Abstract
The noticeable increase in the occurrence of multidrug-resistant Klebsiella pneumoniae strains separated from different hospitals in Taif city, (Saudi Arabia) demonstrates the limitation of antibiotics used for bacterial eradication. The aim of the present study is to detect the virulence genes in some K. pneumoniae isolates that collected from different hospitals in Taif governorate in Saudi Arabia. A total of 134 clinical samples were used to isolate about twenty three K. pneumoniae strains from various clinical specimens throw six months. They were identified by microbiological method as K. pneumoniae and confirmed with 16S rRNA sequencing analysis. The antimicrobial susceptibility of K. pneumoniae isolates was determined. The existence of virulence genes (AcrAB, tolC, arb, OmpK35, RmpA, fimH-1, entB, K2, irP-1 and Mdtk) were performed by PCR. The multidrug-resistant strains were detected in 16 (69.5%), that showed the presence of the most virulence genes. The multidrug-resistant isolates showed resistance against Ampicillin (96%), Amox-Clav (90%), Cephalothin (90%), Cefuroxime (90%), Ceftriaxone (85%), Aztreonam (87%), Cefepime (80%), Ceftazidime (80%), and Trim-Sulf (82%). Molecular diversity between K. pneumoniae isolates was determined using Rep-PCR markers technique. Thirty eight bands were resulted from the rep-PCR primers. Out of them, 31 bands were polymorphic with a polymorphism average of 81.6%. Total loci detected for each primer varied from 11 to 15 loci, and the loci size ranging from 200 to 2000 bp. These data may present novel epidemiological information regarding the clonal nature of K. pneumoniae separated from Taif governorate hospitals, Saudi Arabia.
Keywords: Klebsiella pneumoniae, Multidrug-resistance stains, Rep-PCR, Taif, KSA
1. Introduction
Gram-negative bacteria are potential causes of both infections acquired in hospital and community. Multiple resistance to broad spectrum antibiotics are one of the most important features. Gram-negative pathogens were successfully defeated using carbapenems, cephalosporins and fluoroquinolones in the 1980s [1]. Globally, Klebsiella pneumoniae is consider one of the most common nosocomial pathogens that lead to pneumoniae, urinary tract infections and bacteremia [2]. The increase incidence of multidrug-resistance (MDR) bacteria, such as K. pneumoniae, in the past few decades was due to the uncontrolled use of antimicrobial drugs as a treatment [2,3]. Gram-negative bacteria, in particular, have developed a number of mechanisms to become more resistant against antibiotics. One of these mechanisms is the horizontal gene transfer for transferring MDR genes between different types of bacteria [4]. For example, the increase incidence of K. oxytoca, K. pneumoniae, Escherichia coli, Salmonella sp and Enterobacter sp were due to the horizontal transfer of the common shared plasmids [4,5]. Another important mechanism and may be the important one for increasing the incidence of MDR bacteria is the efflux pump systems [6]. Such system in K. pneumoniae includes the functions of mdtK and AcrAB systems that belong to the multi antimicrobial extrusion family and resistance nodulation division of efflux pumps, respectively [6]. Also, AcrAB-TolC is another system for multidrug efflux pump which consisted of an outer-membrane channel (TolC), a secondary transporter situated in the periplasmic component (AcrA) and finally inner membrane (AcrB) [7]. This pump is important for the resistance against chloramphenicol, quinolones and tetracyclines in different MDR strains. Additionally, porins components, such as OmpK36 and OmpK35, are important for the diffusion of the antibiotics into the cells and for the sensitivity against carbapenems and cephalosporins [8].
Molecular diversity and detection of virulence genes of clinical isolates are useful assets that can reveal insight MDR K. pneumonia infections. Several studies have repeatedly used a PCR vertical component like repetitive element palindromic PCR (rep-PCR), which targets REP palindromic sequences, to compare diversity of bacterial genome [5]. In rep-PCR DNA fingerprinting, PCR amplification is achieved among heterogeneous contiguous elements to obtain DNA fingerprints that can be analyzed easily using a software program to identify patterns. Previously studies chose the rep-PCR technique because of their simplify, differentiate and cheap technique [9,10]. The rep-PCR has been used successfully to characterize K. pneumoniae and E. coli isolates [11]. Despite the fact that K. pneumoniae strains convey virulence related genes that may encode capsules, adhesions, lipopolysaccharides, iron acquisition systems and other harmfulness factors [12], it is not clear how these genes are linked with infection forms or antibiotic resistance.
The main objective of the present research is screening for different virulence genes like AcrAB, tolC, arb, OmpK36, RmpA, fimH, entB, K2, irP-1 and Mdtk in twenty three K. pneumoniae isolates that collected from various clinical specimens throw six months from different hospitals in Taif governorate in Saudi Arabia. Additionally, the molecular diversity between these isolates were accomplished by using Rep-PCR technique.
2. Materials and methods
2.1. Bacterial strains
Out of 134 bacterial isolates, twenty three K. pneumoniae isolates were obtained from inpatients of different hospital in Taif governorate, Saudi Arabia. The bacterial species were identified using the fully automated VITEK-2 COMPACT microbiology system (BioMérieux, Inc., Durham, NC, USA). The K. pneumoniae strain number ATCC 700603 (from ATCC, USA) was used as a positive control for the antimicrobial susceptibility test. The susceptibility test of K. pneumoniae against 20 types of antibiotics including amikacin, amox-Clav, ampicillin, aztreonam, cefepime, cefoxitin, ceftazidime, ceftriaxone, cefuroxime, cephalothin, ciprofloxacin, ertapenem, gentamicin, imipenem, levofloxacin, meropenem, nitrofurantoin, pipe-tazo, tigecycline, trim-sulf was determined using the recommended clinical standard (CLSI) as previously described [4].
2.2. The 16S rRNA gene sequencing
The genomic DNA was isolated by DNA extraction kit (Gena Bioscience, Germany) from all K. pneumoniae isolates according to the manufacturer’s instructions. For each isolate, one fragment of the specific gene (about 1465 bp) was amplified using primers as documented previously [4]. The specific band was punctuated by QIA quick PCR purification kit (QIAGEN, Valencia, CA, USA) and sequenced in DNA Analyzer 3146 (Applied Biosystems, USA). The sequencing results were edited and compiled using DNASTAR software (Laser gene, Madison, WI, USA). BLAST searches were performed using NCBI server (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
2.3. Data analysis
The raw sequencing data of 16S rRNA was gathered using ABI software data collection version 3.1 (ABI, Applied Biosystems). To avoid sequence mismatching, 100 bp from both ends of the 16S rRNA gene was omitted. ClustalW aligned a uniform length of about 1365 bp in MEGA 7.0 program [13]. Genetic diversity and phylogenetic tree analysis was carried out using Maximum Likelihood method [14] applied in MEGA 7.0 program package [13].
2.4. Isolation of virulence genes
Ten virulence genes (AcrAB, tolC, arb, OmpK36, RmpA, fim-H-1, entB, K2, irP-1 and Mdtk) were checked in all strains of K. pneumoniae. Primers sequences (Table 1) and PCR conditions were achieved as previously reported [15]. Expected sizes of the amplicons were verified by electrophoresis in 1.5% agarose gel using 100-bp DNA ladder (MBI, Fermentas, Lithuania, USA).
Table 1.
Primer sequences and amplicon sizes of virulence genes in K. pneumoniae.
| Primer Name | Primer Sequence (5′→3′) | Product Size (bp) |
|---|---|---|
| AcrAB-F | ATCAGCGGCCGGATTGGTAAA | 312 |
| AcrAB-R | CGGGTTCGGGAAAATAGCGCG | |
| tolC-F | ATCAGCAACCCCGATCTGCGT | 527 |
| tolC-R | CCGGTGACTTGACGCAGTCCT | |
| arb-F | TGGGGCAAAGAGGCGCTG GAG | 636 |
| arb-R | CAGCCAGCGACACGGATTCTC | |
| OmpK35-F | CTCCAGCTCTAACCGTAGCG | 241 |
| OmpK35-R | GGTCTGTACGTAGCCGATGG | |
| RmpA-F | ACTGGGCTACCTCTGCTTCA | 535 |
| RmpA-R | CTTGCATGAGCCATCTTTCA | |
| fimH-1F | GCCAACGTCTACGTTAACCTG | 180 |
| fimH-1R | ATATTTCACGGTGCCTGAAAA | |
| entB-F | CTGCTGGGAAAAGCGATTGTC | 385 |
| entB-R | AAGGCGACTCAGGAGTGGCTT | |
| K2-F | GGATTATGACAGCCTCTCCT | 908 |
| K2-R | CGACTTGGTCCCAACAGTTT | |
| irP-1-F | TGAATCGCGGGTGTCTTATGC | 238 |
| irP-1-R | TCCCTCAATAAAGCCCACGCT | |
| Mdtk-F | GCGCTTAACTTCAGCTCA | 543 |
| Mdtk-R | GATGATAAATCCACACCAGAA |
2.5. Rep-PCR analysis
Three repetitive sequence primers (REP, ISSR12 and ISSR29) were used to perform the molecular diversity of K. pneumoniae isolates according to Wasfi et al. [15]. Rep-PCR results were obtained with the used of the following primers: REP 1R (5′-IIIICGICGICATCI GGC-3′) and 2I (5′-ICGICTTATCIGGCCTAC-3′); ISSR12 (AG)9T; and ISSR29 (GA)8A.
2.6. Statistical analysis
Similarity matrix and cluster analysis was achieved using an unweighted pair group method for arithmetic mean and the dendrogram was generated by NTSYS-PC program version 2.01 [16].
3. Results and discussion
3.1. Antimicrobial susceptibility
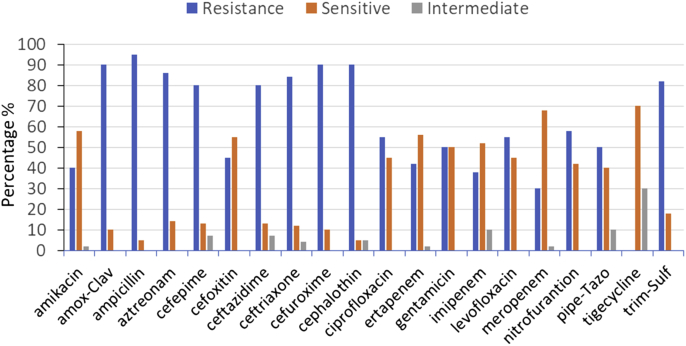
Twenty three K. pneumoniae isolates were obtained from patients of different hospitals in Taif governorate in Saudi Arabia and tested for its susceptibility against 20 types of antibiotics. The overall susceptibility, intermediate and resistance were determined and the results are showing in Fig. 1. Most K. pneumoniae strains showed a high percentage of resistance against Ampicillin (96%), Amox-Clav (90%), Cephalothin (90%), Cefuroxime (90%), Ceftriaxone (85%), Aztreonam (87%), Cefepime (80%), Ceftazidime (80%), and Trim-Sulf (82%). A moderate susceptible were against Tigecycline (70%), Meropenem (68%), Amikacin (57%), Imipenem (52%), Gentamicin (50%), Ertapenem (58%), and Cefoxitin (55%). On other hand, intermediate resistances were identified against Tigecycline (30%). It was reported that intermediate resistance is a compromise between resistance and sensitivity [17,18]. In this case, patients can take high doses of antibiotics. Antibiotics also play an important role in reducing the disease or death in human caused by bacterial infections. However, the uncontrolled use of antibiotics was the main cause of the emergence and spread of MDR bacterial strains [1,19].
Fig. 1.
Antimicrobial resistance profiles of twenty three K. pneumoniae isolates against twenty antibiotics. R = resistance, S = sensitive and I = intermediate.
3.2. Detection of virulence genes in MDR K. pneumoniae
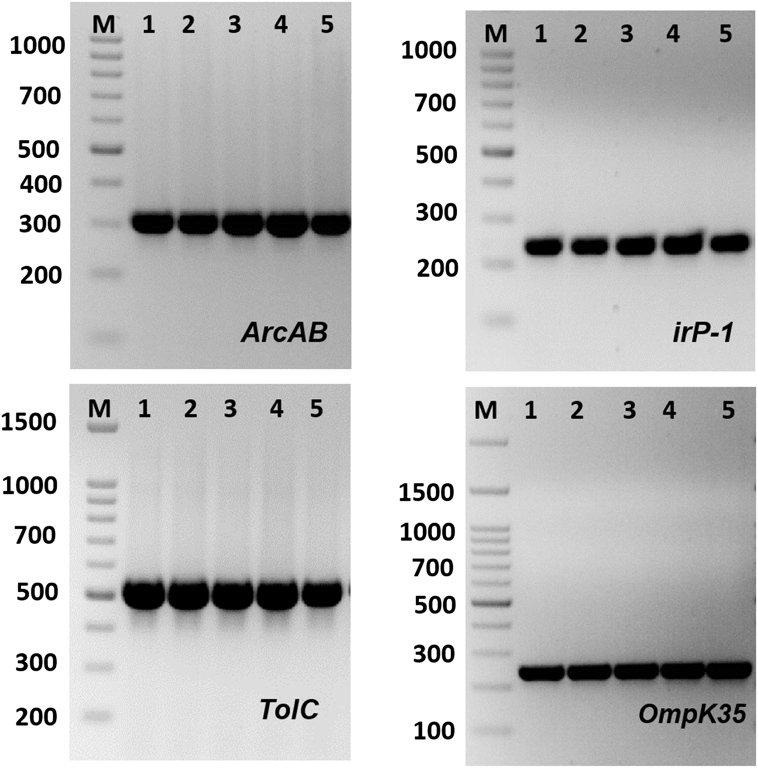
The existence of MDR genes are shown in Fig. 2 and Table 2. The enterobactin biosynthesis (entB), Yersinibactin biosynthesis (irp-1), tolC, AcrAB,fumH-1,Mdtk and Ompk35 genes were exist in all of MDR K. pneumoniae isolates (Table 2). K2 gene which responsible for the formation of capsule K genotypes was found in four isolates only (17.4%). Interestingly, Klebsiella-22 and Klebsiella-23 isolates found to have all tested virulence genes. Both OmpK35 and OmpK36 play a role in K. pneumoniae infection and virulence [8]. Deletion of OmpK35/OmpK36 can result in the decrease in virulence of greatly virulent strains and can rise their susceptibility to neutrophil phagocytosis [20,21]. In the present study, Ompk 35 porin-coding gene was identified in all the isolates of K. pneumoniae. A straight relationship between virulence of infectious bacteria and efflux pumps was previously described by Padilla et al. [22]. The intracellular attack of pathogenic bacteria were decreased in mutant bacterial strains that was lacking in acrAB-tolC efflux pumps [23]. Most strains of Enterobacteriaceae family, especially the genus of Klebsiella, have genes encoding iron uptake systems, including aerobactin or enterochelin systems [24]. These siderophores have dual functions as they can enhance iron uptake and inhibit T cell proliferation. The yersinibactin biosynthesis gene (irp-1) and enterobactin biosynthesis gene (entB) were identified in all MDR K. pneumoniae isolates. Highly pathogenic Yersinia strains have high-pathogenicity island (HPI) that include the gene Irp-1. This HPI is also widespread in Klebsiella genus especially K. pneumonia, K. oxytoca and other enterbocateria family such as and E. coli, Enterobacter species and Citrobacter species [3,25].
Fig. 2.
Amplification of some specific genes producing in some K. pneumoniae isolates by single PCR. (A) ArcAB gene with size about of 312 bp. (B) irP gene with size about of 238 bp. (C) TolC gene with size about of 527 bp. (D) OmpK35 gene with size about of 241 bp. First lane on each panel is 100 bp molecular weight markers.
Table 2.
Virulence genes patterns among pathogenic K. pneumoniae isolates.
| Isolates | Present and absent of virulence genes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AcrAB | tolC | arb | OmpK36 | RmpA | fimH | entB | K2 | irP-1 | Mdtk | |
| Klebsiella-1 | + | + | ــ | + | + | + | + | + | + | + |
| Klebsiella-2 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-3 | + | + | ــ | + | + | + | + | ــ | + | + |
| Klebsiella-4 | + | + | + | + | + | + | + | ــ | + | + |
| Klebsiella-5 | + | + | + | + | ــ | + | + | ــ | + | + |
| Klebsiella-6 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-7 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-8 | + | + | ــ | + | + | + | + | ــ | + | + |
| Klebsiella-9 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-10 | + | + | ــ | + | + | + | + | ــ | + | + |
| Klebsiella-11 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-12 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-13 | + | + | ــ | + | + | + | + | + | + | ــ |
| Klebsiella-14 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-15 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-16 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-17 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-18 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-19 | + | + | ــ | + | ــ | + | + | ــ | + | + |
| Klebsiella-20 | + | + | + | + | ــ | + | + | ــ | + | + |
| Klebsiella-21 | + | + | + | + | ــ | + | + | ــ | + | + |
| Klebsiella-22 | + | + | + | + | + | + | + | + | + | + |
| Klebsiella-23 | + | + | + | + | + | + | + | + | + | + |
3.3. Molecular genotyping of K. pneumoniae isolates according to 16S-rRNA gene
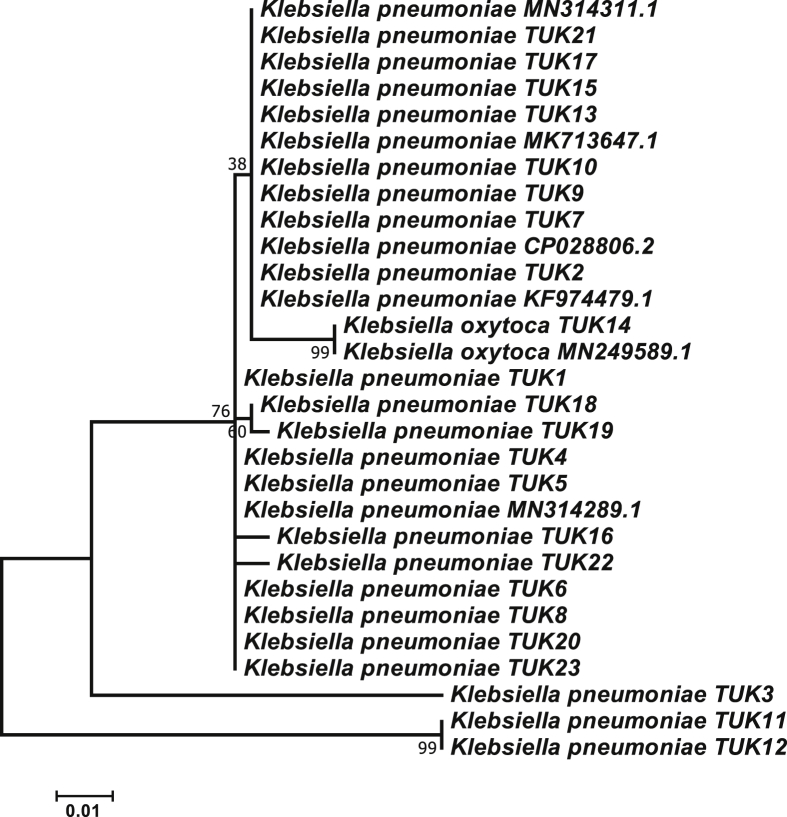
We amplified and sequenced the 16S-rRNA gene of all K. pneumoniae isolates and then the specific fragments were aligned and compare with the available 16S-rRNA sequences for other K. pneumoniae isolates at the NCBI database [26]. The BLAST results showed that the partial 16S rRNA sequences are more similar to other sequences of K. pneumoniae strains with about 98% of similarity matrix with K. pneumoniae strain MN-314311 and 97% similarity matrix with K. pneumoniae strain MK713647 (Fig. 3). The 16S rRNA sequence contains characteristics that make it appropriate methods as a global indicator of evolution. In addition, the 16S rRNA gene sequence is a useful method for bacterial identification, where nucleotide sequences are identified in this region and compared to the available sequences of databases to obtain homogeneous matches, allowing bacterial identification of the target samples [[27], [28], [29]]. Therefore, 16S rRNA has been comprehensively used to reconstruct phylogenetic evolution of microorganisms. The genetic proximity between K. pneumoniae strains was also observed by various researchers [4,30] using sequencing of approximately 1200 bp of the 16S rRNA.
Fig. 3.
Phylogenetic tree analysis of the twenty three isolates of K. pneumoniae with other K. pneumoniae strains that blasted from NCBI based on the 1365 bp of 16s rRNA gene sequences using the Maximum Likelihood method [14]. Numbers by nodes designate Maximum Likelihood bootstrap. The bootstrap consensus tree inferred from 1000 replicates.
3.4. Rep-PCR and genetic distances analysis
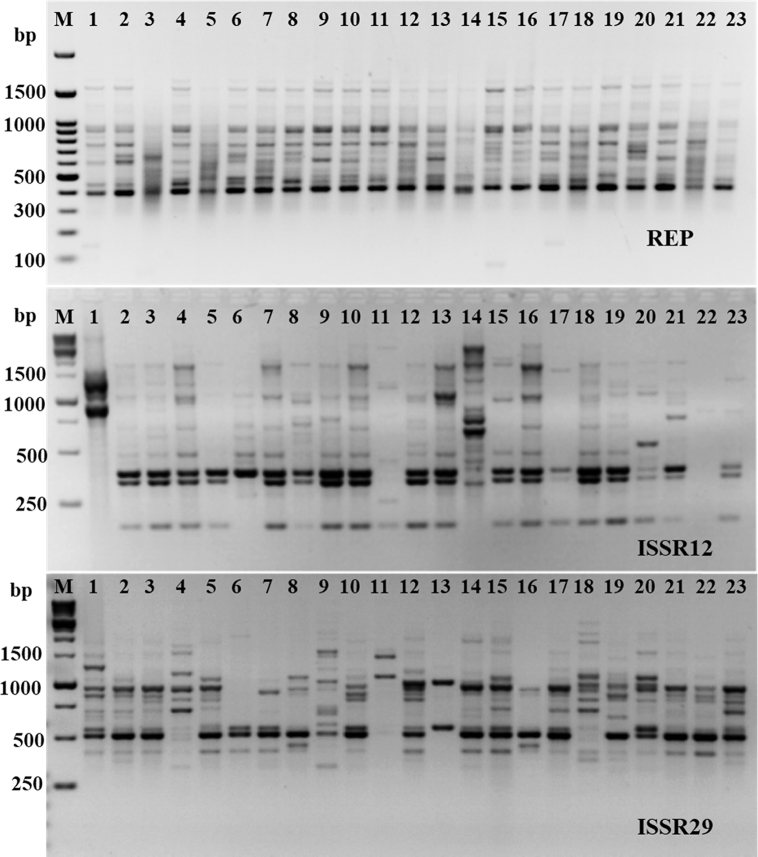
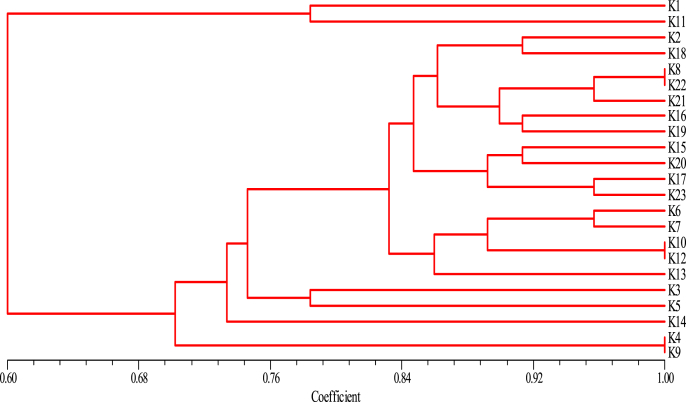
Rep-PCR markers are effective technique for molecular characterization and correlation estimation through DNA fingerprinting [25]. The rep-PCR results were summarized in Table 3 and shown in Fig. 4. The monomorphic and polymorphic loci were produced from the PCR amplification. About 38 bands were resulted from the rep-PCR primers. Out of them, 31 bands were polymorphic bands with a polymorphism average of 81.6%. The number of total bands varied from 11 for primer ISSR12 to 15 bands for Rep-10 (Table 3). The loci sizes ranging from 200 to 2000 bp (Fig. 4). The highest polymorphism among K. pneumoniae isolates was produced using ISSR12 primer (100%), followed by ISSR29 primer (86.7%). On other hand, the lowest polymorphism was produced from primer REP with 54.5% (Table 3). Overall, 38 fragments produced from rep-PCR analysis were sufficient for determination of genetic similarities and constructing the phylogenetic tree for all K. pneumoniae isolates using UPGMA method according to Jaccard’s similarity coefficients (Fig. 5). All K. pneumoniae isolates were assembled into two different groups with about 40% of genetic similarity. The first group contains isolates numbers 1 and 11 only. The second group divided into two clusters, first one includes K. pneumoniae isolates 4 and 9, while, the second cluster includes the remaining isolates of K. pneumoniae (Fig. 5). The highest genetic similarity was between K. pneumoniae isolates 10 and 12 (Fig. 5).
Table 3.
Polymorphic bands of each genetic primers and percentage of polymorphism in twenty three K. pneumoniae isolates based on the five rep-PCR primers.
| Primers | Total Bands | No. of Monomorphic Bands | No. Polymorphic Bands | % Monomorphic bands | % Polymorphic bands |
|---|---|---|---|---|---|
| REP | 11 | 5 | 6 | 45.5 | 54.5 |
| ISSR12 | 12 | 0 | 12 | 0.0 | 100 |
| ISSR29 | 15 | 2 | 13 | 13.3 | 86.7 |
| Total | 38 | 7 | 31 | 18.4 | 81.6 |
Fig. 4.
Rep-PCR profile of 23 K. pneumoniae isolates generated with three primers. First lane on each panel is DNA molecular weight markers.
Fig. 5.
Dendrogram analysis among twenty three K. pneumoniae isolates (K1 to K23) generated with three rep-PCR primers.
4. Conclusion
The screening of antimicrobial resistance of K. pneumoniae would give a better understanding for choosing the appropriate type of antimicrobial agents and avoid the existence of increasingly antimicrobial-resistant K. pneumoniae strains. The present data propose a temperately high predominance of antibiotic resistance in K. pneumoniae strains towards ampicillin, cephalothin, and cefuroxime. The present data will be very worthy to scope of antibiotics to which resistance has been gained after some time. Therefore, it better to incorporate new developing antibiotics that will be used for treatment of K. pneumoniae contamination.
Funding
None.
Declaration of competing interest
The author declare that there is no conflict of interest regarding the publication of this paper.
Acknowledgments
None.
References
- 1.Aarestrup F.M., Wegener H.C., Collignon P. Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti Infect. Ther. 2008;5:733–750. doi: 10.1586/14787210.6.5.733. [DOI] [PubMed] [Google Scholar]
- 2.Pitout J.D., Laupland K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 2008;8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 3.Alzahrani A.K., Farag M.M., Abbadi S.H., Hassan M.M., Gaber A., Abdel-Moneima A.S. Antibiotic resistance profile and random amplification typing of β-lactamase-producing Enterobacteriaceae from the local area of Al-Taif and nearby cities in Saudi Arabia. Asian Biomed. 2016;10:219–228. [Google Scholar]
- 4.Alsanie W.F., Felemban E.M., Farid M.A., Hassan M.M., Sabry A., Gaber A. Molecular identification and phylogenetic analysis of multidrug-resistant bacteria using 16S rDNA sequencing. J. Pure Appl. Microbiol. 2018;12(2):489–496. [Google Scholar]
- 5.Hassan M.M., Gaber A., El-Hallous E.I. Molecular and morphological characterization of Trichoderma harzianum from different Egyptian soils. Wulfenia J. 2014;21:80–96. [Google Scholar]
- 6.Meletis G., Exindari M., Vavatsi N., Sofianou D., Diza E. Mechanisms responsible for the emergence of carbapenem resistancein Pseudomonas aeruginosa. Hippokratia. 2012;16:303–307. [PMC free article] [PubMed] [Google Scholar]
- 7.Du D., Wang Z., James N.R. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi W., Li K., Ji Y. Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 beta-lactamase production. Braz. J. Microbiol. 2013;44:435–442. doi: 10.1590/S1517-83822013000200015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lessa S., Paes R., Santoro P., Mauro R., Vieira-daMotta O. Identification and antimicrobial resistance of microflora colonizing feral pig of Brazilian Pantanal. Braz. J. Microbiol. 2012;42(2):740–749. doi: 10.1590/S1517-838220110002000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alharthi A., Gaber A., Hassan M.M. Molecular characterization of mecA and SCCmec genes in pathogenic Staphylococcus spp. collected from hospitals in Taif region, KSA. Biotechnology. 2016;15:26–34. [Google Scholar]
- 11.Priscilla E.D., Ann L., Johnson K., Zimmerley S.T., Sadowsky M.J. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 2011;66:2572–2577. doi: 10.1128/aem.66.6.2572-2577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candan E.D. N Aksöz, Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 2015;62(4):867–874. doi: 10.18388/abp.2015_1148. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 15.Wasfi R., Elkhatib W.F., Ashour H.M. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci. Rep. 2016;6:38929. doi: 10.1038/srep38929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohlf F.J. Exeter Software; Setauket, New York (NY): 2000. NTSYS-PC Numerical Taxonomy and Multivariate Analysis System, Version 2.1. [Google Scholar]
- 17.Bazzaz B.S., Naderinasab M., Mohamad A.H., Farshadzadeh Z., Ahmadi S., Yousefi F. Escherichia coli and Klebsiella pneumoniae among clinical isolates from a general hospital in Iran. Acta Microbiol. Immunol. Hung. 2009;56(1):89–99. doi: 10.1556/AMicr.56.2009.1.7. [DOI] [PubMed] [Google Scholar]
- 18.Adzitey F. Antibiotic resistance of Escherichia coli isolated from beef and its related samples in Techiman Municipality of Ghana. Asian J. Anim. Sci. 2015;9:233–240. [Google Scholar]
- 19.Krishnamurthy V., Vijaykumar G.S., Kumar M.S., Prashanth H.V., Prakash R., Nagaraj E.R. Phenotypic and genotypic methods for detection of extended spectrum β lactamase producing Escherichia coli and Klebsiella pneumoniae isolated from ventilator associated pneumonia. J. Clin. Diagn. Res. 2013;7(9):1975–1978. doi: 10.7860/JCDR/2013/6544.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J.H., Siu L.K., Fung C.P. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2010;65:986–990. doi: 10.1093/jac/dkq056. [DOI] [PubMed] [Google Scholar]
- 21.Tsai Y.K., Fung C.P., Lin J.C. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2011;55(4):1485–1493. doi: 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padilla E., Llobet E., Doménech-Sánchez A., Martínez-Martínez L., Bengoechea J.A., Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2010;54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webber M.A., Randall L.P., Cooles S., Woodward M.J., Piddock L.J.V. Triclosan resistance in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2008;62:83–91. doi: 10.1093/jac/dkn137. [DOI] [PubMed] [Google Scholar]
- 24.Carbonetti N.H., Williams P.H. A cluster of five genes specifying the aerobactin iron uptake system of plasmid ColV-K30. Infect. Immun. 1984;46:7–12. doi: 10.1128/iai.46.1.7-12.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan M.M., Belal E.B. Antibiotic resistance and virulence genes in Enterococcus strains isolated from different hospitals in Saudi Arabia. Biotechnol. Biotechnol. Equip. 2016;30:726–732. [Google Scholar]
- 26.Haiwen L., Freder M., Vinson S.B., Coates J.C. Isolation, characterization and molecular identification of bacteria from the red imported fire ant (Solenopsisinvicta) midgut. J. Invertebr. Pathol. 2005;89:203–209. doi: 10.1016/j.jip.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Salman V., Amann R., Shub D.A., Schulz-Vogt H.N. Multiple self-splicing introns in the 16S rRNA genes of giant sulfur bacteria. Proc. Natl. Acad. Sci. 2012;109:4203–4208. doi: 10.1073/pnas.1120192109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabir J.S.M., Abo-Aba E.M., Sabry A., Hussein R.H., Bahieldin A., Baeshen N.A. Isolation, identification and comparative analysis of 16S rRNA of Bacillus subtilis grown around Rhazya stricta roots. Life Sci. J. 2013;10:980–986. [Google Scholar]
- 29.Chandran A., Mazumder A. Pathogenic potential, genetic diversity and population structure of Escherichia coli strains isolated from a forest-dominated watershed (comox lake) in British Columbia, Canada. Appl. Environ. Microbiol. 2015;81(5):1788–1798. doi: 10.1128/AEM.03738-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lade H., Paul D., Kweon J.H. Isolation and molecular characterization of biofouling bacteria and profiling of quorum sensing signal molecules from membrane bioreactor activated sludge. Int. J. Mol. Sci. 2014;15:2255–2273. doi: 10.3390/ijms15022255. [DOI] [PMC free article] [PubMed] [Google Scholar]