Abstract
Accumulating evidence has demonstrated that circular RNAs (circRNAs) play important roles in regulating gene expression involved in tumor development. However, the role of circRNAs in modulating the radiosensitivity of oral squamous cell carcinoma (OSCC) and its potential mechanisms have not been documented. We performed high-throughput RNA sequencing (RNA-seq) to investigate the circRNA expression profile in OSCC patients and discovered that the circATRNL1 expression was significantly downregulated and closely related to tumor progression. The circATRNL1 was structurally validated via Sanger sequencing, RNase R treatment, and specific convergent and divergent primer amplification. Importantly, the expression levels of circATRNL1 decreased after irradiation treatment, and upregulation of circATRNL1 enhanced the radiosensitivity of OSCC through suppressing proliferation and the colony survival fraction, inducing apoptosis and cell-cycle arrest. Moreover, we observed that circATRNL1 could directly bind to microRNA-23a-3p (miR-23a-3p) and relieve inhibition for the target gene PTEN. In addition, the tumor radiosensitivity-promoting effect of circATRNL1 overexpression was blocked by miR-23a-3p in OSCC. Further experiments also showed that PTEN can reverse the inhibitory effect of OSCC radiosensitivity triggered by miR-23a-3p. We concluded that circANTRL1 may function as the sponge of miR-23a-3p to promote PTEN expression and eventually contributes to OSCC radiosensitivity enhancement. This study indicates that circANTRL1 may be a novel therapeutic target to improve the efficiency of radiotherapy in OSCC.
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common cancer in head and neck squamous cell carcinoma and is characterized by highly metastatic and invasive malignancy in the oral cavity, accounting for more than 300,000 newly diagnosed cancer cases annually worldwide.1,2 Despite advances in research and therapy, the 5-year survival rate has shown little improvement in recent decades.3 Radiotherapy is the primary nonsurgical approach for OSCC patients; however, the outcomes remain unsatisfactory due to tumor radioresistance.4,5 Further, the specific molecules underlying radioresistance in OSCC have been poorly elucidated. Therefore, it is urgent for us to clarify the molecular mechanisms of OSCC radioresistance and to provide novel therapeutic targets for OSCC patients.
Circular RNA (circRNA), another class of non-coding RNAs (ncRNAs), is a closed-loop structure with back-splicing without 3′ and 5′ ends, which differs from the typical linear RNAs which have 5′ caps and 3′ tails.6 Compared with their linear counterparts, circRNAs are extensively expressed and are generally stable and conserved in eukaryotic cells.7 It has been well-established that circRNAs may play a significant role in physiology and pathological processes and also regulate multiple diseases.8,9 Lately, increasing evidence has demonstrated that circRNAs were generally dysregulated in various cancers and involved in cancer progression, implying that circRNAs may be a new kind of potential biomarker for cancers.10, 11, 12 Moreover, recent studies have demonstrated that circRNAs could serve as competing endogenous RNA (ceRNA) by competitive binding to microRNA (miRNA) response elements (MREs) to regulate gene transcription.13 Moreover, certain kinds of circRNAs have been confirmed by function as a ceRNA mechanism in breast cancer, bladder cancer, and ovarian cancer.14, 15, 16 However, there are currently no reports describing the role of circRNAs and their potential mechanisms in modulating the radiosensitivity of OSCC.
In this study, we analyzed the expression profiles of circRNAs in OSCC tissues and identified a circRNA derived from ATRNL1, termed “circATRNL1,” which was significantly downregulated and positively correlated with OSCC progression. More importantly, we found that circATRNL1 overexpression may act as a ceRNA for miR-23a-3p to regulate phosphatase and the tensin homolog deleted on chromosome ten (PTEN) expression and consequently improved tumor radiosensitivity in OSCC. Our findings established a strong connection between circRNAs and OSCC radiosensitivity and revealed that circATRNL1 may serve as a highly attractive target to radiosensitize OSCC.
Results
Dysregulated circRNAs and Decreased circATRNL1 in OSCCs
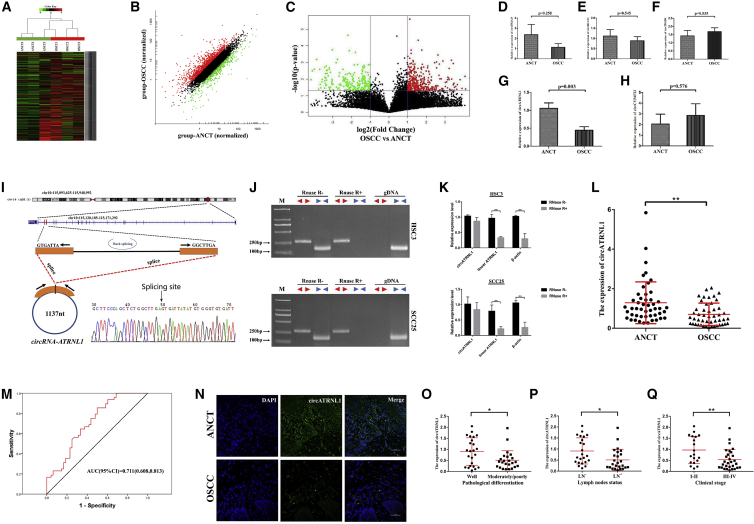
To investigate the role of circRNAs in OSCC tissues, we collected three pairs of OSCC tissues and matched adjacent non-cancerous tissue (ANCT) and screened them for dysregulation using circRNA high-throughput sequencing analysis. The expression profiles of these circRNA transcripts demonstrated that a series of circRNAs was aberrantly expressed in OSCC and ANCT (Figure 1A). The scatterplots present the variations of circRNA expression between OSCC and ANCT specimens (Figure 1B). In total, 474 differentially expressed circRNAs with fold change >2.0 and p <0.05 were identified, among which 267 were upregulated and 207 downregulated (Figure 1C). Through expression intensity sorting, the cluster heatmap demonstrated the five significantly increased and decreased circRNAs in OSCC tissues compared with ANCTs (Figure S1). To screen the key differentially expressed circRNA, we selected the expression of five mostly changed circRNAs from another nine patients to validate their expression. Among them, the expression of circATRNL1 was consistently and significantly decreased in OSCC tissues compared with matched controls (Figures 1D–1H). circATRNL1 is spliced from the ATRNL1 gene on chr10:115120185–115171292. Subsequently, we determined the head-to-tail splicing of circATRNL1 in the RT-PCR product of circATRNL1 by Sanger sequencing and also confirmed its sequence and genomic size (Figure 1I). However, to rule out the possibility that such head-to-tail splicing products could be the result of trans-splicing and genomic rearrangements, we specially designed divergent and convergent primers and discovered that circATRNL1 could be detected only in cDNA even under RNase R treatment, since no products were detected in the genomic DNA (gDNA) in OSCC cells (Figure 1J). Additionally, to confirm the stability of circATRNL1, we performed a quantitative real-time PCR assay, which indicated that circATRNL1 can resist RNase R, while linear-ATRNL1 and β-actin can be degraded by RNase R (Figure 1K). Furthermore, circATRNL1 expression was then confirmed in another 48 patients, and the results of the quantitative real-time PCR assay showed that its level was significantly decreased in OSCC tissues (Figure 1L). ROC curves were highly discriminative between OSCC patients and ANCT, with an area-under-the-ROC-curve (AUC) of 0.711 (95% confidence interval [CI], 0.608–0.813) (Figure 1M). Fluorescence in situ hybridization (FISH) analysis further indicated that circATRNL1 was localized in the cytoplasm, and its expression was also meaningfully downregulated in OSCC tissues compared with ANCTs (Figure 1N). Moreover, to identify the relationship between circATRNL1 expression and OSCC development, we conducted a correlation analysis which showed that circATRNL1 expression was closely associated with clinicopathology, including pathological differentiation, lymph node metastasis, and clinical stage (Figures 1O–1Q; Table 1). Thus, we focused on the expression and the role of circATRNL1 in OSCC.
Figure 1.
circRNA Expression Profile and Decreased circATRNL1 Expression in OSCCs
(A) Heatmap analyses of RNA-seq showing differentially expressed circRNAs with fold changes >2.0. Each column represents a tissue specimen, and each row represents the expression level of a circRNA. The intensity increased from low to high (green to red). (B) Scatterplot of circRNAs in OSCC and ANCT samples. The circRNAs above the top red line and below the green line indicated more than 2-fold changes of up- and downregulation. (C) Volcano plot visualizes the dysregulated circRNAs between OSCC and ANCT groups. The vertical purple lines correspond to 2.0-fold up and down, respectively, and the horizontal purple line represents a p value of 0.05. The red and green points separately represent significantly up- and downregulated circRNAs in OSCC. (D–H) Quantitative real-time PCR was used to examine the expression of five mostly changed circRNAs in OSCC and matched ANCT from nine patients. These five circRNAs included the two mostly increased circRNAs (circITGAD, D; circBCAT1, E) and the three mostly decreased circRNAs (circTHSD4, F; circATRNL1, G; and circCYP4F22, H). (I) Schematic illustration indicates the formation of circATRNL1. The head-to-tail splicing of circATRNL1 in the circATRNL1 RT-PCR products was validated by Sanger sequencing. (J) The existence of circATRNL1 in the presence or absence of RNase R supplementation in cDNA and gDNA from OSCC cells was detected by RT-PCR, and divergent primers could produce circRNAs in cDNA but not in genomic DNA (gDNA). (K) The circATRNL1, rather than linear ATRNL1 or β-actin, could resist digestion by RNase R followed by quantitative real-time PCR assay. (L) The differential expression of circATRNL1 was evaluated by quantitative real-time PCR in 48 pairs of OSCC and matched ANCT. (M) ROC curve analysis revealed that circATRNL1 was capable of distinguishing OSCC patients from healthy controls. (N) The expression of circATRNL1 in OSCC and ANCT specimens was evaluated for detection by FISH. Scale bar, 50 μm. (O–Q) Vertical scatterplots show relative levels of circATRNL1 in OSCC with different pathological differentiation (O), lymph node metastasis (P), and clinical stage (Q). Values are presented as mean ± SD. *p < 0.05; **p < 0.01.
Table 1.
Association between circATRNL1 Expression and Clinicopathological Features in Patients with OSCC (n = 48)
| Clinicopathologic Features | Number of Cases | circATRNL1 Expression |
p Value∗ | |
|---|---|---|---|---|
| Higha (n) | Low (n) | |||
| Gender | ||||
| Male | 33 | 18 | 15 | 0.350 |
| Female | 15 | 6 | 9 | |
| Age | ||||
| ≥55 | 18 | 8 | 10 | 0.551 |
| <55 | 30 | 16 | 14 | |
| Differentiation | ||||
| Well | 25 | 16 | 9 | 0.043 |
| Moderate+ poor | 23 | 8 | 15 | |
| T Stage | ||||
| TI-II | 32 | 19 | 13 | 0.066 |
| TIII-IV | 16 | 5 | 11 | |
| Clinical Stage | ||||
| I-II | 19 | 13 | 6 | 0.039 |
| III-IV | 29 | 11 | 18 | |
| LN Metastasis | ||||
| N- | 23 | 15 | 8 | 0.043 |
| N+ | 25 | 9 | 16 | |
The median expression level was used as the cutoff.
p < 0.05
circATRNL1 Overexpression Improves the Radiosensitivity of OSCC Cells
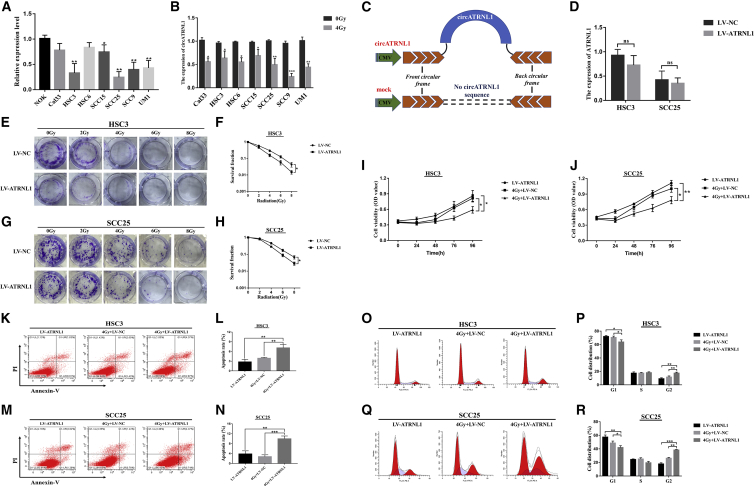
The endogenous expression levels of circATRNL1 in a panel of normal oral keratinocyte (NOK) cells and seven oral cancer cell lines were further examined. Quantitative real-time PCR analysis showed that circATRNL1 expression was decreased in seven OSCC cell lines compared with that of NOK cells, and HSC3 and SCC25 cells were selected for overexpression of circATRNL1 because of their lower circATRNL1 expression (Figure 2A). To evaluate the role of circATRNL1 in radiosensitivity in OSCC cells, we treated seven OSCC cell lines with 4Gy, and the results showed that the expression levels of circATRNL1 were significantly decreased after the 24-h incubation after irradiation treatment (Figure 2B). Next, we constructed circATRNL1 overexpression with stable transfection (lentiviral vector [LV]-circATRNL1) and found that its expression could be significantly upregulated compared with control groups (LV-NC) in HSC3 and SCC25 cells by quantitative real-time PCR (Figure 2C; Figure S2). Moreover, to evaluate whether any linear ATRNL1 could be produced during circATRNL1 overexpression, we performed a quantitative real-time PCR assay, and the results showed that there was no significant difference in linear ATRNL1 between LV-NC and LV-circATRNL1 in OSCC cells (Figure 2D).
Figure 2.
Effect of circATRNL1 Overexpression on Radiosensitivity of OSCC Cells
(A) Relative expression levels of circATRNL1 in a panel of seven OSCC cell lines and NOK. (B) Expression of circATRNL1 in seven OSCC cell lines was detected by quantitative real-time PCR before and after 4 Gy irradiation. (C) The illustration of vector for overexpression of circATRNL1 (pCD-circATRNL1) was presented. (D) The linear ATRNL1 expression was detected by quantitative real-time PCR between LV-NC and LV-ATRNL1 groups in OSCC cells. (E–H) The colony survival rate was determined by colony-formation assay in HSC3 (E and F) and SCC25 (G and H) cells treated with different doses of irradiation. (I and J) MTT assay for cell growth vitality was evaluated in HSC3 (I) and SCC25 (J) cells transfected with LV-NC or LV-ATRNL1 under 4 Gy irradiation or non-irradiated. (K–N) Apoptosis was determined by flow cytometry after transfection in OSCC cells. 4Gy+LV-ATRNL1 groups significantly increased the apoptosis rates compared with the 4Gy+LV-NC groups and LV-ATRNL1 groups in HSC3 (K and L) and SCC25 cells (M and N). (O–R) Calculated cell-cycle distribution and representative charts in circATRNL1 overexpression and the control groups under 4 Gy irradiation or non-irradiated. It demonstrated that cells at the G2 phase were dramatically increased in 4Gy+LV-ATRNL1 after irradiation in HSC3 (O and P) and SCC25 cells (Q and R). Data are presented as mean ± SD of three experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
The colony-formation assay was used to explore the influence of circATRNL1 on radiosensitivity in OSCC cells. The results showed that circATRNL1 overexpression caused a significant dose-dependent radiosensitization, and the dose-survival curves were acquired in OSCC cells (Figures 2E–2H). Next, after cells were exposed to 4 Gy irradiation treatment, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) results revealed that a combination of circATRNL1 overexpression and irradiation led to inhibited proliferation of OSCC cells compared with either circATRNL1 overexpression or radiation therapy alone (Figures 2I and 2J). Furthermore, the results of the apoptosis assay indicated that 4 Gy+LV-ATRNL1 groups dramatically increased the apoptosis rates of OSCC cells compared with the 4 Gy+LV-NC groups and LV-ATRNL1 groups (Figures 2K–2N). Cell-cycle analysis also demonstrated that cells at the G2 phase were significantly increased in 4 Gy+LV-ATRNL1, suggesting that the cycle arrested the G2 phase after irradiation (Figures 2O–2R). These results demonstrated that circATRNL1 overexpression enhanced OSCC cell radiosensitivity by suppressing cell survival and inducing apoptosis.
circATRNL1 Directly Binds to miR-23a-3p and Regulates Its Expression
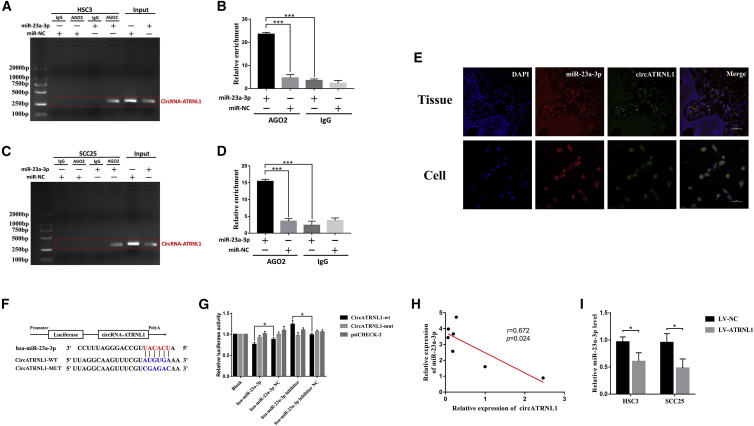
circRNAs can function mainly as miRNA sponges and suppress gene activity. The circATRNL1/miRNA interaction network was predicted by TargetScan and MiRanda, and the results showed that six miRNAs were predicted and listed as the potential targets of circATRNL1 (Table S2). To identify the functional miRNAs that may interact with circATRNL1 in OSCC cells, we used the quantitative real-time PCR technique and found that miR-23a-3p was significantly negatively correlated with circATRNL1 expression, whereas the other miRNAs did not change (Figure S3). Next, anti-AGO2 RNA immunoprecipitation (RIP) was conducted in HSC3 and SCC25 cells transfected with miR-23a-3p mimics or miR-NC to identify circATRNL1. We found significant enrichment of circATRNL1 and miR-23a-3p as compared with the controls (Figures 3A–3D). Furthermore, FISH analysis not only demonstrated that circATRNL1 and miR-23a-3p were colocalized in OSCC tissues and cells (Figure 3E), but also confirmed the direct interaction between circATRNL1 and miR-23a-3p in OSCC cells. Luciferase reporter results showed that the luciferase intensity of wild-type circATRNL1 sequence was decreased or increased by transfection with miR-23a-3p mimics or inhibitors but was restored by miR-NC or miR-inhibitor NC respectively, suggesting that miR-23a-3p can directly target the circATRNL1 (Figures 3F and 3G). Moreover, we measured the basic expression levels between circATRNL1 and miR-23a-3p in OSCC cell lines using quantitative real-time PCR analysis, and a negative correlation between the levels of circATRNL1 and miR-23a-3p was demonstrated in seven cell lines (Figure 3H). More importantly, miR-23a-3p was significantly downregulated by circATRNL1 overexpression in OSCC cells (Figure 3I). Analysis of these data implied that circATRNL1 may serve as a ceRNA for the miR-23a-3p in OSCC cells.
Figure 3.
circANTRL1 Acts as a Sponge for miR-23a-3p in OSCC Cells
(A–D) Anti-AGO2 RIP was measured in OSCC cells transfected with miR-23a-3p mimics or miR-23a-3p NC followed by RT-PCR. It found significant enrichment of circATRNL1 and miR-23a-3p as compared with the controls, and the relative enrichment of circATRNL1 was determined by quantitative real-time PCR in HSC3 (A and B) and SCC25 cells (C and D). (E) miR-23a-3p colocalized with circATRNL1 in OSCC tissues and cells was detected by FISH. (F and G) Schematic model of circATRNL1 wild-type (WT) and mutant (MUT) luciferase reporter vectors (F). Luciferase intensity was confirmed in 293T cells transfected with circATRNL1 (wild-type/mutant), miR-23a-3p (mimics/NC/inhibitor/inhibitor NC), and luciferase reporter vectors psiCHECK-2 (G). (H) Relationship between circATRNL1 expression and miR-23a-3p expression levels in different OSCC cell lines was analyzed by Spearman correlation. (I) The expression of miR-23a-3p was evaluated by quantitative real-time PCR in OSCC cells transfected with LV-circATRNL1 or mock vector, respectively. Values are shown as mean ± SD of triplicate experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
circATRNL1 Enhances OSCC Cell Radiosensitivity by Sponging miR-23a-3p
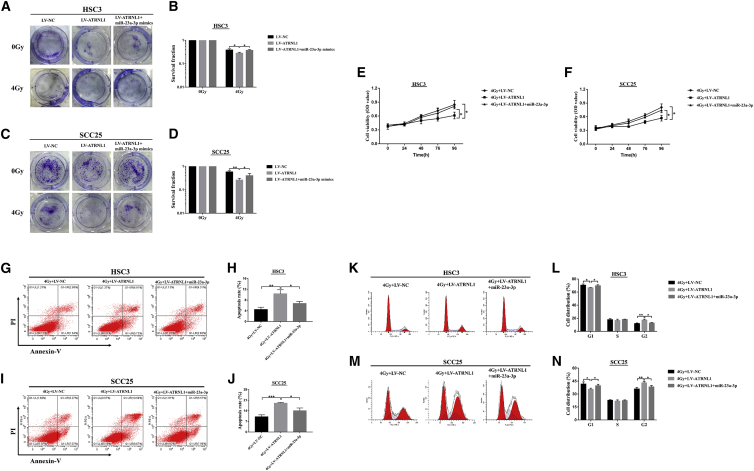
Considering that circATRNL1 could suppress the expression of miR-23a-3p, we further investigated whether circATRNL1 exerted its function through modulating miR-23a-3p expression. HSC3 and SCC25 cells, transfected with LV-NC or LV-circATRNL1 or co-transfected with LV-circATRNL1 and miR-23a-3p mimics, were exposed to 4 Gy irradiation. Clonogenic results revealed that circATRNL1 overexpression repressed colony-formation rates of OSCC cells, while the inhibiting effects of circATRNL1 were apparently rescued by miR-23a-3p mimics (Figures 4A–4D). Additionally, we examined the effect of miR-23a-3p only on colony-forming ability under 4 Gy treatment and found that mR-23a-3p mimics could improve colony-forming ability compared with that of the miR-23a-3p NC groups (Figure S4). Moreover, MTT results indicated that upregulation of circATRNL1 resulted in a significant cell proliferation decrease, while miR-23a-3p mimics completely reversed this effect in OSCC cells (Figures 4E and 4F). Furthermore, apoptosis assay and cell-cycle analysis detected by flow cytometry demonstrated that miR-23a-3p mimics restored the apoptosis and cell-cycle arrest induced by circATRNL1 overexpression in OSCC cells (Figures 4G–4N). Taken together, these results suggest that upregulation of circATRNL1 enhanced the radiosensitivity of OSCC cells by negatively regulating miR-23a-3p expression.
Figure 4.
circATRNL1 Enhanced Radiosensitivity through Modulating miR-23a-3p in OSCC Cells
(A–D) The clonogenic survival ability of transfected HSC3 (A and B) and SCC25 (C and D) cells was calculated by colony-formation assay after 4 Gy irradiation. (E and F) MTT assay was performed to determine the cell viability in transfected HSC3 (E) and SCC25 (F) cells under 4 Gy irradiation. (G–J) Flow cytometry analysis was carried out to measure the apoptosis and found that 4Gy+LV-ATRNL1 + miR-23a-3p groups significantly decreased the apoptosis rates compared with the 4G + LV-ATRNL1 groups in HSC3 (G and H) and SCC25 cells (I and J). (K–N) The analysis of cell-cycle distribution showed that cells at the G2 phase were dramatically decreased in 4Gy+LV-ATRNL1 + miR-23a-3p after irradiation in HSC3 (K and L) and SCC25 cells (M and N). All results are given as mean ± SD obtained from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
circATRNL1 Regulates PTEN Expression and Inactivates the AKT Signaling Cascade through Sequestering miR-23a-3p
To further identify the potential mechanism of circATRNL1 involved in OSCC cell radiosensitivity, we found that both circATRNL1 and eight mRNAs (FKBP5, MAP3K3, PHLPP1, RAP1A, PTEN, SMAD3, PHLPP2, and TGFBR3) contain mutual MREs to miR-23a-3p according to TargetScan and the MiRanda program (Figure S5). Upregulation of circATRNL1 could significantly increase PTEN mRNA expression but not that of the other mRNAs (Figure S6), suggesting that circATRNL1 and PTEN may be interrelated in OSCC cells. We hypothesized that circATRNL1 exerts its biological function as a miR-23a-3p sponge to regulate PTEN expression. For confirmation of whether PTEN is a direct target of miR-23a-3p, the wild-type 3′ UTR sequence and the mutant 3′ UTR sequence of PTEN were transfected into 293T cells with miR-23a-3p mimics or miR-23a-3p NC in a luciferase reporter assay. The results revealed that miR-23a-3p mimics reduced the luciferase activity of PTEN-WT compared with the control group, but no influence was observed on the luciferase activity of PTEN-MUT in OSCC cells (Figures 5A and 5B). Furthermore, using western blot, we found that circATRNL1 overexpression significantly increased the levels of PTEN in OSCC cells (Figures 5C and 5D). In contrast, miR-23a-3p mimics obviously reduced the PTEN mRNA and protein expression in OSCC cells, whereas this effect was abolished by circATRNL1 overexpression (Figures 5E–5H). In addition, to examine whether the Akt signal pathway was involved in the process, we performed western blot analysis and demonstrated that circATRNL1 overexpression meaningfully decreased phosphorylation levels of AKT (Figure S7). Furthermore, transfection of miR-23a-3p mimics significantly enhanced the p-AKT molecules, while co-overexpression of miR-23a-3p and circATRNL1 abrogated these effects in OSCC cells (Figure S8). Analysis of these data, taken together, implied that circATRNL1 could function as a ceRNA for miR-23a-3p to positively regulate PTEN expression and the inactive AKT signaling pathway.
Figure 5.
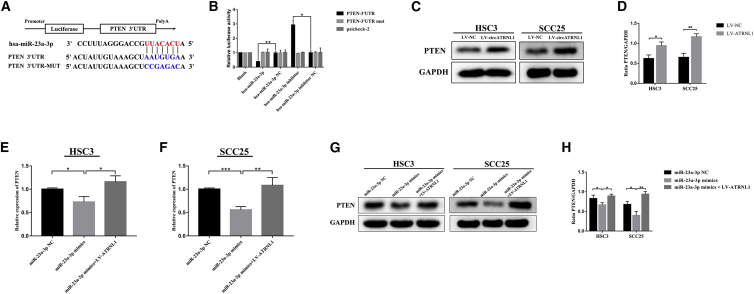
circATRNL1 Regulates PTEN Expression through Serving as a Sponge for miR-23a-3p
(A and B) Schematic of PTEN 3′ UTR wild-type (WT) and mutant (MUT) luciferase reporter vector (A). The relative luciferase activities were evaluated in 293T cells co-transfected with miR-23a-3p or miR-23a-3p NC, miR-23a-3p inhibitor, or miR-23a-3p inhibitor NC and luciferase reporter vectors psiCHECK2 (B). (C and D) The PTEN protein expression levels were revealed by western blot after circATRNL1 overexpression (C). Quantitative data were estimated as a ratio to the GAPDH levels in OSCC cells (D). (E–H) The effects of miR-23a-3p mimics or co-transfected with LV-ATRNL1 on the expression of PTEN were detected by quantitative real-time PCR (E and F) and western blot (G and H). All values are presented as mean ± SD, n = 3. *p < 0.05; **p < 0.01; ***p < 0.001.
Inhibition of miR-23a-3p Promotes Radiosensitivity of OSCC Cells by Increasing PTEN
For further confirmation of the effect of interaction between miR-23a-3p and PTEN on OSCC radiosensitivity, rescue experiments were performed in OSCC cells transfected with miR-23a-3p inhibitor NC (inh-NC), miR-23a-3p inhibitor (inh-23a-3p), or combined with the si-PTEN+miR-23a-3p inhibitor under 4 Gy irradiation. Colony-formation results demonstrated that the miR-23a-3p inhibitor led to an inhibition of cell-survival fractions compared with those in inh-NC groups, which was reversed by PTEN knockdown (Figures 6A–6D). Subsequently, MTT assay showed that downregulation of miR-23a-3p inhibited the proliferation of OSCC cells, but co-transfection with si-PTEN and miR-23a-3p inhibitor could abrogate this effect (Figures 6E and 6F). Furthermore, flow cytometry analysis illustrated that apoptosis and cell-cycle arrest were induced by the inhibition of miR-23a-3p compared with the vector transfection group; however, PTEN knockdown exhibited a restorative role in OSCC cells (Figures 6G–6N). These results suggest that miR-23a-3p could attenuate OSCC radiosensitivity through targeting PTEN.
Figure 6.
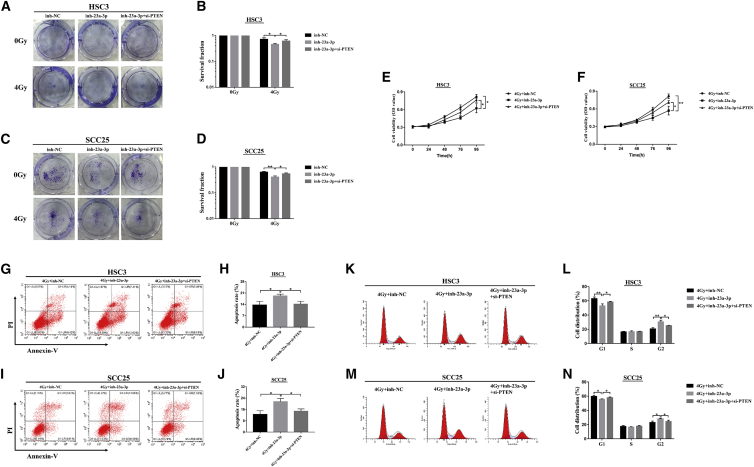
miR-23a-3p Downregulation Enhanced Radiosensitivity through Targeting PTEN in OSCC Cells
(A–D) The colony-formation assay detected the colony survival rate in HSC3 (A and B) and SCC25 (C and D) cells transfected with inh-23a-3p or simultaneous inh-23a-3p+si-PTEN under 4 Gy irradiation. (E and F) The growth curves of cells were analyzed after transfection with the indicated vectors by MTT assay combined with 4 Gy irradiation in HSC3 (E) and SCC25 cells (F). (G–N) The percentages of apoptotic cells (G–J) and cell-cycle distribution (K–N) were measured by flow cytometry in OSCC cells transfected with miR-23a-3p inhibitor alone or co-transfected with the si-PTEN after 4 Gy irradiation. 4Gy+inh-23a-3p+si-PTEN groups significantly decreased the apoptosis rates and the G2 phase compared with the 4Gy+inh-23a-3p groups in HSC3 (G and H; K and L) and SCC25 cells (I and J; M and N). Data are shown as mean ± SD, n = 3. *p < 0.05; **p < 0.01.
PTEN Activation Is Essential for circATRNL1-Mediated Enhancement of OSCC Radiosensitivity
The results above indicate that the overexpression of circATRNL1 increases the radiosensitivity of OSCC by inhibiting miR-23a-3p and downregulation of PTEN by miR-23a-3p decreases OSCC radiosensitivity. We then decided to identify the functional role of PTEN in circATRNL1-mediated OSCC radiosensitization. As shown in Figures 7A–7D, downregulation of PTEN can significantly increase colony-forming ability compared with the si-NC groups, whereas this effect can be markedly restored by upregulation of circATRNL1 under 4 Gy irradiation. The proliferation of HSC3 and SCC25 cells was increased by PTEN inhibition, while the addition of circATRNL1 overexpression partially suppressed the cell viability upon 4 Gy exposure (Figures 7E and 7F). Flow cytometry of apoptosis and the cell cycle also confirmed that PTEN knockdown inhibited apoptosis and decreased cell-cycle arrest at the G2 stage, while circATRNL1 overexpression induced apoptosis and promoted cell-cycle arrest progression followed by irradiation exposure (Figures 7G–7N). Analysis of these data confirmed that PTEN regulation by circATRNL1 was crucial for the radiosensitization of OSCC.
Figure 7.
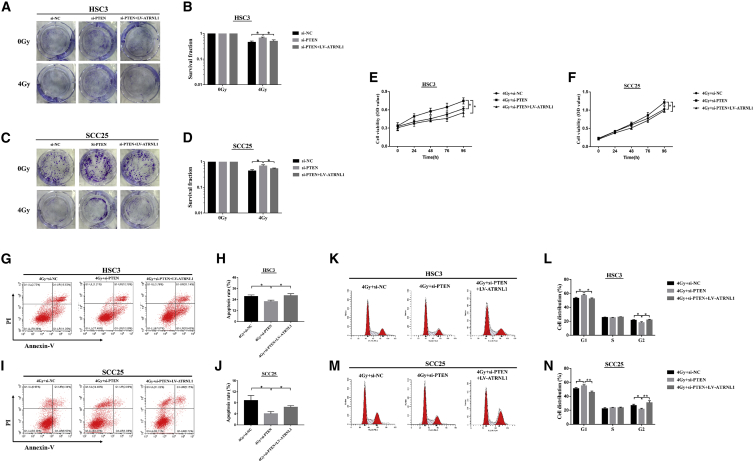
PTEN Knockdown Decreased the Radiosensitivity of OSCC Cells and Restored It by circATRNL1 Overexpression
(A–D) The clonogenic survival rates of transfected OSCC cells were determined by colony-formation assay after 4 Gy irradiation in HSC3 (A and B) and SCC25 (C and D) cells. (E and F) MTT assay was conducted to measure the cell viability in transfected HSC3 (E) and SCC25 (F) cells when exposed to 4 Gy irradiation. (G–J) Flow cytometry of apoptosis showed that PTEN knockdown inhibited apoptosis and circATRNL1 overexpression exhibited a restorative role in HSC3 (G and H) and SCC25 cells (I and J) followed by irradiation exposure. (K–N) The analysis of cell-cycle distribution showed that cells at the G2 phase were significantly increased in 4Gy+si-PTEN+LV-ATRNL1 after irradiation in HSC3 (K and L) and SCC25 cells (M and N). All results are given as mean ± SD, n = 3. *p < 0.05; **p < 0.01.
Discussion
Radiotherapy is a major modality in tumor combinational treatment and is widely used for the treatment of inoperable cancer, including OSCC at advanced stages.17,18 Nevertheless, resistance to irradiation remains an obstacle, resulting in relapse and treatment failure for patients with OSCC.19 Recent studies have reported that some ncRNAs may emerge as key regulators in OSCC radiosensitivity.20,21 However, no research has reported whether circRNAs play a role in OSCC radiosensitivity. This is the report defining the regulatory function role of circRNAs in the radiosensitivity of OSCC.
Since the first circRNA was observed by Hsu and Coca-Prados in eukaryotic cells, increasingly aberrant expression of circRNAs in cancer development has attracted much attention because of their potential biological functions.22,23 Considering that circRNAs have high stability and a specific loop structure, they may be regarded as promising potential biomarkers for cancer prognosis and diagnosis.24,25 Zhang et al. indicated that circLARP4 expression was downregulated and represented an independent prognostic biomarker in patients with gastric cancer.26 Meanwhile, Chen et al.27 showed that dysregulated circRNAs might participate in cell responses to irradiation and that downregulation of hsa_circ_0071410 resulted in the attenuation of irradiation-induced hepatic stellate cell activation. Wang et al.28 also found that inhibition of hsa_circ_0001313 could induce radiosensitivity of colon cancer by negatively regulating miR-338-3p. However, the effects of circRNAs in response to OSCC irradiation have not yet been reported. In this study, we analyzed the aberrantly expressed circRNAs between OSCC and ANCTs through the RNA-sequencing (RNA-seq) technique and focused on a novel circular RNA, termed “circATRNL1,” which formed a ring structure by connecting the 3′ and 5′ splice sites, even under treatment with RNase R. We also confirmed its splice junction, genomic size, and sequences followed by Sanger sequencing. Moreover, the expression levels of circATRNL1 were significantly downregulated and closely related to clinicopathology in OSCC. We obtained an efficient risk score (AUC) of circATRNL1 (0.711) in distinguishing OSCC and ANCT. Although the exact applicable value still needs to be validated in large-scale studies, the ROC curve of circATRNL1 expression is part of the diagnostic prediction for OSCC patients. Interestingly, the levels of circATRNL1 were significantly downregulated after irradiation. We found that, functionally, circATRNL1 overexpression could inhibit cell proliferation and colony formation and induce apoptosis and cell-cycle arrest in OSCC cells under irradiation. Next, we further investigated the underlying mechanisms for circATRNL1 function in OSCC radiosensitivity.
In 2011, Pandolfi et al.29 presented a ceRNA hypothesis proposing that mRNA, long non-coding RNA (lncRNA), and pseudogene transcripts could be enriched with MREs and play a role in the miRNA sponge to remove the suppressive effect of miRNA on its target genes. In earlier studies, the ceRNA interaction among RNA transcripts was found largely in mRNAs.30 Further research, however, has confirmed that circRNAs were not simply byproducts of splicing errors, but rather were considered highly effective ceRNAs, forming the mechanism of the circRNA-miRNA-mRNA axis.31 For example, a circRNA identified as ciRS-7 was first reported to act as ceRNA, containing 70 binding sites for miR-7 in neuronal tissues.32 circHIPK3 can promote human cancer cell growth by sponging miR-124.33 Additionally, the circNRIP1/miR-149-5p/AKT1 regulatory axis was verified as promoting metastasis in gastric cancer.34 The circRAPGEF5/miR-198/FGFR1 pathway played a vital role in modulating the progression of papillary thyroid cancer.35 These findings suggested that numerous circRNAs can competitively bind with miRNAs and consequently interfere in target gene expression at the post-transcription level. It has been proposed that circular RNAs may function as sponges that reduce the number of freely available miRNA transcripts and lead to a decrease in miRNAs expression. For instance, decreasing CDR1as in cell lines resulted in downregulation of miR-7 target mRNAs, presumably because miR-7 was no longer sequestered.36 Furthermore, some studies found that some endogenous or viral transcripts can control miRNAs through target RNA-directed miRNA degradation (TDMD) mechanism and also result in miRNA degradation.37 However, whether circRNAs mediated miRNA degradation through TDMD remains largely unknown and needs to be clarified in the future. In our study, to identify the potential miRNA of circATRNL1, using TargetScan and MiRanda, we predicted a putative circATRNL1-miRNAs interaction network according to the complementary matching sequence. We screened and found that circATRNL1 functioned as an endogenous sponge for miR-23a-3p in OSCC. This prediction subsequently verified the true and definite interaction between circATRNL1 and miR-23a-3p by means of the luciferase reporter assay, the RIP approach, FISH analysis, and quantitative real-time PCR. miR-23a-3p, serving as a regulator involved in tumor progression, has been described in prostate cancer, breast cancer, and mucosal melanoma.38, 39, 40 miRNAs play a critical role in the modulation of radiosensitivity through interplay with the key irradiation-related genes in tumor cells.41 However, whether miR-23a-3p could affect cancer radiosensitivity has not been reported.
In our present studies, we discovered that miR-23a-3p not only promoted cell proliferation and inhibited apoptosis and cell-cycle G2 arrest, but it also attenuated the colony formation of OSCC cells in the presence of irradiation. The G2 arrest of the cell cycle is the most sensitive phase and can be activated to stop cell-cycle progression, allowing DNA double-strand breaks time to repair, which aids cancer cell survival.42,43 Moreover, miR-23a-3p rescued the function of circATRNL1 overexpression, further demonstrating that circATRNL1 enhanced OSCC radiosensitivity by sponge activity on miR-23a-3p. To explore in-depth the regulatory mechanisms within circATRNL1 and miR-23a-3p, we screened and analyzed eight mRNAs according to bioinformatics analysis prediction and discovered that circATRNL1 overexpression could positively improve PTEN activity. The luciferase reporter assay, quantitative real-time PCR, and western blot analysis confirmed that miR-23a-3p had definitive binding sites with PTEN and could negatively regulate its expression.
PTEN plays an essential role in apoptosis and cell-cycle arrest and functions as a tumor suppressor to inhibit tumor metastasis of various cancers.44,45 Although some researchers have reported that PTEN downregulated by miRNAs can decrease the radiosensitivity of human cancers,46,47 the functional role of the miR-23a-3p/PTEN axis in OSCC radiosensitivity requires further investigation. It has been demonstrated that circATRNL1 overexpression could positively improve PTEN activity. Functionally, PTEN knockdown could abolish the effects of circATRNL1 overexpression and miR-23a-3p suppression on cell proliferation and clone inhibition, apoptosis, and cell-cycle arrest in OSCC cells when exposed to irradiation, implying that PTEN participates in radiosensitivity regulation. ATM/ATR, which are the sensors of DNA damage, can mediate activation of the tumor suppressor p53 and lead to cell-cycle arrest in response to irradiation.48 Although PTEN has no direct effect on ATM/ATR activation, it can form a complex with p53 and that protects p53 from MDM-2-mediated degradation.49,50 However, the relationship between PTEN and ATM/ATR or p53 remains unclear in OSCC and needs to be further clarified in the later experiments. It is well-known that PTEN can dephosphorylate PI3K, generating phosphatidylinositol triphosphate (PIP3) and negatively regulating the Akt signaling pathway, which is closely associated with cancer radiation resistance.51,52 Blocking the activation of the PI3K/Akt signaling pathway can improve the sensitivity to radiation therapy in head and neck cancer.53 Therefore, we examined the expression levels of the Akt signaling pathway after circATRNL1 overexpression and confirmed that it was involved in the process. However, whether circATRNL1 influences the radiosensitivity of OSCC through mediating the Akt signal after irradiation is still unclear and requires further investigation. In summary, our findings revealed that circATRNL1 was significantly decreased in OSCC tissues and cell lines and closely correlated with OSCC progression. Moreover, upregulation of circATRNL1 contributed to the improved radiosensitivity of OSCC cells by reducing cell colony formation and cell growth and inducing apoptosis and cell-cycle arrest via sponging miR-23a-3p, which could halt the endogenous suppressive effect on target gene PTEN. Analysis of our data suggested that targeting the circATRNL1/miR-23a-3p/PTEN axis may be a novel therapeutic intervention for OSCC radioresistance.
Materials and Methods
Patients and Tissue Specimens
The tissue specimens of OSCC and matched adjacent non-cancerous tissue were randomly obtained from the Hospital of Stomatology, Sun Yat-Sen University, between April 2017 and April 2018. All tissues were collected with the consent of the patients, and the clinicopathology of the tumors was determined according to the TNM classification system of UICC. All samples were instantly frozen in liquid nitrogen after resection and stored at −80°C until use. This study was approved by the ethics committee of the Hospital of Stomatology of Sun Yat-Sen University.
Cell Culture
The human OSCC cell lines HSC3, HSC6, and CAL27 along with 293T cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, CA, USA). The UM1, SCC9, SCC15, and SCC25 cells were grown in DMEM-F12 (GIBCO) with the addition of 10% FBS and 0.4 μg /mL glucocorticoid. Human NOKs were cultured in keratinocyte serum-free medium (K–SFM) containing bovine pituitary extract and human recombinant epidermal growth factor (GIBCO). All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
circRNA Expression Profile Analysis
Total RNA was extracted from three pairs of OSCC and adjacent normal tissues by means of Trizol reagent (Invitrogen). Briefly, the linear RNA was digested by RNase R (Epicenter Technologies, Madison, WI, USA). Following purification, the poly(A)− or poly(A)+ RNA fractions were fragmented into small pieces under elevated temperatures. Then the cleaved RNA fragments were reverse-transcribed to create the final cDNA library following the protocol of the NEBNext Ultra Directional RNA Liberary Prep Kit (NEB, Beverly, MA, USA). Briefly, after the ribosome RNA depletion and RNA fragmentation, the second-strand cDNA synthesis was performed by adding a dUTP mix. Then the ends of the double-stranded cDNA fragments were repaired and the base A was added to the 3′ end of the DNA fragments. Purified cDNA was subjected to 12 cycles of PCR amplification, followed by library analysis using a Bioanalyser 2100 system (Agilent, Santa Clara, CA, USA). Then the cDNA was sequenced in a HiSeq 4000 (Illumina, San Diego, CA, USA) on a 150 bp paired end run. circRNAs indicating fold changes of ≥2, and p values of <0.05 were considered as significantly differentially expressed. Moreover, Sanger sequencing following all PCR products was performed for validation of the head-to-tail splicing of circATRNL1.
Irradiation
Cells were x-irradiated with Rs2000 equipment (Radsource, Brentwood, TN, USA) at a dose rate of 3 Gy/min and different doses of irradiation treatment (0, 2, 4, 6, and 8 Gy). We selected the optimized irradiation dose (4 Gy) based on the time-course irradiation experiment and then collected cells at the indicated times in each experiment.
Transfection, miRNA Mimic, and Inhibitor
Stable lentiviral transfection was used to increase circATRNL1 expression in HSC3 and SCC25 cells. Briefly, the LVs were constructed by GeneCopoeia and were packaged into pseudoviral particles. Lentivirus infection of cell lines was decided in the presence of 0.5 μg/mL puromycin (Sigma, St. Louis, MO, USA). The three different sequences of LV-RNA-circATRNL1 were tested to validate its expression by quantitative real-time PCR for optimal overexpression. RNA transient transfection was used to knock down PTEN expression with the siRNA transfection kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. miR-23a-3p mimic, inhibitor, and negative control were purchased from RiboBio and then transfected into OSCC cells with lipofectamine RNAimax regents (Thermo Fisher, Waltham, MA, USA) following the manufacturer’s recommendations. The cells were harvested at 24 h after transfection for further experiments.
FISH
FISH was carried out for evaluation of the expression level of circATRNL1 with a Cy5-labeled probe (5′-CCACATATAATCACTCAAGCCAGAGCTGGG-3′) and of miR-23a-3p with a Cy3-labeled probe (5′-GCGGAACTTAGCCACTGTGAA-3′), respectively. Briefly, samples were fixed on glass slides with 4% paraformaldehyde. Hybridization was performed at 37°C overnight with specific probes after dehydration. Then the slides were washed in formamide/2× SSC and were incubated with anti-DIG-HRP (PerkinElmer, Boston, MA, USA) at 4°C overnight, followed by the addition of TSA fluorescent signal reaction solution (PerkinElmer), then sealed with tablets containing DAPI. The images were acquired by fluorescence microscopy (Leica, Heidelberg, Germany).
RIP
After transfection with miR-23a-3p mimics or miR-NC, RIP assay was performed with a Magna RIP RNA binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. HSC3 and SCC25 cells at 85% confluence were harvested and subjected to lysis in complete RIP lysis buffer, after which the cell extracts were separately incubated with RIP immunoprecipitation buffer containing magnetic beads conjugated with human anti-Argonaute2 (AGO2) antibody (Cell Signaling Technology, Beverly, MA, USA) and negative control normal mouse immunoglobulin G (IgG) (Millipore). Samples were incubated with Proteinase K, and then the RNA immunoprecipitation fraction was purified. Finally, the isolated RNAs were detected by quantitative real-time PCR to demonstrate the presence of circATRNL1.
Luciferase Reporter Assay
Luciferase reporter assay was performed for the detection of direct binding between circATRNL1 and miR-23a-3p and between miR-23a-3p and PTEN. pmir-GLO dual-luciferase vectors (GenePharma, Shanghai, China) containing Renilla luciferase gene and firefly luciferase gene were applied in this experiment. Sequences of circATRNL1, miR-23a-3p, and PTEN were separately cloned into psiCHECK-2 vectors, and mutations were achieved in the binding sites. After 48 h of co-transfection, the luciferase activity was assessed by the dual-luciferase reporter assay (Promega, Madison, WI, USA). Relative firefly luciferase activity was normalized to the corresponding Renilla luciferase internal control.
Quantitative Real-Time PCR
Total RNA was extracted from the tissue and cells with Trizol reagent (Invitrogen), and RNA concentration was measured by Nanodrop (Thermo Fisher). In addition, all RNAs were incubated for 20 min at 37°C with or without RNase R (Epicenter Technologies). Reverse transcription to cDNA was then performed with the cDNA synthesis kit (TaKaRa, Tokyo, Japan) and cDNA amplification with the LightCycler 480 Real-Time System (Roche, Mannheim, Germany) with SYBR Green I Master Mix Reagent kit (Roche) according to the manufacturer’s protocol. To confirm the specificity of the circATRNL1 PCR products, we separated the PCR products amplified by divergent or convergent primers on an agarose gel. Samples were normalized to β-actin followed by its control, and finally the relative expression was calculated by the 2-ΔΔCt method. The sequences of primers are listed in Table S1.
Western Blot Analysis
Total proteins were extracted by means of the lysis buffer containing proteinase inhibitor (CWBIO, Beijing, China). After protein concentration was determined, equal amounts of protein were separated by SDS-PAGE gels and transferred into polyvinylidene fluoride (PVDF) membranes (Millipore). Following hybridization with specific primary antibodies at 4°C overnight, the membranes were probed with secondary antibodies at room temperature for 1 h. Finally, the immunoreactive bands were visualized by enhanced chemiluminescence (Millipore) according to the manufacturer’s instructions. The primary antibodies used were as follows: PTEN, p-AKT, AKT, and GAPDH (Cell Signaling, Danvers, MA, USA).
Colony-Formation Assay
The cells were seeded in six-well culture plates. After 24 h of incubation, adhesive cells were exposed to different doses of x-irradiation (0, 2, 4, 6, and 8 Gy) at a dose rate of 3 Gy/min. After an incubation period of 10 days, the cells were fixed with paraformaldehyde and stained with crystal violet. Colonies of at least 50 cells were counted as survivors by microscopy. Each experiment was performed three times independently, and each time there was a control for comparison.
Cell Proliferation Assay
Cell growth rate was determined by MTT assay (KeyGen Biotech, Nanjing, China). Briefly, cells were seeded in 96-well plates and incubated overnight. After transfection, cells were exposed to 4 Gy doses of x-irradiation followed by incubation for indicated times (0, 24, 48, 72, and 96 h). Subsequently, MTT reagent was added to each well for an additional 4 h of incubation, and then the DMSO was supplemented to dissolve the formazan crystals. The absorbance was measured at 450 nm by means of a micro-plate reader (Bio-Rad, Winooski, VT, USA). Experiments were repeated three times.
Flow Cytometry for Apoptosis and Cell-Cycle Analysis
In brief, cells were seeded in 6-well plates and treated with 4 Gy of irradiation. After 24 h of incubation, apoptotic cells were collected and examined by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) with Annexin V and the PI Apoptosis Detection Kit (Invitrogen). The apoptosis rates were calculated by the sum of Annexin V+/PI+ cells and Annexin V+ cells. For cell-cycle assays, cells were treated under the same conditions as described previously. The cells were then added to RNase A and stained with PI at room temperature. Subsequently, the cell cycle was analyzed by the same flow cytometry as above and with ModFit LT software (BD). Each experiment was conducted in triplicate.
Statistical Analyses
All statistical analyses were performed with SPSS 20.0 (Chicago, IL, USA). Fisher’s exact test was used to determine the correlation between circATRNL1 expression and the clinicopathological characteristics of OSCC patients. Two-group comparisons were assessed by Student’s t test, and multiple-group comparisons were analyzed by one-way analysis of variance (ANOVA). Person correlation analyses were conducted to confirm the correlation between circATRNL1 and miR-23a-3p expression. Data were presented as means ± SD, and a p value <0.05 was considered a statistically significant difference.
Author Contributions
D.Y. and W.Z. conceived the experiments and provide all financial support. G.C. and Y.L. performed the experiments. Y.H. and B.Z. analyzed and interpreted the data. C.Y. and C.W. wrote and revised the paper. X.Z. prepared the figures. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant numbers 81873711 and 81974146) and the Guangdong Science and Technology Project (grant number 2016A020216007).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.12.031.
Contributor Information
Wei Zhao, Email: zhaowei3@mail.sysu.edu.cn.
Dongsheng Yu, Email: yudsh@mail.sysu.edu.cn.
Supplemental Information
References
- 1.Chi A.C., Day T.A., Neville B.W. Oral cavity and oropharyngeal squamous cell carcinoma--an update. CA Cancer J. Clin. 2015;65:401–421. doi: 10.3322/caac.21293. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Rivera C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015;8:11884–11894. [PMC free article] [PubMed] [Google Scholar]
- 4.Mazeron R., Tao Y., Lusinchi A., Bourhis J. Current concepts of management in radiotherapy for head and neck squamous-cell cancer. Oral Oncol. 2009;45:402–408. doi: 10.1016/j.oraloncology.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Hu C., Eisbruch A. Organ-sparing radiation therapy for head and neck cancer. Nat. Rev. Clin. Oncol. 2011;8:639–648. doi: 10.1038/nrclinonc.2011.106. [DOI] [PubMed] [Google Scholar]
- 6.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rong D., Tang W., Li Z., Zhou J., Shi J., Wang H., Cao H. Novel insights into circular RNAs in clinical application of carcinomas. OncoTargets Ther. 2017;10:2183–2188. doi: 10.2147/OTT.S134403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li P., Chen S., Chen H., Mo X., Li T., Shao Y., Xiao B., Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Qin M., Liu G., Huo X., Tao X., Sun X., Ge Z., Yang J., Fan J., Liu L., Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 12.Gu C., Zhou N., Wang Z., Li G., Kou Y., Yu S., Feng Y., Chen L., Yang J., Tian F. circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol. Ther. Nucleic Acids. 2018;13:633–641. doi: 10.1016/j.omtn.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Yang R., Xing L., Zheng X., Sun Y., Wang X., Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol. Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Zhong Z., Huang M., Lv M., He Y., Duan C., Zhang L., Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q., Zhang J., He Y., Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol. Ther. Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joubert A., Vogin G., Devic C., Granzotto A., Viau M., Maalouf M., Thomas C., Colin C., Foray N. [Radiation biology: major advances and perspectives for radiotherapy] Cancer Radiother. 2011;15:348–354. doi: 10.1016/j.canrad.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Rajapakshe R.M., Pallegama R.W., Jayasooriya P.R., Siriwardena B.S., Attygalla A.M., Hewapathirana S., Weerasinghe J.U., Dias D.K., Tilakaratne W.M. A retrospective analysis to determine factors contributing to the survival of patients with oral squamous cell carcinoma. Cancer Epidemiol. 2015;39:360–366. doi: 10.1016/j.canep.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Schiegnitz E., Kämmerer P.W., Rode K., Schorn T., Brieger J., Al-Nawas B. Growth differentiation factor 15 as a radiation-induced marker in oral carcinoma increasing radiation resistance. J. Oral Pathol. Med. 2016;45:63–69. doi: 10.1111/jop.12323. [DOI] [PubMed] [Google Scholar]
- 20.Shiiba M., Shinozuka K., Saito K., Fushimi K., Kasamatsu A., Ogawara K., Uzawa K., Ito H., Takiguchi Y., Tanzawa H. MicroRNA-125b regulates proliferation and radioresistance of oral squamous cell carcinoma. Br. J. Cancer. 2013;108:1817–1821. doi: 10.1038/bjc.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng J.H., Yu C.C., Lee Y.C., Lin C.W., Chang W.W., Kuo Y.L. miR-494-3p induces cellular senescence and enhances radiosensitivity in human oral squamous carcinoma cells. Int. J. Mol. Sci. 2016;17:1092. doi: 10.3390/ijms17071092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene J., Baird A.M., Brady L., Lim M., Gray S.G., McDermott R., Finn S.P. Circular RNAs: biogenesis, function and role in human diseases. Front. Mol. Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 24.Wang K., Sun Y., Tao W., Fei X., Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao K.Y., Lin Y.C., Gupta S.K., Chang N., Yen L., Sun H.S., Tsai S.J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Liu H., Hou L., Wang G., Zhang R., Huang Y., Chen X., Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Yuan B., Wu Z., Dong Y., Zhang L., Zeng Z. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene. 2017;629:35–42. doi: 10.1016/j.gene.2017.07.078. [DOI] [PubMed] [Google Scholar]
- 28.Wang L., Peng X., Lu X., Wei Q., Chen M., Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: Effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol. Res. Pract. 2019;215:689–696. doi: 10.1016/j.prp.2018.12.032. [DOI] [PubMed] [Google Scholar]
- 29.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tay Y., Kats L., Salmena L., Weiss D., Tan S.M., Ala U., Karreth F., Poliseno L., Provero P., Di Cunto F. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu S., Zhong Y., Shang R., Zhang X., Song W., Kjems J., Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X., Wang S., Wang H., Cao J., Huang X., Chen Z., Xu P., Sun G., Xu J., Lv J., Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W., Zhao J., Jin M., Zhou M. circRAPGEF5 contributes to papillary thyroid proliferation and metastatis by regulation miR-198/FGFR1. Mol. Ther. Nucleic Acids. 2019;14:609–616. doi: 10.1016/j.omtn.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 37.Kato M. Target RNA-directed microRNA degradation; which controls which? Non-coding RNA Investig. 2018;2:1243–1255, E7. doi: 10.21037/ncri.2018.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen Y.C., Lee W.J., Tan P., Yang S.F., Hsiao M., Lee L.M., Chien M.H. By inhibiting snail signaling and miR-23a-3p, osthole suppresses the EMT-mediated metastatic ability in prostate cancer. Oncotarget. 2015;6:21120–21136. doi: 10.18632/oncotarget.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarmo E.L., Havunen R., Häyrynen S., Penkki S., Ketolainen J., Nykter M., Kallioniemi A. Bone morphogenetic protein 4 regulates microRNA expression in breast cancer cell lines in diverse fashion. Genes Chromosomes Cancer. 2016;55:227–236. doi: 10.1002/gcc.22324. [DOI] [PubMed] [Google Scholar]
- 40.Ma M., Dai J., Tang H., Xu T., Yu S., Si L., Cui C., Sheng X., Chi Z., Mao L. MicroRNA-23a-3p inhibits mucosal melanoma growth and progression through targeting adenylate cyclase 1 and attenuating cAMP and MAPK pathways. Theranostics. 2019;9:945–960. doi: 10.7150/thno.30516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metheetrairut C., Slack F.J. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr. Opin. Genet. Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang A., Muschel R.J. Radiation and the G2 phase of the cell cycle. Radiat. Res. 1998;150(5, Suppl):S52–S59. [PubMed] [Google Scholar]
- 43.Sancar A., Lindsey-Boltz L.A., Unsal-Kaçmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 44.Lee Y.R., Chen M., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 2018;19:547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 45.Hollander M.C., Blumenthal G.M., Dennis P.A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qu C., Liang Z., Huang J., Zhao R., Su C., Wang S., Wang X., Zhang R., Lee M.H., Yang H. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Zheng L., Ding Y., Li Q., Wang R., Liu T., Sun Q., Yang H., Peng S., Wang W., Chen L. MiR-20a induces cell radioresistance by activating the PTEN/PI3K/Akt signaling pathway in hepatocellular carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2015;92:1132–1140. doi: 10.1016/j.ijrobp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 49.Ming M., He Y.Y. PTEN in DNA damage repair. Cancer Lett. 2012;319:125–129. doi: 10.1016/j.canlet.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayo L.D., Dixon J.E., Durden D.L., Tonks N.K., Donner D.B. PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J. Biol. Chem. 2002;277:5484–5489. doi: 10.1074/jbc.M108302200. [DOI] [PubMed] [Google Scholar]
- 51.Cully M., You H., Levine A.J., Mak T.W. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 52.Zhao L., Lu X., Cao Y. MicroRNA and signal transduction pathways in tumor radiation response. Cell. Signal. 2013;25:1625–1634. doi: 10.1016/j.cellsig.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tonlaar N., Galoforo S., Thibodeau B.J., Ahmed S., Wilson T.G., Yumpo Cardenas P., Marples B., Wilson G.D. Antitumor activity of the dual PI3K/MTOR inhibitor, PF-04691502, in combination with radiation in head and neck cancer. Radiother. Oncol. 2017;124:504–512. doi: 10.1016/j.radonc.2017.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.