Abstract
Background
Long non‐coding RNA (lncRNA) has been regarded as crucial regulator for cancer progression. Roles of KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) in cancers including osteosarcoma and colon cancer have been previously reported. However, its role in ovarian cancer (OC) remains unclear.
Methods
Expression level of KCNQ1OT1 on OC cells and normal cell was analyzed with quantitative real‐time PCR. Gain and loss‐of‐function experiments were performed to analyze the biological roles of KCNQ1OT1 in OC. Moreover, whether KCNQ1OT1 functions its role via mediating MICRORNA‐142‐5p (MIR‐142‐5p)/calpain 10 (CAPN10) axis was analyzed. In addition, effects of KCNQ1OT1, MIR‐142‐5p, and CAPN10 on overall survival of OC patients were analyzed at Kaplan–Meier plotter website.
Results
We showed KCNQ1OT1 was elevated expression in OC cells and indicated poorer overall survival of OC patients. Besides, we found KCNQ1OT1 could promote OC cell proliferation and migration in vitro. Moreover, MIR‐142‐5p was found reduced expression, while CAPN10 was found elevated expression in OC cells compared with normal cell. Kaplan–Meier curve analysis showed low MIR‐142‐5p or high CAPN10 expression were indicators for poorer overall survival of OC patients. At length, we showed KCNQ1OT1 could regulate OC development via MIR‐142‐5p/CAPN10 axis.
Conclusions
Taken together, KCNQ1OT1 upregulates CAPN10 expression via sponging MIR‐142‐5p, thus promoting the proliferation and migration of OC.
Keywords: CAPN10, KCNQ1OT1, MIR‐142‐5p, ovarian cancer
KCNQ1OT1 expression was revealed highly expressed in GC cells. High KCNQ1OT1 expression was a predictor for poor overall survival of cancer patients.

1. INTRODUCTION
Ovarian cancer (OC) is a common occurred cancer type in female reproductive system (Siegel, Miller, & Jemal, 2018). The 5‐year overall survival for OC patients is about 30% due to approximately 75% of cancer patients are diagnosed at late stages (Siegel, Miller, & Jemal, 2017; Sorensen, Schnack, Karlsen, & Hogdall, 2015). Therefore, it is imperative to explore the novel targets which can be used for tumor‐targeted therapy.
Emerging evidence showed OC carcinogenesis is correlated with aberrantly expressed of long non‐coding RNAs (lncRNAs) (Wang, Lu, & Chen, 2019). Even though lncRNA lacks the ability to code proteins, it was reported to regulate various cellular processes and tumorigenesis (Bartonicek, Maag, & Dinger, 2016). For instance, lncRNA FLVCR1 antisense RNA 1 (FLVCR1‐AS1) was reported to overexpressed in OC tissues, serum, and cells (Yan et al., 2019). The overexpression of FLVCR1‐AS1 was revealed to promote cancer proliferation, metastasis, and epithelial to mesenchymal transition via regulating Yes associated protein 1 (606608) expression via modulating MICRORNA‐513, while the knockdown of FLVCR1‐AS1 caused opposite effects on OC cell behaviors (Yan et al., 2019). Moreover, lncRNA growth arrest specific 5 (GAS5, 608280) was identified to be elevated expression in epithelial ovarian cancer tissues and cells (Long, Xiong, et al., 2019). Mechanism study showed that GAS5 could recruit transcription factor enhancer of zeste 2 polycomb repressive complex 2 subunit (601573)to regulate map kinase signal pathway (Long, Song, et al., 2019).
Potassium Voltage‐Gated Channel Subfamily Q Member 1 opposite strand/antisense transcript 1 (KCNQ1OT1, 604115) is a long non‐coding RNA located at chromosome 11p15.5 (Mitsuya et al., 1999). KCNQ1OT1 was revealed to be elevated expression in colon and rectal adenocarcinoma (Zhang, Yan, Yi, Rui, & Hu, 2019). Also, high KCNQ1OT1 was found to be a predictor for poorer overall survival and recurrence‐free survival of colon adenocarcinoma patients (Zhang et al., 2019). Moreover, KCNQ1OT1 was found could regulate the response of cancer cells to chemo‐reagents. For example, KCNQ1OT1 was found overexpressed in methotrexate resistant colorectal cancer cells (Xian & Zhao, 2019). Also, they showed KCNQ1OT1 was able to affect the chemosensitivity of colorectal cancer cell via targeting protein phosphatase 1 regulatory inhibitor subunit 1B (604399) through sponging MIR‐760 (Xian & Zhao, 2019). In colon cancer, KCNQ1OT1 was also identified to be elevated expression and correlated with poor overall survival of cancer patients (Li et al., 2019). Moreover, it was found knockdown of KCNQ1OT1 inhibits cell proliferation but promotes cell apoptosis through MIR‐34a (611172)/autophagy‐related 4B cysteine peptidase (611338) (Li et al., 2019). Besides that, KCNQ1OT1 was shown to regulate the osteosarcoma cell behaviors and its response to cisplatin via regulating Kcnq1/DNA methyltransferase 1 mediated KCNQ1 expression (Li et al., 2019). However, its expression and roles of KCNQ1OT1 in OC remains to be explored.
In this study, expression of KCNQ1OT1 in OC tissues and cell lines was explored. Moreover, a series of loss and gain‐of‐function experiments were performed to investigate the biological roles of KCNQ1OT1. Moreover, the underlying mechanism of KCNQ1OT1 in regulating OC cell behaviors was explored.
2. MATERIALS AND METHODS
2.1. Cell culture
The Roswell Park Memorial Institute‐1640 (RPMI‐1640) and fetal bovine serum (FBS) purchased from Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to incubate OC cells (SKOV3, OVCAR3) and normal ovarian epithelial cell line (IOSE80). The incubation condition was maintained at 37°C and contained 5% of CO2. All these three cells were obtained from American Type Culture Collection (ATCC).
2.2. Cell transfection
For manipulating the expression of KCNQ1OT1, small‐interfering RNA (siRNA) for KCNQ1OT1 (si‐KCNQ1OT1, 5ʹ‐GGTAGAATAGTTCTGTCTT‐3ʹ), overexpression construct for KCNQ1OT1 (pKCNQ1OT1) or calpain 10 (605286, pCAPN10), and the relative controls (siR‐NC, 5ʹ‐ATCTATGATTGGTCTATGG‐3ʹ and pcDNA3.1) was synthesized by GenScript. MIR‐140‐5p mimic (5ʹ‐CAGUGGUUUUACCCUAUGGUAG‐3ʹ) and the corresponding negative control (miR‐NC, 5ʹ‐GGUCAUCGGAUUGUAUCUCGUA‐3ʹ) were designed by GenePharm. Cells were transfected using Lipofectamine 2000 (Invitrogen) until incubated to approximately 80% confluence according to the standard protocols.
2.3. Quantitative real‐time PCR (qRT‐PCR)
To explore relative gene expression level, qRT‐PCR was performed on ABI 7,500 system (Applied Biosystem). RNA from cells was extracted using Trizol reagent (Invitrogen) and then reverse transcribed into complementary DNA using First‐Strand cDNA Synthesis kit (Invitrogen). qRT‐PCR was performed using SYBR Green Mix (Takara, Dalian, Liaoning, China) with the following procedure: 95°C for 10 min, 40 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 30 s. Relative expression level was calculated with the 2−ΔΔCt method. Primers used were as follows: KCNQ1OT1: forward: 5ʹ‐CCTCCCTCACTGAGCTTTGG‐3ʹ, reverse: 5ʹ‐GTGCGGACCCTATACGGAAG‐3ʹ; CAPN10: forward: 5ʹ‐GCTGGCTGGTGACATCAGTG‐3ʹ, reverse: 5ʹ‐TCAGGTTCCATCTTTGGGCCAG‐3ʹ; glyceraldehyde‐3‐phosphate dehydrogenase (138400): forward: 5ʹ‐GCTGCTGAGTATGTCGTGGAGT‐3ʹ, reverse: 5ʹ‐AGTCTTCTGGGTGGCAGTGAT‐3ʹ; MIR‐142‐5p: forward: 5ʹ‐CAUAAAGUAGAAAGCACUACU‐3ʹ, reverse: 5ʹ‐UAGUGCUUUCUACUUUAUGUU‐3ʹ; U6 snRNA: forward: 5ʹ‐CTCGCTTCGGCAGAC‐3ʹ, reverse: 5ʹ‐AACGCTTACGAATTT‐3ʹ. The procedures were as follows: 95°C for 3 min, 40 cycles of 95°C for 10 s, and 58°C for 30 s.
2.4. Cell counting kit‐8 (CCK‐8) assay
Cell proliferation ability was analyzed with CCK‐8 (Beyotime, Haimen, Jiangsu, China) assay according to the supplier's instructions. Cell proliferation rate was analyzed at 0, 1, 2, and 3 days after incubation. At these time points, CCK‐8 reagent was added to each well and then the optical density at 450 nm was measured using microplate reader.
2.5. Colony formation assay
Eight hundred cells were seeded into 6‐well plate and incubated for 14 days. Then, methanol was added to the plate to fix the colonies. Colonies were further stained with crystal violet and counted under microscope.
2.6. Transwell invasion assay
Cell invasion ability was analyzed with transwell invasion assay. 2 × 105 in serum‐free medium was added into the upper chamber of 8 μm transwell unit (Corning, NY, USA) that pre‐coated with Matrigel. RPMI‐1640 supplemented with FBS was filled into the lower chamber. After incubation for 48 hr, invasive cells were fixed and stained. At length, the invasive cell numbers were counted under microscope.
2.7. Luciferase reporter assay
lncBase was used to explore the potential miRNA target of KCNQ1OT1, and we identified MIR‐142‐5p was a potential target. TargetScan was utilized to investigate the potential target of MIR‐142‐5p, and we showed CAPN10 was a putative target. The wild‐type sequence of KCNQ1OT1 and CAPN10 was inserted into pmirGLO to generate KCNQ1OT1‐wt and CAPN10‐wt. Site‐direct mutagenesis kit was used to generate mutant luciferase vectors (KCNQ1OT1‐wt and CAPN10‐wt). Cells were co‐transfected with luciferase vectors or miRNAs using Lipofectamine 2000. After 48 hr of transfection, relative luciferase activity was analyzed with Dual‐luciferase reporter assay system (Promega, Madison, WI, USA).
2.8. Prognostic values of KCNQ1OT1, MIR‐142‐5p, and CAPN10 on OC patients
The effects of KCNQ1OT1, MIR‐142‐5p, and CAPN10 on OC patients were explored at Kaplan‐Meier plotter website (http://kmplot.com/analysis/index.php?p=background, Nagy, Lánczky, Menyhárt, & Győrffy, 2018).
2.9. Statistical analysis
Results obtained from in vitro experiments were analyzed at Graphpad prism 7 (San Diego, CA, USA) and then presented as mean ± standard deviation (SD). Differences among groups were analyzed using Student's t‐test or one‐way Analysis of Variance and Tukey post‐hoc test. p < .05 was believed as statistically significant.
3. RESULTS
3.1. Expression of KCNQ1OT1 was elevated in OC
qRT‐PCR revealed that KCNQ1OT1 levels were significant higher in OC cell lines than in normal cell line (Figure 1a). Survival analysis revealed that patients with high KCNQ1OT1 expression level tend to have worse prognosis compared to those with low KCNQ1OT1 expression level (Figure 1b).
Figure 1.
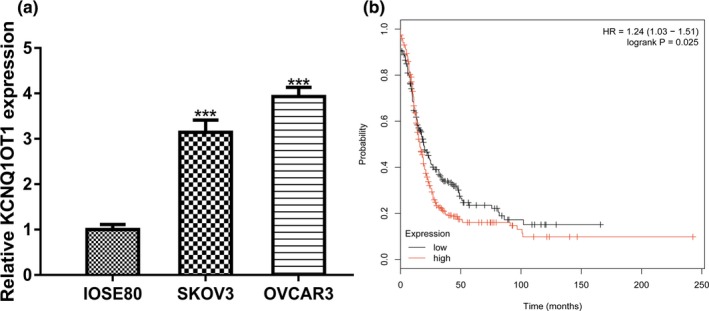
KCNQ1OT1 expression and its impact on overall survival of OC patients. (a) Expression of KCNQ1OT1 in OC cells and normal cell line. (b) Effect of KCNQ1OT1 on the overall survival of OC patients. KCNQ1OT1, Potassium Voltage‐Gated Channel Subfamily Q Member 1 opposite strand/antisense transcript 1; OC, ovarian cancer
3.2. KCNQ1OT1 overexpression promotes OC cell proliferation and invasion in vitro
The SKOV3 cell line was selected for gain‐of‐function experiments. qRT‐PCR revealed that KCNQ1OT1 levels could be significantly elevated by pKCNQ1OT1 (Figure 2a). CCK‐8 assay and colony formation assay revealed that KCNQ1OT1 overexpression promoted OC cell growth (Figure 2b,c). Furthermore, transwell invasion assay showed the cell invasion of OC cell was promoted after KCNQ1OT1 overexpression (Figure 2d).
Figure 2.
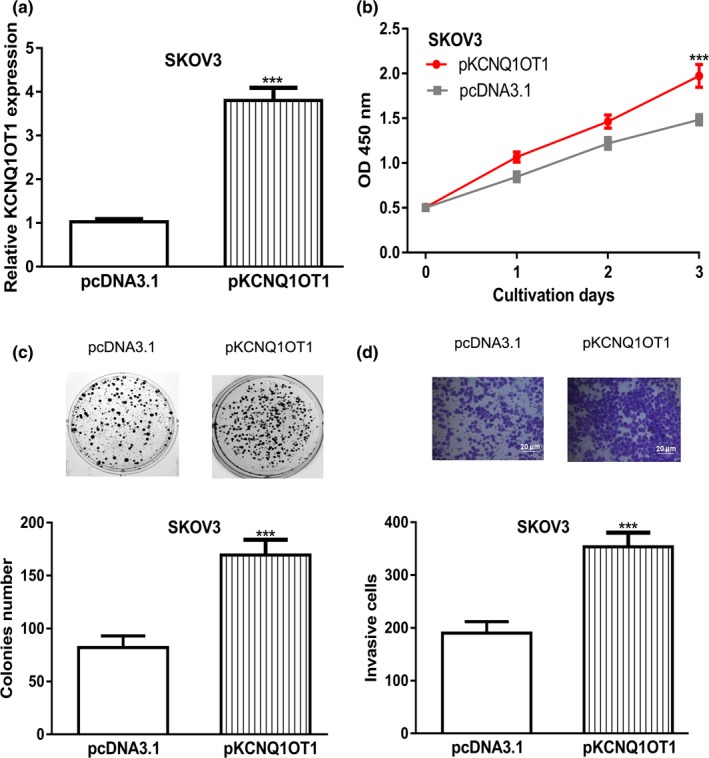
KCNQ1OT1 overexpression promoted cell proliferation, colony formation, and invasion. (a) KCNQ1OT1 expression was detected following pKCNQ1OT1 transfection. (b) CCK‐8 assay to detect proliferation of cells transfected with pKCNQ1OT1. (c) Colony formation of cells after transferring of pKCNQ1OT1. (d) Cell invasion rate of cells with pKCNQ1OT1 transfection was measured. CCK‐8: cell counting kit‐8; KCNQ1OT1, Potassium Voltage‐Gated Channel Subfamily Q Member 1 opposite strand/antisense transcript 1; OC, ovarian cancer
3.3. Knockdown of KCNQ1OT1 inhibits OC cell proliferation and invasion in vitro
OVCAR3 cell line was used for loss‐of‐function experiments. si‐KCNQ1OT1 introduction significantly decreased the levels of KCNQ1OT1 (Figure 3a). In vitro experiments revealed that knockdown the expression KCNQ1OT1 was able to inhibit OC cell proliferation, colony formation, and cell invasion (Figure 3b‐d).
Figure 3.
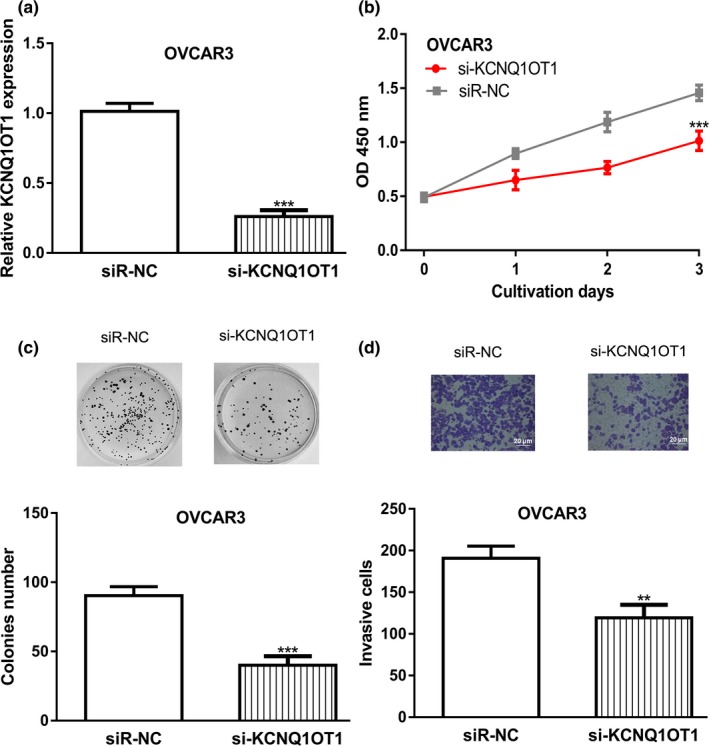
KCNQ1OT1 knockdown inhibited cell proliferation, colony formation, and invasion. (a) KCNQ1OT1 expression was detected following si‐KCNQ1OT1 transfection. (b) CCK‐8 assay to detect proliferation of cells transfected with si‐KCNQ1OT1. (c) Colony formation of cells after transferring of si‐KCNQ1OT1. (d) Cell invasion rate of cells with si‐KCNQ1OT1 transfection was measured. CCK‐8: cell counting kit‐8; KCNQ1OT1, Potassium Voltage‐Gated Channel Subfamily Q Member 1 opposite strand/antisense transcript 1; OC, ovarian cancer; siR‐NC, negative control small interfering RNA
3.4. Interaction of KCNQ1OT1 and MIR‐142‐5p in OC
lncBase showed that MIR‐142‐5p was a putative target for KCNQ1OT1 (Figure 4a). MIR‐142‐5p expression level was revealed to be decreased expression in OC cells compared with normal cell line (Figure 4b). Meanwhile, we showed that low MIR‐142‐5p was a predictor for poor overall survival of OC patients (Figure 4c). Luciferase activity reporter assay revealed that overexpression of MIR‐142‐5p inhibited the luciferase activity of cells transfected with KCNQ1OT1‐wt (Figure 4d).
Figure 4.
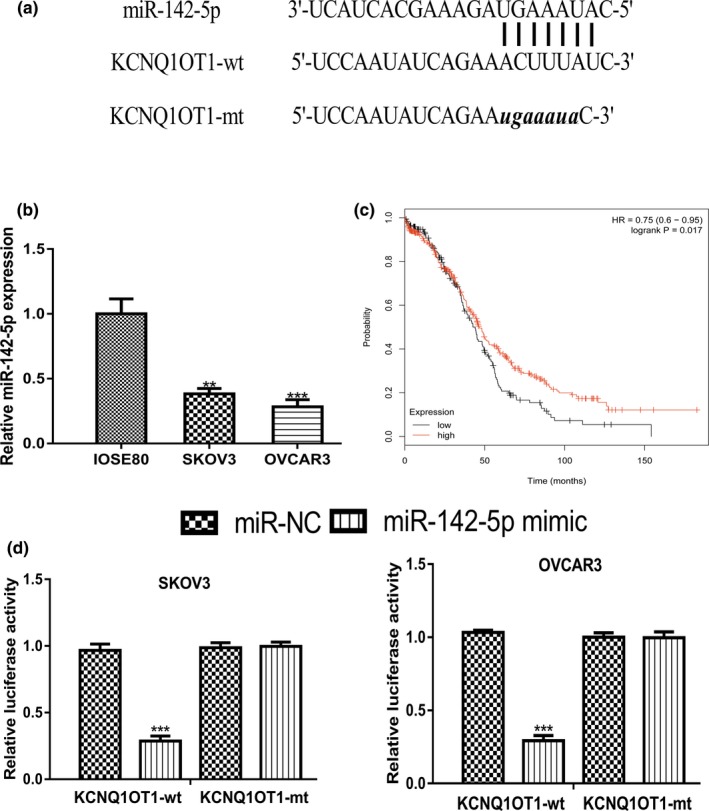
KCNQ1OT1 functioned as a sponge for MIR‐142‐5p. (a) Binding model of KCNQ1OT1 and MIR‐142‐5p. (b) Expression of MIR‐142‐5p in OC cells and normal cell line. (c) Effect of MIR‐142‐5p on the overall survival of OC patients. (d) Luciferase activity in cells with luciferase vectors and miRNAs transfection. KCNQ1OT1, Potassium Voltage‐Gated Channel Subfamily Q Member 1 opposite strand/antisense transcript 1; MIR‐142‐5p, MICRORNA‐142‐5p; miR‐NC, negative control microRNA; mt: mutant; OC, ovarian cancer; wt: wild‐type
3.5. Interaction of MIR‐142‐5p and CAPN10 in OC
Furthermore, TargetScan showed that CAPN10 was a putative target for MIR‐142‐5p (Figure 5a). qRT‐PCR revealed that CAPN10 expression level in OC cells was higher than in normal cell line (Figure 5b). Kaplan–Meier plotter showed that high CAPN10 level indicates a poor prognosis of OC cancer patients (Figure 5c). Luciferase activity reporter assay indicated that overexpression of MIR‐142‐5p inhibits the relative luciferase activity of cells transfected with CAPN10‐wt (Figure 5d).
Figure 5.
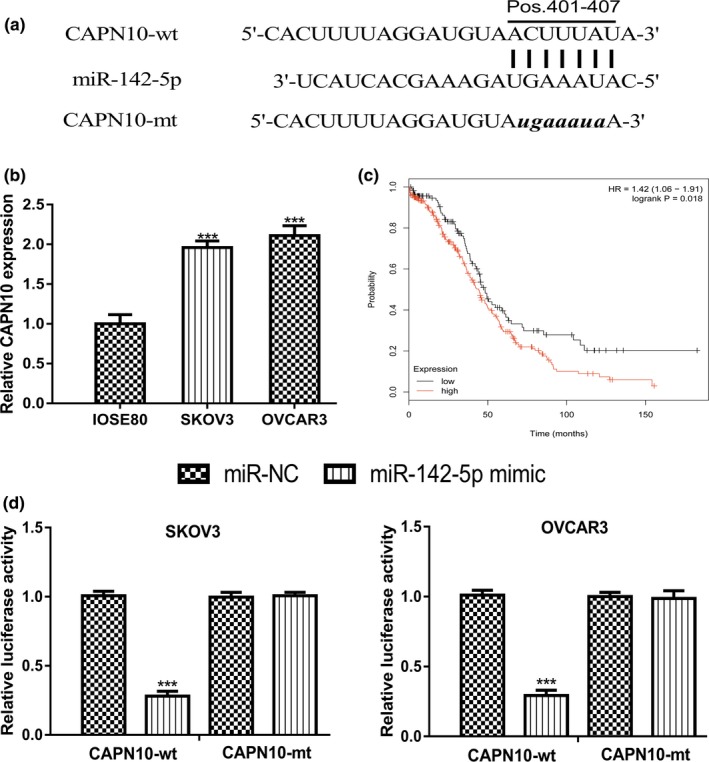
MIR‐142‐5p directly target CAPN10. (a) Binding model of CAPN10 and MIR‐142‐5p. (b) Expression of CAPN10 in OC cells and normal cell line. (c) Effect of CAPN10 on the overall survival of OC patients. (d) Luciferase activity in cells with luciferase vectors and miRNAs transfection. CAPN10, calpain 10; MIR‐142‐5p, MICRORNA‐142‐5p; miR‐NC, negative control microRNA; mt, mutant; OC, ovarian cancer; wt, wild‐type
3.6. KCNQ1OT1 exerts its function through MIR‐142‐5p/CAPN10
qRT‐PCR revealed that CAPN10 expression was significantly increased by pCAPN10 (Figure 6a). CCK‐8 assay, colony formation, and transwell invasion assay showed that CAPN10 overexpression promotes OC cell growth and invasion (Figure 6b‐d). In the meantime, we showed overexpression of MIR‐142‐5p inhibits OC cell proliferation, colony formation, and invasion (Figure 6b‐d). Importantly, we showed KCNQ1OT1 targeting the MIR‐142‐5p/CAPN10 axis to regulate OC cell behaviors through rescue experiments (Figure 6b‐d).
Figure 6.
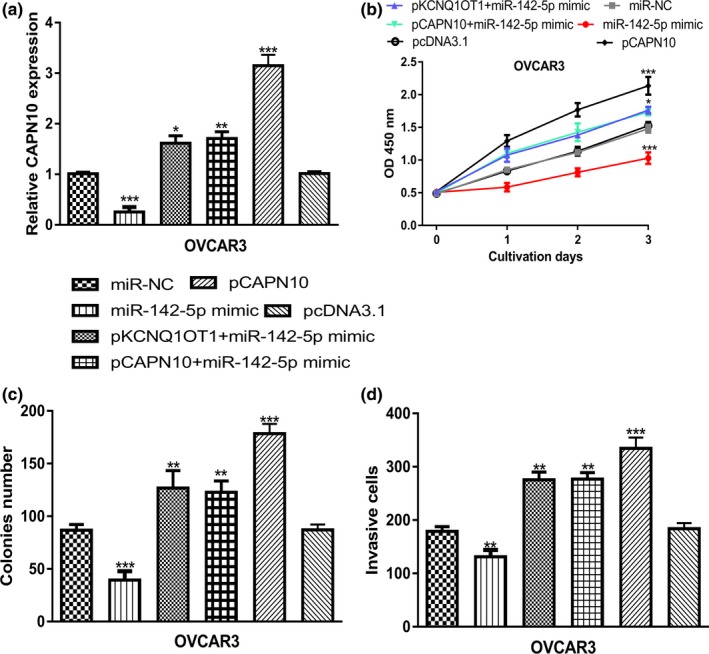
KCNQ1OT1 regulated OC progression via regulating MIR‐142‐5p/CAPN10 axis. (a) CAPN10 expression was detected following miRNA or overexpression construct transfection. (b) CCK‐8 assay to detect proliferation of cells transfected with miRNA or overexpression construct. (c) Colony formation of cells after transferring of miRNA or overexpression construct. (d) Cell invasion rate of cells with miRNA or overexpression construct transfection was measured. CAPN10, calpain 10; CCK‐8, cell counting kit‐8; KCNQ1OT1, Potassium Voltage‐Gated Channel Subfamily Q Member 1 opposite strand/antisense transcript 1; MIR‐142‐5p: MICRORNA‐142‐5p; miR‐NC, negative control microRNA; OC, ovarian cancer
4. DISCUSSION
LncRNA and miRNAs are RNA molecules that play crucial roles in multiplies cellular processes including cell growth, metastasis, and drug resistant (Li & Zhan, 2019; Long, Song, et al., 2019; Qi et al., 2019). The competitive RNA (ceRNA) theory has connected lncRNA and miRNA together (Salmena, Poliseno, Tay, Kats, & Pandolfi, 2011). A recent work conducted by Zhao et al. revealed several lncRNA/miRNA/mRNA triplets that may contribute to the cisplatin‐resistant in epithelial OC (Zhao, Tang, Zuo, Zhang, & Wang, 2019). Hence, the investigations on the aberrantly expressed molecules contributed to OC initiation and progression may help to better control OC in the future.
Previous studies have suggested that lncRNA KCNQ1OT1 plays crucial roles in cancer biology (Li et al., 2019; Xian & Zhao, 2019). In this work, we showed expression levels of lncRNA KCNQ1OT1 in OC cells were higher than in normal cell line. Through Kaplan–Meier plotter analysis, we showed high KCNQ1OT1 expression was correlated with poor overall survival of OC patients. Moreover, after KCNQ1OT1 overexpression, we showed that cell proliferation, colony formation, and cell invasion of OC cells were significantly promoted. Besides that, after KCNQ1OT1 knockdown, cell proliferation, colony formation, and cell invasion of OC cells were significantly inhibited. Collectively, our work indicated that KCNQ1OT1 may have an oncogenic role in OC.
Bioinformatic analysis showed MIR‐142‐5p was a possible target for KCNQ1OT1. MIR‐142‐5p was reported to be a decreased expression in non‐small cell lung cancer tissues and cells (Wang, Liu, Fang, & Yang, 2017). Overexpression of MIR‐142‐5p suppressed non‐small cell lung cancer progression in vitro and in vivo via regulating phosphatidylinositol‐4,5‐bisphosphate 3‐kinase, catalytic subunit alpha (171,834) (Wang et al., 2017). Moreover, MIR‐142‐5p was revealed to function as a bridge between KCNQ1OT1 and cyclin‐dependent kinase 5 (123,831) to regulate lung adenocarcinoma cell migration, invasion, and epithelial mesenchymal transition (Jia et al., 2019). In this work, we showed MIR‐142‐5p expression was decreased in OC cells and correlated with poor overall survival of OC patients. Luciferase activity reporter assay revealed the direct interaction of MIR‐142‐5p and KCNQ1OT1. Moreover, by exploring the target of MIR‐142‐5p, we identified a direct connection of MIR‐142‐5p and CAPN10. CAPN10 was found highly expressed in OC cells and predicted poor overall survival of cancer patients. CAPN10 belongs to the mitochondrial calpain system and reported to be regulated by oncogene GAEC1 (612,130) to promote tumor progression (Chan et al., 2013). One drawback of this work was the lack of in vivo experiments. Hence, we will perform animal experiments in the near future to validate the KCNQ1OT1/MIR‐142‐5p/CAPN10 axis in OC and to confirm their roles in regulating cancer progression.
To sum up, our results revealed that MIR‐142‐5p, sponged by KCNQ1OT1, target CAPN10 to affect the progression of OC. Our study provided novel insights on the understanding of OC progression and the potential to use KCNQ1OT1 as a target for OC therapy. However, further studies are needed to understand the clinical significance of KCNQ1OT1/MIR‐142‐5p/CAPN10 in OC.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Liu H, Chen R, Kang F, Lai H, Wang Y. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR‐142‐5p/CAPN10 axis. Mol Genet Genomic Med. 2020;8:e1077 10.1002/mgg3.1077
Liu and Chen authors contributed equally to this work.
Funding information
This work was supported by Fujian Natural Science Fund Project (No. 2012D050).
REFERENCES
- Bartonicek, N. , Maag, J. L. , & Dinger, M. E. (2016). Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Molecular Cancer, 15(1), 43 10.1186/s12943-016-0530-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D. , Tsoi, M. Y. , Liu, C. D. , Chan, S. H. , Law, S. Y. , Chan, K. W. , … Tang, J. C. (2013). Oncogene GAEC1 regulates CAPN10 expression which predicts survival in esophageal squamous cell carcinoma. World Journal of Gastroenterology, 19(18), 2772–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y. , Duan, Y. , Liu, T. , Wang, X. , Lv, W. , Wang, M. , … Liu, L. (2019). LncRNA TTN‐AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging MIR‐142‐5p to regulate CDK5. Cell Death & Disease, 10(8), 573 10.1038/s41419-019-1811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , & Zhan, X. (2019). Identification of clinical trait‐related lncRNA and mRNA biomarkers with weighted gene co‐expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA Journal, 10(3), 273–290. 10.1007/s13167-019-00175-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Li, C. , Li, D. , Yang, L. , Jin, J. , & Zhang, B. (2019). lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR‐34a/ATG4B pathway. OncoTargets and Therapy, 12, 2649–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J. , Xiong, J. , Bai, Y. I. , Mao, J. , Lin, J. , Xu, W. , … Zhao, H. (2019). Construction and investigation of a lncRNA‐associated ceRNA regulatory network in cholangiocarcinoma. Frontiers in Oncology, 9, 649 10.3389/fonc.2019.00649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, X. , Song, K. , Hu, H. , Tian, Q. I. , Wang, W. , Dong, Q. , … Di, W. (2019). Long non‐coding RNA GAS5 inhibits DDP‐resistance and tumor progression of epithelial ovarian cancer via GAS5‐E2F4‐PARP1‐MAPK axis. Journal of Experimental & Clinical Cancer Research, 38(1), 345 10.1186/s13046-019-1329-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuya, K. , Meguro, M. , Lee, M. P. , Katoh, M. , Schulz, T. C. , Kugoh, H. , … Oshimura, M. (1999). LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Human Molecular Genetics, 8(7), 1209–1217. 10.1093/hmg/8.7.1209 [DOI] [PubMed] [Google Scholar]
- Nagy, A. , Lánczky, A. , Menyhárt, O. , & Győrffy, B. (2018). Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports, 8, 9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X. U. , Yu, X.‐J. , Wang, X.‐M. , Song, T.‐N. , Zhang, J. , Guo, X.‐Z. , … Shao, M. (2019). Knockdown of KCNQ1OT1 Suppresses Cell Invasion and Sensitizes Osteosarcoma Cells to CDDP by Upregulating DNMT1‐Mediated Kcnq1 Expression. Molecular Therapy ‐ Nucleic Acids, 17, 804–818. 10.1016/j.omtn.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Salmena, L. , Poliseno, L. , Tay, Y. , Kats, L. , & Pandolfi, P. P. (2011). A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell, 146, 353–358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R. L. , Miller, K. D. , & Jemal, A. (2017). Cancer statistics, 2017. CA: A Cancer Journal for Clinicians, 67(1), 7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- Siegel, R. L. , Miller, K. D. , & Jemal, A. (2018). Cancer statistics, 2018. CA: A Cancer Journal for Clinicians, 68(1), 7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- Sorensen, R. D. , Schnack, T. H. , Karlsen, M. A. , & Hogdall, C. K. (2015). Serous ovarian, fallopian tube and primary peritoneal cancers: A common disease or separate entities ‐ a systematic review. Gynecologic Oncology, 136(3), 571–581. 10.1016/j.ygyno.2015.01.534 [DOI] [PubMed] [Google Scholar]
- Wang, J. Y. , Lu, A. Q. , & Chen, L. J. (2019). LncRNAs in ovarian cancer. Clinica Chimica Acta, 490, 17–27. 10.1016/j.cca.2018.12.013 [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Liu, Z. , Fang, X. , & Yang, H. (2017). MIR‐142‐5p suppresses tumorigenesis by targeting PIK3CA in non‐small cell lung cancer. Cellular Physiology and Biochemistry, 43(6), 2505–2515. 10.1159/000484459 [DOI] [PubMed] [Google Scholar]
- Xian, D. , & Zhao, Y. (2019). LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR‐760/PPP1R1B via the cAMP signalling pathway. Journal of Cellular and Molecular Medicine, 23(6), 3808–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. , Li, H. , Silva, M. A. , Guan, Y. , Yang, L. I. , Zhu, L. , … Ren, C. (2019). LncRNA FLVCR1‐AS1 mediates miR‐513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer. Journal of Experimental & Clinical Cancer Research, 38(1), 356 10.1186/s13046-019-1356-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Yan, J. , Yi, B. , Rui, Y. , & Hu, H. (2019). High KCNQ1OT1 expression might independently predict shorter survival of colon adenocarcinoma. Future Oncology, 15(10), 1085–1095. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Tang, D. Y. , Zuo, X. , Zhang, T. D. , & Wang, C. (2019). Identification of lncRNA‐miRNA‐mRNA regulatory network associated with epithelial ovarian cancer cisplatin‐resistant. Journal of Cellular Physiology, 234(11), 19886–19894. 10.1002/jcp.28587 [DOI] [PubMed] [Google Scholar]


