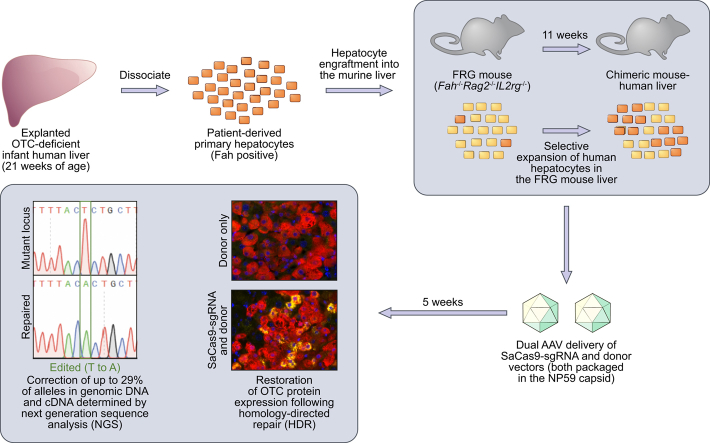
Abstract
Background & Aims
Genome editing technology has immense therapeutic potential and is likely to rapidly supplant contemporary gene addition approaches. Key advantages include the capacity to directly repair mutant loci with resultant recovery of physiological gene expression and maintenance of durable therapeutic effects in replicating cells. In this study, we aimed to repair a disease-causing point mutation in the ornithine transcarbamylase (OTC) locus in patient-derived primary human hepatocytes in vivo at therapeutically relevant levels.
Methods
Editing reagents for precise CRISPR/SaCas9-mediated cleavage and homology-directed repair (HDR) of the human OTC locus were first evaluated against an OTC minigene cassette transposed into the mouse liver. The editing efficacy of these reagents was then tested on the native OTC locus in patient-derived primary human hepatocytes xenografted into the FRG (Fah-/-Rag2-/-Il2rg-/-) mouse liver. A highly human hepatotropic capsid (NP59) was used for adeno-associated virus (AAV)-mediated gene transfer. Editing events were characterised using next-generation sequencing and restoration of OTC expression was evaluated using immunofluorescence.
Results
Following AAV-mediated delivery of editing reagents to patient-derived primary human hepatocytes in vivo, OTC locus-specific cleavage was achieved at efficiencies of up to 72%. Importantly, successful editing was observed in up to 29% of OTC alleles at clinically relevant vector doses. No off-target editing events were observed at the top 10 in silico-predicted sites in the genome.
Conclusions
We report efficient single-nucleotide correction of a disease-causing mutation in the OTC locus in patient-derived primary human hepatocytes in vivo at levels that, if recapitulated in the clinic, would provide benefit for even the most therapeutically challenging liver disorders. Key challenges for clinical translation include the cell cycle dependence of classical HDR and mitigation of unintended on- and off-target editing events.
Lay summary
The ability to efficiently and safely correct disease-causing mutations remains the holy grail of gene therapy. Herein, we demonstrate, for the first time, efficient in vivo correction of a patient-specific disease-causing mutation in the OTC gene in primary human hepatocytes, using therapeutically relevant vector doses. We also highlight the challenges that need to be overcome for this technology to be translated into clinical practice.
Keywords: OTC deficiency, primary human hepatocytes, CRISPR-Cas9, homology-directed repair, genome editing, recombinant AAV, humanised FRG mice, NP59 capsid, synthetic capsid
Abbreviations: AAV, adeno-associated virus; BrdU, bromodeoxyuridine; FRG, Fah-/-Rag2-/-Il2rg-/-; HDR, homology-directed repair; InDels, insertions and deletions; ITR, inverted terminal repeats; LSP1, liver-specific promoter; 7 NGS, next-generation sequencing; NHEJ, non-homologous end joining; pA, bovine growth hormone polyadenylation signal sequence; PAM, protospacer adjacent motif; PRE, mutant form of the Woodchuck hepatitis virus posttranscriptional regulatory element; rAAV, recombinant adeno-associated virus; RTA, Real Time Analysis; SaCas9, Staphylococcus aureus Cas9 nuclease; sgRNA, single guide RNA; SV40 pA, SV40 polyadenylation signal sequence; TBG, human thyroxine binding globulin promoter; U6, RNA polymerase III promoter for human U6 snRNA; WT, wild-type
Graphical abstract
Highlights
-
•
Therapeutically relevant levels of single-nucleotide repair of the human OTC locus were achieved in vivo.
-
•
Single-nucleotide editing of primary human hepatocytes was facilitated by a highly hepatotropic bioengineered AAV capsid.
-
•
A novel human minigene platform proved highly effective for evaluation of human liver-specific genome editing reagents.
Introduction
The demonstrated therapeutic efficacy of recombinant adeno-associated virus (rAAV) vectors in the treatment of haemophilia B1 has intensified interest in exploiting this vector system to treat more demanding genetic/metabolic liver disease phenotypes2 and other hepatic pathologies caused by viral infection,3 hepatotoxin exposure4 and neoplasia.5 Challenges include the need to efficiently target a greater proportion of the hepatic cell mass,6 overcome the limitations imposed by the loss of rAAV episomes that occurs in concert with hepatocellular replication7,8 and develop preclinical models that better predict the performance of AAV-based gene addition and editing technologies in the human liver. We have previously used healthy primary human hepatocytes xenografted and expanded in the FRG (Fah-/-Rag2-/-Il2rg-/-) mouse liver to select and functionally characterise novel AAV capsid variants with unprecedented human hepatocyte tropism.9,10 The most recently developed and potent of these capsid variants, NP59, is capable of transducing in excess of 95% of primary human hepatocytes in chimeric mouse-human livers, offering the prospect of dramatically extending the therapeutic reach of AAV vectors.
To date, there have been relatively few studies using genome editing for successful homology-directed repair (HDR) in vivo, none of which have used human-specific guide RNAs or donor templates. These studies have all employed murine models where either a selective growth advantage is conferred on the corrected cells, such as models of hereditary tyrosinemia type I,[11], [12], [13] or where the correction of only a small percentage of cells can result in phenotypic improvements, as is the case for haemophilia B.14 Alternatively, studies targeting primary hepatocytes in vivo have exploited the more active non-homologous end joining (NHEJ) pathway for locus-specific disruption of the murine Pcsk915,16 or human PCSK917 genes. Only a single study reports the use of dual AAV-based editing reagents in vivo to treat ornithine transcarbamylase (OTC) deficiency, in a murine model with mild disease.18 Although this work provides important proof-of-concept data, sequence differences between mouse and man prevent the clinical translation of these reagents, and the effectiveness of this approach in patient-derived human cells remains to be demonstrated. In contrast, here we utilise a mouse model comprising chimeric livers engrafted and expanded with explanted patient-derived primary human hepatocytes to evaluate editing reagents in vivo.
In order to reserve precious OTC-deficient patient-derived hepatocytes for critical experiments in the current study, human-specific editing reagents were first evaluated in the murine liver against a novel minigene cassette containing the patient-specific target locus to recapitulate the intracellular environment of bona fide hepatocytes in vivo. This was achieved by using a novel hybrid AAV/PiggyBac transposase system to introduce the human minigene, containing a point mutation that causes severe OTC deficiency in humans, into murine hepatocytes. This human locus-specific target sequence was then used to evaluate AAV-based CRISPR-Cas9 guide strands and repair templates in vivo that could be directly translated to the human setting. We then explored the utility of these validated AAV editing vectors, this time pseudo-serotyped with the NP59 capsid, for genome editing of OTC-deficient primary human hepatocytes carrying a missense mutation in exon 9 (c.905A>T), dissociated from an explanted male paediatric liver. Importantly, we demonstrate successful in vivo correction of a disease-causing mutation in patient-derived primary hepatocytes at rates that would provide therapeutic benefit if achieved in humans.
Materials and methods
Minigene design and plasmid construction
The human OTC gene from a patient with OTC deficiency was sequenced. Briefly, genomic DNA was isolated from primary hepatocytes using QIAamp Blood mini kit, as per manufacturer instructions (Qiagen, Hilden, Germany). The genomic DNA was then used as a template to amplify the human OTC gene by PCR with Phusion polymerase (New England Biolabs, Ispswich, MA). Amplicons were then cloned into pGEM-T Easy cloning vector (Promega, Madison, WI). Following ligation and transformation of JM109 chemically competent cells, plasmid DNA was isolated from selected clones using ISOLATE II Plasmid Mini Kit (Bioline, London, UK) and sent for Sanger sequencing at the Australian Genome Research Facility (AGRF; Sydney, Australia). The disease-causing mutation is located in exon 9 (c.905A>T; p.H302L).
The minigene version of the human OTC gene (NCBI Reference Sequence NG_008471.1) was designed by including all the 10 exons and retaining only truncated versions of the introns flanking exon 9. Two different versions of the minigene were created: wild-type and mutant (containing the c.905A>T point mutation). The minigene sequence was synthesised (Genscript, Piscataway, NJ) and cloned into a recipient pAm construct (pAAV2-pBRsh-LSP2) containing the AAV inverted terminal repeats (ITRs), short piggyBac transposon ITRs 5′TRsh (67 bp) and 3′TRsh (40 bp) and a liver-specific enhancer/promoter (hAPO-HCR-hAAT) to produce pAAV2.pBRsh.LSP2.huOTCmini. The piggyBac transposase vector (pAAV2-LSP1.PB) has been described previously.19
For the CRISPR-Cas9 nuclease system, guides were designed by selecting five 21-nt target sequences next to a 5′NNGRRT protospacer adjacent motif (PAM) around the mutation in exon 9. Oligonucleotides for each guide (Table S1) were annealed and cloned into the pX602 plasmid after digestion with BsaI (New England Biolabs). pX602-AAV-TBG::NLS-SaCas9-NLS-HA-OLLAS-bGHpA;U6::BsaI single guide (sg)RNA was a gift from Feng Zhang (Addgene plasmid #61593). pX602 contains the Cas9 nuclease from Staphylococcus aureus (SaCas9) under the control of a liver-specific thyroxine binding globulin (TBG) promoter in an AAV backbone.16
To generate the repair vectors, a 2,597 bp wild-type homology region was cloned into a recipient AAV backbone, with or without a reporter gene expression cassette. The GFP expression cassette was amplified from pAAV.ApoE.short-hAAT.eGFP.SV40poly, kindly donated by Professor Amit Nathwani (University College London, UK). Versions of the repair vectors with a mutated PAM sequence were created to be used in conjunction with sgRNA2 to prevent re-cutting of the locus post-correction. All AAV plasmid vectors were propagated in SURE2 cells (Stratagene, San Diego, CA) and validated by Sanger sequencing. Recombinant AAV vectors are in the single-stranded configuration unless otherwise indicated. Plasmids and their sequences are available upon request.
Cell culture
Human embryonic kidney 293 cells (HEK-293) were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% (v/v) foetal bovine serum (Bovogen Biologicals Pty Ltd, VIC, Australia) at 37 °C in a humidified 5% CO2-air atmosphere. Cells were mycoplasma free.
AAV vector production
Recombinant AAV vectors were produced in HEK-293 cells by triple-transfection as previously described.7 Vectors were packaged in capsid serotypes 8 and rh10 using plasmids constructs p5E18-VD2/8 and pAAVRh/10, respectively. The transposase vector (rAAV2/8-LSP1.PB) and the minigene vector (rAAV2/8-pBRsh-LSP2-huOTCmini) were packaged into capsid AAV8, while the SaCas9-guide and repair vectors were pseudo-serotyped with capsid rh10 to reduce immunoreactivity with vector re-administration. Vector particles were purified by standard caesium chloride (CsCl) gradient centrifugation from the cell lysate, or from cell lysate and supernatant. The cell lysate was processed by using ammonium sulphate precipitation as described.7 Vector particles from the supernatant were precipitated using 40% (w/v) PEG-8000 in 2.5 M NaCl. Vector genomes were titered by real-time quantitative PCR (qPCR) using sequence-specific primers (Table S1).
Mouse studies
All animal care and experimental procedures were evaluated and approved by the Animal Ethics Committee of the Children's Medical Research Institute and The Children's Hospital at Westmead. Otc-deficient Spfash mice (C57BL/6/C3H-F1 background) were bred in-house from breeding pairs originally purchased from The Jackson Laboratory (Bar Harbor, ME). Animals were housed in standard boxes in a temperature-controlled environment and had free access to water and mouse chow containing 18.9% (w/w) protein (Glen Forrest Stockfeeders, Glen Forrest, WA, Australia). All vectors were administered by intraperitoneal injection. Neonatal male mice (3-4 days old) received 1.5×1011 vg transposase vector (rAAV2/8.LSP1.PB) and 1×1011 vg minigene vector (rAAV2/8.pBRsh.LSP2.huOTCmini). Three weeks later, mice were injected with 2×1011 vg SaCas9-guide strand vector (rAAV2/rh10.SaCas9.sgRNA) and/or 5×1011 vg repair vector (rAAV2/rh10.Repair-eGFP or rAAV2/rh10.Repair). At 5 weeks of age, mice were euthanized by CO2 inhalation. Livers were harvested for molecular analysis and immunohistochemistry. Experiments were conducted in an unblinded manner.
FRG mice breeding pairs were obtained from the laboratory of Professor Markus Grompe (Oregon Stem Cell Center, Oregon Health and Science University, Portland, Oregon) and bred in-house. As FRG mice are immunodeficient, they were housed in individually ventilated filter cages. Mice had free access to drinking water supplemented with 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione (NTBC) before engraftment. Eight to 12-week-old female mice were engrafted with human hepatocytes, isolated from paediatric donors or purchased from Lonza (Basel, Switzerland), as described previously.20 Hepatocytes from the paediatric donors were obtained after informed consent and approval by the Sydney Children's Hospitals Network Human Ethics Committee. Engrafted mice were cycled on and off NTBC to promote liver repopulation. Blood was collected every 2 weeks and at the end of the experiment to measure the levels of human albumin, used as a marker to estimate the level of engraftment, in serum by ELISA (Bethyl Laboratories, Inc., Montgomery, TX). Eleven weeks after engraftment, mice were treated with the editing vectors packaged in NP59, a highly human hepatotropic AAV capsid.10 The following vectors were administered by intraperitoneal injection: rAAV2/NP59.SaCas9.sgRNA and/or rAAV2/NP59.Repair. After vector injection, mice were cycled for another 5 weeks before being euthanized.
Vector copy number analysis
Sections collected for molecular analysis were snap-frozen in liquid nitrogen. Genomic DNA was extracted from snap-frozen liver samples by standard phenol/chloroform and ethanol precipitation methods. Vector copy analysis was performed by qPCR using SYBR Green (Takara Bio Inc., Kusatsu, Japan) and primers specific for each vector. The results were normalized to murine Gapdh levels, as previously described.21
Minigene expression analysis and RT-qPCR
Total RNA was isolated from snap-frozen liver sections using RNeasy Kit (Qiagen) and reverse transcribed with SuperScript III First-Strand Synthesis kit (Invitrogen, Carlsbad, CA) as per the manufacturer’s instructions. qPCR to quantify minigene expression was performed using sequence-specific primers. The results were normalized to murine β-actin expression.
Molecular quantification of guide strand efficiency and HDR
To quantify the efficiency of each guide strand, the Surveyor® nuclease assay (Integrated DNA Technologies, Coralville, IO) was performed as previously described.22 Briefly, an 869-bp PCR product was produced using Q5 polymerase (New England Biolabs) and primers Surveyor-F Surveyor-R (Table S1). The purified PCR products were denatured and re-annealed for heteroduplex formation. Finally, the annealed heteroduplexes were digested with Nuclease S + enhancer and loaded into an agarose gel for visualisation. As an alternative method for assessing guide strand efficiency, PCR products were sent to the AGRF for Sanger Sequencing and analysed using TIDE.23 Additionally, the PCR products from 1 mouse per guide were cloned and individually sequenced to confirm the results obtained by Surveyor® and TIDE analysis.
Targeted deep sequencing
PCR products were created by using 2 primers outside of the homology region: AA_74R and AA_75F for minigene mice, hOTC intron8_F2 and hOTC intron9_R2 for humanised FRG mice (Table S1). PCR was carried out with a standard protocol for Q5 polymerase (New England Biolabs) and the products loaded into a 1% (w/v) agarose gel. Amplicons were purified from the gel using the Wizard PCR purification kit (Promega) and used as template in a second PCR reaction to produce 318-bp nested PCR products for targeted deep sequencing, using primers AA_82F and AA_83R (Table S1). Following purification, amplicons were sent to the AGRF or Genewiz (Suzhou, China) for library preparation and MiSeq paired-read sequencing (Illumina, San Diego, CA). Image analysis was performed in real time by the MiSeq Control Software (MCS) v2.6.1.1 and Real Time Analysis (RTA) v1.18.54. RTA performs real-time base calling on the MiSeq instrument computer. The Illumina bcl2fastq 2.19.0.316 pipeline was then used to generate the sequence data. The next-generation sequencing (NGS) data was analysed using CRISPResso.24
In silico off-target predictions for sgRNA4 were identified using Benchling (Table S2). Primers were designed for the top scoring sites (Table S3) and blasted against the human and mouse genomes. Amplicons for each predicted off-target site were generated using the high-fidelity Q5 polymerase and standard PCR conditions and subsequently pooled for deep sequence analysis. Pooled amplicons were sent to Genewiz for library preparation and MiSeq paired-read sequencing (Illumina).
Analysis of NGS data
NGS data was analysed using CRISPResso (version 1.0.8).24 Indel profile histograms were generated in GraphPad Prism 7.02 using indel size data extracted from CRISPResso. Insertions were classified depending on size as short (<10 bp) or large (≥10 bp). Large insertion sequences were extracted from CRISPResso output and aligned to vector sequences using blastn (version 2.6.0+).25 CRISPRessoPooled was used for the analysis of NGS data from pooled amplicons.
Immunofluorescence, immunohistochemistry and OTC enzyme activity staining
Liver samples collected for immunofluorescence and OTC staining were fixed in 4% (w/v) paraformaldehyde overnight at 4°C, cryoprotected through a sucrose gradient (10%, 20% and 30% (w/v) sucrose in PBS-/-) and frozen in optimal cutting temperature O.C.T. (Tissue-Tek; Sakura Finetek USA, Torrance, CA), as previously described.7 Frozen sections (5 μm) were permeabilized in methanol at −20°C. After blocking in 13% (v/v) donkey serum (Sigma-Aldrich), 8.7% (v/v) FBS in PBS-/-, sections were incubated with either a rabbit monoclonal antibody to human GAPDH conjugated with Alexa Fluor 647 (Abcam, Cambridge, UK; Cat# ab215227, clone EPR6256, Lot# GR293665-1; 1:800 dilution, room temperature for 1 h), a rabbit polyclonal antibody to human OTC (Sigma-Aldrich; Cat# HPA000243, Lot# E105743; 1:100 dilution, 4°C overnight) or a rabbit monoclonal antibody to Ki-67 [SP6] (Abcam, Cat# ab16667, Lot# GR3244218-6; 1:250 dilution, 4°C overnight) followed by a polyclonal donkey anti-rabbit Alexa Fluor 594 (Invitrogen; Cat# A21207, Lot# 857218; 1:800 dilution, room temperature for 1 h). OTC enzyme activity staining in frozen liver sections was performed as described.26 Histological analysis using immunofluorescence microscopy was performed using a Zeiss Axio Imager.M1 with ZEN 2 software (ZEN 2 version 2.0.0.0). Samples collected for immunohistochemistry were fixed in 10% formalin (Sigma-Aldrich) and then transferred to 70% (v/v) ethanol. Paraffin embedding and sectioning was performed by Virginia James of the Westmead Institute for Medical Research (Westmead, NSW, Australia).
Bromodeoxyuridine (BrdU) staining was performed as previously described using a rabbit anti-BrdU primary antibody (Abcam; ab1893, 1:100 dilution, room temperature for 45 min).27 A rabbit anti-GAPDH primary antibody (Abcam; ab128915, EPR6256, 1:250 dilution, room temperature for 45 min) was used to label the human hepatocytes in paraffin sections. Sections were incubated for 45 min at room temperature with biotin-conjugated secondary antibodies: donkey anti-sheep IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, 713-065-147, 1:600 dilution) or donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, 711-065-152, 1:800 dilution). Antigens were visualised by chromogenic detection using RTU Elite ABC reagent (Vector Laboratories, Burlingame, CA) and subsequent incubation with one the following substrates: ImmPACT™ DAB peroxidase substrate (Vector Laboratories) or ImmPACT™ VIP (VectorLabs) for BrdU or GAPDH, respectively.
Western blot analysis
Western blot was performed on liver lysates as previously described.28 The blot was incubated with a polyclonal rabbit anti-human OTC antibody (Abcam; Cat# Ab91418, Lot# GR133033-16; 1:1,000 dilution, overnight at 4°C) and monoclonal mouse anti-vinculin antibody (Sigma-Aldrich; Cat# V9131, clone hVIN-1, Lot# 073M4755V; 1:10,000 dilution, overnight at 4°C). Bound primary antibody was detected using a polyclonal goat anti-rabbit secondary antibody (Santa Cruz Biotechnology, Dallas, TX; Cat# Sc-2004, polyclonal, B2615, 1:5,000, room temperature for 1 h) and goat anti-mouse secondary antibody (Dako, Santa Clara, CA; Cat# P0447, polyclonal, Lot# 20025121, 1:10,000, room temperature for 1 h). Protein detection was carried out by adding SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific) to the membrane and imaging in a Fujifilm Luminescent Image Analyzer LAS-4000.
Statistical analyses
Statistical significance was examined using the Mann-Whitney non-parametric test using GraphPad Prism 7.02. p values <0.05 were considered significant.
Data availability
The NGS datasets generated and analyzed during the current study are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), accession number PRJNA543707.
Results
Development of a novel transposon-based minigene platform to evaluate human-specific editing reagents in vivo
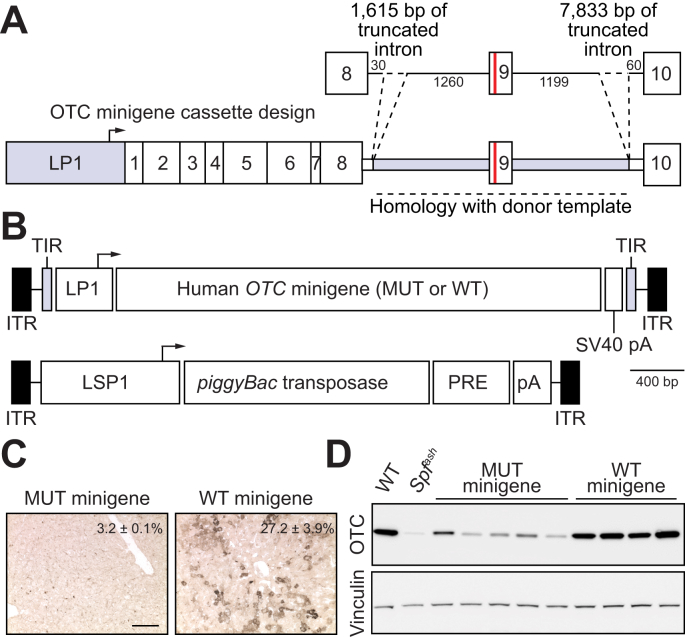
Given the experimentally limiting supply of patient-derived primary human hepatocytes, we first sought to functionally evaluate target locus-specific editing reagents (including CRISPR-SaCas9 constructs,16 containing candidate sgRNAs and donor templates for HDR) against a minigene cassette containing the target human locus in the murine liver (Fig. 1A). This was achieved by transposing patient-specific mutant and positive control wild-type human OTC minigene expression cassettes into the livers of newborn Otc-deficient Spfash mice29 using a novel hybrid AAV/piggyBac transposase vector system19 pseudo-serotyped with the AAV8 capsid (Fig. 1B). Analysis of OTC expression at 4 to 5 weeks of age confirmed the functionality of the wild-type minigene cassette and, as expected, minimal expression from the mutant minigene above background levels in the Spfash mouse liver, consistent with the severe neonatal phenotype conferred by the missense mutation in the donor infant (Fig. 1C,D and Fig. S1A-D).
Fig. 1.
Functional analysis of human minigenes.
(A) Schematic representation of the human OTC minigene containing a point mutation in exon 9 (c.905A>T; p.H302L; red line) that causes severe OTC deficiency with neonatal presentation. Minigene expression is driven by a liver-specific promoter/enhancer (LP1) comprising an ApoE enhancer and short form of the hAAT promoter.47 Intronic sequences flanking exon 9 are shaded in grey. (B) Configuration of the hybrid AAV/piggyBac transposase system used in the study with the structure of the cloned minigene indicated in panel (A) above. ITRs are indicated by the black rectangular boxes and the piggyBac transposase is expressed from the LSP1 promoter containing two copies of the ApoE enhancer and long form of the hAAT promoter. Minigenes were delivered to newborn Spfash mice by intraperitoneal injection using the hybrid AAV/piggyBac transposase vector system. Animals received 1.5×1010 vg transposase and 1×1011 vg minigene vectors (both packaged in the AAV8 capsid) and were harvested 4 weeks later (n = 5 for the mutant and n = 4 for the WT minigenes, respectively). OTC activity (percent WT, mean ± SEM) and expression levels were determined in (C) liver sections (scale bar = 50 μm) and (D) by Western blot analysis. Lysates from WT and Spfash mice were included as controls.
AAV, adeno-associated virus; ApoE, apolipoprotein E; hAAT, human alpha 1-antitrypsin; ITRs, inverted terminal repeats; LP1 and LSP1, liver-specific promoters;7 pA, bovine growth hormone polyadenylation signal sequence; PRE, mutant form of the Woodchuck hepatitis virus posttranscriptional regulatory element; SV40 pA, SV40 polyadenylation signal sequence; WT, wild-type.
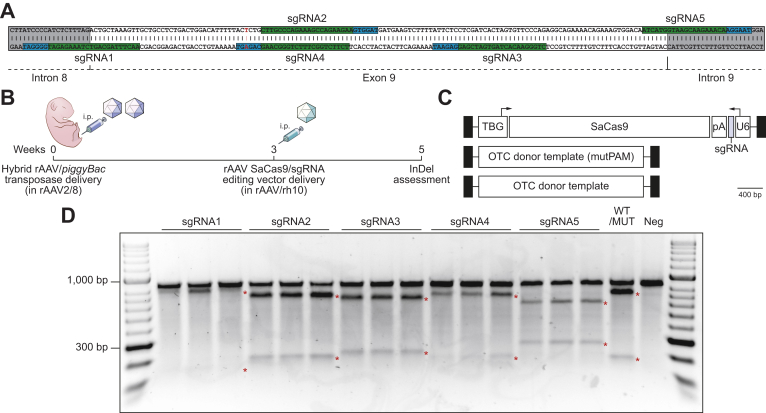
Evaluation of human-specific sgRNAs for SaCas9 in the murine liver in vivo
Five different sgRNAs exploiting PAMs in close proximity to the exon 9 mutation (Fig. 2A) were evaluated against the transposed minigene target using vector pseudo-serotyped with the AAVrh10 capsid to reduce potential immune responses following earlier exposure to the AAV8 capsid (Fig. 2B,C).30 Genome editing reagents were delivered to 3-week-old animals, providing time for the episomal clearance of transposase vectors, which has been shown to be almost complete 2 weeks after neonatal AAV delivery.7
Fig. 2.
Functional analysis of rAAV editing reagents.
(A) The location of sgRNA and SaCas9 PAM sequences are indicated (highlighted in green and blue, respectively). Intronic sequences are shaded in grey and the patient-specific c.905A>T mutation is indicated by the nucleotide shown in red. (B) Overview and timing of intraperitoneal rAAV delivery for CRISPR-SaCas9-mediated gene disruption in Spfash mice. Newborn animals (n = 5 per treatment group) received 5×1010 vg transposase and 1×1011 vg minigene vector (packaged in the AAV8 capsid) and 3 weeks later, 2×1011 of a SaCas9 with sgRNA (1 to 5) rAAV vector (packaged in the rh10 capsid). (C) Configuration of the dual rAAV genome editing vectors used in this study. (D) Functional analysis of sgRNAs used to disrupt the human OTC locus in newborn Spfash mice (n = 5 per treatment group). The presence of small InDels was confirmed by using Surveyor® nuclease. An equal mix of PCR products from WT and mutant minigenes (WT/MUT) and mutant only (neg) were included as controls for the Surveyor® reaction. Red asterisks indicate expected cleavage fragments. InDels, insertions and deletions; pA, bovine growth hormone polyadenylation signal sequence; PAM, protospacer adjacent motif; rAAV, recombinant adeno-associated virus; SaCas9, Staphylococcus aureus Cas9 nuclease; sgRNA, single guide RNA; TBG, human thyroxine binding globulin promoter; U6, RNA polymerase III promoter for human U6 snRNA; WT, wild-type.
Analysis of small insertions and deletions (InDels) in the target human OTC minigene, indicative of repair by non-homologous end joining (NHEJ) was performed using Surveyor® nuclease (Fig. 2D), Decomposition (TIDE) analysis23 (Fig. S2A) and sequencing of at least 24 individual amplicon clones per sgRNA. This confirmed functionality, but variable efficiency, among the 5 sgRNAs tested, with sgRNA2 giving the highest InDel frequency (Fig. S2B). Notably, sgRNA4 contains the patient mutation within the target PAM, which upon correction abolishes the PAM sequence and prevents recleavage by the SaCas9 nuclease (Fig. S2C).
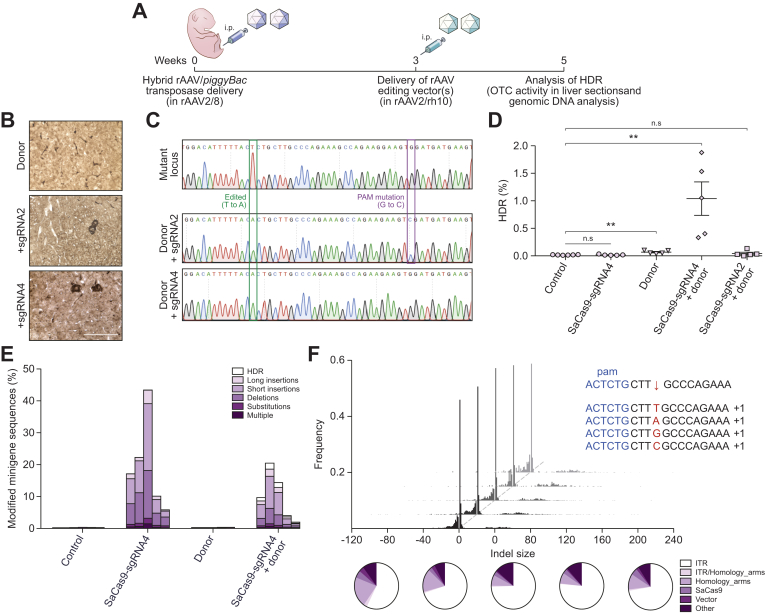
In vivo correction of the patient-specific human minigene by homology-directed repair
Two SaCas9/sgRNA combinations (sgRNA2 and sgRNA4) were then selected for co-delivery with a donor template for correction of the transposed mutant human OTC minigene by HDR (Fig. 3A). Analyses of host mouse livers revealed correction of the mutant minigene with both sgRNAs as evidenced by the presence of murine hepatocytes expressing functional OTC enzyme (Fig. 3B) and the expected corrective T to A substitution, and additional PAM modification when sgRNA2 was used (Fig. 3C). Deep sequencing of minigene amplicons (Fig. S3A) was used to determine HDR frequency (Fig. 3D), InDel frequency and nature (Fig. 3E) and InDel size distribution and insert origin (Fig. 3F, Fig. S3B). Maximal HDR rates of approximately 1% were achieved with sgRNA4, while sgRNA2 was 20-fold less efficient with HDR rates only marginally above those achieved with template alone. Template configuration, whether single-stranded (with or without an eGFP expression cassette) or self-complementary had little effect on HDR frequency (Fig. S4). InDel rates were substantially higher than the rate of HDR, with up to 43% of target minigene cassettes being successfully cut and repaired by NHEJ (Fig. 3E). Notably long insertions were predominately fragments of the AAV vector constructs used for liver-targeted delivery of the editing machinery, particularly AAV ITR-derived sequences (Fig. 3F and Fig. S3B).
Fig. 3.
Homology-directed repair of the transposed human OTC locus in Spfashmice.
(A) Experimental overview and timing of intraperitoneal rAAV delivery for CRISPR-SaCas9-mediated HDR in Spfash mice. Newborn animals received 5×1010 vg transposase and 1×1011 vg mutant minigene vectors (packaged in rAAV2/8 capsid) and 3 weeks later, 2×1011 SaCas9/sgRNA and/or 5×1011 donor vectors (packaged in rAAV2/rh10 capsid). Mutant minigenes were delivered (intraperitoneal) to newborn spfash mice using the hybrid AAV/piggyBac transposase vector system and 3 weeks later animals received genome editing vectors; n = 5 per treatment group. HDR was confirmed 2 weeks later by (B) the presence of functional OTC activity in liver sections (scale bar = 50 μm) and (C) by Sanger sequencing analysis of cloned OTC amplicons amplified using primers with binding sites outside the region of homology with the donor vector. Next-generation Illumina® sequencing was performed across the transposed OTC human minigene locus to more accurately (D) quantitate the HDR rates and (E) characterise the unintended modifications found at the SaCas9 cleavage site. Data are plotted as mean ± SEM. and significance evaluated using the Mann-Whitney non-parametric test (**p <0.005). Control samples represent PCR amplicons amplified from Spfash mice that did not receive genome editing vectors following minigene delivery. (F) In addition to the expected InDels, with a 1 bp insertion the most commonly detected event (insert, top right), large insertions (>10 bp) were observed at the cleavage site. Notably, insertions containing ITR sequences were over-represented (indicated by the white region of the pie diagrams). Each histogram and pie diagram represent an individual animal that received 2×1011 vg SaCas9-sgRNA4 and 5×1011 vg donor vectors. AAV, adeno-associated virus; HDR, homology-directed repair; InDels, insertions and deletions; ITR, inverted terminal repeat; SaCas9, Staphylococcus aureus Cas9 nuclease; sgRNA, single guide RNA; rAAV, recombinant AAV.
Efficient in vivo correction of OTC-deficient patient-derived primary human hepatocytes
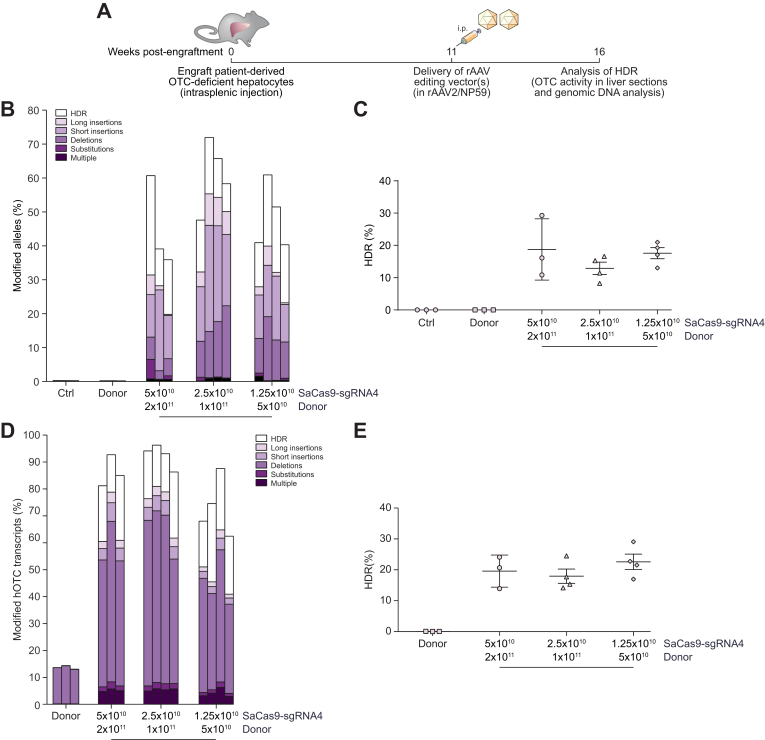
Using these validated reagents packaged into the highly human hepatotropic NP59 capsid,10 we next tested the ability to perform targeted single nucleotide genome editing in patient-derived primary human hepatocytes in vivo. Cells were engrafted and selectively expanded in FRG mice.20 Eleven weeks post-engraftment, single-stranded donor and SaCas9-sgRNA4 vectors (Fig. 2C) were co-delivered via intraperitoneal injection and chimeric livers analysed a further 5 weeks later (Fig. 4A). Deep sequencing of amplicons generated across the SaCas9 target site from genomic DNA was indicative of high rates of locus-specific cleavage of up to 72% (Fig. 4B). Most importantly up to 29% of amplicons carried the desired HDR-mediated corrective single nucleotide editing event (Fig. 4C). While values varied among treated mice, the proportion of cleaved alleles undergoing repair was even higher, ranging up to 48% (Fig. 4B). Analysis of editing events at the mRNA level by deep sequencing of amplicons generated from cDNA provided evidence for rates of locus-specific cleavage higher than those observed in genomic DNA (Fig. 4D). Interestingly, the observation of apparent deletions in donor only control animals proved to be the consequence of aberrant splicing induced by the patient's mutation (Fig. S5). Nevertheless, even considering this effect, in excess of 80% of human OTC transcripts showed evidence of locus-specific cleavage, with up to 29% carrying the desired single nucleotide editing event (Fig. 4E).
Fig. 4.
Efficient correction of OTC-deficient patient-derived primary human hepatocytes in vivo.
(A) Experimental overview. Cells were engrafted into FRG mice and repopulation established by cycling on and off the drug 2-(2-nitro-4-fluoromethylbenzoyl)-1,3-cyclohexanedione.20 Eleven weeks later, when the mice were on day 11 of a 21-day water cycle, animals received 5×1010 vg/mouse SaCas9-sgRNA4 and 2×1011 vg/mouse donor vectors, 2.5×1010 vg/mouse SaCas9-sgRNA4 and 1×1011 vg/mouse donor vectors or 1.25×1010 vg/mouse SaCas9-sgRNA4 and 5×1010 vg/mouse donor vectors packaged in the NP59 capsid via intraperitoneal delivery (n = 3 per treatment group for controls and highest vector dose, and n = 4 for 2 lower vector doses). Livers were analyzed 5 weeks following vector delivery. Next-generation Illumina® sequencing was performed across the OTC locus from (B, C) genomic DNA and (D, E) cDNA isolated from FRG mice transplanted with patient-derived human hepatocytes to quantitate the HDR rates and characterise the unintended modifications found at the SaCas9 cleavage site. Data are plotted as mean ± SEM. Control samples represent PCR amplicons from engrafted FRG mice that did not receive AAV vector treatment. AAV, adeno-associated virus; FRG, Fah-/-Rag2-/-Il2rg-/; HDR, homology-directed repair; InDels, insertions and deletions; ITR, inverted terminal repeat; SaCas9, Staphylococcus aureus Cas9 nuclease; sgRNA, single guide RNA; rAAV, recombinant AAV.
Cellular replication in the FRG mouse model
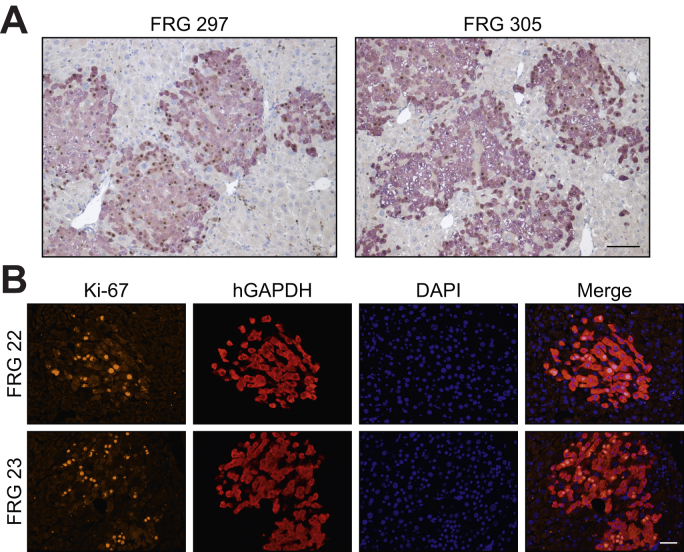
Given that classical HDR occurs only in cycling cells,31 we next sought to determine the basal level of cellular replication occurring in primary human hepatocytes in chimeric mouse-human livers. This was assessed using 2 independent methods, BrdU incorporation into newly synthesised genomic DNA and immunohistochemical staining for Ki67. At week 11 post-transplantation, mice received twice daily injections of BrdU for 1 week while on a water cycle, after which time the livers were harvested for immunohistochemical analysis. Of the total number of primary human hepatocytes, 52.2% ± 5.5% (mean ± SEM, n = 5 animals) were found to contain BrdU-positive nuclei (Fig. 5A). Similarly, the level of Ki-67 labelling in primary human hepatocytes was evaluated 11 weeks post-transplantation when animals were on a water cycle. In 2 chimeric mouse-human livers we observed 28.1% and 39.6% of primary human hepatocytes staining positive for Ki-67 (Fig. 5B). Taken together, these data confirm the supraphysiological levels of proliferation of the human hepatocytes when under positive selection in this model.
Fig. 5.
Evaluation of cellular replication in primary human hepatocytes in chimeric FRG livers.
(A) Quantitation of BrdU-positive human hepatocytes in engrafted FRG mice following vector treatment. Eleven weeks post-engraftment, mice received BrdU injections twice daily following vector delivery and livers were harvested 1 week later for immunohistochemical analysis. The number of BrdU-positive (brown) hepatocyte nuclei within human GAPDH positive clusters were counted across liver sections from engrafted FRG mice (n = 5 animals; 10 images per mouse). Representative images from 2 mice (FRG 297 and FRG 305) are shown. Scale bar = 100 μm. (B) Ki-67 labelling was determined in human GAPDH positive clusters at 11 weeks post-engraftment (n = 2 animals; 4 images per mouse). Representative images from the 2 mice (FRG 22 and FRG 23) are shown. Scale bar = 50 μm. BrdU, bromodeoxyuridine; FRG, Fah-/-Rag2-/-Il2rg-/.
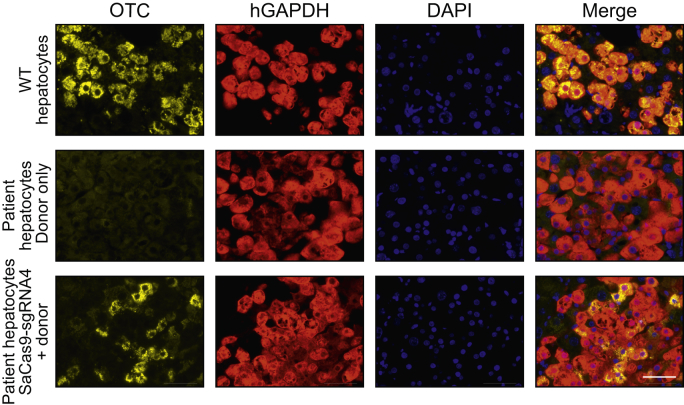
OTC protein expression is restored following in vivo correction
Molecular evidence of human OTC locus-specific correction at the DNA and mRNA level was confirmed by direct immunohistochemical staining of chimeric liver sections (Fig. 6). Clear restoration of OTC expression was observed in up to 36% of human hepatocytes in individual clusters, but with considerable inter-cluster and inter-mouse variation (mean ± SEM; 14.6% ± 7.7%). The intensity of OTC staining also differed among individual hepatocytes, reminiscent of the natural variation observed across the hepatic lobule, a phenomenon known as metabolic zonation.32 Collectively these data confirm unprecedented rates of HDR-mediated locus-specific single nucleotide repair, that could provide unequivocal therapeutic efficacy in many disease contexts. Finally, no off-target cleavage was observed at the top 10 in silico-predicted sites in the human genome (Fig. S6).
Fig. 6.
Restoration of OTC expression in patient-derived primary human hepatocytes in vivo.
Representative immunohistochemistry images of FRG mice engrafted with patient-derived human hepatocytes 5-weeks post-treatment. Sections were labelled with antibodies against the OTC (yellow) and human GAPDH (red) proteins in liver sections. Nuclei were stained with DAPI. Top panels show representative images from an FRG control mice that was engrafted with WT human hepatocytes but did not receive vector treatment, middle and bottom panels are representative images from engrafted patient-derived OTC-deficient hepatocytes treated with donor only (middle panels) or SaCas9-sgRNA4 and donor vectors (lower panels), respectively, at a dose of 1×1011 vg/mouse of donor or 2.5×1010 vg/mouse SaCas9-sgRNA4 and 1 × 1011 vg/mouse of donor. Scale bar = 50 μm. FRG, Fah-/-Rag2-/-Il2rg-/; SaCas9, Staphylococcus aureus Cas9 nuclease; sgRNA, single guide RNA.
Discussion
Using OTC deficiency as a model of genetic/metabolic liver disease, we report, for the first time, precise in vivo locus-specific single-nucleotide correction of a disease-causing mutation in patient-derived primary human hepatocytes. Using an AAV vector-based approach involving CRISPR-saCas9-mediated cleavage and HDR, unprecedented rates of correction, in up to 29% of human hepatocytes, were achieved that if recapitulated in the clinic would provide unequivocal therapeutic benefit. Even higher rates of locus-specific cleavage (up to 72%) were achieved, indicating that not all cleaved loci underwent HDR. This cleavage efficiency is substantially above the highest previously reported rate achieved in vivo, without attempted HDR, in primary human hepatocytes targeting the PCSK9 locus using adenovirus-mediated delivery (≤47%).17 The editing rates achieved in the current study, at clinically relevant vector doses, are likely due to unparalleled gene delivery efficiency facilitated by the use of the highly human liver tropic NP59 capsid.10 In vivo validation and selection of editing reagents in our novel transposon-based minigene system was also a contributing factor.
The results presented herein describe, for the first-time, in vivo editing of primary human hepatocytes with high efficiency. However, several issues need to be addressed before there is a clear research trajectory towards human clinical translation. These include i) the prime importance of highly efficient liver-targeted gene transfer technology, which is currently undergoing rapid development, ii) the critical need to configure gene repair approaches that are cell cycle independent, particularly in the adult liver, iii) the necessity of avoiding unintended harm to hypermorphic target alleles, and iv) the challenge of developing editing strategies that are universally applicable, rather than mutation specific.
Vectors based on AAV have emerged as the leading system for liver-targeted gene transfer and editing applications.1,16,33,34 The AAV capsid, which determines tropism in a target cell type and species-specific manner, is currently an area of dynamic research activity that is continuing to improve and expand the clinical potential of the AAV vector system.9,10,[35], [36], [37] Naturally occurring capsid variants, such as AAV2 and AAV8, which have been used for gene transfer to the murine liver and in human clinical trials have been shown to transduce only a minority of human hepatocytes engrafted into the FRG mouse liver.9 In contrast, LK03, a synthetic capsid with high tropism for human hepatocytes, was created by directed evolution using selection in the same clinically predictive xenograft model.9 This capsid has been successfully deployed in early phase clinical trials for haemophilia A38 and is currently being evaluated in phase III trials, with promising results to date. More recently, the highly human hepatotropic capsid NP59, used in this study, which is 3-fold more efficient than LK03, was also identified by directed capsid evolution in the FRG model.10 This synthetic capsid is capable of transducing in excess of 95% of primary human hepatocytes in vivo at clinically relevant vector doses. This level of in vivo delivery efficiency will be mandatory in future clinical applications given the two-hit kinetics required to deliver all elements of the editing machinery to individual hepatocytes at the rates required to achieve therapeutic benefit. Ongoing developments in capsid technology can be expected to further improve liver-targeted gene delivery efficiency and specificity, allowing for lower vector doses to be used and reducing the probability of patients being excluded from AAV-based trials based on anticapsid seroreactivity in the general population. This is the consequence of exposure to natural infection by AAV2, which is prevalent in human populations.
Genome editing strategies that use classical HDR require cell division for successful correction.39 Therefore, an important, but surmountable, caveat is that human hepatocytes in their native hepatic context will have lower patient age-dependent rates of replication with correspondingly lower rates of HDR. This requirement also likely explains the lower rates of HDR observed in the transposed minigene target locus in murine hepatocytes in the Otc-deficient Spfash mouse liver. Of note, this replication-dependence of HDR favours targeting the paediatric liver, particularly in the early months of life, during which hepatocytes are highly proliferative leading to a doubling of liver mass by 8 months of age.40 To further explore the impact of cellular turnover in the FRG model system, we used BrdU incorporation to define the level of replication occurring in engrafted primary human hepatocytes at the time of vector delivery. We observed a close approximation between the number of cells containing BrdU-positive nuclei after 1 week and rates of HDR, consistent with the dependence of HDR on cell division. With this dependency in mind, it is conceivable that editing rates might still be therapeutically useful in the relatively quiescent human liver for conditions with a low therapeutic threshold, including non-cell-autonomous diseases, such as haemophilia, or where the corrected cells have a selective advantage.
Another important consideration in the clinical translation of genome editing strategies is the risk of converting hypomorphic alleles to null alleles through the occurrence of undesired DNA repair events at the target locus (see Fig. 4B,D). For example, hepatocytes from the donor infant in this study had 2.6% residual OTC enzymatic activity which is sufficient to confer clinically relevant levels of ureagenic activity. Accordingly, inadvertent conversion of such alleles to a null state has the potential for therapeutic harm if there is not simultaneous conversion of a sufficient number of target alleles to a fully functional state. This critical balance will vary among disease phenotypes and be particularly important in cell autonomous diseases, such as urea cycle defects, where significantly subphysiological levels of enzymatic activity in a large number of hepatocytes is predicted to confer greater therapeutic benefit than physiological or supraphysiolocal activity in a smaller number of hepatocytes.41 In the current study, analyses of the target locus following saCas9-mediated cleavage and classical HDR-mediated template repair (Fig. 4B), not only revealed unprecedented rates of correction with restoration of OTC expression, but also a background pattern of small InDels, single nucleotide substitutions, and large insertions (defined as ≥10 bp; Fig. S7). Interestingly, the vast majority of insertions were of vector origin with a dominant contribution from the ITR sequences, an observation already reported by others.42 Irrespective of the nature of these events, the key consequence is the likely disruption of the reading frame and resultant loss of residual OTC function. In the context of OTC deficiency, this balance of intended and undesired DNA repair events would almost certainly be of clear net therapeutic benefit if achieved in the clinic. One class of undesired DNA repair events that would not have been detected in our study with the PCR-based strategy employed is large deletions removing one or both primer binding sites lying 147 bp and 171 bp 5' and 3' to the SaCas9 cleavage site, respectively. However, the relatively close approximation of the observed HDR rates and recovery of OTC expression in our study are consistent with large undetected deletions making a minimal contribution to all undesired DNA repair events that would have resulted in a null allele. This finding is also consistent with other recent studies examining repair events following double-strand breaks, where large deletions were found to be relatively infrequent and represent a minority of deletion events.43,44 Taken together, these data highlight the need to balance justified enthusiasm with appropriate caution in moving editing strategies to the clinic.
A further challenge in moving to the clinic is that strategies based on HDR will require a customised set of editing reagents for each mutation being targeted, namely a donor vector with homology arms flanking the targeted site and a vector containing the CRISPR/Cas9 machinery, including validated sgRNAs with target sequences adjacent to the mutation site. This will limit the therapeutic potential of this approach to disease phenotypes with frequent mutations at defined genomic locations because the development and manufacture of multiple mutation-specific reagents will make commercialization difficult.
The broadly applicable minigene system described in this study has the potential to ensure that only the most effective reagents be evaluated in humanised models, where the availability of disease-specific patient-derived primary cells is limiting. Here we demonstrate that HDR was at least 1 log lower in mice that received sgRNA2 than sgRNA4 in combination with a donor vector, despite this guide being identified as the most efficient at targeting the transposed human locus among the panel tested (Fig. 2D and Fig. S2A). It is noteworthy that an additional nucleotide change at the PAM site had to be introduced when using sgRNA2 to prevent re-cutting of the target sequence after correction of the mutation (Fig. S2C), which may have limited the rate of HDR. In addition to evaluating guide strand efficiency, a comparison of different rAAV donor template vector configurations was performed in the minigene model. Interestingly, no significant differences in HDR rates were detected between mice cohorts treated with single-stranded or self-complementary rAAV. Moreover, the inclusion of a reporter cassette adjacent to the template for HDR did not significantly reduce repair rates in the minigene model (Fig. S4). Importantly, this novel minigene system can also be used to evaluate HDR-independent editing approaches.
The key strategic insights arising from this study are the need to further explore editing strategies that are cell cycle independent, relatively mutation-agnostic and that carry a low risk of damaging hypomorphic alleles. An example of an editing approach that addresses these considerations is homology independent targeted insertion as recently reported by Suzuki and colleagues.39 Avoidance of cell cycle dependence is most relevant in the more quiescent adult human liver and for disease phenotypes, including urea cycle disorders, where a relatively large number of hepatocytes must be successfully edited to confer therapeutic benefit. Finally, systems designed to deliver Cas9 transiently are also being investigated in order to increase the safety profile and reduce unwanted off-target effects resulting from prolonged Cas9 expression.11,45,46 Irrespective of the editing strategy used, the results reported and near-term prospect of human clinical translation remains critically dependent upon the ongoing development and use of AAV capsids, such as NP59 used herein,10 with the capacity to target primary human hepatocytes in vivo with unprecedented efficiency.
Financial support
This work was supported by a project grant from the National Health and Medical Research Council of Australia (1080330). AKA was supported by a University of Sydney International Scholarship and Children's Medical Research Institute PhD stipend.
Conflict of interest
The authors declare no competing financial interests. Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
S.L.G. and A.K.A. conceived, designed and performed experiments, analyzed data and wrote the manuscript; S.H.Y.L. and E.Z. performed experiments and analyzed data; M.L. analyzed data; C.V.H. designed and performed experiments; S.C.C. and K.D. performed experiments; G.J.L. conceived experiments; S.S.T. designed experiments; A.J.C. provided data analysis insights; H.A.P. supervised research; M.G. and L.L. conceived experiments; and I.E.A. jointly supervised the research, conceived and designed experiments, analyzed data and wrote the manuscript.
Acknowledgements
We thank Mark A. Kay (Stanford University School of Medicine, CA) for providing the NP59 capsid.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2019.100065.
Supplementary data
References
- 1.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander I.E., Kok C., Dane A.P., Cunningham S.C. Gene therapy for metabolic disorders: an overview with a focus on urea cycle disorders. J Inherit Metab Dis. 2012;35:641–645. doi: 10.1007/s10545-012-9467-0. [DOI] [PubMed] [Google Scholar]
- 3.Berraondo P., Di Scala M., Korolowicz K., Thampi L.M., Otano I., Suarez L. Liver-directed gene therapy of chronic hepadnavirus infection using interferon alpha tethered to apolipoprotein A-I. J Hepatol. 2015;63:329–336. doi: 10.1016/j.jhep.2015.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satishchandran A., Ambade A., Rao S., Hsueh Y.C., Iracheta-Vellve A., Tornai D. MicroRNA 122, regulated by GRLH2, protects livers of mice and patients from ethanol-induced liver disease. Gastroenterology. 2018;154:238–252.e7. doi: 10.1053/j.gastro.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santiago-Ortiz J.L., Schaffer D.V. Adeno-associated virus (AAV) vectors in cancer gene therapy. J Control Release. 2016;240:287–301. doi: 10.1016/j.jconrel.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginn S.L., Alexander I.E. 2016. Gene Therapy; pp. 1–19. Wiley StatsRef: Statistics Reference Online. [Google Scholar]
- 7.Cunningham S.C., Dane A.P., Spinoulas A., Logan G.J., Alexander I.E. Gene delivery to the Juvenile mouse liver using AAV2/8 vectors. Mol Ther. 2008;16:1081–1088. doi: 10.1038/mt.2008.72. [DOI] [PubMed] [Google Scholar]
- 8.Wang L., Bell P., Lin J., Calcedo R., Tarantal A.F., Wilson J.M. AAV8-mediated hepatic gene transfer in infant rhesus monkeys (Macaca mulatta) Mol Ther. 2011;19:2012–2020. doi: 10.1038/mt.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–28610. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borel F., Tang Q., Gernoux G., Greer C., Wang Z., Barzel A. Survival advantage of both human hepatocyte xenografts and genome-edited hepatocytes for treatment of alpha-1 antitrypsin deficiency. Mol Ther. 2017;25:2477–2489. doi: 10.1016/j.ymthe.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao Y., Wang L., Guo N., Wang S., Yang L., Li Y. Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats. J Biol Chem. 2018;293:6883–6892. doi: 10.1074/jbc.RA117.000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohmori T., Nagao Y., Mizukami H., Sakata A., Muramatsu S.I., Ozawa K. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci Rep. 2017;7:4159. doi: 10.1038/s41598-017-04625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Q., Strong A., Patel K.M., Ng S.L., Gosis B.S., Regan S.N. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Raghavan A., Chen T., Qiao L., Zhang Y., Ding Q. CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo-brief report. Arterioscler, Thromb Vasc Biol. 2016;36:783–786. doi: 10.1161/ATVBAHA.116.307227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham S.C., Siew S.M., Hallwirth C.V., Bolitho C., Sasaki N., Garg G. Modeling correction of severe urea cycle defects in the growing murine liver using a hybrid recombinant adeno-associated virus/piggyBac transposase gene delivery system. Hepatology. 2015;62:417–428. doi: 10.1002/hep.27842. [DOI] [PubMed] [Google Scholar]
- 20.Azuma H., Paulk N., Ranade A., Dorrell C., Al Dhalimy M., Ellis E. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logan G.J., Dane A.P., Hallwirth C.V., Smyth C.M., Wilkie E.E., Amaya A.K. Identification of liver-specific enhancer-promoter activity in the 3′ untranslated region of the wild-type AAV2 genome. Nat Genet. 2017;49:1267–1273. doi: 10.1038/ng.3893. [DOI] [PubMed] [Google Scholar]
- 22.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye X., Robinson M.B., Batshaw M.L., Furth E.E., Smith I., Wilson J.M. Prolonged metabolic correction in adult ornithine transcarbamylase-deficient mice with adenoviral vectors. J Biol Chem. 1996;271:3639–3646. doi: 10.1074/jbc.271.7.3639. [DOI] [PubMed] [Google Scholar]
- 27.Dane A.P., Cunningham S.C., Kok C.Y., Logan G.J., Alexander I.E. Transient suppression of hepatocellular replication in the mouse liver following transduction with recombinant adeno-associated virus. Gene Ther. 2015;22:917–922. doi: 10.1038/gt.2015.66. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham S.C., Kok C.Y., Dane A.P., Carpenter K., Kizana E., Kuchel P.W. Induction and prevention of severe hyperammonemia in the spfash mouse model of ornithine transcarbamylase deficiency using shRNA and rAAV-mediated gene delivery. Mol Ther. 2011;19:854–859. doi: 10.1038/mt.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M.X., Nakajima T., Fukushige T., Kobayashi K., Seiler N., Saheki T. Aberrations of ammonia metabolism in ornithine carbamoyltransferase-deficient spf-ash mice and their prevention by treatment with urea cycle intermediate amino acids and an ornithine aminotransferase inactivator. Biochim Biophys Acta. 1999;1455:1–11. doi: 10.1016/s0925-4439(99)00048-4. [DOI] [PubMed] [Google Scholar]
- 30.Kok C.Y., Cunningham S.C., Carpenter K.H., Dane A.P., Siew S.M., Logan G.J. Adeno-associated virus-mediated rescue of neonatal lethality in argininosuccinate synthetase deficient mice. Mol Ther. 2013;21:1823–1831. doi: 10.1038/mt.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hustedt N., Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 32.Haussinger D., Lamers W.H., Moorman A.F. Hepatocyte heterogeneity in the metabolism of amino acids and ammonia. Enzyme. 1992;46:72–93. doi: 10.1159/000468779. [DOI] [PubMed] [Google Scholar]
- 33.Barzel A., Paulk N.K., Shi Y., Huang Y., Chu K., Zhang F. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander I.E., Russell D.W. The potential of AAV-mediated gene targeting for gene and cell therapy applications. Curr Stem Cell Rep. 2015;1:16–22. [Google Scholar]
- 35.Cabanes-Creus M., Ginn S.L., Amaya A.K., Liao S.H.Y., Westhaus A., Hallwirth C.V. Codon-optimization of wild-type adeno-associated virus capsid sequences enhances DNA family shuffling while conserving functionality. Mol Ther Methods Clin Dev. 2019;12:71–84. doi: 10.1016/j.omtm.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ojala D.S., Sun S., Santiago-Ortiz J.L., Shapiro M.G., Romero P.A., Schaffer D.V. In vivo selection of a computationally designed SCHEMA AAV library yields a novel variant for infection of adult neural stem cells in the SVZ. Mol Ther. 2018;26:304–319. doi: 10.1016/j.ymthe.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm D., Zolotukhin S. E pluribus unum: 50 years of research, millions of viruses, and one goal–tailored acceleration of AAV evolution. Mol Ther. 2015;23:1819–1831. doi: 10.1038/mt.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan S.K., George L.A., Ragni M.V., Samelson-Jone B., Cuker A., Chen Y. SPK-8011: preliminary results from a phase 1/2 trial of investigational gene therapy for hemophilia confirm transgene derived increases in FVIII activity that are persistent and stable beyond eight months. Cell Press. 2018;26(suppl):350. [Google Scholar]
- 39.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stocker J.T., Dehner L.P., editors. Pediatric Pathology. Lippincott, Williams and Wilkins; Philadelphia: 2002. [Google Scholar]
- 41.Kok C.Y., Cunningham S.C., Kuchel P.W., Alexander I.E. Insights into gene therapy for urea cycle defects by mathematical modeling. Hum Gene Ther. 2019;30:1385–1394. doi: 10.1089/hum.2019.053. [DOI] [PubMed] [Google Scholar]
- 42.Hanlon K.S., Kleinstiver B.P., Garcia S.P., Zaborowski M.P., Volak A., Spirig S.E. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun. 2019;10:4439. doi: 10.1038/s41467-019-12449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson C.E., Wu Y., Gemberling M.P., Oliver M.L., Waller M.A., Bohning J.D. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 47.Nathwani A.C., Gray J.T., Ng C.Y., Zhou J., Spence Y., Waddington S.N. Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver. Blood. 2006;107:2653–2661. doi: 10.1182/blood-2005-10-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS datasets generated and analyzed during the current study are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), accession number PRJNA543707.