Abstract
Background & Aims
A DNA methylation (DNAm) signature derived from 353 CpG sites (the Horvath clock) has been proposed as an epigenetic measure of chronological and biological age. This epigenetic signature is accelerated in diverse tissue types in various disorders, including non-alcoholic steatohepatitis, and is associated with mortality. Here, we assayed whole blood DNAm to explore age acceleration in patients with primary sclerosing cholangitis (PSC).
Methods
Using the MethylationEPIC BeadChip (850K) array, DNAm signatures in whole blood were analyzed in 36 patients with PSC enrolled in a 96-week trial of simtuzumab (Ishak F0-1, n = 13; F5-6, n = 23). Age acceleration was calculated as the difference between DNAm age and chronological age. Comparisons between patients with high and low age acceleration (≥ vs. < the median) were made and Cox regression evaluated the association between age acceleration and PSC-related clinical events (e.g. decompensation, cholangitis, transplantation).
Results
Age acceleration was significantly higher in patients with PSC compared to a healthy reference cohort (median, 11.1 years, p <2.2 × 10-16). In PSC, demographics, presence of inflammatory bowel disease, and ursodeoxycholic acid use were similar between patients with low and high age acceleration. However, patients with high age acceleration had increased serum alkaline phosphatase, gamma glutamyltransferase, alanine aminotransferase, enhanced liver fibrosis test scores, and greater hepatic collagen and α-smooth muscle actin expression on liver biopsy (all p <0.05). Moreover, patients with high age acceleration had an increased prevalence of cirrhosis (89% vs. 39%; p = 0.006) and greater likelihood of PSC-related events (hazard ratio 4.19; 95% CI 1.15–15.24).
Conclusion
This analysis of blood DNAm profiles suggests that compared with healthy controls, patients with PSC – particularly those with cirrhosis - exhibit significant acceleration of epigenetic age. Future studies are required to evaluate the prognostic implications and effect of therapies on global methylation patterns and age acceleration in PSC.
Lay summary
An epigenetic clock based on DNA methylation has been proposed as a marker of age. In liver diseases such as non-alcoholic steatohepatitis, age acceleration based on this epigenetic clock has been observed. Herein, we show that patients with primary sclerosing cholangitis have marked age acceleration, which is further accentuated by worsening fibrosis. This measure of age acceleration could be a useful marker for prognostication or risk stratification in primary sclerosing cholangitis.
Keywords: Aging, biomarker, inflammatory bowel disease, primary sclerosing cholangitis, prognosis, ursodeoxycholic acid
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; BMI, body mass index; DNAm, DNA methylation; ELF, enhanced liver fibrosis; FDR, false discovery rate; GGT, gamma-glutamyltransferase; IBD, inflammatory bowel disease; IL, interleukin; LOXL2, lysyl oxidase-like-2; NASH, non-alcoholic steatohepatitis; PSC, primary sclerosing cholangitis; SMA, smooth muscle actin; UDCA, ursodeoxycholic acid
Graphical abstract
Highlights
-
•
A peripheral blood DNA methylation (DNAm) score identifies age acceleration in PSC patients vs. healthy controls.
-
•
PSC patients with high age acceleration had significantly more PSC-related events than those with low age acceleration.
-
•
These findings may enable stratification of at-risk PSC patients based on a DNAm score from peripheral blood.
Introduction
Primary sclerosing cholangitis (PSC) is a complex cholestatic liver disease characterized by inflammation and scarring of the intra and/or extrahepatic bile ducts, with a markedly elevated risk of developing several malignancies, most notably cholangiocarcinoma. The pathogenesis of PSC remains poorly understood and currently no pharmacological therapy is available.1 The histological appearance is marked by a chronic inflammatory infiltrate and fibro-obliterative destruction of the bile ducts. Assessment of liver injury by biopsy is a standard method of assessing chronic injury, but the regional heterogeneity characteristic of PSC accentuates sampling variability; moreover, liver biopsy is restricted to evaluation of smaller intrahepatic bile ducts. Although liver biopsy is not currently recommended by practice guidelines,2 histological staging likely has prognostic value and may represent a key endpoint in clinical trials.3,4 Beyond liver biopsy, novel surrogate markers of fibrosis (e.g. the enhanced liver fibrosis [ELF™] test) that provide a global assessment may also provide meaningful prognostic information.5
Epigenetic-based estimates of biological age using a DNA methylation (DNAm) signature (i.e. DNAm age) assayed from whole blood or tissue are a promising new technique to ascertain a biological ‘snapshot’ of aging. One such ‘epigenetic clock’ accurately predicts an individual's age based on methylation levels at 353 CpG sites. This epigenetic clock has been validated in multiple cohorts and has demonstrated predictive utility across different tissue sites including the liver.[6], [7], [8] Cases where a person's epigenetic age exceeds their chronological age represent a state of age acceleration with consequences for developing overt manifestations of disease.7,8
Determination of age acceleration may have practical consequences. For instance, successful liver transplantation from chronologically old, but biologically ‘fit’ donors reflects the clear distinction between biological vs. chronological age.9 The converse is also true as the intrinsic rate of the DNAm clock can be altered by diseases that involve the liver. HIV and obesity predispose to increased liver injury, and both accelerate the epigenetic clock more than would be expected from age-matched control specimens.8,10,11 Our group previously reported that patients with non-alcoholic steatohepatitis (NASH) and moderate to severe fibrosis demonstrate age acceleration compared to their healthy counterparts based on a DNAm signature from whole blood.12 In this setting, age acceleration was associated with hepatic fibrosis, the only independent predictor of adverse liver-related outcomes in NASH.13,14 In other conditions, age acceleration has been associated with poorer performance on a range of physical and cognitive assessments, and higher overall mortality even after adjusting for known risk factors.15,16
Whether age acceleration is a reflection of the fibrogenic process across different liver diseases is unknown. If this were indeed the case, then age acceleration in patients with NASH would be comparable to that of patients with PSC with similar fibrosis severity. In the current study, we confirmed the hypothesis that patients with PSC have higher age acceleration than a control population. Moreover, in patients with PSC, age acceleration reflects the severity of hepatic fibrosis and is associated with an increased risk of liver-related complications. These findings support the use of a novel, non-invasive method (based on a peripheral blood DNAm signature) to assess the biological fitness of patients with PSC and stratify them according to their risk of clinical events.
Materials and methods
Study population
The PSC study population was derived from a phase IIb, placebo-controlled trial of simtuzumab, a LOXL2 inhibitor, as described elsewhere.17 Since simtuzumab demonstrated no evidence of efficacy in this trial for clinical or histologic endpoints, both placebo- and simtuzumab-treated patients were included in the current analysis. Centrally read liver biopsies were obtained at baseline and fibrosis was staged according to the Ishak classification. For the purpose of this analysis, the study population was restricted to patients with no-to-mild fibrosis (Ishak F0-1) or cirrhosis (F5-6).
The healthy reference samples were chosen from a publicly available DNAm database18 such that the age and sex distribution of the chosen 50 samples matched the PSC dataset. Specifically, each reference sample was assigned a weight based on the age and sex distribution of the PSC cohort, such that the reference samples with greater weights were more like the PSC cohort than samples with lower weights. Fifty reference samples were then chosen at random using a method that was biased toward choosing samples with greater weight.
Sample collection and methylation analysis
DNA extracted from PBMCs was assayed for cytosine methylation using the Infinium Methylation Assay (850k platform), as described by the manufacturer. DNA was treated with sodium bisulfite to convert unmethylated cytosines to uracil, leaving methylated cytosines unchanged. The treated DNA sample was then denatured, neutralized, and isothermally amplified. The amplified DNA was fragmented, precipitated with isopropanol and re-suspended prior to hybridization onto BeadChips. The converted and non-converted amplified DNAs were hybridized to their corresponding probes, and excess DNA was washed away. Hybridized DNAs then underwent single-base extension and staining for labeling, followed by scanning on an Illumina iScan instrument for detection. Scanned images were then analyzed using Illumina Genome Studio (version 2.0) software. DNAm data were processed using the minfi R Bioconductor package version 1.2,19 and wateRmelon package version 1.18.20 DNAm data were quantile normalized.
Epigenetic age and determination of differential DNAm
Epigenetic age, based on the Horvath model, was calculated from the methylation beta values using the minfi's agep function. Age acceleration was calculated as the difference between an individual's DNAm age and chronological age. Differential DNAm analysis, using methylation M values, was restricted to include DNAm probes that do not overlap known single nucleotide polymorphisms (SNPs) and are not thought to be cross-reactive, as described previously.21,22 This analysis was implemented in rmSNPandCH function (part of the DMRcate package23), which resulted in removal of 51,695 probes. Tests for differential methylation were carried out using the limma R package version 3.3.24 A false discovery rate (FDR)-adjusted p value of 0.1 was used to identify differentially methylated probes. Probe to gene mapping was achieved using a processed version of an array manufacturer's microarray annotation file.25
Ingenuity Pathway Analysis
Genomic DNAm and the relationship between different methylation of genes was analyzed using Ingenuity® Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA). Networks of methylated genes were algorithmically generated based on their connectivity and assigned a score. Scores were used to rank signaling and developmental pathway networks according to their methylation content.
Statistical analyses
The PSC cohort was categorized as having low or high age acceleration based on the median age acceleration value in the cohort (<11.1 years vs. ≥11.1 years). Comparisons between groups were made using Wilcoxon rank-sum tests. In addition, time to first PSC-related clinical event (defined as ascending cholangitis, hepatic decompensation [e.g. ascites, variceal hemorrhage, hepatic encephalopathy], cholangiocarcinoma, hepatocellular carcinoma, liver transplantation, and death) was assessed using Kaplan-Meier and Cox proportional hazards regression analysis using the survival R package version 2.41.26
Study approval
The study was approved by the Institutional Review Boards of contributing institutions to this study. Written informed consent was obtained from each individual prior to his or her participation in the study.
Results
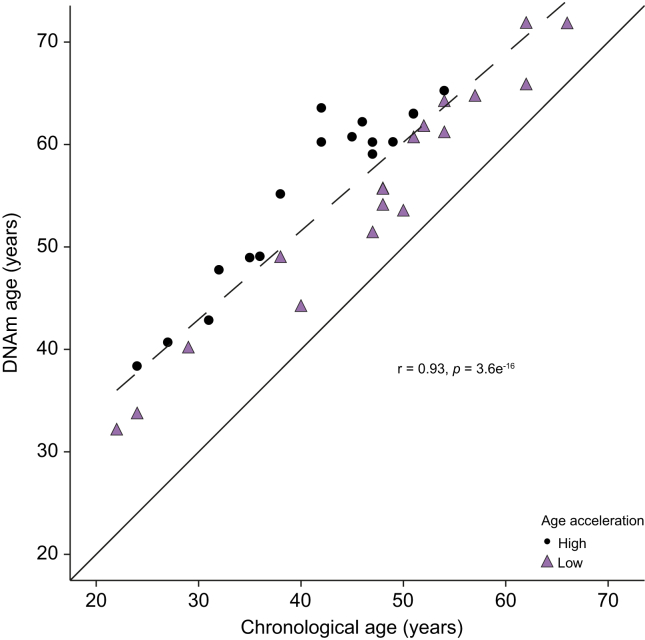
The study population included 36 patients with PSC enrolled in a 96-week, phase IIb study of simtuzumab, a monoclonal antibody directed against lysyl oxidase-like-2 (LOXL2).17 The median age of the PSC cohort was 47 years; median body mass index (BMI) was 25 kg/m2, and the majority (78%) were male. Additional demographic and clinical characteristics of the PSC cohort are included in Table 1. In this cohort, the DNAm age and chronological age were highly correlated (r = 0.93; p = 3.6 × 10-16; Fig. 1). Importantly, in every patient with PSC, the DNAm age was greater than the chronological age, with a median difference between DNAm and chronological age (age acceleration) of 11.1 years (range 3.4–21.6). In contrast to patients with PSC, the median age acceleration of an age/sex-matched healthy cohort18 (Table S1) was not significantly different from 0 (median age acceleration = 0.3 years; p = 0.9) (Fig. S1).
Table 1.
Demographic and baseline characteristics of patients with PSC.
| Total (n = 36) | Low age acceleration (n = 18) | High age acceleration (n = 18) | p value | |
|---|---|---|---|---|
| Demographics | ||||
| Age, years | 47 (38–51) | 49 (42–54) | 43.5 (35–48) | 0.10 |
| European descent | 30 (83.3) | 15 (83.3) | 15 (83.3) | 0.57 |
| African American | 4 (11.1) | 2 (11.1) | 2 (11.1) | |
| Native American or Alaska native | 1 (2.8) | 1 (5.6) | 0 (0) | |
| Other race | 1 (2.8) | 0 (0) | 1 (5.6) | |
| Female | 8 (22.2) | 4 (22) | 4 (22) | 1.0 |
| BMI, kg/m2 | 24.8 (23–30) | 24.8 (23–28) | 26.3 (23–31) | 0.70 |
| IBD | 17 (47.2) | 9 (50) | 8 (44.4) | 1.0 |
| UDCA use | 24 (66.7) | 12 (66.7) | 12 (66.7) | 1.0 |
| Liver biochemistry | ||||
| ALT, U/L | 52.5 (27–107) | 29 (21–52) | 83 (56–110) | 0.013 |
| ALP, U/L | 250 (119–459) | 125.5 (92–196) | 365.5 (261–599) | 0.0008 |
| GGT, U/L | 171.5 (69–459) | 84.5 (50–162) | 373 (178–521) | 0.006 |
| Liver histology | ||||
| F0-1 fibrosis | 13 (36.1) | 11 (61.1) | 2 (11.1) | 0.006 |
| F5-6 fibrosis | 23 (63.9) | 7 (38.9) | 16 (88.9) | |
| Hepatic collagen, % | 4.5 (2–9) | 2.7 (2–6) | 8.2 (4–14) | 0.005 |
| α-SMA expression, % | 4.6 (1–12) | 1.3 (1–5) | 11.4 (4–14) | 0.003 |
| Serum markers | ||||
| ELF | 9.9 (9–11) | 9.23 (8–11) | 10.9 (10–12) | 0.024 |
| IL-8, pg/ml | 29.5 (14–69) | 15.5 (11–30) | 53 (29–134) | 0.0007 |
| Total bile acids, ng/ml | 5,409 (2,249–9,973) | 4,390 (1,494–5,409) | 9,207 (6,575–43,978) | 0.11 |
Comparisons between groups were made using Wilcoxon rank-sum tests. The chi-squared test was used to compare categorical frequencies.
ALP, alkaline phosphatase; ALT, alanine aminotransferase; IL-8, interleukin-8; GGT, gamma-glutamyltransferase; PSC, PSC, primary sclerosing cholangitis; SMA, smooth muscle actin; UDCA, ursodeoxycholic acid.
Fig. 1.
Relationship between DNAm and chronological age.
DNAm age (y-axis) is presented as a function of chronological age (x-axis) for individuals with PSC. The regression line is presented as a dashed line. The identity line is presented as a solid line. Vertical distance between each point and the identity line is representative of age acceleration. Patients with high age acceleration (circles) have age acceleration at or above the median value, whereas patients with low age acceleration (triangles) have age acceleration below the median value. The scatterplot is annotated with the Pearson correlation coefficient and its p value between DNAm and chronological age. DNAm, DNA methylation; PSC, primary sclerosing cholangitis.
Association between age acceleration and hepatic fibrosis in PSC
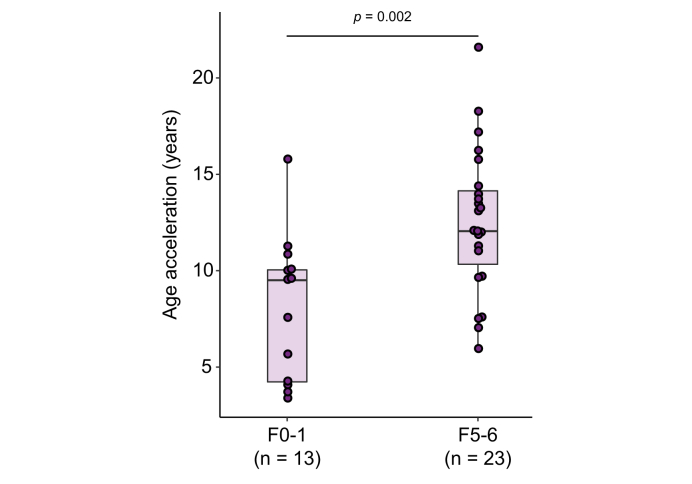
Next, we determined whether age acceleration is associated with the severity of liver fibrosis in patients with PSC. Specifically, we evaluated whether patients with PSC and no-to-mild fibrosis (Ishak stages F0-1 [n=13]) have less age acceleration compared to those with cirrhosis (Ishak stages F5-6 [n=23]). Our results indicate that patients with PSC and cirrhosis exhibit significantly more age acceleration than those with F0-1 fibrosis (median age acceleration, 12.1 vs. 9.5 years; p = 0.002) (Fig. 2). We also categorized patients with PSC into 2 groups, those at or above vs. below the median value of age acceleration (11.1 years) (Table 1). Compared with patients with low age acceleration, those with high age acceleration had greater hepatic collagen content (2.7% vs. 8.2%; p = 0.005) and α-smooth muscle actin (α-SMA) expression (1.3% vs. 11.4%; p = 0.003) on liver biopsy, and higher ELF scores (9.2 vs. 10.9; p = 0.024). As previously noted in patients with NASH-related liver fibrosis, these data support an association between hepatic fibrosis and age acceleration in patients with PSC.
Fig. 2.
Age acceleration in patients with PSC.
Age acceleration, the difference between DNAm and chronological age, plotted for patients with PSC based on Ishak fibrosis stage at baseline. Age acceleration is significantly greater in patients with Ishak F5-6 fibrosis vs. F0-1 fibrosis (p = 0.002). DNAm, DNA methylation; PSC, primary sclerosing cholangitis.
Associations between age acceleration and other baseline characteristics
No significant differences in sex, race, BMI, or use of ursodeoxycholic acid (UDCA) were observed between patients with PSC and low vs. high age acceleration (Table 1). Consistent with previous findings,27 the presence of inflammatory bowel disease (IBD) was not associated with age acceleration. However, median age was non-significantly lower in patients with high age acceleration (44 vs. 49 years, p = 0.10). Moreover, compared with patients with low age acceleration, those with high age acceleration had greater median serum alkaline phosphatase (ALP) (125.5 vs. 365.5 U/L; p <0.001), gamma-glutamyltransferase (GGT) (84.5 vs. 373 U/L; p = 0.006), and alanine aminotransferase (29 vs. 83 U/L; p = 0.013). In addition, serum levels of interleukin (IL)-8, which have been associated with disease progression, were higher in patients with high vs. low age acceleration (53 vs. 15.5 pg/ml; p = 0.0007).
Association between age acceleration and PSC-related clinical events
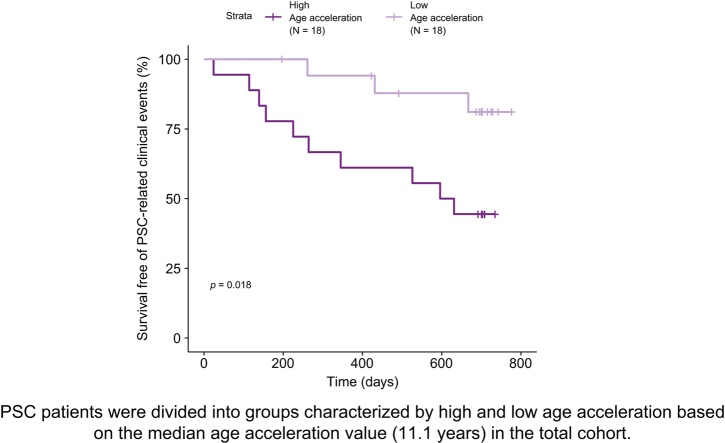
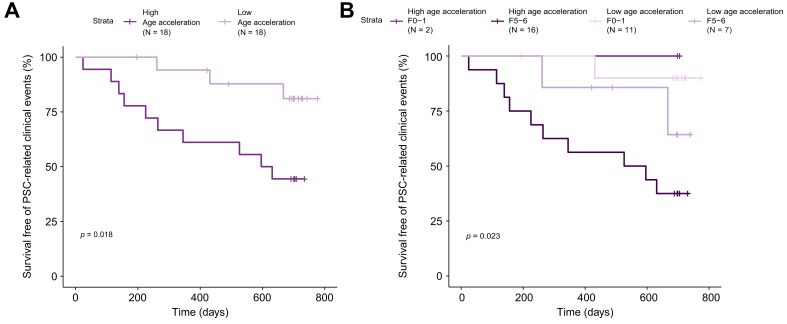
We next asked whether patients with PSC and high age acceleration had a greater risk of PSC-related clinical events (i.e. ascending cholangitis, hepatic decompensation, liver transplantation, cholangiocarcinoma) compared to the low age acceleration group. During a median follow-up of 23 months (range 1–26), 13 individuals (36%) developed PSC-related clinical events. In total, 10/18 (56%) patients with high age acceleration vs. 3/18 (17%) with low age acceleration had an event (Table 2). In Kaplan-Meier analysis, survival free of PSC-related clinical events was reduced in patients with PSC and high age acceleration (log-rank p = 0.018) (Fig. 3A). Compared to patients with low age acceleration, those with high age acceleration had an approximately 4-fold higher risk of PSC-related events (hazard ratio 4.19; 95% CI 1.15–15.24; p = 0.03). In patients with cirrhosis (F5-6) specifically, the presence of high age acceleration also conferred a greater likelihood of clinical disease progression (Fig. 3B). These results suggest that a greater difference between DNAm-based age and chronological age is associated with an increased risk of disease progression in PSC.
Table 2.
PSC-related clinical events according to age acceleration.
| Low age acceleration (n = 18) | High age acceleration (n = 18) | |
|---|---|---|
| All PSC progression events | 3 | 10 |
| Ascites | 0 | 1 |
| Bilirubin elevated | 0 | 1 |
| Ascending cholangitis | 3 | 3 |
| Hepatic encephalopathy | 0 | 2 |
| Esophageal varices | 0 | 3 |
PSC, primary sclerosing cholangitis.
Fig. 3.
Survival free of PSC-related clinical events stratified by age acceleration and fibrosis severity.
(A) Patients with PSC were divided into groups characterized by high and low age acceleration based on the median age acceleration value (11.1 years) in the total cohort. (B) Patients with PSC were divided into high and low age acceleration groups based on the median age acceleration value (11.1 years) for the cohort and by Ishak fibrosis stage. p value calculated using log-rank test. PSC, primary sclerosing cholangitis.
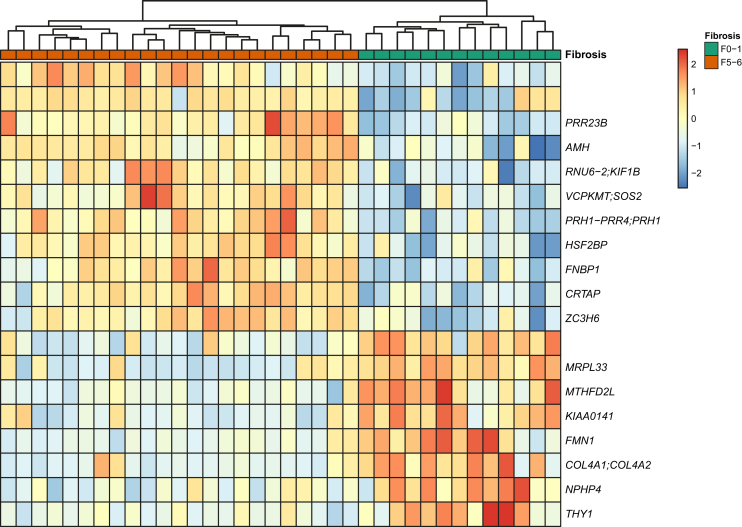
Differential methylation patterns according to fibrosis stage
To assess whether global DNAm patterns exist that distinguish patients with PSC and no-to-mild (F0-1) fibrosis from those with cirrhosis (F5-6), we performed differential DNAm analysis on the PSC cohort. After correcting for multiple testing (FDR cut-off of 0.1), we identified 19 differentially methylated probes between the low and high fibrosis groups that map to 20 genes (Fig. 4). A pathway enrichment analysis identified collagen formation as the top enriched pathway (FDR q-value = 0.003) based on the presence of 3 genes in the differentially methylated list: COL4A1 (collagen, type IV, alpha 1), COL4A2 (collagen, type IV, alpha 2), and CRTAP (cartilage associated protein).
Fig. 4.
Differentially methylated regions between patients with PSC and no-to-mild (Ishak F0-1) fibrosisvs.cirrhosis (F5-6).
Heatmap where rows are DNA methylation sites and columns are patients with PSC. PSC, primary sclerosing cholangitis.
Discussion
The current study identified age acceleration in patients with PSC enrolled in a controlled clinical trial. In contrast to other chronic diseases such as NASH or HIV infection, where age acceleration has been determined to be in the range of 5 to 7 years, far greater age acceleration was observed in the current PSC cohort.10,12,28 Specifically, PSC patients with minimal fibrosis or cirrhosis demonstrated median age acceleration of approximately 10 and 12 years, respectively, greater than a control population. These divergent levels of age acceleration, despite the presence of advanced fibrosis in both the NASH and PSC cohorts, suggest distinct epigenetic evidence of aging in these 2 liver diseases. Moreover, greater fibrosis on liver biopsy and higher serum markers of cholestasis were associated with additional age acceleration, thereby allowing for potential risk stratification from a peripheral blood sample.
The distinction between chronological and biological age is a useful one in chronic conditions such as PSC where the latency period between the initial diagnosis and clinical manifestations can be decades. Determination of the epigenetic clock and its deviation from chronological age could help gauge the unique impact of a disease on a particular individual. From a practical standpoint, the current study found that despite being 6 years younger chronologically than the low age acceleration group, the group with high age acceleration had more fibrosis, more severe cholestasis, and an approximately 4-fold increased risk of PSC-related clinical events. Notably, all portal hypertension-related clinical events were in the high age acceleration group, whereas only episodes of ascending cholangitis were observed in the low age acceleration group. Despite the small sample set of this study, the use of biological age assessed by this peripheral blood DNAm signature rather than chronological age may provide a novel method to risk stratify patients with this disease.
The cause(s) of the marked age acceleration observed in this PSC cohort is unclear; however, obesity, the use of UDCA, and concomitant IBD do not appear to play a role. The absence of an association between age acceleration and UDCA use is consistent with the lack of clear clinical benefit of UDCA in PSC. While the lack of association with concomitant IBD could be explained by the relatively quiescent intestinal disease in this cohort, additional study is warranted based on the prognostic impact of IBD on the progression of PSC.29,30 One potential explanation for our findings may be the presence of cellular senescence that has been reported in cholangiopathies including PSC. Sasaki et al. described the presence of markers of cholangiocyte senescence in PSC livers and found increased senescent cholangiocytes compared to controls.31 Subsequent studies confirmed the presence of senescence markers in PSC in the setting of intact telomere sequences, consistent with a non-replicative, stress-induced senescent phenotype.32,33 This secretory phenotype is marked by the significant expression and production of pro-inflammatory cytokines IL-6 and IL-8.32 The latter cytokine has particular relevance in PSC, as higher serum levels of IL-8 have been associated with an increased risk of clinical events.34 In the current study, compared to patients with PSC and low age acceleration, those with high age acceleration had greater serum levels of IL-8 and an increased risk of PSC-related complications. Thus, a link between cholangiocyte senescence, an inflammation-associated secretory phenotype, and elevated IL-8 levels associated with clinical decompensation are consistent with an overall global picture of organ dysfunction in PSC.34,35
Our data showing significant associations between age acceleration with serum ALP and GGT, as well as a trend to increased serum bile acids in patients with high age acceleration, suggest a link between the severity of cholestasis and accelerated aging in PSC. Indeed, prior studies have observed direct effects of bile acids on DNA methylation.36,37 The composition of specific bile acid species may also contribute to the inflammation-related senescent phenotype. A previous study found that bile acids such as deoxycholic acid contribute to the senescent and inflammatory characteristics of hepatic stellate cells, which combined with high fat feeding leads to fibrosis and hepatocarcinogenesis.38 Identification of specific bile acid species as a driver of liver disease is consistent with previous work that noted the development of liver cancer with deletion of the farnesoid X receptor, the major transcriptional regulator of bile acid homeostasis.39,40 Whether alterations in bile acids alone contribute to age acceleration or merely represent one notable feature of organ dysfunction brought on by age acceleration is unclear. However, the current experimental data suggests that the biologic clock represents an epigenetic maintenance system that may be distinct from the process of senescence.41 It would therefore be important to determine whether age acceleration in PSC occurs in the earliest stages of PSC, prior to the onset of the senescent phenotype and its associated inflammatory phenotype.
In conclusion, we have identified marked age acceleration in a PSC cohort with varying stages of fibrosis compared with a healthy control population based on a peripheral blood DNAm signature. In patients with PSC, age acceleration is associated with increased hepatic fibrosis, biochemical markers of cholestasis, and a higher risk of PSC-related clinical events. These findings provide a potentially novel and useful approach to risk stratification in PSC.
Financial support
This study was supported by Gilead Sciences.
Conflicts of interest
Michael Trauner has served as a consultant for Albireo, BiomX, Falk, Genfit, Gilead, Intercept, MSD, Novartis, Phenex and Regulus and received speaker's honoraria from Falk, Gilead, Intercept, MSD, Novartis and Roche. He further received travel grants from Abbvie, Falk, Roche and Gilead and unrestricted research grants from Albireo, Cymabay, Falk, Gilead, Intercept and MSD. He is also coinventor of a patent on the medical use of nor-UDCA filed by the Medical University of Graz. Yevgeniy Gindin, Zhaoshi Jiang, Chuhan Chung, G. Mani Subramanian, and Robert Myers are employees of and hold stock interest in Gilead Sciences. Aliya Gulamhusein has received speaking honoraria from AbbVie and Intercept and has served on advisory boards for Intercept. Kris Kowdley has received research support from Gilead, HighTide and Intercept, has served as a consultant and speaker for Gilead and Intercept, has served on advisory boards for Gilead, and has received royalties from Up-To-Date. Zachary Goodman has received research grant support from Gilead, Intercept, Novartis, Bristol-Myers Squibb, Allergan, and Merck. Michael Manns has received research support, served as a consultant, received lecture fees, and received travel support from Gilead, Intercept, Falk Pharma, and Novartis. Andrew Muir has received research grants from Gilead, Intercept, and NGM Pharmaceuticals, and has served on advisory boards for Gilead. Christopher Bowlus has served on advisory boards for BiomX, Intercept and GlaxoSmithKline and has received research grants from BiomX, Gilead, Intercept, CymaBay, Takeda, Bristol-Myers Squibb, GlaxoSmithKline, Tobira, Merck, TaiwanJ, Eli Lilly, Novartis, and Target Pharmasolutions.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
Yevgeniy Gindin, Chuhan Chung, G. Mani Subramanian, and Robert P. Myers designed this study and provided study oversight. Michael Trauner, Aliya Gulamhusein, Kris V. Kowdley, Cynthia Levy, Zachary Goodman, Michael P. Manns, Andrew J. Muir, and Christopher L. Bowlus served as study investigators and collected data. Michael Trauner, Yevgeniy Gindin, Chuhan Chung, and Zhaoshi Jiang provided data analysis and interpretation. Yevgeniy Gindin, Chuhan Chung, and Rob P. Myers drafted the manuscript. All authors reviewed and approved the manuscript prior to submission.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2019.11.004.
Supplementary data
References
- 1.Lazaridis K.N., LaRusso N.F. Primary sclerosing cholangitis. N Engl J Med. 2016;375(25):2501–2502. doi: 10.1056/NEJMc1613273. [DOI] [PubMed] [Google Scholar]
- 2.Chapman R., Fevery J., Kalloo A., Nagorney D.M., Boberg K.M., Shneider B. Diagnosis and management of primary sclerosing cholangitis. Hepatology. 2010;51(2):660–678. doi: 10.1002/hep.23294. [DOI] [PubMed] [Google Scholar]
- 3.de Vries E.M., de Krijger M., Farkkila M., Arola J., Schirmacher P., Gotthardt D. Validation of the prognostic value of histologic scoring systems in primary sclerosing cholangitis: an international cohort study. Hepatology. 2017;65(3):907–919. doi: 10.1002/hep.28963. [DOI] [PubMed] [Google Scholar]
- 4.de Vries E.M., Verheij J., Hubscher S.G., Leeflang M.M., Boonstra K., Beuers U. Applicability and prognostic value of histologic scoring systems in primary sclerosing cholangitis. J Hepatol. 2015;63(5):1212–1219. doi: 10.1016/j.jhep.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 5.de Vries E.M.G., Farkkila M., Milkiewicz P., Hov J.R., Eksteen B., Thorburn D. Enhanced liver fibrosis test predicts transplant-free survival in primary sclerosing cholangitis, a multi-centre study. Liver Int. 2017;37(10):1554–1561. doi: 10.1111/liv.13402. [DOI] [PubMed] [Google Scholar]
- 6.Chen B.H., Marioni R.E., Colicino E., Peters M.J., Ward-Caviness C.K., Tsai P.C. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging. 2016;8(9):1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath S., Erhart W., Brosch M., Ammerpohl O., von Schonfels W., Ahrens M. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacalini M.G., Franceschi C., Gentilini D., Ravaioli F., Zhou X., Remondini D. Molecular aging of human liver: an epigenetic/transcriptomic signature. J Gerontol A Biol Sci Med Sci. 2019;74(1):1–8. doi: 10.1093/gerona/gly048. [DOI] [PubMed] [Google Scholar]
- 10.Horvath S., Levine A.J. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212(10):1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine A.J., Quach A., Moore D.J., Achim C.L., Soontornniyomkij V., Masliah E. Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol. 2016;22(3):366–375. doi: 10.1007/s13365-015-0406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loomba R., Gindin Y., Jiang Z., Lawitz E., Caldwell S., Djedjos C.S. DNA methylation signatures reflect aging in patients with nonalcoholic steatohepatitis. JCI Insight. 2018;3(2) doi: 10.1172/jci.insight.96685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., Bjornsson E.S., Charatcharoenwitthaya P. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149(2):389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhala N., Angulo P., van der Poorten D., Lee E., Hui J.M., Saracco G. The natural history of nonalcoholic fatty liver disease with advanced fibrosis or cirrhosis: an international collaborative study. Hepatology. 2011;54(4):1208–1216. doi: 10.1002/hep.24491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christiansen L., Lenart A., Tan Q., Vaupel J.W., Aviv A., McGue M. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15(1):149–154. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marioni R.E., Shah S., McRae A.F., Ritchie S.J., Muniz-Terrera G., Harris S.E. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44(4):1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muir A.J., Levy C., Janssen H.L.A., Montano-Loza A.J., Shiffman M.L., Caldwell S. Simtuzumab for primary sclerosing cholangitis: phase 2 study results with insights on the natural history of the disease. Hepatology. 2019;69(2):684–698. doi: 10.1002/hep.30237. [DOI] [PubMed] [Google Scholar]
- 18.Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pidsley R., Y Wong C.C., Volta M., Lunnon K., Mill J., Schalkwyk L.C. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters T.J., Buckley M.J., Statham A.L., Pidsley R., Samaras K., V Lord R. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen KD. IlluminaHumanMethylationEPICanno.ilm10b2.hg19: Annotation for Illumina's EPIC methylation arrays. 0.6 ed.
- 26.Therneau TM. A Package for Survival Analysis in S. version 2.38 ed2015.
- 27.Ventham N.T., Kennedy N.A., Adams A.T., Kalla R., Heath S., O'Leary K.R. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun. 2016;7:13507. doi: 10.1038/ncomms13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross A.M., Jaeger P.A., Kreisberg J.F., Licon K., Jepsen K.L., Khosroheidari M. Methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA. Mol Cell. 2016;62(2):157–168. doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weismuller T.J., Trivedi P.J., Bergquist A., Imam M., Lenzen H., Ponsioen C.Y. Patient age, sex, and inflammatory bowel disease phenotype associate with course of primary sclerosing cholangitis. Gastroenterology. 2017;152(8):1975–1984. doi: 10.1053/j.gastro.2017.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muir A.J., Levy C., Janssen H.L.A., Montano-Loza A.J., Shiffman M.L., Caldwell S. Simtuzumab for primary sclerosing cholangitis: phase 2 study results with insights on the natural history of the disease. Hepatology. 2019;69(2):684–698. doi: 10.1002/hep.30237. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki M., Ikeda H., Haga H., Manabe T., Nakanuma Y. Frequent cellular senescence in small bile ducts in primary biliary cirrhosis: a possible role in bile duct loss. J Pathol. 2005;205(4):451–459. doi: 10.1002/path.1729. [DOI] [PubMed] [Google Scholar]
- 32.Tabibian J.H., O'Hara S.P., Splinter P.L., Trussoni C.E., LaRusso N.F. Cholangiocyte senescence by way of N-ras activation is a characteristic of primary sclerosing cholangitis. Hepatology. 2014;59(6):2263–2275. doi: 10.1002/hep.26993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng L., Quezada M., Levine P., Han Y., McDaniel K., Zhou T. Functional role of cellular senescence in biliary injury. Am J Pathol. 2015;185(3):602–609. doi: 10.1016/j.ajpath.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vesterhus M., Holm A., Hov J.R., Nygard S., Schrumpf E., Melum E. Novel serum and bile protein markers predict primary sclerosing cholangitis disease severity and prognosis. J Hepatol. 2017;66(6):1214–1222. doi: 10.1016/j.jhep.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 35.Zweers S.J., Shiryaev A., Komuta M., Vesterhus M., Hov J.R., Perugorria M.J. Elevated interleukin-8 in bile of patients with primary sclerosing cholangitis. Liver Int. 2016;36(9):1370–1377. doi: 10.1111/liv.13092. [DOI] [PubMed] [Google Scholar]
- 36.Bajpai M., Kessel R., Bhagat T., Nischal S., Yu Y., Verma A. High resolution integrative analysis reveals widespread genetic and epigenetic changes after chronic in-vitro acid and bile exposure in Barrett's epithelium cells. Genes Chromosomes Cancer. 2013;52(12):1123–1132. doi: 10.1002/gcc.22106. [DOI] [PubMed] [Google Scholar]
- 37.Baptissart M., Sedes L., Holota H., Thirouard L., Martinot E., de Haze A. Multigenerational impacts of bile exposure are mediated by TGR5 signaling pathways. Sci Rep. 2018;8(1):16875. doi: 10.1038/s41598-018-34863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimoto S., Loo T.M., Atarashi K., Kanda H., Sato S., Oyadomari S. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 39.Degirolamo C., Modica S., Vacca M., Di Tullio G., Morgano A., D'Orazio A. Prevention of spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice by intestinal-specific farnesoid X receptor reactivation. Hepatology. 2015;61(1):161–170. doi: 10.1002/hep.27274. [DOI] [PubMed] [Google Scholar]
- 40.Kim I., Morimura K., Shah Y., Yang Q., Ward J.M., Gonzalez F.J. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28(5):940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowe D., Horvath S., Raj K. Epigenetic clock analyses of cellular senescence and ageing. Oncotarget. 2016;7(8):8524–8531. doi: 10.18632/oncotarget.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.