use of the starBase database,21 we found that there was a targeting relationship between lncRNA H19 and miR-152-3p (Figure 3A). Since expression of lncRNA H19 has been detected in the cytoplasm and nucleus before, RNA-fluorescence in situ hybridization (FISH) was applied for verification (Figure 3B). The dual-luciferase reporter gene assay clearly displayed that relative luciferase activity of H19-WT was significantly weakened by miR-152-3p mimic (p < 0.01), whereas no similar effects were observed in relation to the luciferase activity of H19-Mut in comparison to mimic-NC treatment (Figure 3C). After miR-152-3p was marked by biotin, RNA pull-down was conducted. Results demonstrated that Bio-miR-152-3p-WT could pull down lncRNA H19 (p < 0.01), but Bio-miR-152-3p-Mut exerted no significant effects on lncRNA H19 (Figure 3D), suggesting that there was a direct interaction between miR-152-3p and lncRNA H19. To explore further the occurrence of the RNA-induced silencing complex when lncRNA H19 bound to miR-152-3p, RNA immunoprecipitation (RIP) assay was performed. Results showed that argonaute2 (Ago2) antibody could significantly enrich lncRNA H19 and miR-152-3p (p < 0.01; Figure 3E), implying that lncRNA H19 can target miR-152-3p by binding to Ago2.
Figure 3.
Overexpressed lncRNA H19 Competitively Binding to miR-152-3p Affects Fibroblast Proliferation, Migration, and Apoptosis
(A) Binding sites between lncRNA H19 and miR-152-3p. (B) Subcellular localization of lncRNA H19 detected by FISH (400×). (C) Luciferase activity of H19-WT and H19-Mut detected by dual-luciferase reporter gene assay. (D) Relative enrichment of lncRNA H19 detected by RNA pull-down. (E) Relative enrichment of Ago2 by lncRNA H19 and miR-152-3p detected by RIP assay. (F) lncRNA H19 and miR-152-3p expression and mRNA expression of PTEN, determined by qRT-PCR. (G) Fibroblast proliferation detected by EdU assay (200×). (H) Fibroblast migration detected by Transwell assay (200×). (I) Fibroblast apoptosis detected by TUNEL assay (200×). **p < 0.01 versus the oe-NC group (fibroblasts treated with oe-NC); ##p < 0.01 versus the sh-NC group (fibroblasts treated with sh-NC). Measurement data were expressed as mean ± SD. Comparison between two groups was conducted using independent sample t test. One-way ANOVA was used for data comparison among multiple groups, followed by Tukey’s post hoc test. The experiment was repeated independently three times.
For verification on whether lncRNA H19 could competitively bind to miR-152-3p, different plasmids were delivered into fibroblasts to determine the expression of miR-152-3p and PTEN. Overexpression of H19 led to lower miR-152-3p expression and higher PTEN expression, whereas higher miR-152-3p expression and lower PTEN expression were induced by short hairpin (sh)-H19 treatment rather than sh-NC treatment, indicating that lncRNA H19 can competitively bind to miR-152-3p to regulate PTEN expression (Figure 3F). Subsequent changes of proliferation (Figure 3G), migration (Figure 3H), and apoptosis (Figure 3I) of fibroblasts were investigated by means of 5-ethynyl-2′-deoxyuridine (EdU), Transwell, and TUNEL assays. Results demonstrated that the transduction of oe-H19 resulted in promoted proliferation and migration along with suppressed apoptosis (p < 0.01). A reasonable conclusion can be drawn that lncRNA H19 facilitated fibroblast proliferation and migration by downregulating miR-152-3p.
Characterization of MSCs and the Derived Exosomes
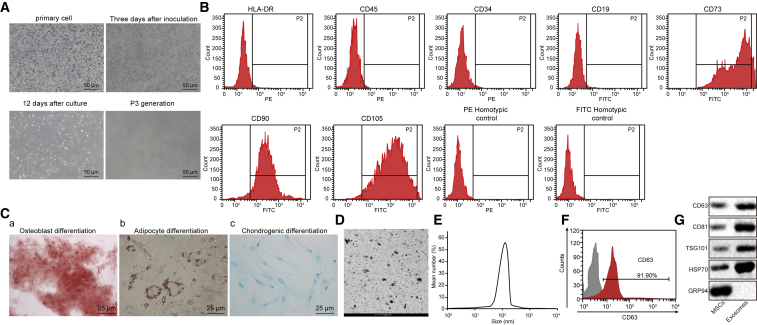
A previous study has reported that MSCs can promote the wound-healing process of DFU.22 Hereby, explorations were carried out regarding the function of exosomes from myeloid-derived MSCs in the wound-healing process of DFU. In order to verify whether MSCs exhibited the ability of multidirectional differentiation into osteoblasts or adipocytes, we observed the cellular morphology, identified the surface antigen by flow cytometry, as well as treated cells with osteogenic induction and adipogenic induction. MSCs were first isolated and cultured. At the 3rd day after culture, cells started to grow adhered to the wall in a fusiform shape. The growth manner tended to be swirling or clustered with clear nucleus. Typical characteristics of MSCs presented (Figure 4A). The expression of MSC surface antigens was analyzed by flow cytometry. As shown in Figure 4B, CD73 was 100%, CD90 was 94.2%, and CD105 was 97.8%, whereas the rest was less than 1%. The results accorded with the biological characteristics of MSCs, suggesting that fibroblasts had specific molecular markers of MSCs. As for results of MSC differentiation, 21 days after osteogenic induction, cells overlapped with each other and formed calcified nodules containing a small amount of mineral salt sediment, suggesting that MSCs had the potential to differentiate into osteoblasts (Figure 4Ca). After 25 days of adipogenic induction, lipid accumulation was observed inside, and lipid droplets gradually became larger or beaded, indicating the adipogenic differentiation potential of MSCs (Figure 4Cb). Alcian blue staining showed the potential of MSCs to differentiate into chondrocytes (Figure 4Cc).
Figure 4.
Characterization of MSCs and MSC-exo
(A) Morphological observation of MSCs (200×). (B) Surface marker molecules of MSCs detected by flow cytometry. (C) Representative images of osteocyte, adipocyte, and chondrocyte differentiation of MSCs analyzed using cytochemical staining with alizarin red (a), oil red O (b), and Alcian blue (c) (400×). (D) Morphology change of MSC-exo detected by TEM (scale bar, 200 nm). (E) The size distribution of the MSC-exo examined using dynamic light scattering. (F) Surface marker (CD63) of exosomes detected by flow cytometry. (G) The positive markers for exosomes, including CD63, CD81, TSG101, and HSP70, were detected in MSC-exo using western blot analysis, whereas the negative marker GRP94 was not detected. The experiment was repeated independently three times.
According to the transmission electron microscope (TEM) observation, the size of MSC-derived exosomes (MSC-exo) varied, ranging from 30 nm to 120 nm, and the morphology was basically the same as round or oval membranous vesicles (Figure 4D). The diameter of exosome granules was measured by dynamic light scattering (Figure 4E). The content of exosome surface marker CD63 was determined by flow cytometry, results of which showed that CD63 content increased remarkably. Western blot analysis was conducted for quantification of CD63, CD81, TSG101, heat shock protein 70 (HSP70), and the negative marker GRP94 expression in exosomes, results of which provided verification that exosomes displayed overexpression of CD63, CD81, TSG101, and HSP70 (Figure 4G). The above-mentioned results demonstrate that fibroblasts have the ability of multidirectional differentiation, and exosomes were successfully extracted from MSCs.
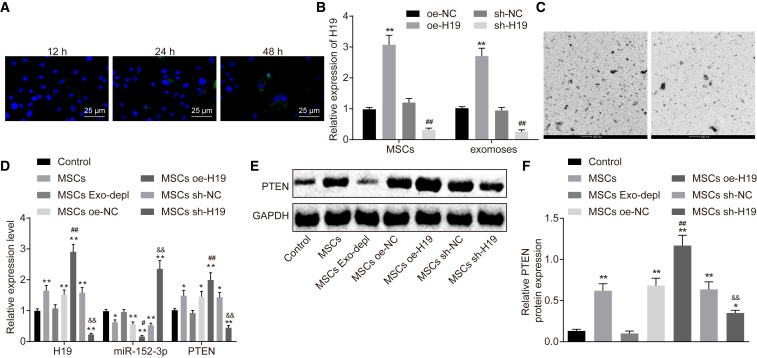
MSCs Regulate Expression of miR-152-3p and PTEN in Fibroblasts by MSC-exo Carrying lncRNA H19
Subsequently, exosomes traced by PKH67 (green) were cultured with fibroblasts. The uptake of exosomes was observed under a confocal fluorescence microscope at the 12th, 24th, and 48th h postculture (Figure 5A). Over the period of coculture, more and more cells showed green fluorescence, suggesting that the number of PKH67 exosomes uptaken by fibroblasts was increased. At 48 h after coculture, the uptake of PKH67 exosomes by fibroblasts was significantly obvious, indicating that exosomes can transfer from donor cells (MSCs) to recipient cells (fibroblasts). To confirm whether lncRNA H19 could be transferred by MSC-exo to fibroblasts, expression of lncRNA H19 was altered in MSCs, and qRT-PCR was performed for quantification of lncRNA H19 in MSCs and MSC-exo. Results showed that lncRNA H19 expression was increased in MSCs and MSC-exo in the presence of oe-H19 but decreased in the presence of sh-H19 (p < 0.01; Figure 5B)
Figure 5.
MSCs Regulate the Expression of miR-152-3p and PTEN in Fibroblasts through MSC-exo Carrying lncRNA H19
(A) The uptake of MSC-exo by fibroblasts observed under an inverted microscope (400×). (B) Expression of lncRNA H19 in MSCs and exosomes, determined by qRT-PCR. (C) Secretion of exosomes in medium with exosome inhibitor added, observed under TEM (scale bar, 200 nm). (D) Expression of lncRNA H19, miR-152-3p, and PTEN, determined by qRT-PCR. (E) Protein bond diagram of PTEN determined by western blot analysis. (F) Relative protein expression of PTEN normalized to GAPDH, determined by western blot analysis. **p < 0.01 versus the control group (fibroblasts without any treatment); ##p < 0.01 versus the MSC oe-NC group (MSCs treated with oe-NC); &&p < 0.01 versus the MSC sh-NC group (MSCs treated with sh-NC). Measurement data were expressed as mean ± SD. One-way ANOVA was used for data comparison among multiple groups, followed by Tukey’s post hoc test. The experiment was repeated independently three times.
To confirm further the role of exosomes, the secretion of exosomes was inhibited by adding GW4869 in MSCs, and the TEM observation illustrated that the secretion was almost blocked (Figure 5C). Investigation was then made to explore whether transfer of lncRNA H19 in vitro could regulate expression of miR-152-3p and PTEN in fibroblasts. qRT-PCR and western blot analysis were conducted to quantify the expression of lncRNA H19, miR-152-3p, and PTEN (Figures 5D–5F). In comparison to fibroblasts without any treatment, expression of lncRNA H19, miR-152-3p, and PTEN did not differ significantly when secretion of exosomes was blocked, suggesting that secretion of exosomes exerted no significant effects on expression of lncRNA H19, miR-152-3p, and PTEN. When MSCs were treated with oe-H19, miR-152 expression was decreased significantly, whereas expression of lncRNA H19 and PTEN was increased (p < 0.01), indicating that transfer of lncRNA H19 in vitro can efficiently downregulate expression of miR-152-3p and upregulate PTEN expression. The aforementioned results shed light on the important role of exosomes in the transfer from MSCs to fibroblasts, revealing that transfer of exogenous lncRNA H19 from MSCs to fibroblasts depends on exosomes.
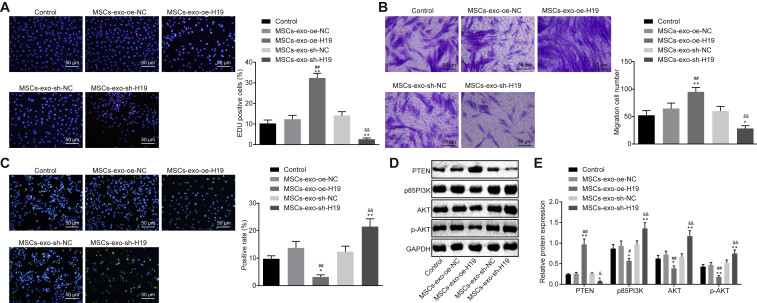
MSC-exo Carrying lncRNA H19 Regulates Fibroblast Proliferation, Migration, and Apoptosis via the PI3K/AKT Signaling Pathway
Since MSCs could carry lncRNA H19 to fibroblasts via exosomes, the regulatory role of exosomes in proliferation, migration, and apoptosis of fibroblasts was explored by allowing MSCs to coculture with fibroblasts. As shown in Figures 6A–6C, oe-H19 delivery led to promoted fibroblast proliferation and migration with suppressed apoptosis, whereas sh-H19 caused opposite changes (p < 0.01), suggesting that MSC-exo carrying lncRNA H19 regulates fibroblast proliferation, migration, and apoptosis. It is acknowledged that PTEN is a critical suppressor for the PI3K/AKT signaling pathway, which plays an important part in mediating cell growth, differentiation, proliferation, apoptosis, and glycometabolism. Herein, a hypothesis was proposed that effects of lncRNA H19/miR-152-3p/PTEN axis on biological functions of fibroblasts were achieved through the PI3K/AKT signaling pathway. The expression of p85 PI3K and AKT, as well as the extent of AKT phosphorylation, was determined. Results revealed that exosomes treated with oe-H19 had significantly higher PTEN expression and lower expression of p85 PI3K and AKT, as well as the extent of AKT phosphorylation. On the contrary, sh-H19 treatment resulted in higher expression of p85 PI3K and AKT, as well as the extent of AKT phosphorylation, from which it could be concluded that expression of p85 PI3K and AKT, as well as the extent of AKT phosphorylation, can be mediated by lncRNA H19 (p < 0.01; Figures 6D and 6E). These findings uncover the participation of the PI3K/AKT signaling pathway in the regulating mechanism of the lncRNA H19/miR-152-3p/PTEN axis in proliferation, migration, and apoptosis of fibroblasts.
Figure 6.
MSC-exo Carrying lncRNA H19 Regulates Fibroblast Proliferation, Migration, and Apoptosis via the PI3K/AKT Signaling Pathway
(A) Fibroblast proliferation detected by EdU assay (200×). (B) Fibroblast migration detected by Transwell assay (200×). (C) Fibroblast apoptosis detected by TUNEL assay (200×). (D) Protein bond diagram of PTEN, p85 PI3K, and AKT, as well as the extent of AKT phosphorylation determined by western blot analysis. (E) Relative protein expression of PTEN, p85 PI3K, and AKT, as well as the extent of AKT phosphorylation normalized to GAPDH, determined by western blot analysis. **p < 0.01 versus the control group (fibroblasts without any treatment); ##p < 0.01 versus the MSC-exo-oe-NC group (MSC-exo treated with oe-NC); &&p < 0.01 versus the MSC-exo-sh-NC (MSC-exo treated with sh-NC). Measurement data were expressed as mean ± SD. Comparison among multiple groups was conducted using one-way ANOVA, followed by Tukey’s post hoc test. The experiment was repeated independently three times.
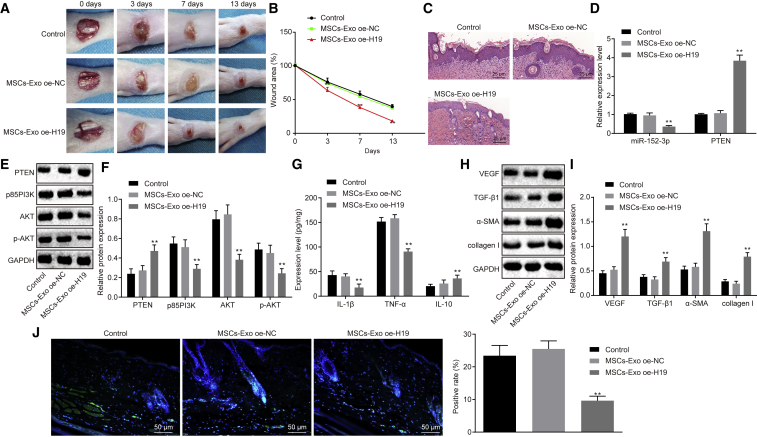
MSC-exo Carrying lncRNA H19 Promotes Wound Healing in Mice with DFU
In order to assess the effect of MSC-exo on the wound-healing process in mice with DFU, we developed a DFU mouse model and injected MSC-exo containing ad-oe-H19 into the surrounding tissues of the wound, followed by H&E staining and immunohistochemical staining for observation on wound healing. It was shown that exosomes containing ad-oe-H19 in vivo could significantly accelerate the wound-healing process (Figures 7A–7C), as evidenced by fewer inflammatory cells and thicker granulation tissues around the wound. qRT-PCR results revealed that miR-152-3p expression was diminished, and PTEN expression was elevated after the treatment of MSC-exo oe-H19 in mice with DFU (Figure 7D). Meanwhile, expression of p85 PI3K and AKT, as well as the extent of AKT phosphorylation, was decreased following the treatment of MSC-exo oe-H19 in mice with DFU (Figures 7E and 7F).
Figure 7.
MSC-exo Carrying lncRNA H19 Promotes Wound Healing in Mice with DFU
(A) Representative images displaying the wound-healing process of mice with DFU. (B) Wound area of mice with DFU. (C) Edge tissues of wound identified by H&E staining (200×). (D) miR-152-3p expression and mRNA expression of PTEN in wound tissues, determined by qRT-PCR. (E) Protein bond diagram of PTEN, p85 PI3K, and AKT, as well as the extent of AKT phosphorylation determined by western blot analysis. (F) Relative protein expression of PTEN, p85 PI3K, and AKT, as well as the extent of AKT phosphorylation normalized to GAPDH, determined by western blot analysis. (G) Levels of inflammatory factors (IL-1β, TNF-α, and IL-10), determined by ELISA. (H and I) Relative protein expression of angiogenesis-related factors (VEGF, TNF-β1, α-SMA, and collagen I) normalized to GAPDH, determined by western blot analysis. (J) Cell apoptosis detected by TUNEL assay (200×). **p < 0.01 versus the control group (n = 12, DFU mice without any treatment). Measurement data were expressed as mean ± SD. Comparison among multiple groups was conducted using one-way ANOVA, followed by Tukey’s post hoc test. Repeated-measures ANOVA was applied for data comparison among multiple groups at different time points, followed by Tukey’s post hoc test (B).
Lastly, an inflammatory response of the wound was detected by quantifying proinflammation factors (interleukin 1β [IL-1β] and tumor necrosis factor α [TNF-α]) and an anti-inflammation factor (IL-10) using ELISA. Results revealed that MSC-exo oe-H19 led to higher level of IL-10 and lower levels of IL-1β and TNF-α (p < 0.01; Figure 7G), suggesting that MSC-exo expressing oe-H19 can ameliorate the inflammation of the wound in mice with DFU. For further verification, western blot analysis was performed to determine the expression of vascular endothelial growth factor (VEGF), TGF-β1, α smooth muscle actin (α-SMA), and collagen I. As displayed in Figures 7H and 7I, injection of MSC-exo oe-H19 significantly increased expression of VEGF, TGF-β1, α-SMA, and collagen I (p < 0.01), indicating that angiogenesis can be enhanced by MSC-exo oe-H19 to accelerate the wound-healing process. TUNEL assay was carried out to detect cell apoptosis further, results of which demonstrated that cell apoptosis was significantly suppressed by MSC-exo oe-H19 in comparison to MSC-exo vector treatment (p < 0.01; Figure 7J). To conclude, an indication is revealed that MSC-exo carrying overexpressed lncRNA H19 is capable of accelerating the wound-healing process of mice with DFU.
Discussion
DFU is a health complication caused by diabetes-induced blood-vessel damage, where high blood glucose levels slowed down the wound-healing process.23 Therefore, the identification of efficient treatment approaches for DFU is a challenging task, which brooks no delay. In the present study, the mechanism of MSC-derived exosomal lncRNA H19 working in tandem with miR-152-3p and PTEN via the PI3K/AKT signaling pathway was probed through a series of in vitro and in vivo experiments. Based on the available evidence, it is reasonable to conclude that MSC-derived exosomes carrying overexpressed lncRNA H19 can be stimulative for the wound-healing process of mice suffering from DFU, which is involved with an interaction with miR-152-3p via PTEN-mediated PI3K/AKT signaling pathway.
The miR-152-3p was verified as the potential regulator of PTEN, based on the starBase database prediction and dual-luciferase reporter gene assay. The targeting relationship between miR-152-3p and PTEN has been previously reported in the therapy candidate investigation for heart failure.14 Additionally, the elevated miR-152-3p expression and diminished PTEN expression were observed in fibroblasts from patients with DFU in this present study. These findings were consistent with a previous study, where miR-12-5p, miR-34a-5p, miR-143-3p, and miR-145-5p were observed to be upregulated in fibroblasts with DFU rather than their counterparts in fibroblasts without DFU, whereas the opposite alternatives induced fibroblast proliferation, differentiation, and migration.17 An in-depth investigation on the changes of cellular behaviors in response to delivery of the miR-152-3p inhibitor and oe-PTEN was performed in this study. The results revealed that proliferation and migration abilities of fibroblasts were strengthened, whereas apoptotic capability was diminished. Likewise, the regulation of inflammatory stress and immune response of dengue viruses by miR-152 was validated. The upregulated miR-152 in suppressing cell proliferation was also proved in similar study.24 Downregulation of PTEN predisposes to diabetic nephropathy by impairing glomerular functions.25
Interestingly, this present study discovered a significant finding, where lncRNA H19 upregulated the expression of PTEN by binding to miR-152-3p. A similar mechanism of lncRNA H19 has been unveiled previously that lncRNA H19 mediated the expression of a connective tissue growth factor by binding to miR-455 in cardiac fibrosis, and the expression of suppresses aquaporin-3 was subjected to the regulation of lncRNA H19-sponging miR-874 in an intestinal barrier.26,27 MSC regenerative therapy is a novel way of tissue regeneration, where MSCs can improve the wound healing of DFU by accelerating wound closure, alleviating clinical parameters, as well as preventing patients from amputation.28 A rat model of DFU has demonstrated that the transplantation of human adipose-derived stem cells promotes wound healing, highlighting the application of exogenous stem cells in clinical practice of DFU.29 According to Zeng et al.,30 a patient with DFU received administration of a placenta-derived MSC hydrogel for the treatment of DFU and displayed desirable outcomes. Moreover, exosomes derived from miR-126-3p-overexpressing synovium MSCs have been documented as a potential drug delivery system to improve the cutaneous wound healing in a diabetic rat model.31 In this study, the mice with DFU were competently recovered from DFU after subsequent injection of MSC-exo oe-H19. The recipient fibroblasts were altered, leading to the increase of proliferation and migration. Moreover, circumstantial evidence has verified that the blocked PI3K/AKT signaling pathway also plays an important role in the mechanism, showing decreased expression of p85 PI3K and AKT, as well as the extent of AKT phosphorylation in the presence of oe-H19. The PI3K/AKT signaling pathway has been reviewed as an axis that is responsible to counteract pain relief or inflammation caused by bone cancer, where the inhibition of this axis may be beneficial to chronic pain management,32 which is consistent with the results of this present study.
Taken together, these promising data allowed us to propose a molecular mechanism underlying DFU treatment by which MSC-released exosomal lncRNA H19 inhibits miR-152-3p and promotes PTEN expression to enhance the proliferation and migration and suppress apoptosis of fibroblasts, thereby alleviating the damage of DFU and accelerating the wound-healing process. In conclusion, MSC-exo lncRNA H19 may function as a promising therapeutic target to enhance the development of therapeutic treatments for DFU. Nevertheless, the current research is still in the preclinical stage, and the investigation on the mechanism of action is insufficient. More experiments are needed to explore the intrinsic mechanisms further.
Materials and Methods
Ethics Statement
The study was approved by the Institutional Review Board of the Fourth Affiliated Hospital of Harbin Medical University in strict accordance with Declaration of Helsinki. Written, informed consent was obtained from each participant. The animal protocol and experiment procedure were approved by the Institutional Animal Care and Use Committee of the Fourth Affiliated Hospital of Harbin Medical University.
Sample Collection
A total of 69 tissue samples of foot ulcer were selected from patients with DFU (age: 30–75 years) in the Fourth Affiliated Hospital of Harbin Medical University. Patients were selected if they fall into grades 1–3 according to Wagner grading,33 without any serious lower extremity arterial diseases, cardiocerebrovascular diseases, or other complications. Another 40 normal foot tissue samples were collected from healthy individuals without DFU.
Isolation, Culture, and Identification of Fibroblasts
Under sterile condition, full-thickness skin graft (1.0 cm × 1.0 cm) was collected from the edge of the patient’s ulcer and placed in DMEM (Gibco by Life Technologies, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco by Life Technologies, Grand Island, NY, USA) in a sterile bottle. On a super-clean bench, the sample was repeatedly rinsed two to three times with PBS containing 100 U/mL penicillin and streptomycin (Gibco by Life Technologies, Grand Island, NY, USA) to remove the adhered serum and fat particles. Then, the subcutaneous adipose tissues were cut into pieces of 0.5–l mm3 and incubated with 2–3 mL DMEM containing 10% FBS in 25 cm culture flasks for a 4-h incubation at 37°C with 5% CO2. The medium was renewed after 3–5 days for the first time, then renewed every 3–4 days afterward. The cellular morphology was observed and photographed. When the cells grew to 80%–90%, the cells were passaged. The cells at passages 3–5 were used for all the experiments.
After the fibroblasts adhered to the coverslip, the fibroblasts were fixed with 4% paraformaldehyde, immersed in Triton X-100 for 10 min, then soaked in 3% hydrogen peroxide (H2O2) for 10 min, and blocked with serum for 10 min. Subsequently, the fibroblasts were incubated with the primary antibody, mouse anti-human antibody to vimentin (1:20, ab3974; Abcam, Cambridge, UK), or keratin (1:1,000, ab8068; Abcam, Cambridge, UK) at 4°C for 3 h and the secondary antibody, goat anti-mouse antibody (1:1,000, ab6789; Abcam, Cambridge, UK), at 37°C for 10 min. After diaminobenzidine (DAB) treatment and hematoxylin staining, the sections were mounted for observation and photographed under an optical microscope.
Establishment of a Mouse Model with DFU
A total of 60 healthy male C57BL/6J mice (age: 5 weeks; weight: 20.88 ± 1.94 g) were purchased from Shanghai Laboratory Animal Center, Chinese Academy of Sciences (Shanghai, China), and housed at a constant temperature and humidity, with a 12:12-h light-dark cycle. All mice were treated under normal feeding condition in separated cages. After 1 week of adaptive feeding, ten mice were randomly selected as the normal control group by injecting normal saline with normal feeding. The remaining fifty mice were used to establish diabetic models by feeding with a high-glucose and high-fat diet for 6 weeks and intraperitoneally injected with 0.45% streptozotocin (STZ; 45 mg/kg, S0130-50MG; Sigma-Aldrich, St. Louis, MO, USA), which was prepared by 0.1 mM sterile sodium citrate buffer solution. After 72 h, the tail blood glucose level was measured by an offline blood glucose monitoring system (Glucotrend 2; Roche Diagnostics, Mannheim, Germany). Mice with a blood glucose level ≥16.7 mM continuously for 10 days were enrolled in our study. The modeling success rate was 74% (37/50).
Two weeks after the diabetes was molded, the mice were anesthetized by intraperitoneal injection of pentobarbital sodium at 50 mg/kg (57-33-0; Shanghai Beizhuo Biotechnology, Shanghai, China). The dorsal area of the mice was shaved, and then a depilatory agent was applied to remove the remaining hair. Full-thickness wounds (10 mm diameter) were made on the back using a sterile punch. Exosomes of MSCs, including MSC-exo vector and MSC-exo carrying oe-H19, were injected into the skin around the wound (twelve mice for each). Another twelve mice remained untreated as controls, and one mouse remained as an alternative. Then, the wound was sterilized with 0.5% iodine daily. Blood glucose, weight, mean amount of drinking water, humidity of padding, and urine output were measured on the 3rd, 7th, and 13th day postinjection. The wound area was determined by tracing the wound edge on the transparent plastic film. Wound tissues and their surrounding areas were harvested at 3, 7, and 13 days. The tissues were fixed in 10% paraformaldehyde, paraffin embedded, and H&E stained. The wound-healing condition was observed under an optical microscope.
H&E Staining
The obtained wound edge tissue of the mice was fixed in 10% neutral formalin for at least 24 h, deparaffinized in xylene, dehydrated through a graded series of ethanol, stained with H&E, and mounted in resin. The images were captured using an optical microscope.
Immunohistochemistry
The paraffin-embedded sections were dehydrated by an ascending series of ethanol and washed with running water for 2 min. Then, the sections were immersed in 3% H2O2 for 20 min, followed by washing with distilled water for 2 min and 0.1 M PBS for 3 min. The samples were blocked by incubation in normal goat serum (C-0005; Shanghai Haoran Biological Technology, Shanghai, China) at room temperature for 20 min. Slides were incubated overnight at 4°C with the primary rabbit anti-human antibody to PTEN (1:100, ab170941; Abcam, Cambridge, UK). Following incubation with the secondary goat anti-rabbit antibody to immunoglobulin G (IgG; 1:1,000, ab6785; Abcam, Cambridge, UK) and a horseradish peroxidase (HRP)-labeled streptavidin working solution (0343-10000U; Imunbio, Beijing, China) at 37°C for 20 min, labeling was visualized using DAB (ST033; Whiga Biotech, Guangzhou, Guangdong, China) with hematoxylin (PT001; Bogoo Biotech, Shanghai, China). The sections were returned blue by adding 1% ammonia, then were dehydrated with gradient alcohol, cleared by xylene, and mounted with neutral resin. The section was observed and photographed under the optical microscope.
RNA Isolation and Quantitation
The total RNA of tissues or cells was extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA). A total of 1 μg RNA was reversely transcribed into cDNA, according to the instructions described in a PrimeScript RT reagent kit with a genomic DNA (gDNA) Eraser kit (RR037A; Takara, Tokyo, Japan). The miRNA-specific complementary DNA was synthesized using miRNA-specific RT primers from a TaqMan MicroRNA Assay and TaqMan MicroRNA RT kit (4366596; Thermo Fisher Scientific, Waltham, CA, USA). The expression of miR-152-3p was determined, according to the instructions described in the TaqMan miRNA Assay (Thermo Fisher Scientific, Waltham, CA, USA). qRT-PCR was performed on an ABI7500 qPCR instrument (Thermo Fisher Scientific, Waltham, CA, USA) using a SYBR Premix Ex Taq (Tli RNaseH Plus) kit (RR820A; Takara, Tokyo, Japan). The relative expression of miRNA or mRNAs was evaluated by the 2−ΔΔCt method and normalized to U6 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH), respectively.34 The primer sequences for qRT-PCR were provided from Shanghai GenePharma (Shanghai, China), as shown in Table 1.
Table 1.
Primer Sequences for qRT-PCR
| Gene | Primer Sequences |
|---|---|
| lncRNA H19 | F: 5′-TACAACCACTGCACTACCTG-3′ |
| R: 5′-TGGAATGCTTGAAGGCTGCT-3′ | |
| miR-152-3p | F: 5′-ACACTCCAGCTGGGTCAGTGCATGACAG-3′ |
| R: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCAAGTT-3′ | |
| PTEN | F: 5′-TTTGAAGACCATAACCCACCAC-3′ |
| R: 5′-ATTACACCAGTTCGTCCCTTTC-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| R: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| GAPDH | F: 5′-AATCCCATCACCATCTTCCA-3′ |
| R: 5′-TGGACTCCACGACGTACTCA-3′ |
lncRNA, long noncoding RNA; miR, microRNA; PTEN, phosphatase and tensin homolog deleted on chromosome ten; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward; R, reverse.
Western Blot Analysis
The total proteins from tissue or cells were lysed by cell lysis buffer containing PMSF and proteinase inhibitor (P0100; Solarbio, Beijing, China) at 4°C for 15 min. After centrifugation at 15,000 rpm for 15 min, the concentration of the protein was quantified by a bicinchoninic acid protein quantification kit (23227; Thermo Fisher Scientific, Waltham, CA, USA). The protein was separated by PAGE and transferred onto the polyvinylidene fluoride membrane. The membrane was then blocked with 5% BSA at room temperature for 1 h and probed with primary antibodies at 4°C overnight. The primary antibodies used in the study included antibodies to PTEN (1:2,000, ab109454), AKT (1:500, ab8805), phosphorylated AKT (p-AKT; 1:1,000, ab38449), p85 PI3K (1:1,000, ab86714), collagen I (1:1,000, ab90395), VEGF (1:2,000, ab32152), TGF-β1 (ab155613), α-SMA (1:2,000, ab108424), CD63 (1:1,000, ab216130), HSP70 (1:2,000, ab2787), CD81 (1:1,000, ab79559), TSG101 (1:1,000, ab83), and GAPDH (1:5,000, ab8245). The membrane was subsequently incubated with the HRP-conjugated goat anti-rabbit secondary antibody to IgG (1:20,000, ab205718) at room temperature for 1.5 h. The antibodies were all purchased from Abcam (Cambridge, UK). Bands were detected by developing solution (NCI4106; Pierce, Rockford, IL, USA) and analyzed with ImageJ 1.48u software (Bio-Rad, Hercules, CA, USA) based on the gray value of target protein and gray ratio of the internal reference (GAPDH).
TUNEL Assay
The de-waxed tissue was incised into five slices. Each slice was incubated with 50 μL 1% protease K dilution at 37°C for 30 min. The activity of endogenous peroxidase (POD) was eliminated by the incubation with 0.3% H2O2-methanol solution at 37°C for 30 min. After that, the sections were treated with TUNEL reaction solution at 37°C for 1 h, 50 μL Converter-POD at 37°C for 30 min, and 2% DAB at room temperature for 15 min. Based on the microscopic observation, after the appearance of a brown-yellow nucleus, distilled water was added to terminate the reaction. Hematoxylin was added for counterstaining, followed by addition of distilled water to terminate the reaction. The sample was then hydrated by gradient ethanol, cleared with xylene, and mounted. The mounted samples were observed under an optical microscope (400 times). Ten fields of view were randomly selected from each slice. The numbers of positive cells and podocytes were counted. The brown-yellow nucleus represented apoptotic-positive cells, whereas blue nucleus represented normal cells. The mean value was obtained. Apoptotic index = the number of positive cells/the number of normal cells.
Isolation, Culture, and Osteogenic, Adipogenic, and Chondrogenic Differentiation of MSCs
The C57BL/6 mice were euthanized and soaked in alcohol for 10 min. The femur and tibia were resected under sterile condition. After the flesh was removed, the femur and tibia were maintained in DMEM (Gibco by Life Technologies, Grand Island, NY, USA) for further use. Both edges of femur and tibia were cut with sterile scissors, and the marrow cells were transferred to a 15-mL centrifuge tube using DMEM, followed by centrifugation at 1,500 rpm for 3 min. Cells were resuspended in DMEM complete medium containing 10% FBS (Gibco by Life Technologies, Grand Island, NY, USA) and 100 U/mL penicillin and streptomycin (Gibco by Life Technologies, Grand Island, NY, USA) and cultured at 37°C with 5% CO2. The culture medium was renewed 3 days later to remove the nonadhered cells. The cellular morphology changes were observed and photographed. When cells were grown to 80%–90% confluency, the cells were passaged in separated bottles. The cells at passage 3 were harvested for further use.
Passage 3 MSCs were detached by Trypsin (Gibco by Life Technologies, Grand Island, NY, USA) and suspended in PBS, and the cell concentration was adjusted to 1 × 106 cells/mL. A total of 200 μL cell suspension was then stained with 5 μL monoclonal antibody, fluorescein isothiocyanate (FITC)-conjugated antibodies (Abcam, Cambridge, UK) to CD19 (ab24936), CD73 (ab202122), CD90 (ab226), and CD105 (ab184667), or polyethyleneglycol (PE)-conjugated antibodies (Bio-Techne China, Shanghai, China) to CD34 (NB600-1071PE), CD45 (NB100-77417PE), and HLA-DR (NBP2-47670PE) at 4°C for 15 min in the dark. Then, the sample was centrifuged at 1,000 rpm for 5 min with 2 mL PBS to remove the unbound antibodies. Then, 400 μL 0.01 M PBS containing 0.5% paraformaldehyde was added for cell resuspension. The IgG antibody was set as an isotype control. Each tube was examined by flow cytometry.
The passage 3 MSCs were trypsinized to prepare a single-cell suspension. The cell concentration was adjusted to 1 × 105 cells/mL in a 6-well plate. To accomplish the differentiation, MSCs from C57BL/6 mice were cultured in MSC osteogenic (MUBMX-90021), adipogenic (MUBMX-90031), or chondrogenic (MUBMX-90041; all from Cyagen, Suzhou, Jiangsu, China) differentiation media, as described by the manufacturer. The osteogenic, adipogenic, and chondrogenic differentiations were identified by alizarin red staining, oil red O staining, and Alcian blue staining, respectively.
Isolation and Identification of MSC-exo
The passage 3 MSCs were cultured until 80%–90% confluence overnight in serum-free DMEM and centrifuged at 2,000 g for 20 min at 4°C to remove cell debris. The obtained supernatant was ultracentrifuged at 10,000 g for 1 h at 4°C. The pellet was suspended in serum-free DMEM containing 25 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonicacid (pH 7.4) and submitted to another ultracentrifugation in the same conditions. The pellet was stored at −80°C until further use.35
TEM was used to identify exosomes. An amount of 30 μL of exosomes was added dropwise on copper mesh and allowed to stand for 1 min. The liquid was blotted from the side using filter paper. Then, 30 μL phosphotungstic acid solution (pH 6.8) was added dropwise for counterstaining at room temperature for 5 min. After drying under an electric incandescent lamp, photographs were acquired using TEM.
The concentration of exosome surface marker CD63 was detected by flow cytometry. MSCs were trypsinized, centrifuged at 1,000 rpm for 5 min, and resuspended with PBS. The cell suspension was evenly aliquoted into ten, 1.5 mL eppendorf (EP) tubes, followed by centrifugation at 1,000 rpm for 5 min. The cells were then treated with 1 mL PBS (containing 1% BSA) and incubated at room temperature for 30 min to block the nonspecific antigens. Following another centrifugation at 1,000 rpm for 5 min, the cells in each EP tube were resuspended with 200 μL PBS and incubated with PE-conjugated CD63 antibody (ab234251; Abcam, Cambridge, UK) at room temperature for 30 min. Blank control was set without addition of any antibody. Isotype control was set with addition of PE-labeled human antibody to IgG. The cells were then centrifuged at 1,000 rpm for 5 min and resuspended in PBS containing 1% BSA. Guava easyCyte system flow cytometry was employed for detection. The expression of miR-152-3p in exosomes was determined by qRT-PCR.
Dual-Luciferase Reporter Gene Assay
Through the starBase database,21 we obtained that there were specific binding regions between the PTEN gene sequence and the miR-152-3p sequence. The WT-PTEN (PmirGLO-PTEN-WT) and Mut-PTEN (PmirGLO-PTEN-Mut) vectors, along with the WT-H19 (PGLO-H19-WT) and Mut-PTEN (PGLO-H19-Mut) vectors were constructed, respectively. The reporter vectors, miR-152-3p mimic plasmid and NC plasmid, were cotransfected into 293T cells, respectively. After 24 h of transfection, the cells were harvested, lysed, and centrifuged at 12,000 rpm for 1 min. The luciferase activity was measured using a Dual-Luciferase Reporter Assay System (E1910; Promega, Madison, WI, USA). Luciferase activity was normalized by the Renilla/firefly luciferase signal in cells.
Plasmid Transfection
Fibroblasts at the logarithmic growth phase were seeded in a 6-well plate at a density of 4 × 105 cells/well. When cells reached about 80%–90% confluency, plasmids of vector (control plasmid), oe-PTEN, miR-152-3p NC, miR-152-3p mimic, miR-152-3p inhibitor NC, and miR-152-3p inhibitor were delivered into the cells following the instructions described in Lipofectamine 2000 (11668-019; Invitrogen, NY, CA, USA). All the above-mentioned plasmids were provided by Shanghai GenePharma (Shanghai, China).
24 h before transfection, the cells were plated at a density of 2 × 105 cells/well using antibiotic-free DMEM medium. When MSCs reached 70%–90% confluency, transfection was performed in accordance with instructions described in TransIT-2020 (MIR5404; Mirus, Madison, WI, USA). After 6 h, the medium was renewed with fresh culture medium, and 48 h after transfection, the cells were harvested and subjected to further analysis.
Coculture of MSCs or Exosomes and Fibroblasts
MSCs were treated with an exosome inhibitor (GW4869) to inhibit the exosomal secretion of MSCs. MSCs were plated in a 6-well plate at a density of 1 × 106 cells/well. When the cells reached 80%–90% confluence, 10% GW4869 (D1692-5MG; Sigma-Aldrich, St. Louis, MO, USA) was introduced to treat MSCs. DMSO was also introduced to treat cells for the control group. The cells and supernatant were harvested after 24 h for further use.
MSCs were plated on the 24-well basolateral chambers of a Transwell chamber at a density of 1 × 104 cells/well, whereas apical chambers were added with fibroblasts. The insertion well size between apical and basolateral chambers was 0.4 um. After coculture for 24 h, the fibroblasts were isolated, and the expression of miR-152-3p and PTEN in each group was determined. Fibroblasts were cocultured with MSCs, MSCs + GW4869, MSCs + oe-H19, and MSCs + oe-NC, respectively. The exosomal secretion from GW4869-treated MSCs was observed under a TEM.
The exosomes of the extracted MSCs were labeled with PKH67 (green) (MINI67-1KT; Sigma-Aldrich, St. Louis, MO, USA) following the suggested instructions. The fluorescence-labeled exosomes were cocultured with the supernatant of fibroblasts with 50%–60% confluence in the 24-well plate for 48 h. Plasmids of oe-NC and oe-H19 were delivered into fibroblasts and MSC-exo, respectively. Fibroblasts were cocultured with MSC-exo-vector and MSC-exo oe-H19, respectively. The inverted fluorescence microscope was used for observation. The expression of lncRNA H19 and miR-152-3p was determined by qRT-PCR, whereas protein expression of PTEN and AKT, as well as the extent of AKT phosphorylation, was determined by western blot analysis.
EdU Cell Proliferation Assay
Fibroblasts were isolated from the coculture system and seeded in a 96-well plate at a density of 5 × 103 cells/well. After 6 h, the cells in each well were incubated with 100 μL EdU medium for 2 h, then fixed with 100 μL 4% paraformaldehyde at room temperature for 30 min, and incubated with 2 mg/mL glycine for 5 min. Following PBS wash, the cells in each well were permeabilized with 100 μL PBS containing 0.5% Triton X-100 for 10 min, stained with 100 μL, one times Apollo staining solution at room temperature for 30 min in the dark, and incubated with 100 μL, one times Hoechst 33342 at room temperature for 30 min in the dark. After addition of 100 μL anti-fluorescence quenching, the cells in each well were observed under a fluorescence microscope. The number of EdU-labeled cells was counted and recorded, where cells with nucleus stained in red were regarded as positive cells. Three fields of view were selected on a random basis. The EdU labeling rate (%) = the number of positive cells/(the number of positive cells + the number of negative cells) × 100%.
Transwell Assay
The migration ability of transfected cells was performed using a Transwell chamber inserted with 8 μm pore-size polycarbonate membranes (Corning, Corning, NY, USA). The apical chambers were added with 600 mL DMEM containing 20% FBS and allowed to stand at 37°C for 1 h. After 48 h of transfection, the fibroblasts were resuspended in FBS-free DMEM medium. The cell suspension at a density of 1 × 106 cells/mL was seeded into apical chambers. After 24 h of cell migration at 37°C with 5% CO2, the cells that had migrated to the lower surface of the membrane were fixed with 5% glutaraldehyde and stained with 0.1% crystal violet for 5 min. The surface cells were then wiped off with a cotton swab and observed under an inverted fluorescence microscope (TE2000; Nikon Instruments, Melville, NY, USA). Five fields of view were randomly selected and photographed. The mean value was regarded as the number of cells that had migrated through chambers.
TUNEL Assay for Cell Apoptosis
The cells in the 24-well plate were fixed with 4% paraformaldehyde and fixed with 0.3% H2O2-formalin prepared at a ratio of 1:99 for 30 min. The 24-well plate was ice bathed with 0.3% Triton X-100 for 2 min. Cell apoptosis was assessed by a TUNEL Apoptosis Detection kit (green fluorescence, C1088; Beyotime Biotechnology, Shanghai, China). After anti-fluorescence quenching, the cells were observed under a fluorescence microscope at the excitation wavelength of 450 nm and at an emission wavelength of 550 nm (green fluorescence).
RNA-FISH
The subcellular localization of lncRNA H19 in fibroblasts was identified using FISH techniques, following the instructions described in a Ribo lncRNA FISH Probe Mix (red) kit (lnc10000; Ribobio, Guangzhou, Guangdong, China). The coverslip was placed onto the wells of the 24-well plate, and the cells were seeded at a density of 6 × 104 cells per well. When the cell reached about 80% confluence, the cells were fixed with 1 mL 4% paraformaldehyde and treated with 2 μg/mL proteinase K, glycine, and acetylation reagent. Then, cells were prehybridized with a 250-μL prehybridization solution at 42°C for 1 h and then hybridized with a 250-μL hybridization solution, which contained 300 ng/mL probe lncRNA H19 at 42°C overnight. Afterward, the nucleus was stained with DAPI, diluted with PBS with Tween 20 at a ratio of 1:800 for 5 min. The cells were lastly mounted with an anti-fluorescence-quenching agent, observed, and photographed under a fluorescence microscope (Olympus, Tokyo, Japan) with five randomly selected visual fields.
RNA Pull-Down Assay
Fibroblasts were lysed in specific lysis buffer (Ambion, Austin, TX, USA). The lysates were incubated with 3 μg biotinylated miR-152-3p probe at room temperature for 2 h, followed by an incubation with precoated M-280 streptavidin beads (S3762; Sigma-Aldrich, St. Louis, MO, USA) at 4°C for 4 h. The samples were washed two times with precooled lysis buffer, then three times with low-salt buffer solution, and one time with high-salt buffer solution. The combined RNA was purified by Trizol, and the enrichment of lncRNA H19 was detected by qRT-PCR.
RIP
The binding between lncRNA H19 and Ago2 was detected using a RIP kit (17-700; Millipore, Temecula, CA, USA). The cells were lysed with an equal volume of radio immunoprecipitation assay lysis buffer (P0013B; Beyotime Biotechnology, Shanghai, China) in an ice bath for 5 min. The cells were centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was then collected. A part of the cell extract was used as an input, whereas the rest was probed with Ago2 antibody (ab32381, 1:50; Abcam, Cambridge, UK) for a coprecipitation reaction. IgG antibody (ab109489, 1:100; Abcam, Cambridge, UK) served as NC. RNA was extracted from the sample and input after protease K detachment, followed by qRT-PCR.
Statistical Analysis
SPSS 21.0 software (IBM, Armonk, NY, USA) was applied for data analysis. The measurement data were expressed as mean ± SD. Comparison between two groups was conducted by independent sample t test. One-way ANOVA was used for data comparison among multiple groups, followed by Tukey’s post hoc test. The correlation between PTEN and miR-152-3p was analyzed using Pearson correlation coefficient. Repeated-measures ANOVA was applied for data comparison among multiple groups at different time points, followed by Tukey’s post hoc test. Values of p < 0.01 were considered statistically significant.
Author Contributions
B.L., S.L., and J.C. designed the study. Y.Z., T.W., and Z.L. collated the data, designed and developed the database, carried out data analyses, and produced the initial draft of the manuscript. B.L., Y.F., C.B., and A.Z. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors thank their colleagues for their helpful comments and technical assistance. This study was supported by the National Natural Science Foundation of China (81501737, 81871562, and 81800739).
Contributor Information
Aixia Zhai, Email: aixiazhai@126.com.
Changlong Bi, Email: bichanglongdr@163.com.
References
- 1.Yazdanpanah L., Nasiri M., Adarvishi S. Literature review on the management of diabetic foot ulcer. World J. Diabetes. 2015;6:37–53. doi: 10.4239/wjd.v6.i1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong D.G., Boulton A.J.M., Bus S.A. Diabetic Foot Ulcers and Their Recurrence. N. Engl. J. Med. 2017;376:2367–2375. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]
- 3.Amin N., Doupis J. Diabetic foot disease: From the evaluation of the “foot at risk” to the novel diabetic ulcer treatment modalities. World J. Diabetes. 2016;7:153–164. doi: 10.4239/wjd.v7.i7.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Squillaro T., Peluso G., Galderisi U. Clinical Trials With Mesenchymal Stem Cells: An Update. Cell Transplant. 2016;25:829–848. doi: 10.3727/096368915X689622. [DOI] [PubMed] [Google Scholar]
- 5.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop Dj., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 6.Frykberg R.G., Banks J. Challenges in the Treatment of Chronic Wounds. Adv. Wound Care (New Rochelle) 2015;4:560–582. doi: 10.1089/wound.2015.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ti D., Hao H., Fu X., Han W. Mesenchymal stem cells-derived exosomal microRNAs contribute to wound inflammation. Sci. China Life Sci. 2016;59:1305–1312. doi: 10.1007/s11427-016-0240-4. [DOI] [PubMed] [Google Scholar]
- 8.Lässer C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin. Biol. Ther. 2012;12(Suppl 1):S189–S197. doi: 10.1517/14712598.2012.680018. [DOI] [PubMed] [Google Scholar]
- 9.Gezer U., Özgür E., Cetinkaya M., Isin M., Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014;38:1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- 10.Gong Y.Y., Peng M.Y., Yin D.Q., Yang Y.F. Long non-coding RNA H19 promotes the osteogenic differentiation of rat ectomesenchymal stem cells via Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:8805–8813. doi: 10.26355/eurrev_201812_16648. [DOI] [PubMed] [Google Scholar]
- 11.Zhang N., Geng T., Wang Z., Zhang R., Cao T., Camporez J.P., Cai S.Y., Liu Y., Dandolo L., Shulman G.I. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight. 2018;3:e120304. doi: 10.1172/jci.insight.120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J., Zhao J., Sun L., Pan Y., Wang H., Zhang W.B. Long non-coding RNA H19 mediates mechanical tension-induced osteogenesis of bone marrow mesenchymal stem cells via FAK by sponging miR-138. Bone. 2018;108:62–70. doi: 10.1016/j.bone.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Luan W., Li R., Liu L., Ni X., Shi Y., Xia Y., Wang J., Lu F., Xu B. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote malignant melanoma progression by sponging miR-152-3p. Oncotarget. 2017;8:85401–85414. doi: 10.18632/oncotarget.19910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., Li Y., Sheng C., Yang C., Chen L., Sun J. Tanshinone IIA inhibits apoptosis in the myocardium by inducing microRNA-152-3p expression and thereby downregulating PTEN. Am. J. Transl. Res. 2016;8:3124–3132. [PMC free article] [PubMed] [Google Scholar]
- 15.Hou S.Q., Ouyang M., Brandmaier A., Hao H., Shen W.H. PTEN in the maintenance of genome integrity: From DNA replication to chromosome segregation. BioEssays. 2017;39:1700082. doi: 10.1002/bies.201700082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X.M., Yao M., Liu S.X., Hao J., Liu Q.J., Gao F. Interplay between the Notch and PI3K/Akt pathways in high glucose-induced podocyte apoptosis. Am. J. Physiol. Renal Physiol. 2014;306:F205–F213. doi: 10.1152/ajprenal.90005.2013. [DOI] [PubMed] [Google Scholar]
- 17.Liang L., Stone R.C., Stojadinovic O., Ramirez H., Pastar I., Maione A.G., Smith A., Yanez V., Veves A., Kirsner R.S. Integrative analysis of miRNA and mRNA paired expression profiling of primary fibroblast derived from diabetic foot ulcers reveals multiple impaired cellular functions. Wound Repair Regen. 2016;24:943–953. doi: 10.1111/wrr.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S., Wang L., Dou L., Guo J., Fang W., Li M., Meng X., Man Y., Shen T., Huang X., Li J. MicroRNA 152 regulates hepatic glycogenesis by targeting PTEN. FEBS J. 2016;283:1935–1946. doi: 10.1111/febs.13713. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J., Li X. Association of PTEN expression with liver function and inflammatory changes in patients with liver cancer after chemotherapy. Oncol. Lett. 2018;16:6633–6637. doi: 10.3892/ol.2018.9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L., Wang Y., He J., Zhang C., Chen J., Shi D. Long non-coding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol Res. 2018;26:1419–1428. doi: 10.3727/096504018X15178768577951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv Y., Ge L., Zhao Y. Effect and mechanism of SHED on ulcer wound healing in Sprague-Dawley rat models with diabetic ulcer. Am. J. Transl. Res. 2017;9:489–498. [PMC free article] [PubMed] [Google Scholar]
- 23.Grennan D. Diabetic Foot Ulcers. JAMA. 2019;321:114. doi: 10.1001/jama.2018.18323. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang X., Jiang X., Gu D., Zhang Y., Kong S.K., Jiang C., Xie W. Dysregulated Serum MiRNA Profile and Promising Biomarkers in Dengue-infected Patients. Int. J. Med. Sci. 2016;13:195–205. doi: 10.7150/ijms.13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J., Shi Y., Peng H., Shen X., Thomas S., Wang Y., Truong L.D., Dryer S.E., Hu Z., Xu J. Loss of PTEN promotes podocyte cytoskeletal rearrangement, aggravating diabetic nephropathy. J. Pathol. 2015;236:30–40. doi: 10.1002/path.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Z., Zhi X., Zhang Q., Yang L., Xu H., Xu Z. LncRNA H19 functions as a competing endogenous RNA to regulate AQP3 expression by sponging miR-874 in the intestinal barrier. FEBS Lett. 2016;590:1354–1364. doi: 10.1002/1873-3468.12171. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z.W., Tian L.H., Yang B., Guo R.M. Long Noncoding RNA H19 Acts as a Competing Endogenous RNA to Mediate CTGF Expression by Sponging miR-455 in Cardiac Fibrosis. DNA Cell Biol. 2017;36:759–766. doi: 10.1089/dna.2017.3799. [DOI] [PubMed] [Google Scholar]
- 28.Cao Y., Gang X., Sun C., Wang G. Mesenchymal Stem Cells Improve Healing of Diabetic Foot Ulcer. J. Diabetes Res. 2017;2017:9328347. doi: 10.1155/2017/9328347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi R., Jin Y., Cao C., Han S., Shao X., Meng L., Cheng J., Zhang M., Zheng J., Xu J., Li M. Localization of human adipose-derived stem cells and their effect in repair of diabetic foot ulcers in rats. Stem Cell Res. Ther. 2016;7:155. doi: 10.1186/s13287-016-0412-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng X., Tang Y., Hu K., Jiao W., Ying L., Zhu L., Liu J., Xu J. Three-week topical treatment with placenta-derived mesenchymal stem cells hydrogel in a patient with diabetic foot ulcer: A case report. Medicine (Baltimore) 2017;96:e9212. doi: 10.1097/MD.0000000000009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao S.C., Guo S.C., Li M., Ke Q.F., Guo Y.P., Zhang C.Q. Chitosan Wound Dressings Incorporating Exosomes Derived from MicroRNA-126-Overexpressing Synovium Mesenchymal Stem Cells Provide Sustained Release of Exosomes and Heal Full-Thickness Skin Defects in a Diabetic Rat Model. Stem Cells Transl. Med. 2017;6:736–747. doi: 10.5966/sctm.2016-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S.P., Zhou Y.Q., Liu D.Q., Zhang W., Manyande A., Guan X.H., Tian Y.K., Ye D.W., Omar D.M. PI3K/Akt Pathway: A Potential Therapeutic Target for Chronic Pain. Curr. Pharm. Des. 2017;23:1860–1868. doi: 10.2174/1381612823666170210150147. [DOI] [PubMed] [Google Scholar]
- 33.Wagner F.W., Jr. The dysvascular foot: a system for diagnosis and treatment. Foot Ankle. 1981;2:64–122. doi: 10.1177/107110078100200202. [DOI] [PubMed] [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Zhang G., Zou X., Miao S., Chen J., Du T., Zhong L., Ju G., Liu G., Zhu Y. The anti-oxidative role of micro-vesicles derived from human Wharton-Jelly mesenchymal stromal cells through NOX2/gp91(phox) suppression in alleviating renal ischemia-reperfusion injury in rats. PLoS ONE. 2014;9:e92129. doi: 10.1371/journal.pone.0092129. [DOI] [PMC free article] [PubMed] [Google Scholar]