Abstract
Functional elimination of p53 is a common feature of a large percentage of human malignancies. Here, we report the development of a pharmacological strategy aimed at restoring p53 function and its use for targeted therapy in p53-deficient mice. Specific inhibition of deubiquitinases ubiquitin-specific peptidase 14 (USP14) resulted in durable tumor regressions of autochthonous lymphomas and sarcomas in p53-deficient mice without affecting normal tissues, and therapeutic response was correlated with an increase in the ubiquitination of constitutive photomorphogenesis 9 (COP9) signalosome subunit 5 (COPS5), a key negative regulatory effector for p53. Inhibition of USP14 resulted in durable tumor regression through COPS5 deubiquitilation and a p53-dependent and -independent regulation mechanism by USP14. This series highlights the utility of proteasome deubiquitinating activity inhibition as a novel treatment paradigm for p53-deficient cancers. In addition, it provides preliminary evidence that inhibition of USP14 resulted in durable tumor regression through COPS5 deubiquitilation and p53-dependent and -independent regulation mechanism by USP14. These findings suggest that the deubiquitinating activity of the 19S regulatory particle is a new anticancer drug target for patients with p53 deficiency.
Keywords: p53, COPS5, USP14, DUB, ubiquitination
Graphical Abstract
Introduction
Wild-type p53 protein is critical for cell function and maintaining integrity of the genome, which suppresses cancer in humans and rodents.1 This reduction in p53 protein, a common feature of a large percentage of human malignancies, results in deficiencies in cell-cycle check-point control and induction of apoptosis.2 Mutational inactivation of the p53 gene is also detected in more than 50% of human cancers, in which cancer cells are more resistant to current cancer therapies due to lack of p53-mediated apoptosis.3
Extensive studies have been conducted to identify small molecules that manipulate p53, including restoration of mutant p53 to wild-type, disruption of murine double minute-2 (Mdm2)-p53 binding to prevent p53 degradation, and an increase in p53 level.4 However, it is not known whether sustained inhibition of proteasome deubiquitinating activity to prevent p53 ubiquitination degradation is required for tumor regression.
To explore this issue, in this study, we utilized p53 knockout mice,5 in which targeted germline disruption of the p53 gene results in the absence of p53 in all tissues of the mouse throughout development and in adulthood to develop therapeutic strategies for tumors with p53 deficiency (Figures S1A and S1B). Our results suggest that inhibition of 19S proteasome deubiquitinating activity could provide a general strategy for preventing p53 deficiency in a wide range of malignancies through the constitutive photomorphogenesis 9 (COP9) signalosome subunit 5 (COPS5) deubiquitilation and p53-dependent and -independent regulation mechanism by ubiquitin-specific peptidase 14 (USP14).
Results
Inhibition of Proteasome Deubiquitinating Activity Leads to Tumor Regression In Vivo
Adult p53−/− mice succumb to cancer death mostly by developing lymphomas at an early age (between 4 and 6 months), and heterozygous p53+/− mice have an increased lifespan and in addition to malignant lymphomas of thymus (MLT), also develop sarcomas (including osteosarcoma [OSA] and soft tissue sarcoma [STS]) and to a much lesser extent, carcinomas (Figure 1A).
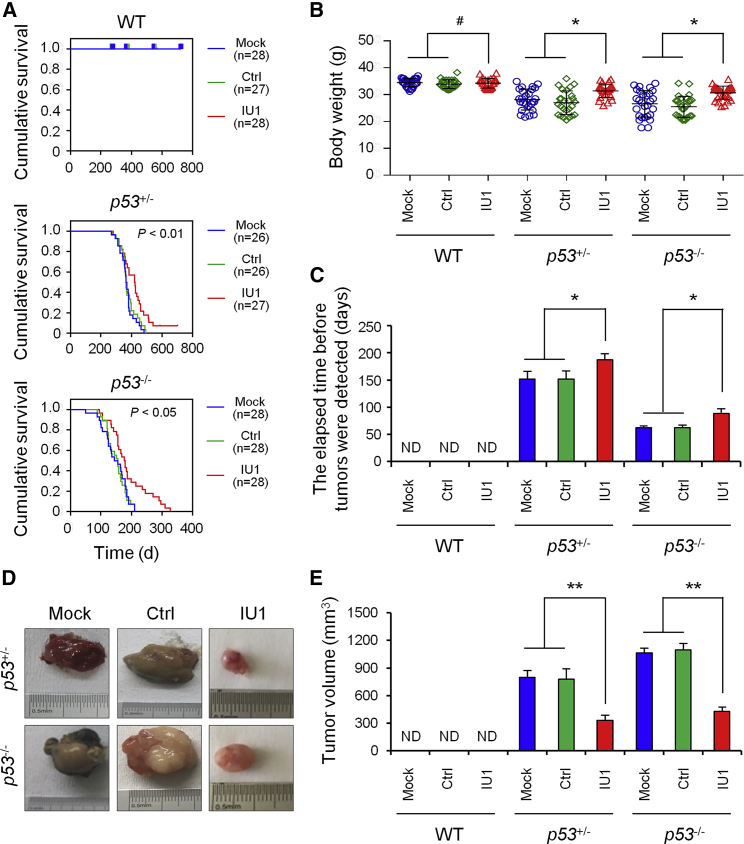
Figure 1.
IU1 Treatment Resulted in Durable Tumor Regressions in p53-Deficient Mice
(A) Kaplan-Meier survival analysis was used to evaluate the treatment effect of IU1 in wild-type (WT), heterozygous p53+/− mice, and homozygous p53−/− mice for OS. (B–E) The effect of IU1 in WT, heterozygous p53+/− mice, and homozygous p53−/− mice on the whole body (B), time to detected tumor (C), tumor volume (D and E). Ctrl, control; Mock, mice without treatment; ND, not detected. Data shown are the mean ± SD. Statistical analyses were performed with one-way ANOVA (*p < 0.05 and **p < 0.01 versus control).
We previously found that an USP14 inhibitor IU1 induced tumor cell apoptosis and inhibits tumor growth in vitro (unpublished data). Here, we investigated the effect of IU1 on tumor growth in the p53 deficiency model in vivo. We administered IU1 to C57BL/6J mice, with or without p53 deficiency (Figure S1C). We observed an obviously prolonged overall survival (OS) in p53+/− and p53−/− mice with IU1 administration (Figure 1A). Weight from control mice significantly decreased. In contrast, IU1-treated mice showed normal body weight (Figure 1B) and main organs’ weights (e.g., liver and lung) (Figures S1D and S1E). Delayed onset of an observable tumor (Figure 1C) and tumor regression (Figures 1D and 1E) was shown in IU1-treated mice compared to the mice in the vehicle-treated group.
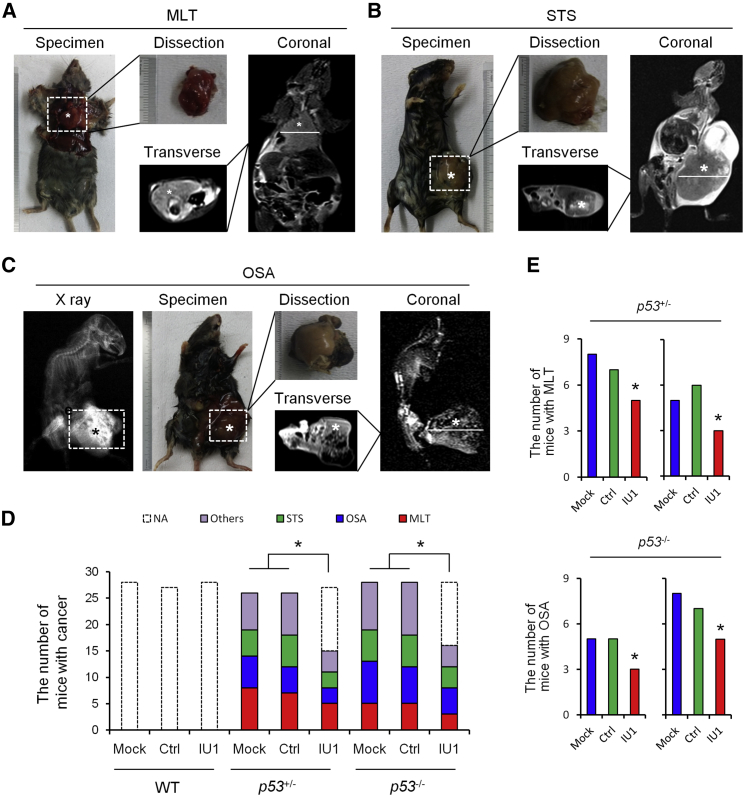
With the comparison of the treatment effect of IU1 on the main type of cancer in p53-deficient mice, including MLT, STS, and OSA (Figures 2A–2C), we verified that the number of mice with MLT obviously decreased in heterozygous p53+/− mice, and the number of mice with OSA decreased in homozygous p53−/− mice (Figures 2D and 2E).
Figure 2.
X-Ray, Micro-CT, and MRI Analysis and Type of Primary Tumors in p53-Deficient Mice
(A–C) X-Ray, micro-CT, and MRI analysis of MLT (A), STS (B), and OSA (C) in p53-deficient mice. (D) The effect of IU1 in WT, heterozygous p53+/− mice, and homozygous p53−/− mice on the number of types of cancer. (E) The number of mice with MLT or OSA in p53+/− and p53−/− mice. Ctrl, control; MLT, malignant lymphomas of thymus; Mock, mice without treatment; NA, not applicable; OSA, osteosarcoma; STS, soft tissue sarcoma; WT, wild-type.
The tissue morphology from IU1-treated mice was restored, which were malignant phenotypes in the control group (Figures S2A–S2C). The mitotic index (Figure S2D) and Ki-67-positive cells (Figure S3A) from IU1-treated mice decreased. Apoptotic (Figure S3B) and senescent cells (Figure S3C) significantly increased, which suggests the mechanisms of tumor inhibition by IU1.
We assessed the p53 levels in response to IU1. In comparison to the control, treatment with IU1 significantly increased the protein levels of p53 (Figure S3D).
Treatment of IU1 Induced Cell Growth Arrest, Apoptosis, and Senescence In Vivo and In Vitro
Recent studies indicate that deubiquitinating enzymes (DUBs) play a pivotal role in the regulation of the cell cycle.6 We found that IU1 induced significant G2/M-phase growth arrest (Figures 3A–3C). In accordance with these findings, IU1 treatment decreased G2/M-phase cell-cycle regulatory proteins’ cell-division cycle 25C (CDC25C) and its downstream protein CDC2, as well as cyclin B1 (Figure 3A). We also examined the effect of IU1 on G1/S-phase marker proteins, including cyclin E1, cyclin D1, and p21. Results showed that IU1 induced p21 without any significant change in cyclin D1 (Figure 3A).
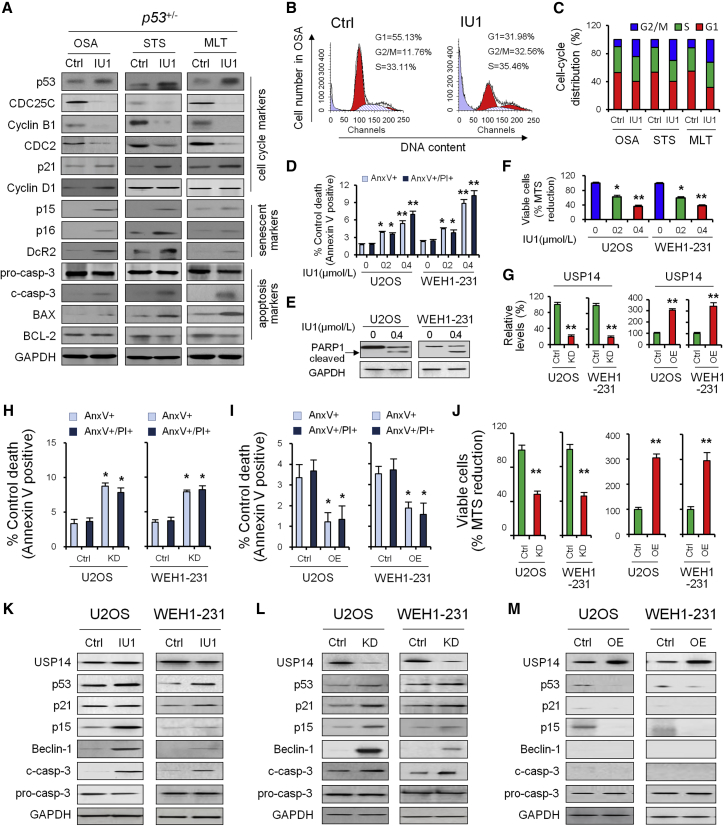
Figure 3.
The Effect of IU1 on the p53-Dependent Regulation Mechanism by USP14
(A) Western blotting was used to measure the protein level of cell-cycle-, senescent-, and apoptosis-associated markers in heterozygous p53+/− mice. (B and C) The effect of IU1 in heterozygous p53+/− mice on cell cycle (B) and distribution (C) was analyzed by flow cytometry. (D) U2OS and WEH1-231 cells were incubated with 0, 0.2, and 0.4 μΜ IU1 for 24 h, as indicated. Bar graphs (mean ± SD) show percentage of Annexin V (AnxV)+ or AnxV+/propidium iodide (PI)+ cells and were derived from three independent experiments. (E) Protein levels of cleaved PARP1 were detected by immunoblotting. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is useful as protein loading control. (F) Viability of U2OS and WEH1-231 cells was measured after the cells were treated with 0, 0.2, and 0.4 μΜ IU1 for 24 h. (G) qRT-PCR was used to validate the effect of overexpression or knockdown of USP14. (H and I) Bar graphs (mean ± SD) show percentage of AnxV+ or AnxV+/PI+ cells before and after USP14 knockdown (H) or overexpression (I). (J) Viability of U2OS and WEH1-231 cells was measured before and after USP14 knockdown or overexpression. (K–M) Western blotting was used to quantify USP14, p53, and p53 target protein level in U2OS and WEH1-231 cells treated with IU1 (K), USP14 knockdown (L), and overexpression (M). Data shown are the mean ± SD. Statistical analyses were performed with one-way ANOVA (*p < 0.05 and **p < 0.01 versus control).
Recent work has led to the identification of a number of proteins whose expression is increased in senescent preneoplastic lesions,7 which include the cyclin-dependent kinase (cdk) inhibitors p15-Ink4b and p16-Ink4a, as well as decoy receptor 2 (DcR2). We examined their expression in different types of cancer derived from p53+/− mice, with or without IU1 treatment (Figure 3A). The OSA from p53+/− mice induced expression of p15-Ink4b, p16-Ink4a, and DcR2 (Figure 3A).
We next examined whether cell apoptosis markers (c-casp-3, BAX, and BCL-2) are similarly induced by p53 restoration in p53+/− mice in vivo. We observed that the proapoptotic effects of IU1 in MLT from p53−/− mice were p53 dependent but weakly exhibited in OSA from p53+/− mice (Figure 3A), suggesting that IU1 inhibits tumor growth in vivo, and different molecular mechanisms are involved in different tumor subtypes with p53 deficiency.
Consistent with the data obtained from p53+/− mice, we noted significant Annexin V positivity and poly(ADP-ribose) polymerase 1 (PARP1) cleavage in primary malignant cells treated with IU1 by 12 h (Figure 3), with >35% undergoing total loss of viability at a concentration of 0.4 μM (Figure 3F).
To investigate the biological role of USP14 in tumor progression, we performed loss- and gain-of-function studies using the U2OS and WEH1-231 cells that moderately expressed p53 and USP14 as models to generate stable USP14 knockdown or overexpression cell lines (Figure 3G). Knockdown of USP14 induced cell apoptosis (Figure 3H) and decreased cell viability (Figure 3J); in contrast, overexpression of USP14 significantly inhibited cell apoptosis (Figure 3I) and increase cell viability (Figure 3J).
Inhibition of USP14 activity by IU1 restored the p53 protein level and function that lead to upregulation of downstream effectors p21, p15, beclin-1, and cleaved caspase 3 (Figure 3K). Knockdown of USP14 led to similar results (Figure 3L); on the contrary, overexpression of USP14 significantly reduced the p53 protein level and downstream effectors (Figure 3M), suggesting the regulatory role of USP14 for p53 protein and its downstream effectors.
IU1 Induces Ubiquitination and Degradation of COPS5
Deubiquitination enzymes interact with and then deubiquitinate substrates to inhibit their protein degradation by a ubiquitination pathway through the 26S proteasome.8 We explored the candidate-interacting proteins and potential substrates of USP14 through mass spectrometry after immunoprecipitation. In addition to several previously well-defined proteasome regulatory proteins, including non-ATPase 13 (RPN13),9 RPN10, proteasome 26S subunit, ATPase 1 (PSMC1),10 PSMA4, and PSMD1,11 COPS5, a critical negative regulator, was identified as an interacting partner of USP14 (Figure 4A).
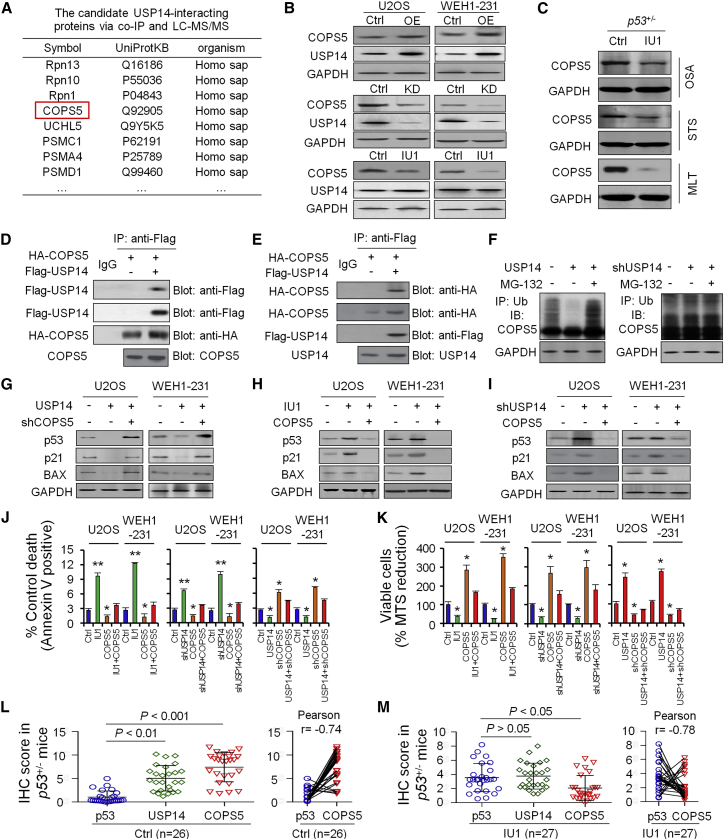
Figure 4.
IU1 Inhibits USP14 Deubiquitinating Activity to Upregulate p53 through COPS5
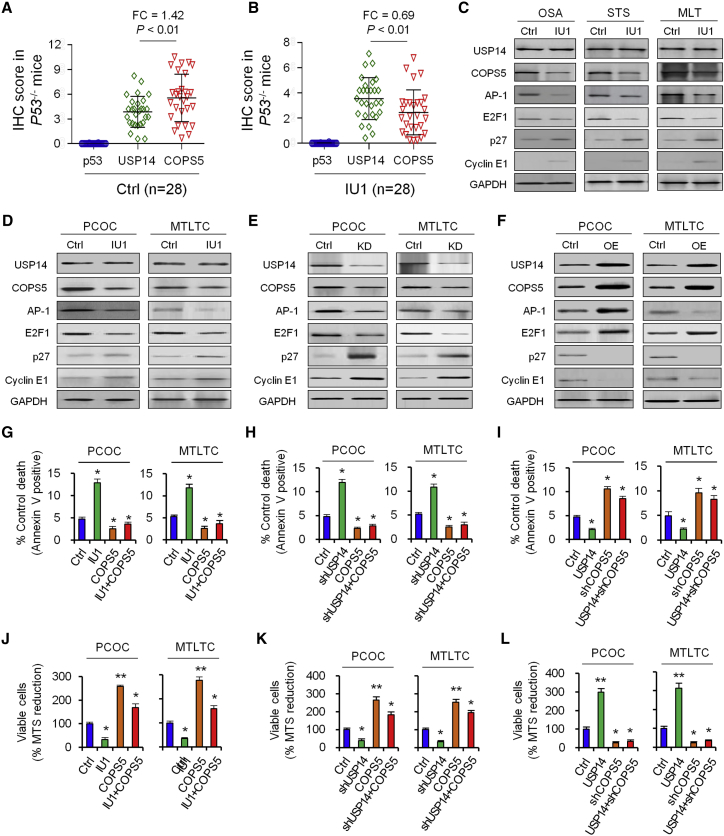
(A) Coimmunoprecipitation (coIP) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to screen the candidate of USP14-interacting proteins. (B) Western blotting was used to quantify COPS5 protein level in U2OS and WEH1-231 cells after IU1 treatment, USP14 knockdown, or overexpression. (C) Western blotting was used to quantify COPS5 protein level in OSA, STS, and MLT tissues from p53+/− mice. (D and E) Immunoprecipitation analysis by either anti-HA (D) or anti-Flag antibody (E) after HA-COPS5, together with Flag-USP14 expression plasmid, was cotransfected in 293T cells. (F) COPS5 ubiquitination level was detected in vitro in 293T cells after USP14 overexpression or treatment with MG-132. (G–I) p53, p21, and BAX protein level was detected in vitro in U2OS and WEH1-231 cells after USP14 overexpression and COPS5 knockdown (G), COPS5 knockdown with IU1 treatment (H), or USP14 knockdown and COPS5 overexpression (I). (J) Bar graphs (mean ± SD) show percentage of AnxV+ cells treated with DMSO (Ctrl), IU1 treatment, knockdown, and overexpression of USP14 or COPS5. (K) Viability was measured in U2OS and WEH1-231 cells treated with DMSO (Ctrl), IU1 treatment, knockdown, and overexpression of USP14 or COPS5. (L and M) Expression and association of p53, USP14, and COPS5 in primary tumor tissues from p53+/− mice treated with DMSO (L; Ctrl, n = 26) or IU1 (M; n = 27).
USP14 significantly upregulated COPS5 protein but did not upregulate other interacting protein candidates, including RPN13, RPN10, PSMC1, and PSMA4 (Figure 4B); in contrast, knockdown of USP14 or treatment with IU1 significantly decreased the COPS5 protein level (Figure 4B). Treatment with IU1 significantly decreased the COPS5 protein level in p53+/− mice (Figure 4C).
Immunoprecipitation analysis by either anti-hemagglutinin (HA) (Figure 4D) or anti-Flag antibody (Figure 4E) confirmed that USP14 associated with COPS5 in 293T cells when both proteins were overexpressed. We also demonstrated that endogenous USP14 interacts with endogenous COPS5 in U2OS cells.
Moreover, USP14 overexpression significantly decreased COPS5 ubiquitination level; in contrast, knockdown of USP14 significantly increased ubiquitination of COPS5 in vitro (Figure 4F).
Inhibition of USP14 Resulted in Durable Tumor Regression through a COPS5-Induced and p53-Dependent Regulation Mechanism in p53+/− Mice
COPS5 mediated degradation of proteins by factors, such as tumor suppressor p53 and cell-cycle inhibitor p27,12 to enhance cell proliferation. Thus, it is possible that IU1 acts as an anti-tumor inhibitor in tumors with p53 deficiency by inducing degradation of p53 and p27 through the COPS5 pathway. Our results showed that knockdown of COPS5 rescued p53 and the downstream p21 and BAX protein level, induced by USP14 overexpression (Figure 4G). Similarly, IU1 treatment (Figure 4H) or knockdown of USP14 (Figure 4I) rescued the p53 protein, induced by COPS5 overexpression, and enhanced the protein levels of p53 downstream target genes p21 and BAX.
COPS5 overexpression rescued cell apoptosis induced by IU1 treatment (Figure 4J) and enhanced cell viability (Figure 4K). Similarly, COPS5 overexpression rescued cell apoptosis and viability induced by knockdown of USP14 (Figures 4J and 4K); in contrast, COPS5 knockdown induced significant cell apoptosis and reduced cell viability, even when USP14 was overexpressed (Figures 4J and 4K).
The COPS5 protein level significantly increased in various types of tumors in p53+/− mice (Figure 4L) and decreased after treatment of IU1, and the changes in the COPS5 protein level showed a remarkable negative correlation with the p53 protein level (Figure 4M).
Inhibition of USP14 Resulted in Durable Tumor Regression through p53-Independent Regulation Mechanism in p53−/− Mice
We further investigated the tumor inhibition mechanism of IU1 through the p53-independent regulation mechanism by USP14 in p53−/− mice.
The COPS5 protein level significantly increased in various types of tumors in p53−/− mice (Figure 5A) and decreased after treatment of IU1, and the changes in COPS5 protein level showed a remarkable negative correlation with the USP14 protein level (Figure 5B).
Figure 5.
Effect of IU1 on COPS5-Induced Downstream Effectors In Vivo and In Vitro
(A and B) Expression and association of p53, USP14, and COPS5 in primary tumor tissues from p53−/− mice treated with DMSO (A; Ctrl, n = 28) or IU1 (B; n = 28). (C) Western blotting was used to measure the protein level of USP14, COPS5, and COPS5 downstream effectors in homozygous p53−/− mice. (D–F) Western blotting was used to measure the protein levels of USP14, COPS5, and COPS5 downstream effectors in PCOC and MTLTC cells with treatment of IU1 (D), knockdown (E), or overexpression (F) of USP14. (G–I) Bar graphs (mean ± SD) show percentage of AnxV+ cells treated with IU1 treatment with or without COPS5 overexpression (G), USP14 knockdown with or without COPS5 overexpression (H), and USP14 overexpression with or without COPS5 knockdown (I). (J–L) Viability was measured in PCOC and MTLTC cells treated with IU1 treatment with or without COPS5 overexpression (J), USP14 knockdown with or without COPS5 overexpression (K), and USP14 overexpression with or without COPS5 knockdown (L). Data shown are the mean ± SD. Statistical analyses were performed with one-way ANOVA (*p < 0.05 and **p < 0.01 versus control).
IU1 treatment in p53−/− mice resulted in tumor regression (Figure 1), increased apoptosis, induced cell senescence, inhibited cell proliferation, and the transition of cell cycle (Figure 3) that could be attributed to COPS5 substrates, including proto-oncogene activator protein 1 (AP-1), E2F transcription factor 1 (E2F1), cell-cycle inhibitor p27, and cyclin E1 (Figure 5C).
Overexpression of USP14 restored COPS5 protein level and led to an upregulation of downstream effectors AP-1 and E2F1 and reduced p27 and cyclin E1 levels (Figure 5D); in contrast, knockdown of USP14 (Figure 5E) or inhibition of USP14 activity by IU1 (Figure 5F) reduced COPS5 protein level and led to downregulation of downstream effectors AP-1 and E2F1 and induced p27 and cyclin E1 levels in primary cultured osteosarcoma cells (PCOCs) and murine thymic lymphoma T cell (MTLTC) lines from p53−/− mice.
COPS5 overexpression rescued cell apoptosis induced by IU1 treatment (Figure 5G) and enhanced cell viability (Figure 5J). Similarly, COPS5 overexpression rescued cell apoptosis (Figure 5H) and enhanced viability (Figure 5K), induced by knockdown of USP14; in contrast, COPS5 knockdown induced significant cell apoptosis (Figure 5I) and reduced cell viability (Figure 5L), even when USP14 was overexpressed.
Thus, our results suggested that IU1 treatment rescued p53 protein level and blocked p53 ubiquitination degradation induced by USP14-dependent COPS5 deubiquitilation and regulation for p53 and other transcription factors. Given that IU1 is already under clinical evaluation in advanced solid malignancies,13 the translation of our preclinical findings could be fast tracked into the clinic for individuals with p53 expression and functional deficiency.
Discussion
Cancer is associated with evading apoptosis and uncontrolled cell cycle and development of apoptosis resistance and insensitivity to anti-growth signal in cancer cells, which are significant contributing factors to the failure of cancer therapies.14, 15, 16 Thus, induction of apoptosis and control of cell-cycle progression would be ideal strategies for effective cancer therapy.17, 18, 19, 20 One promising approach to achieving this is the modulation of p53 or the components of the p53 signaling pathways.21, 22, 23, 24 The p53 protein acts biochemically as a transcription factor and biologically as a tumor suppressor, exemplified by the fact that many cancers selectively inactivate p53 and/or the p53 pathway.25, 26, 27 Loss of p53 functions accelerates tumorigenesis compared to mice expressing at least one wild-type Trp53 allele.1,26,28,29
In some cancer cells that bear a wild-type p53, p53 is nonfunctional, either by being excluded from the nucleus where p53 acts as a transcription factor or by being targeted for degradation by E3 ubiquitin ligase, including Mdm2 and COPS5.4,30, 31, 32, 33 COPS5, also known as Jab1 or CSN5, is implicated in regulating p53 activity and is overexpressed in various tumors.34 However, the precise roles of COPS5 in the p53 network and in tumorigenesis are not well characterized.
In order to develop therapeutic strategies for tumors with p53 deficiency in vivo, we have utilized a mouse strain carrying a germline disruption of the gene, which eliminates synthesis of the p53 protein and is highly predisposed to malignancy. We show that USP14, a DUB associated with the proteasome in mammalian cells and reversibly associated with the 19S regulatory particle, can deubiquitilate and upregulate COPS5 to promote its nuclear transport. COPS5 was found to interact with p53, and COPS5 expression leads to p53 degradation, facilitating Mdm2-mediated p53 ubiquitination and promoting p53 nuclear export. Additionally, COPS5 can antagonize the transcriptional activity of p53, which demonstrates that USP14 is a pivotal regulator for COPS5 and p53. Thus, our studies may pave the way for targeting USP14 for anti-cancer drug development.
To restore wild-type p53 conformation in p53-deficient cancer cells, a small molecule compound, 19S regulatory particle inhibitor IU1, was utilized. IU1 specifically and selectively blocks USP14 deubiquitilating activity without inhibiting proteasome activity and can block growth and induce apoptosis. It shows antitumor activity of b-AP15, a novel, small molecule inhibitor of deubiquitilating enzyme USP14 and ubiquitin carboxyl-terminal hydrolase isozyme L5 (UCHL5), in a number of tumor models, including multiple myeloma, and it overcomes bortezomib resistance.35 Tumor cells are rapidly committed to apoptosis/cell death after exposure to IU1 through binding to USP14 and react with intracellular nucleophiles, such as cysteine thiolates, to inhibit proteasome function. Thus, IU1 targets the proteasome-associated USP14 and inhibits its activities, resulting in a rapid accumulation of protein-ubiquitin conjugates but without inhibiting the proteolytic activities of 20S proteasomes.36
Our previous results showed that IU1 exhibits cytotoxic effects against various cancer cell lines in vitro, selectively kills cancer cells from hepatocellular carcinoma (HCC) patients ex vivo, and efficiently inhibits the growth of non-small cell lung carcinoma xenografts in nude mice.37 In the current study, in vivo studies show that IU1 is well tolerated, inhibits tumor growth, and prolongs survival. Moreover, IU1 induces cell-cycle arrest, decreases viability, and induces apoptosis in cultured cell lines and patient-derived primary cells.
The 26S proteasome complex, which degrades ubiquitinated proteins, contains the 20S core particle and a 19S regulatory particle necessary for binding protein substrates.38, 39, 40, 41, 42 The mammalian 19S cap contains three DUBs that unfold and deubiquitinate proteins prior to their entry into the proteasomal core.43, 44, 45, 46, 47 Of the three, USP14 and UCHL5 reversibly associate with the proteasome through scaffolding proteins RPN1 and RPN13, respectively.48 Suppression of either DUB or scaffolding protein individually via RNA interference partly upregulates proteasomal catalytic activity and accumulation of polyubiquitinated proteins.49, 50, 51, 52, 53 The combined inhibition of both UCHL5 and USP14 results in lethality, indicates their nonredundancy, and suggests their role in maintaining cancer cell survival, which partly explains the finding that b-AP15, which selectively disrupts both USP14 and UCHL5 activity, was shown to significantly increase cancer cell apoptosis in vitro and to inhibit tumor progression, as well as exhibit robust antitumor activity.54, 55, 56
Anti-cancer activity of IU1 is associated with growth arrest through inhibition of deubiquitilating activity of USP14, downregulation of COPS5, and upregulation of p53-dependent p21, p15, and beclin-1 and p53-independent COPS5 downstream effects AP-1, E2F1, p27, and cyclin E1, as well as induction of caspase-dependent apoptosis. Additionally, the effects of IU1 were shown to be independent of p53 status, as well as the expression of BCL-2, both of which can influence the response to bortezomib therapy.
Conclusions
Our preclinical data, showing efficacy of USP14 in p53-deficient disease models, validates targeting DUBs in the ubiquitin proteasomal cascade and provides the new anticancer drug target and framework for clinical evaluation of the USP14 inhibitor to improve outcome for patients with p53 deficiency.
Methods
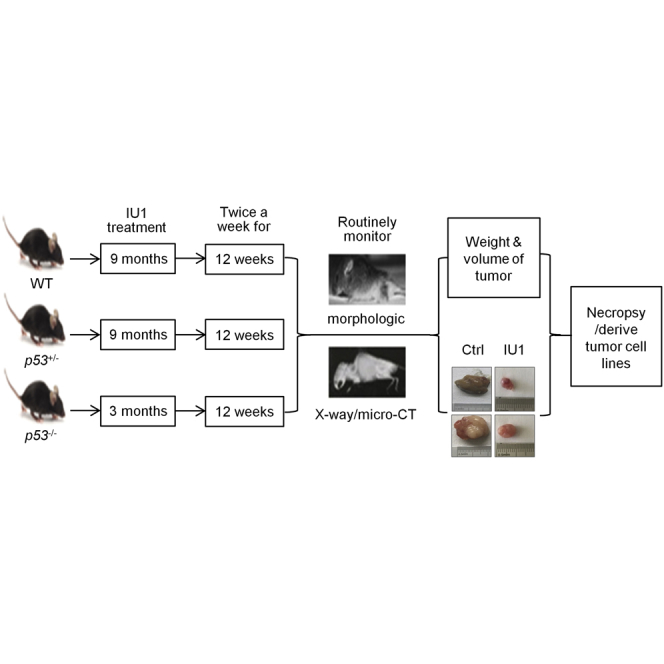
Animal Studies
All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) guidelines at Tongji University School of Medicine (SYDW-19-215). Experiments were performed in 9-month-old wild-type and p53+/− mice and 3-month-old p53−/− mice. Genomic DNA from tail biopsies was genotyped by polymerase chain reaction (PCR).57 IU1 (5 mg/kg) was administered intraperitoneally (i.p.) weekly for the number of days indicated.58 All mice were monitored by X-ray, magnetic resonance imaging (MRI), or micro-computed tomography (CT) diagnosis for tumor phenotypes, three times a week.59 Supplemental Methods contains detailed information.
Cell Treatments
HEK293T, U2OS, and WEH1-231 cells were cultured in DMEM media and supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin for viability, flow cytometry, immunofluorescence, immunoprecipitation, and functional analysis. Derivation of MTLTC lines and primary cultured osteosarcoma cell (PCOC) lines was obtained from p53−/− mice and cultured.60,61
Plasmid Construction and Transfection
Gene overexpression was performed using the pMSCV retroviral plasmid.62 Knockdown cell lines were generated using short hairpin RNAs and retroviral transduction.63 All constructs were confirmed by PCR and Sanger sequencing. The transfected cells were harvested at 48 h post-transfection for further assays.
RNA Isolation, cDNA Synthesis, and qRT-PCR
RNA was isolated using TRIzol reagent. qPCR was conducted using the TaqMan Universal PCR Kit (Life Technologies, Carlsbad, CA, USA).
Immunoprecipitation and Immunoblotting
Cells were pretreated with MG132 or IU1 for the indicated time periods. Immunoprecipitations were performed using the indicated primary antibody and protein A/G agarose beads. For immunoblotting, total proteins were extracted from cells following the standard protocol.64
Histology and Immunohistochemistry (IHC) Analysis
Standard IHC and H&E staining were used to evaluate protein expression levels in tumor samples.65 Tissues from mice were flushed and fixed in 4% formaldehyde in PBS for 24 h. Samples were then dehydrated and embedded in paraffin, sectioned at 5 μM, and processed for H&E staining. Serial sections were stained in parallel with the primary antibody replaced by PBS as controls.
Mass Spectrometry
Pellets of U2OS cell expressing Flag-UCH37 from two, 150-mm plates were lysed, and digestion with trypsin was performed for liquid chromatography-tandem mass spectrometry analysis.66
Statistical Analysis
Data were expressed as mean ± SD. Categorical data were reported as numbers and percentages. Statistical significance was assessed by Student’s t test or one-way ANOVA with Tukey test using the SPSS 20.0 software program or Prism 6.0 GraphPad. The level of significance was set as p <0.05.
Author Contributions
Y.-S.M., X.-F.W., Y.-J.Z., and D.F. designed the study. Y-.S.M., X.-F.W., Y.-J.Z., P.L., H.-D.L., H.-Q.Y., R.-T.X., C.-Y.J., and D.F. performed the cytological experiments. Y.-S.M., X.-F.W., Y.-J.Z., P.L., H.-D.L., L.L., G.-X.L., Z.-Y.C., J.-J.Z., S.-B.X., Q.X., and D.F. performed the animal experiments. Y.-S.M., Y.-J.Z., H.-D.L., and D.F. performed the statistical analyses and interpreted the data. Y.-S.M., X.-F.W., Z.-W.L., F.Y., Q.X., and D.F. contributed to study materials and consumables. Y.-S.M., X.-F.W., Y.-J.Z., and D.F. wrote the manuscript. All authors contributed to the final version of the manuscript and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We would like to thank LetPub (https://www.letpub.com) for providing linguistic assistance during the preparation of this manuscript. The datasets supporting the conclusions of this article are included within the article. This study was supported partly by grants from the National Natural Science Foundation of China (81972214, 81772932, 81472202, 81201535, 81302065, 81671716, 81301993, 81372175, and 81472209); Fundamental Research Funds for the Central Universities (22120170212 and 22120170117); Shanghai Natural Science Foundation (12ZR1436000 and 16ZR1428900); Shanghai Municipal Commission of Health and Family Planning (201540228 and 201440398); Construction of Clinical Medical Center for Tumor Biological Samples in Nantong (HS2016004); and Wu Jieping Medical Foundation (320.6750.14326).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.12.013.
Supplemental Information
References
- 1.Mastropasqua F., Marzano F., Valletti A., Aiello I., Di Tullio G., Morgano A., Liuni S., Ranieri E., Guerrini L., Gasparre G. TRIM8 restores p53 tumour suppressor function by blunting N-MYC activity in chemo-resistant tumours. Mol. Cancer. 2017;16:67. doi: 10.1186/s12943-017-0634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P., Boeckx B., Wijnhoven P.W., Radaelli E., Vermi W. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 3.Wu S.D., Ma Y.S., Fang Y., Liu L.L., Fu D., Shen X.Z. Role of the microenvironment in hepatocellular carcinoma development and progression. Cancer Treat. Rev. 2012;38:218–225. doi: 10.1016/j.ctrv.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Mancini F., Teveroni E., Di Conza G., Monteleone V., Arisi I., Pellegrino M., Buttarelli M., Pieroni L., D’Onofrio M., Urbani A. MDM4 actively restrains cytoplasmic mTORC1 by sensing nutrient availability. Mol. Cancer. 2017;16:55. doi: 10.1186/s12943-017-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 6.Sun H., Zhang Q., Jing Y.Y., Zhang M., Wang H.Y., Cai Z., Liuyu T., Zhang Z.D., Xiong T.C., Wu Y. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat. Commun. 2017;8:15534. doi: 10.1038/ncomms15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar D., Gorain M., Kundu G., Kundu G.C. Therapeutic implications of cellular and molecular biology of cancer stem cells in melanoma. Mol. Cancer. 2017;16:7. doi: 10.1186/s12943-016-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y., Fu D., Shen X.Z. The potential role of ubiquitin C-terminal hydrolases in oncogenesis. Biochim. Biophys. Acta. 2010;1806:1–6. doi: 10.1016/j.bbcan.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K.J., Finley D., Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T., Duan W., Yang H., Lee M.K., Bte Mustafa F., Lee B.H., Teo T.S. Identification of two proteins, S14 and UIP1, that interact with UCH37. FEBS Lett. 2001;488:201–205. doi: 10.1016/s0014-5793(00)02436-4. [DOI] [PubMed] [Google Scholar]
- 11.Huang H., Liu N., Liao Y., Liu N., Cai J., Xia X., Guo Z., Li Y., Wen Q., Yin Q. Platinum-containing compound platinum pyrithione suppresses ovarian tumor proliferation through proteasome inhibition. J. Exp. Clin. Cancer Res. 2017;36:79. doi: 10.1186/s13046-017-0547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R., Hu X., Zhou J., Sun J., Zhu A.Z., Xu X., Zheng H., Gao X., Wang X., Jin H. COPS5 amplification and overexpression confers tamoxifen-resistance in ERα-positive breast cancer by degradation of NCoR. Nat. Commun. 2016;7:12044. doi: 10.1038/ncomms12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B., Jiang S., Li M., Xiong X., Zhu M., Li D., Zhao L., Qian L., Zhai L., Li J. Proteome-wide analysis of USP14 substrates revealed its role in hepatosteatosis via stabilization of FASN. Nat. Commun. 2018;9:4770. doi: 10.1038/s41467-018-07185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X., Nowicka U., Sridharan V., Liu F., Randles L., Hymel D., Dyba M., Tarasov S.G., Tarasova N.I., Zhao X.Z. Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets. Nat. Commun. 2017;8:15540. doi: 10.1038/ncomms15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y.S., Huang T., Zhong X.M., Zhang H.W., Cong X.L., Xu H., Lu G.X., Yu F., Xue S.B., Lv Z.W., Fu D. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol. Cancer. 2018;17:139. doi: 10.1186/s12943-018-0890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F., Long Q., Fu D., Zhu D., Ji Y., Han L., Zhang B., Xu Q., Liu B., Li Y. Targeting SPINK1 in the damaged tumour microenvironment alleviates therapeutic resistance. Nat. Commun. 2018;9:4315. doi: 10.1038/s41467-018-06860-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajaraman S., Canjuga D., Ghosh M., Codrea M.C., Sieger R., Wedekink F., Tatagiba M., Koch M., Lauer U.M., Nahnsen S. Measles Virus-Based Treatments Trigger a Pro-inflammatory Cascade and a Distinctive Immunopeptidome in Glioblastoma. Mol. Ther. Oncolytics. 2018;12:147–161. doi: 10.1016/j.omto.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong B., Muili K., Bolyard C., Russell L., Lee T.J., Banasavadi-Siddegowda Y., Yoo J.Y., Yan Y., Ballester L.Y., Bockhorst K.H., Kaur B. Suppression of HMGB1 Released in the Glioblastoma Tumor Microenvironment Reduces Tumoral Edema. Mol. Ther. Oncolytics. 2018;12:93–102. doi: 10.1016/j.omto.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mooney R., Majid A.A., Batalla-Covello J., Machado D., Liu X., Gonzaga J., Tirughana R., Hammad M., Lesniak M.S., Curiel D.T., Aboody K.S. Enhanced Delivery of Oncolytic Adenovirus by Neural Stem Cells for Treatment of Metastatic Ovarian Cancer. Mol. Ther. Oncolytics. 2018;12:79–92. doi: 10.1016/j.omto.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bressy C., Hastie E., Grdzelishvili V.Z. Combining Oncolytic Virotherapy with p53 Tumor Suppressor Gene Therapy. Mol. Ther. Oncolytics. 2017;5:20–40. doi: 10.1016/j.omto.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Echchannaoui H., Petschenka J., Ferreira E.A., Hauptrock B., Lotz-Jenne C., Voss R.H., Theobald M. A Potent Tumor-Reactive p53-Specific Single-Chain TCR without On- or Off-Target Autoimmunity In Vivo. Mol. Ther. 2019;27:261–271. doi: 10.1016/j.ymthe.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh A., Bhattacharyya N., Srivastava A., Pruett N., Ripley R.T., Schrump D.S., Hoang C.D. MicroRNA-215-5p Treatment Suppresses Mesothelioma Progression via the MDM2-p53-Signaling Axis. Mol. Ther. 2019;27:1665–1680. doi: 10.1016/j.ymthe.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vile R.G. The Immune System in Oncolytic Immunovirotherapy: Gospel, Schism and Heresy. Mol. Ther. 2018;26:942–946. doi: 10.1016/j.ymthe.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toh T.B., Lim J.J., Chow E.K. Epigenetics in cancer stem cells. Mol. Cancer. 2017;16:29. doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ylä-Herttuala S. CRISPR/Cas9 and p53: An Odd Couple Requiring Relationship Management. Mol. Ther. 2018;26:2711. doi: 10.1016/j.ymthe.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y.S., Yu F., Zhong X.M., Lu G.X., Cong X.L., Xue S.B., Xie W.T., Hou L.K., Pang L.J., Wu W. miR-30 family reduction maintains self-renewal and promotes tumorigenesis in NSCLC-initiating cells by targeting oncogene TM4SF1. Mol. Ther. 2018;26:2751–2765. doi: 10.1016/j.ymthe.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi M., Jiao D., Xu H., Liu Q., Zhao W., Han X., Wu K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer. 2018;17:129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meister M.T., Boedicker C., Klingebiel T., Fulda S. Hedgehog signaling negatively co-regulates BH3-only protein Noxa and TAp73 in TP53-mutated cells. Cancer Lett. 2018;429:19–28. doi: 10.1016/j.canlet.2018.04.025. [DOI] [PubMed] [Google Scholar]
- 30.Lee C.C., Yang W.H., Li C.H., Cheng Y.W., Tsai C.H., Kang J.J. Ligand independent aryl hydrocarbon receptor inhibits lung cancer cell invasion by degradation of Smad4. Cancer Lett. 2016;376:211–217. doi: 10.1016/j.canlet.2016.03.052. [DOI] [PubMed] [Google Scholar]
- 31.Thiem A., Hesbacher S., Kneitz H., di Primio T., Heppt M.V., Hermanns H.M., Goebeler M., Meierjohann S., Houben R., Schrama D. IFN-gamma-induced PD-L1 expression in melanoma depends on p53 expression. J. Exp. Clin. Cancer Res. 2019;38:397. doi: 10.1186/s13046-019-1403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z., Yi L., Gao P., Zhang R., Li J. The cornerstone of integrating circulating tumor DNA into cancer management. Biochim. Biophys. Acta Rev. Cancer. 2019;1871:1–11. doi: 10.1016/j.bbcan.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Subburayan K., Thayyullathil F., Pallichankandy S., Rahman A., Galadari S. Par-4-dependent p53 up-regulation plays a critical role in thymoquinone-induced cellular senescence in human malignant glioma cells. Cancer Lett. 2018;426:80–97. doi: 10.1016/j.canlet.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Wang X., Stafford W., Mazurkiewicz M., Fryknäs M., Brjnic S., Zhang X., Gullbo J., Larsson R., Arnér E.S., D’Arcy P., Linder S. The 19S Deubiquitinase inhibitor b-AP15 is enriched in cells and elicits rapid commitment to cell death. Mol. Pharmacol. 2014;85:932–945. doi: 10.1124/mol.113.091322. [DOI] [PubMed] [Google Scholar]
- 35.Tian Z., D’Arcy P., Wang X., Ray A., Tai Y.T., Hu Y., Carrasco R.D., Richardson P., Linder S., Chauhan D., Anderson K.C. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo C.L., Goldberg A.L. Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc. Natl. Acad. Sci. USA. 2017;114:E3404–E3413. doi: 10.1073/pnas.1701734114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu F., Liu J.B., Wu Z.J., Xie W.T., Zhong X.J., Hou L.K., Wu W., Lu H.M., Jiang X.H., Jiang J.J. Tumor suppressive microRNA-124a inhibits stemness and enhances gefitinib sensitivity of non-small cell lung cancer cells by targeting ubiquitin-specific protease 14. Cancer Lett. 2018;427:74–84. doi: 10.1016/j.canlet.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 38.Dheilly E., Moine V., Broyer L., Salgado-Pires S., Johnson Z., Papaioannou A., Cons L., Calloud S., Majocchi S., Nelson R. Selective Blockade of the Ubiquitous Checkpoint Receptor CD47 Is Enabled by Dual-Targeting Bispecific Antibodies. Mol. Ther. 2017;25:523–533. doi: 10.1016/j.ymthe.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou H., Chen H., Zhou Z., Wan Y., Liu Z. ATXN3 promotes breast cancer metastasis by deubiquitinating KLF4. Cancer Lett. 2019;467:19–28. doi: 10.1016/j.canlet.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Xia X., Liao Y., Huang C., Liu Y., He J., Shao Z., Jiang L., Dou Q.P., Liu J., Huang H. Deubiquitination and stabilization of estrogen receptor α by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 2019;465:118–128. doi: 10.1016/j.canlet.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Cui H., Wang Q., Lei Z., Feng M., Zhao Z., Wang Y., Wei G. DTL promotes cancer progression by PDCD4 ubiquitin-dependent degradation. J. Exp. Clin. Cancer Res. 2019;38:350. doi: 10.1186/s13046-019-1358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X., Liu M., Zhu H., Wang J., Dai W., Li J., Zhu D., Tang W., Xiao Y., Lin J. Ubiquitin-specific protease 3 promotes cell migration and invasion by interacting with and deubiquitinating SUZ12 in gastric cancer. J. Exp. Clin. Cancer Res. 2019;38:277. doi: 10.1186/s13046-019-1270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin N.T., Bell J.C. Oncolytic Virus Combination Therapy: Killing One Bird with Two Stones. Mol. Ther. 2018;26:1414–1422. doi: 10.1016/j.ymthe.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim J.A., Sun B., Puertollano R., Raben N. Therapeutic Benefit of Autophagy Modulation in Pompe Disease. Mol. Ther. 2018;26:1783–1796. doi: 10.1016/j.ymthe.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu J., Mao W., Ren W., Xu F., Zhu Q., Lu C., Lin Z., Zhang Z., Chu Y., Liu R., Ge D. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett. 2019;443:125–134. doi: 10.1016/j.canlet.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 46.Han H.G., Moon H.W., Jeon Y.J. ISG15 in cancer: Beyond ubiquitin-like protein. Cancer Lett. 2018;438:52–62. doi: 10.1016/j.canlet.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Zhou S., Lu J., Li Y., Chen C., Cai Y., Tan G., Peng Z., Zhang Z., Dong Z., Kang T., Tang F. MNAT1 is overexpressed in colorectal cancer and mediates p53 ubiquitin-degradation to promote colorectal cancer malignance. J. Exp. Clin. Cancer Res. 2018;37:284. doi: 10.1186/s13046-018-0956-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Hillert E.K., Brnjic S., Zhang X., Mazurkiewicz M., Saei A.A., Mofers A., Selvaraju K., Zubarev R., Linder S., D’Arcy P. Proteasome inhibitor b-AP15 induces enhanced proteotoxicity by inhibiting cytoprotective aggresome formation. Cancer Lett. 2019;448:70–83. doi: 10.1016/j.canlet.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Hotani T., Mizuguchi H., Sakurai F. Systemically Administered Reovirus-Induced Downregulation of Hypoxia Inducible Factor-1α in Subcutaneous Tumors. Mol. Ther. Oncolytics. 2018;12:162–172. doi: 10.1016/j.omto.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing F., Wang S., Zhou J. The expression of microRNA-598 inhibits ovarian cancer cell proliferation and metastasis by targeting URI. Mol. Ther. Oncolytics. 2018;12:9–15. doi: 10.1016/j.omto.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Wei W.S., Chen X., Guo L.Y., Li X.D., Deng M.H., Yuan G.J., He L.Y., Li Y.H., Zhang Z.L., Jiang L.J. TRIM65 supports bladder urothelial carcinoma cell aggressiveness by promoting ANXA2 ubiquitination and degradation. Cancer Lett. 2018;435:10–22. doi: 10.1016/j.canlet.2018.07.036. [DOI] [PubMed] [Google Scholar]
- 52.Gu Y., Lv F., Xue M., Chen K., Cheng C., Ding X., Jin M., Xu G., Zhang Y., Wu Z. The deubiquitinating enzyme UCHL1 is a favorable prognostic marker in neuroblastoma as it promotes neuronal differentiation. J. Exp. Clin. Cancer Res. 2018;37:258. doi: 10.1186/s13046-018-0931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma J., Chang K., Peng J., Shi Q., Gan H., Gao K., Feng K., Xu F., Zhang H., Dai B. SPOP promotes ATF2 ubiquitination and degradation to suppress prostate cancer progression. J. Exp. Clin. Cancer Res. 2018;37:145. doi: 10.1186/s13046-018-0809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parasramka M., Yan I.K., Wang X., Nguyen P., Matsuda A., Maji S., Foye C., Asmann Y., Patel T. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol. Cancer. 2017;16:22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castro I., Ekinci E., Huang X., Cheaito H.A., Ahn Y.H., Olivero-Verbel J., Dou Q.P. Proteasome-associated cysteine deubiquitinases are molecular targets of environmental optical brightener compounds. J. Cell. Biochem. 2019;120:14065–14075. doi: 10.1002/jcb.28682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D’Arcy P., Brnjic S., Olofsson M.H., Fryknäs M., Lindsten K., De Cesare M., Perego P., Sadeghi B., Hassan M., Larsson R., Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 57.Chen K., Qian W., Jiang Z., Cheng L., Li J., Sun L., Zhou C., Gao L., Lei M., Yan B. Metformin suppresses cancer initiation and progression in genetic mouse models of pancreatic cancer. Mol. Cancer. 2017;16:131. doi: 10.1186/s12943-017-0701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia X., Huang C., Liao Y., Liu Y., He J., Guo Z., Jiang L., Wang X., Liu J., Huang H. Inhibition of USP14 enhances the sensitivity of breast cancer to enzalutamide. J. Exp. Clin. Cancer Res. 2019;38:220. doi: 10.1186/s13046-019-1227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perales-Puchalt A., Wojtak K., Duperret E.K., Yang X., Slager A.M., Yan J., Muthumani K., Montaner L.J., Weiner D.B. Engineered DNA Vaccination against Follicle-Stimulating Hormone Receptor Delays Ovarian Cancer Progression in Animal Models. Mol. Ther. 2019;27:314–325. doi: 10.1016/j.ymthe.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jinadasa R., Balmus G., Gerwitz L., Roden J., Weiss R., Duhamel G. Derivation of thymic lymphoma T-cell lines from Atm(-/-) and p53(-/-) mice. J. Vis. Exp. 2011;50:2598. doi: 10.3791/2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma O., Cai W.W., Zender L., Dayaram T., Shen J., Herron A.J., Lowe S.W., Man T.K., Lau C.C., Donehower L.A. MMP13, Birc2 (cIAP1), and Birc3 (cIAP2), amplified on chromosome 9, collaborate with p53 deficiency in mouse osteosarcoma progression. Cancer Res. 2009;69:2559–2567. doi: 10.1158/0008-5472.CAN-08-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma Y.S., Yu F., Zhong X.M., Lu G.X., Cong X.L., Xue S.B., Xie W.T., Hou L.K., Pang L.J., Wu W. miR-30 Family Reduction Maintains Self-Renewal and Promotes Tumorigenesis in NSCLC-Initiating Cells by Targeting Oncogene TM4SF1. Mol. Ther. 2018;26:2751–2765. doi: 10.1016/j.ymthe.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma Y.S., Lv Z.W., Yu F., Chang Z.Y., Cong X.L., Zhong X.M., Lu G.X., Zhu J., Fu D. MicroRNA-302a/d inhibits the self-renewal capability and cell cycle entry of liver cancer stem cells by targeting the E2F7/AKT axis. J. Exp. Clin. Cancer Res. 2018;37:252. doi: 10.1186/s13046-018-0927-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma Y.S., Wu Z.J., Zhang H.W., Cai B., Huang T., Long H.D., Xu H., Zhao Y.Z., Yin Y.Z., Xue S.B. Dual regulatory mechanisms of expression and mutation involving metabolism-related genes FDFT1 and UQCR5 during CLM. Mol. Ther. Oncolytics. 2019;14:172–178. doi: 10.1016/j.omto.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin F., Yang R., Wei Y., Wang D., Zhu Y., Wang X., Lu Y., Wang Y., Zen K., Li L. HIF-1α-induced miR-23a∼27a∼24 cluster promotes colorectal cancer progression via reprogramming metabolism. Cancer Lett. 2019;440-441:211–222. doi: 10.1016/j.canlet.2018.10.025. [DOI] [PubMed] [Google Scholar]
- 66.Simon T., Pinioti S., Schellenberger P., Rajeeve V., Wendler F., Cutillas P.R., King A., Stebbing J., Giamas G. Shedding of bevacizumab in tumour cells-derived extracellular vesicles as a new therapeutic escape mechanism in glioblastoma. Mol. Cancer. 2018;17:132. doi: 10.1186/s12943-018-0878-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.