Abstract
Cryptosporidium spp., Giardia spp. and microsporidia are important intestinal protozoa responsible for diarrhea in humans and other mammals. China is a major chicken-raising country, and studies on these protozoa in chickens have important public health significance. Here, we investigated the prevalence and genetic characterization of these parasites in chickens from Ezhou City, Hubei Province, China. In total, 206 stool specimens were collected from chickens in four villages of Ezhou between July 2014 and February 2015. Genomic DNA of each specimen was tested by nested PCR based on the Cryptosporidium small subunit rRNA gene, the Giardia intestinalis triose phosphate isomerase gene, and the internal transcribed spacer of the Enterocytozoon bieneusi rRNA gene, respectively. The public health significance of G. intestinalis and E. bieneusi identified in our study was evaluated via phylogenetic analysis. The infection rates were determined to be 2.43% (5/206), 8.25% (17/206), and 1.94% (4/206) for Cryptosporidium, G. intestinalis, and E. bieneusi, respectively. One sample showed coinfection with G. intestinalis and E. bieneusi. Meanwhile, sequence analysis of the PCR-positive samples showed that the Cryptosporidium was C. baileyi, G. intestinalis was assemblage C, and E. bieneusi was genotype D and novel genotype EZ0008. This is the first report of zoonotic G. intestinalis assemblage C in chickens in the world, and the first report of zoonotic E. bieneusi genotype D in chickens in China. These findings indicate new transmission dynamics and molecular epizootiology.
Keywords: Cryptosporidium, Giardia intestinalis, Enterocytozoon bieneusi, assemblage C, genotype D
Introduction
Cryptosporidium spp., Giardia spp. and microsporidia are important intestinal protozoa of humans, livestock, and wild animals, which cause acute or self-limiting diarrhea (1–3). The diseases caused by infection with these parasites are distributed worldwide (4, 5). Cryptosporidium spp. often infect immunodeficient patients (especially HIV-infected persons), children and the elderly, which can be fatal (6, 7). Giardia intestinalis (G. intestinalis), which is the etiologic agent of giardiasis, usually appears in tourists, resulting in diarrhea called “Traveler's Diarrhea.” Zoonotic giardiasis is one of the ten principal parasitoses threatening human health worldwide (1). Microsporidia is mainly associated with immunocompromised individuals, causing wasting syndrome. One of the most frequently identified microsporidian species in fecal samples of clinical patients and domestic animals worldwide is Enterocytozoon bieneusi (E. bieneusi) (8). Many outbreaks among humans have been caused by these parasites (9–13), for example, the massive Cryptosporidium-associated waterborne outbreak in Milwaukee, Wisconsin in 1993 (14). Such outbreaks pose significant challenges to public health. In the United States, it is estimated that 748,000 cryptosporidiosis cases occur every year (15). According to the World Health Organization, annually, there are 500,000 emerging giardiasis cases globally. The prevalence of human microsporidiosis ranges from 0 to 50% depending on the geographical region (16).
Epidemiological data on human cryptosporidiosis, giardiasis and microsporidiosis has confirmed their common occurrence in China (6, 17). For example, the occurrence rates of these three parasitoses were 13.49, 6.75, and 13.49% respectively in outpatients suffering from diarrhea in Shanghai, China in 2014 (6) and 2.0, 1.4, and 0.2% respectively in children with a history of diarrhea in Hubei province, China in 2017 (13).
Poultry play a significant role in the agricultural economy of China. As a major chicken-raising country, the total population of layer chicken was 2.6 billion in 2013, accounting for 37.3% of the total number in the world (http://kids.fao.org/glipha/). Up to now, there is limited information regarding the distribution and molecular characterization of Cryptosporidium spp., G. intestinalis, and E. bieneusi in chickens in China. Due to transmission by these parasites is the fecal-oral route, often through direct contact with feces from infected animals or people, contaminated food and/or water (18), together with the close contact between chickens and humans in rural areas, knowledge of parasitic species in chickens has important public health significance.
In the present study, we examined the occurrence of Cryptosporidium spp., G. intestinalis and E. bieneusi in chickens from four villages in Ezhou City, Hubei Province, China, identified the species/genotypes of these intestinal protozoa, and assessed their potential for zoonotic transmission. Furthermore, the public health significance of G. intestinalis and E. bieneusi identified in our study was evaluated via phylogenetic analysis.
Materials and Methods
Specimen Collection
Between July 2014 and February 2015, 206 fresh stool specimens were collected from chickens on four different farms from villages in Ezhou City, Hubei Province, China. Animal details, including their location, age and sampling time, were recorded. The chickens ranged in age from 2 months to 1 year old. On these farms, they were kept separately in individual cages, and fresh fecal excretion was collected from cages with care, avoiding contamination from other cages. Each sample was >5 g.
All specimens were taken to laboratory in a cool box at 4°C, registered and stored at −20°C until DNA extraction.
DNA Extraction
Sufficient specimens (200–300 mg of each stool specimen) were used for DNA extraction and purification using a QIAamp® Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany), following the manufacturer-recommended procedures. The extracted genomic DNA samples were stored at −20°C before polymerase chain reaction (PCR).
Parasite Identification in Animal Samples
Cryptosporidium spp., G. intestinalis, and E. bieneusi in the fecal specimens were detected using individual nested PCR and the sequences analyzed were of the small subunit (SSU) rRNA gene, the triose phosphate isomerase (tpi) gene, and the internal transcribed spacer (ITS) of the rRNA gene, respectively. The primers for each parasite referred to previous descriptions (6, 19–21).
The PCR was conducted in a 25 μl reaction mixture including 12.5 μl Taq mix (2×), 11.3 μl nuclease-free water, 1 μl genomic DNA template (20–60 ng/μl), 0.1 μl of sense and antisense primers each (100 μM). For the nested PCR, 1 μl of the first PCR product was used as the template. The genes from Cryptosporidium spp. and G. intestinalis were amplified using GoTaq® Green Master Mix (Promega, code no. M7123, Madison, WI, USA), while the gene from E. bieneusi was amplified using Premix Taq® (Takara, code no. RR901M, Dalian, China).
The PCR cycling condition for Cryptosporidium spp. was as follows: denaturation at 94°C for 1 min, followed by 35 cycles (94°C for 10 s, 55°C for 30 s and 72°C for 1 min) and a final extension step at 72°C for 10 min. The secondary reaction was carried out similarly. The cycling condition for G. intestinalis was: denaturation at 94°C for 5 min, followed by 35 cycles (94°C for 45 s, 57.5°C for 45 s, and 72°C for 1 min) and a final extension step at 72°C for 7 min. For E. bieneusi, the cycling condition was the same to G. intestinalis, except the annealing temperature was 55°C. The secondary reactions were identical to the primary PCR cycling conditions.
Each DNA sample was analyzed three times to ensure the reliability of results with positive (templates were positive nucleic acids of Cryptosporidium, G. intestinalis and E. bieneusi stored in our laboratory) and negative (template was nuclease-free water) controls in each PCR. All final secondary PCR products were visualized by electrophoresis in 2% agarose gels after ethidium bromide staining.
Sequencing of Positive Genes
Positive PCR products were treated by a Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, USA), and sequenced in both directions on an ABI 3730 DNA analyzer (Applied Biosystems), using the secondary primers. Sequencing was performed by the Shanghai Sunny Biotechnology Co., Ltd. (Shanghai, China).
DNA Sequences and Statistical Analysis
ContigExpress was used to evaluate the wave peak and assemble the sequences. Each sequence was compared against sequences in the NCBI database and analyzed using Clustal X 1.83 and MEGA 5, determining the species/genotypes of parasites. To assess the phylogenetic relationships of the sequences identified in our study and the known ones, the neighbor-joining analyses of G. intestinalis assemblages at the tpi locus and E. bieneusi genotypes at the ITS locus were calculated by the Kimura two-parameter model, and 1,000 replicates were used. All statistical analyses were performed using SPSS version 17.0 (SPSS Inc., Chicago, IL). Association of age category and parasitic infections was analyzed using the chi-square test, and P < 0.05 was considered statistically significant.
Results
Occurrence of Cryptosporidium spp., G. intestinalis, and E. bieneusi
In chickens (n = 206), the positive rates of Cryptosporidium, G. intestinalis, and E. bieneusi determined using nested PCR were 2.43% (5/206), 8.25% (17/206), and 1.94% (4/206), respectively (Table 1). Polyparasitism was observed in one specimen from village A, which was coinfected with G. intestinalis and E. bieneusi. Specimens positive for Cryptosporidium and E. bieneusi were not restricted to a particular village, whereas G. intestinalis was only detected in village A (Table 2). No obvious age-associated difference in parasitic infections was observed in the chickens (P > 0.05).
Table 1.
Occurrence of Cryptosporidium, Giardia, and microsporidia.
| Genus | Number of positive specimens (n%) | Species | Genotype |
|---|---|---|---|
| Cryptosporidium | 5 (2.43%) | C. baileyi | – |
| Giardia | 17 (8.25%) | G. intestinalis | Assemblage C |
| Enterocytozoon | 4 (1.94%) | E. bieneusi | Genotype D (n = 2)/EZ0008 (n = 2) |
| Total specimens | 206 |
Table 2.
Distribution of positive samples in different villages.
| Village | Number of specimens | Cryptosporidium | G. intestinalis | E. bieneusi |
|---|---|---|---|---|
| A | 151 | – | 17 (11.3%) | 1 (0.66%) |
| B | 20 | 4 (20.0%) | – | – |
| C | 19 | – | – | 2 (10.5%) |
| D | 16 | 1 (6.25%) | – | 1 (6.25%) |
Samples were analyzed by nested PCR.
Molecular Analyses of the Parasites
Five Cryptosporidium-positive specimens were identified by nested PCR. DNA sequencing followed by alignment of the SSU rRNA gene fragments revealed these isolates belonged to C. baileyi. Of them, two sequences (four cases: KY448454, KY448455, KY448457, and KY448458) showed 100% homology with that previously reported, and one novel sequence (KY448456) was found.
Seventeen specimens were identified as G. intestinalis-positive by nested PCR. Sequence analysis of the tpi gene indicated that these isolates belonged to assemblage C. Among them, five sequences (13 cases: KY448449, KY448450, KY448460-KY448469, and KY448471) were 100% identical to that previously reported, and two novel sequences (four cases: KY448459/KY448448 and KY448447/KY448470) were found.
DNA sequencing and analysis of the ITS gene by nested PCR showed that the two E. bieneusi-positive specimens (KY448446 and KY448451) were identical to zoonotic genotype D. One novel genotype (two cases: KY448452 and KY448453) was found and named EZ0008.
Phylogenetic Analysis
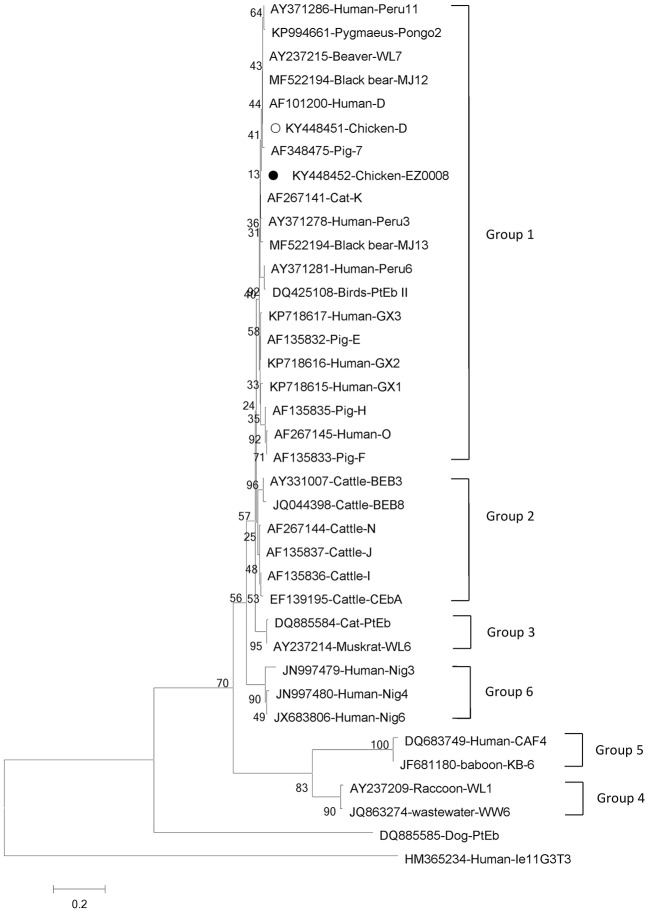
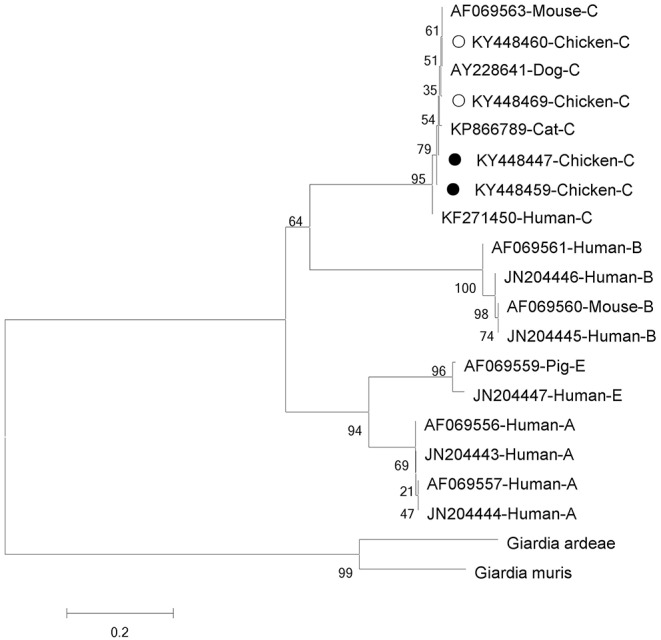
Phylogenetic analyses of G. intestinalis and E. bieneusi were performed to understand the relationships on the basis of published nucleotide sequences. The two novel sequences of G. intestinalis belong to assemblage C, which can infect mice, dogs, cats, and humans (Figure 1). The novel genotype of E. bieneusi was phylogenetically related to Group 1, which includes most of the human pathogenic genotypes (Figure 2).
Figure 1.
Phylogenetic relationships between assemblages of Giardia intestinalis. The relationships were inferred using neighbor-joining analysis of triose phosphate isomerase (tpi) sequences based on genetic distance calculated by the Kimura two-parameter model. Each sequence is marked with its accession number, host origin and assemblage. The numbers on the branches are percentage bootstrapping values from 1,000 replicates. Open and solid circles, respectively, indicate previously known and novel sequences identified in this study.
Figure 2.
Phylogenetic relationships between genotype groups of Enterocytozoon bieneusi. The relationships were inferred using neighbor-joining analysis of internal transcribed spacer (ITS) sequences based on genetic distance calculated by the Kimura two-parameter model. Each sequence is marked with its accession number, host origin and genotype designation. The numbers on the branches are percentage bootstrapping values from 1,000 replicates. Open and solid circles, respectively, indicate previously known and novel sequences identified in this study.
Discussion
To date, 38 species of Cryptosporidium have been identified in diverse hosts, totaling more than 40 subtypes infecting mammals (22–24). Among these, C. hominis and C. parvum are the main pathogens infecting humans and responsible for approximately 90% of human cryptosporidiosis (25). The reported main species of Cryptosporidium in chickens include C. meleagridis, C. baileyi (26, 27), C. parvum (28), C. galli (29), and avian genotype II (30), and C. meleagridis, C. parvum are zoonotic. In this study, only C. baileyi was identified in chickens, and the occurrence rate was 2.43%, which was in accord with previous reports (0–33.3% in China, 0.7% in Iran, 4.8% in Jordan) (27, 31, 32). One novel sequence (KY448456) was found, which has a high homology with isolate GU377273 from ostrich, based on sequence alignment.
Based on genetic analysis, G. intestinalis has been grouped into 8 assemblages (A–H) (33). Assemblages A and B are the major genotypes infecting humans, and assemblages C (6), E (34), and F (35) also infect people. Only zoonotic genotypes of assemblages A and B have been reported in chickens (36). Interestingly, all G. intestinalis-positive chickens on farm A in this study were found to be infected with assemblage C. Assemblage C of G. intestinalis usually occurs in dogs (4, 37), and occasionally in cats (38), coyotes (39), and humans (6). To our knowledge, this is the first report of assemblage C in chickens. Two novel sequences (four cases: KY448459/KY448448 and KY448447/KY448470) were identified, which, based on sequence alignment and phylogenetic analysis, have high homology with isolate KP866789 from cat, and also have homology with isolate KF271450 from human, indicating public health significance. A survey of diarrhea patients in Shanghai, China, in 2013 indicated that the infection rate with assemblage C was up to 6.35% (6). Thus, G. intestinalis-infected chickens might pose a great risk of human infection in this area. More specimens and deeper study should be undertaken to understand the transmission dynamics.
E. bieneusi has about 11 genotype groups, and Group 1 contains most zoonotic genotypes; the other groups contain mostly host-adapted genotypes (40). Several genotypes of E. bieneusi in chickens have been reported, including genotype J in Germany (41), genotype Peru8 in Brazil (42), genotypes Henan-IV, and CC-1 in China (43) and genotypes Peru6, Peru11, Type IV, and D in Brazil (44), which are zoonotic except for CC-1. E. bieneusi genotype D belongs to Group 1 and is the most common zoonotic genotype, which has been found in humans, dogs, cats, rhesus monkeys, some livestock and wild mammals (18, 21). The genotype is also widespread in wastewater (45). The first report of genotype D infecting chickens came from Minas Gerais, Brazil (9.27%) (44). However, only one report has been published concerning E. bieneusi infection in chickens in China, with genotypes Henan-IV and CC-1 (43). In our study, genotype D and novel EZ0008 were identified for the first time in chickens in China, suggesting multi-genotype infections of E. bieneusi in chickens in this country. The novel genotype (two cases: KY448452 and KY448453) was related to Group 1 and has high homology with isolate AF348475 from pig and isolate AF101200 from human, indicating public health significance. Our results imply that humans in rural areas are at significant risk of infection by E. bieneusi because of intimate contact with livestock.
Overall, chickens from Ezhou were determined to carry Cryptosporidium, G. intestinalis, and E. bieneusi. Among the parasites detected, species of G. intestinalis assemblage C and E. bieneusi genotype D are zoonotic, and can be transmitted through water or food, resulting in giardiasis and microsporidiosis, respectively. Presently, prevention is the predominant measure to control these diseases. Knowledge of the parasite genetic profile, source of infection, mode of transmission and susceptible population is beneficial for effective control. Considering these zoonoses in Ezhou, Hubei, China, peasants should prevent direct/indirect contact with chicken feces, and also avoid discharging these feces to nearby water, because contamination of water sources is the principal cause of outbreaks and prevalence of these parasitic diseases (46). Zoonotic species of G. intestinalis and E. bieneusi/EZ0008 were identified in villages A, C, and D in this study (Table 2); in particular, we found numerous examples of G. intestinalis assemblage C in village A. Although no zoonotic parasites were detected in village B in our study, their presence cannot be excluded because the sample number was small. In future, we will investigate the occurrence rates of these three parasites in humans (especially diarrhea patients) and waters in Ezhou, to clarify their patterns of transmission and help with risk control. In summary, the present study provides important reference data on the epidemiology of Cryptosporidium, G. intestinalis, and E. bieneusi infections in chickens.
Data Availability Statement
The nucleotide sequences generated in present study were submitted to the NCBI GenBank with accession numbers KY448446 to KY448471.
Ethics Statement
Ethical clearance for the collection and examination of chicken feces samples was obtained from the Guide for the Care and Use of Laboratory Animals of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention. The protocol was approved by the Laboratory Animal Welfare & Ethics Committee (LAWEC), National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention, China (reference no. 2012-12). Before beginning our work, we contacted the farm owners and obtained their permission. No specific permits were required for the described field studies. We directly collected fecal specimens from the cages using plastic bags, requiring very little contact with the chickens. The chickens were not harmed in any way during the procedure.
Author Contributions
YS, SC, and JC conceived and designed the experiments. SC, YJ, HL, ZY, and LS performed the experiments. MX and SC analyzed the data. SC wrote the manuscript. YS and JC revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Zu'an He, Qiong Zhang, Jing Wang (Hubei Provincial Center for Disease Control and Prevention), and Yu Luo, Meihong Yang, Shun Liu, Qiang Sun, Conggang Zhou, Shaobing Yan (Ezhou Center for Disease Control and Prevention) for their assistance in specimen collection.
Footnotes
Funding. This work was supported by the Chinese Special Program for Scientific Research of Public Health (Grant No. 201502021), the Fourth Round of Three-Year Public Health Action Plan of Shanghai, China (Grant No. 15GWZK0101) and the Shanghai Municipal Commission of Health and Family Planning (Grant Nos. 20164Y0085 and 20164Y0225).
References
- 1.Feng YY, Xiao LH. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. (2011) 24:110–40. 10.1128/CMR.00033-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao LH. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. (2010) 124:80–9. 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 3.Mathis A, Weber R, Deplazes P. Zoonotic potential of the microsporidia. Clin Microbiol Rev. (2005) 18:423–45. 10.1128/CMR.18.3.423-445.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao LH, Fayer R. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol. (2008) 38:1239–55. 10.1016/j.ijpara.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 5.Henriques-Gil N, Haro M, Izquierdo F, Fenoy S, del Aguila C. Phylogenetic approach to the variability of the microsporidian Enterocytozoon bieneusi and its implications for inter- and intrahost transmission. Appl Environ Microbiol. (2010) 76:3333–42. 10.1128/AEM.03026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Shen YJ, Yin JH, Yuan ZY, Jiang YY, Xu YX, et al. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect Dis. (2014) 14:25. 10.1186/1471-2334-14-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Current WL, Garcia LS. Cryptosporidiosis. Clin Microbiol Rev. (1991) 4:325–58. 10.1128/cmr.4.3.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Didier ES. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. (2005) 94:61–76. 10.1016/j.actatropica.2005.01.010 [DOI] [PubMed] [Google Scholar]
- 9.Karanis P, Kourenti C, Smith H. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J Water Health. (2007) 5:1–38. 10.2166/wh.2006.002 [DOI] [PubMed] [Google Scholar]
- 10.Giangaspero A, Berrilli F, Brandonisio O. Giardia and Cryptosporidium and public health: the epidemiological scenario from the Italian perspective. Parasitol Res. (2007) 101:1169–82. 10.1007/s00436-007-0598-4 [DOI] [PubMed] [Google Scholar]
- 11.Chalmers RM, Elwin K, Hadfield SJ, Robinson G. Sporadic human cryptosporidiosis caused by Cryptosporidium cuniculus, United Kingdom, 2007–2008. Emerg Infect Dis. (2011) 17:536–8. 10.3201/eid1703.100410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng YY, Wang L, Duan LP, Gomez-Puerta LA, Zhang LX, Zhao XK, et al. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg Infect Dis. (2012) 18:312–4. 10.3201/eid1802.110666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Fan YY, Koehler AV, Ma GX, Li T, Hu M, et al. First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect Genet Evol. (2017) 51:127–31. 10.1016/j.meegid.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 14.Hoxie NJ, Davis JP, Vergeront JM, Nashold RD, Blair KA. Cryptosporidiosis-associated mortality following a massive waterborne outbreak in Milwaukee, Wisconsin. Am J Public Health. (1997) 87:2032–5. 10.2105/AJPH.87.12.2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florescu DF, Sandkovsky U. Cryptosporidium infection in solid organ transplantation. World J Transplant. (2016) 6:460–71. 10.5500/wjt.v6.i3.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghoyounchi R, Ahmadpour E, Spotin A, Mahami-Oskouei M, Rezamand A, Aminisani N, et al. Microsporidiosis in Iran: A systematic review and meta-analysis. Asian Pac J Trop Med. (2017) 10:341–50. 10.1016/j.apjtm.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 17.Xu HL, Jin Y, Wu WX, Li P, Wang L, Li N, et al. Genotypes of Cryptosporidium spp., Enterocytozoon bieneusi and Giardia duodenalis in dogs and cats in Shanghai, China. Parasit Vectors. (2016) 9:121. 10.1186/s13071-016-1409-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zambrano LD, Levy K, Menezes NP, Freeman MC. Human diarrhea infections associated with domestic animal husbandry: a systematic review and meta-analysis. Trans R Soc Trop Med Hyg. (2014) 108:313–25. 10.1093/trstmh/tru056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao LH, Escalante L, Yang CF, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. (1999) 65:1578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. (2003) 9:1444. 10.3201/eid0911.030084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulaiman IM, Fayer R, Yang CF, Santin M, Matos O, Xiao LH. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol Res. (2004) 92:328–34. 10.1007/s00436-003-1049-5 [DOI] [PubMed] [Google Scholar]
- 22.Jiang YY, Ren JH, Yuan ZY, Liu AQ, Zhao H, Liu H, et al. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect Dis. (2014) 14:555. 10.1186/s12879-014-0555-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunha FS, Peralta RHS, Peralta JM. New insights into the detection and molecular characterization of Cryptosporidium with emphasis in Brazilian studies: a review. Rev Inst Med Trop São Paulo. (2019) 61:e28. 10.1590/S1678-9946201961028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan U, Zahedi A, Paparini A. Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol. (2016) 38:535–47. 10.1111/pim.12350 [DOI] [PubMed] [Google Scholar]
- 25.Elwin K, Hadfield SJ, Robinson G, Chalmers RM. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000-2008. Epidemiol Infect. (2012) 140:673–83. 10.1017/S0950268811000860 [DOI] [PubMed] [Google Scholar]
- 26.Wang RJ, Jian FC, Sun YP, Hu QS, Zhu JJ, Wang F, et al. Large-scale survey of Cryptosporidium spp. in chickens and Pekin ducks (Anas platyrhynchos) in Henan, China: prevalence and molecular characterization. Avian Pathol. (2010) 39:447–51. 10.1080/03079457.2010.518314 [DOI] [PubMed] [Google Scholar]
- 27.Liao C, Wang T, Koehler AV, Fan YY, Hu M, Gasser RB. Molecular investigation of Cryptosporidium in farmed chickens in Hubei Province, China, identifies 'zoonotic' subtypes of C. meleagridis. Parasit Vectors. (2018) 11:484. 10.1186/s13071-018-3056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helmy YA, Krucken J, Abdelwhab EM, Samson-Himmelstjerna G, Hafez HM. Molecular diagnosis and characterization of Cryptosporidium spp. in turkeys and chickens in Germany reveals evidence for previously undetected parasite species. PLoS ONE. (2017) 12:e0177150. 10.1371/journal.pone.0177150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassouha M, Soukkarieh C, Alkhaled A. First genotyping of Cryptosporidium spp. in pre-weaned calves, broiler chickens and children in Syria by PCR-RFLP analysis. Vet Parasitol. (2016) 225:86–90. 10.1016/j.vetpar.2016.06.009 [DOI] [PubMed] [Google Scholar]
- 30.Wang LM, Xue X, Li JQ, Zhou QJ, Yu YC, Du AF. Cryptosporidiosis in broiler chickens in Zhejiang Province, China: molecular characterization of oocysts detected in fecal samples. Parasite. (2014) 21:36. 10.1051/parasite/2014035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamidinejat H, Jalali MHR, Jafari RA, Nourmohammadi K. Molecular determination and genotyping of Cryptosporidium spp. in fecal and respiratory samples of industrial poultry in Iran. Asian Pac J Trop Med. (2014) 7:517–20. 10.1016/S1995-7645(14)60086-9 [DOI] [PubMed] [Google Scholar]
- 32.Hijjawi N, Mukbel R, Yang RC, Ryan U. Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet Parasitol. (2016) 228:116–20. 10.1016/j.vetpar.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 33.Monis PT, Andrews RH, Mayrhofer G, Ey PL. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect Genet Evol. (2003) 3:29–38. 10.1016/S1567-1348(02)00149-1 [DOI] [PubMed] [Google Scholar]
- 34.Foronda P, Bargues MD, Abreu-Acosta N, Periago MV, Valero MA, Valladares B, et al. Identification of genotypes of Giardia intestinalis of human isolates in Egypt. Parasitol Res. (2008) 103:1177–81. 10.1007/s00436-008-1113-2 [DOI] [PubMed] [Google Scholar]
- 35.Gelanew T, Lalle M, Hailu A, Pozio E, Caccio SM. Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. (2007) 102:92–9. 10.1016/j.actatropica.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 36.Berrilli F, D'Alfonso R, Giangaspero A, Marangi M, Brandonisio O, Kaboré Y, et al. Giardia duodenalis genotypes and Cryptosporidium species in humans and domestic animals in Côte d'Ivoire: occurrence and evidence for environmental contamination. Trans R Soc Trop Med Hyg. (2012) 106:191–5. 10.1016/j.trstmh.2011.12.005 [DOI] [PubMed] [Google Scholar]
- 37.Thompson RCA. The zoonotic significance and molecular epidemiology of Giardia and giardiasis. Vet Parasitol. (2004) 126:15–35. 10.1016/j.vetpar.2004.09.008 [DOI] [PubMed] [Google Scholar]
- 38.Read CM, Monis PT, Thompson RCA. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. (2004) 4:125–30. 10.1016/j.meegid.2004.02.001 [DOI] [PubMed] [Google Scholar]
- 39.Trout JM, Santin M, Fayer R. Giardia and Cryptosporidium species and genotypes in coyotes (Canis latrans). J Zoo Wildl Med. (2006) 37:141–4. 10.1638/05-06TYM-123005.1 [DOI] [PubMed] [Google Scholar]
- 40.Li W, Feng YY, Santin M. Host Specificity of Enterocytozoon bieneusi and Public Health Implications. Trends Parasitol. (2019) 35:436–51. 10.1016/j.pt.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 41.Reetz J, Rinder H, Thomschke A, Manke H, Schwebs M, Bruderek A. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens). J Clin Microbiol. (2002) 32:785–7. 10.1128/JCM.02624-10 [DOI] [PubMed] [Google Scholar]
- 42.Feng YY, Li N, Dearen T, Lobo ML, Matos O, Cama V, et al. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Appl Environ Microbiol. (2011) 77:4822–8. 10.1128/AEM.02803-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, Tao W, Jiang YX, Diao RN, Yang JP, Xiao LH. Genotypic distribution and phylogenetic characterization of Enterocytozoon bieneusi in diarrheic chickens and pigs in multiple cities, China: potential zoonotic transmission. PLoS ONE. (2014) 9:e108279. 10.1371/journal.pone.0108279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Cunha MJ, Cury MC, Santin M. Widespread presence of human-pathogenic Enterocytozoon bieneusi genotypes in chickens. Vet Parasitol. (2016) 217:108–12. 10.1016/j.vetpar.2015.12.019 [DOI] [PubMed] [Google Scholar]
- 45.Li N, Xiao LH, Wang L, Zhao SM, Zhao XK, Duan LP, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl Trop Dis. (2012) 6:1809. 10.1371/journal.pntd.0001809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimoto A, Kunikane S, Hirata T. Prevalence of Cryptosporidium oocysts and Giardia cysts in the drinking water supply in Japan. Water Res. (2002) 36:519–26. 10.1016/S0043-1354(01)00279-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequences generated in present study were submitted to the NCBI GenBank with accession numbers KY448446 to KY448471.