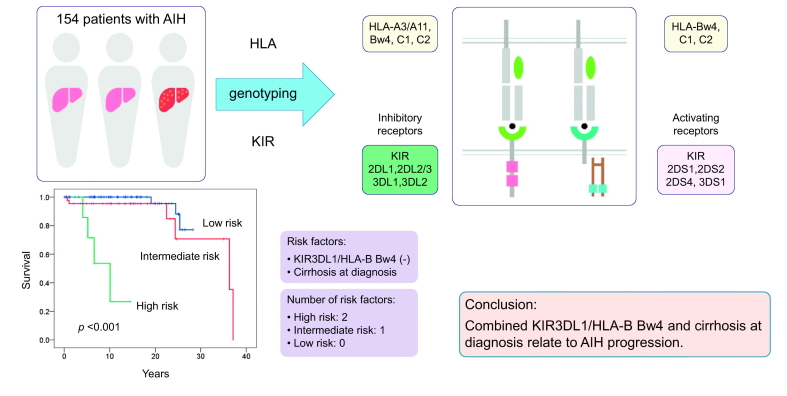
Background & Aims
Natural killer (NK) cells are key participants in the innate immune response. Killer cell immunoglobulin-like receptors (KIRs) are involved in the activation and inhibition of NK cells through the recognition of human leukocyte antigen (HLA) class I molecules. We investigated the impact of KIR/HLA combinations on susceptibility and long-term clinical outcome in Japanese patients with type 1 autoimmune hepatitis (AIH).
Methods
A total of 154 cases of AIH were recruited at Shinshu University Hospital between 1974 and 2018. KIR genes and HLA class I and II alleles were genotyped in all patients along with 201 healthy individuals. Associations between KIR/HLA pairs and clinical outcomes (liver decompensation and liver-related death) were evaluated using the Cox proportional hazards model with stepwise method.
Results
After a median follow-up period of 11.1 years, 12% of patients experienced liver decompensation and 8% died from liver disease. KIR3DL1/HLA-B Bw4-80Ile (p = 0.0062) and the HLA-DRB1*04:05-DQB1*04:01 haplotype (p ≪0.001) were significantly associated with AIH. Conversely, significant protective associations were found for KIR3DL1/HLA-B Bw4-80Thr (p = 0.0092) and KIR2DL1/HLA-C2 (p = 0.0025). The KIR3DL1/HLA-B Bw4-positive phenotype was strongly associated with a favorable clinical outcome (liver decompensation: hazard ratio [HR] 0.37, p = 0.037; liver-related death: HR 0.26, p = 0.038). Cirrhosis was detected in 16 (10%) patients at diagnosis and was significantly related to poor survival (HR 17.87, p ≪0.001) and progression to liver decompensation (HR 9.00, p ≪0.001).
Conclusions
This study revealed the impact of specific KIR/HLA pairs in AIH susceptibility and progression in Japanese patients. KIR3DL1/HLA-B Bw4-negative patients with AIH and cirrhosis at diagnosis are at high risk of adverse outcomes and require careful surveillance.
Lay summary
Autoimmune hepatitis (AIH) is a disease of the liver that can present in acute or chronic hepatitis. We examined whether KIR/HLA pairs were associated with AIH susceptibility or disease progression. KIR3DL1/HLA-B Bw4 was a novel KIR/HLA pair related to a favorable clinical outcome, while cirrhosis at the initial diagnosis was a risk factor for poor prognosis. Thus, frequent and careful surveillance is advised for KIR3DL1/HLA-B Bw4-negative patients with AIH and cirrhosis.
Keywords: HLA, Killer cell immunoglobulin-like receptors, AIH, NK cell, outcome
Graphical abstract
Highlights
-
•
KIR3DL1/HLA-B Bw4-80Ile is significantly associated with autoimmune hepatitis.
-
•
KIR2DL1/HLA-C2 and KIR3DL1/HLA-B Bw4-80Thr have protective associations with autoimmune hepatitis.
-
•
KIR3DL1/HLA-B Bw4 is a novel KIR/HLA pair related to a favorable outcome in autoimmune hepatitis.
-
•
Combined KIR3DL1/HLA-B Bw4 and cirrhosis at diagnosis relate to autoimmune hepatitis progression.
Introduction
Autoimmune hepatitis (AIH) is characterized by an autoimmune reaction towards hepatocytes.1 The nature of AIH is complex and may depend on interactions between genetic susceptibility and environmental triggers.2 Understanding the exact mechanisms involved in AIH development remains elusive. There is considerable evidence supporting a strong association of type 1 AIH with human leukocyte antigen (HLA) (Table S1). In particular, the HLA DRB1*04:05-DQB1*04:01 haplotype is related to type 1 AIH in the Japanese.3 In northern Europe and North America, the DRB1*03:01-DQB1*02:01 and DRB1*04:01-DQB1*03:01 haplotypes are seen at an increased frequency in patients with AIH, while DRB1*15:01-DQB1*06:02 is observed at a lower frequency.4 In Latin Americans, DRB1*13:01-DQB1*06:03 correlates significantly with the disease.5
Natural killer (NK) cells play crucial roles in the innate immune response to viruses, bacteria, and tumor cells.6 NK cell-mediated responses have also been implicated in the pathogenesis of AIH and other autoimmune diseases.[7], [8], [9] NK cells express a wide array of inhibitory and activating killer cell immunoglobulin-like receptors (KIRs) that detect the expression levels of MHC class I ligands on normal and diseased cells in humans.10 The inhibitory signals that are transduced from KIR binding to MHC class I molecules allow NK cells to acquire their full cytotoxic potential.11 KIR2DL1 recognizes HLA-C group 2 (HLA-C2) allotypes that share lysine at position 80, while KIR2DL2 and KIR2DL3 are specific for HLA-C group 1 (HLA-C1) allotypes that have asparagine at position 80.12 KIR3DL1 binds with high affinity to HLA-B Bw4 molecules containing isoleucine at position 80 (Bw4-80Ile) compared with Bw4 molecules containing threonine at that position (Bw4-80Thr).13 KIR3DL2 interacts with HLA-A3 and -A11.14,15 Although KIR gene complexes are important regulators of innate and adaptive immune responses as well as pathogenetic factors in several autoimmune diseases, there remain few reports on KIRs in AIH susceptibility16,17 and none addressing disease progression. We therefore sought to determine whether KIR genes, HLA class I molecules, and KIR/HLA pairs were associated with AIH susceptibility or disease progression in a Japanese population.
Patients and methods
Patients
Between January 1979 and June 2018, a total of 255 Japanese patients were diagnosed as having type 1 AIH at Shinshu University Hospital or its affiliated institutions. Among them, 101 patients were excluded for the following reasons: (1) no stored DNA samples (n = 76); (2) other chronic liver disease, such as hepatitis B, hepatitis C, or non-alcoholic steatohepatitis (n = 5); (3) incomplete medical records (n = 8); (4) follow-up ≪6 months (n = 2); and (5) AIH-primary biliary cholangitis overlap syndrome (n = 10). The remaining 154 patients with type 1 AIH were retrospectively included in this study along with 201 volunteer healthy blood donors.18 The diagnosis of type 1 AIH was made according to the scoring system of the International Autoimmune Hepatitis Group (IAIHG) revised criteria.19 Anti-nuclear antibody and anti-smooth muscle antibody were determined as reported previously.20 The antibody titers to soluble liver antigen (SLA) were determined using ELISA kits (QUANTA Lite® Inova Diagnostics, San Diego, CA), whereby a titer of ≥25 U was interpreted as a positive finding according to the manufacturer’s protocol and instructions for all assays. Cirrhosis was diagnosed by histological examination and/or imaging studies. Hepatocellular carcinoma was diagnosed by imaging studies, and liver decompensation was identified by bleeding from esophageal varices, ascites, and hepatic encephalopathy. Each patient was initially treated with prednisolone at an oral dosage of 20–50 mg/day that was gradually reduced and then maintained at 5–10 mg/day as reported previously.21 Azathioprine at a dose of 50 mg/day was added for only 3 patients whose alanine aminotransferase (ALT) level did not normalize or in whom severe adverse effects were seen from prednisolone therapy, since it and other immunomodulatory drugs were not covered by national health insurance in Japan. Complete biochemical remission was defined as the normalization of aminotransferases and IgG levels.22 If patients exhibited an increase in ALT ≫3 times the upper limit of normal as defined as a biochemical relapse, prednisolone therapy was reinstituted.23 All individuals were negative for hepatitis B surface antigen, antibodies to hepatitis B core antigen, hepatitis C virus, and the human immunodeficiency virus. No patient had a history of excess alcohol consumption. This study was reviewed and approved by the Institutional Review Board of Shinshu University School of Medicine in Matsumoto, Japan (approval number: 524). Written informed consent was obtained from all participating patients. The investigation was conducted according to the principals of the Declaration of Helsinki.
HLA and KIR genotyping
Genomic DNA from all participants was extracted from whole-blood samples using QuickGene-800 assays (Fujifilm, Tokyo, Japan). We genotyped HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 as reported previously.24 HLA-A3 and -A11, HLA-Bw4, HLA-B Bw4-80Ile, HLA-B Bw4-80Thr, HLA-C1, and HLA-C2 KIR ligands were assigned based on the amino acid residues of the HLA-A, HLA-B, and HLA-C alleles, as described previously.25,26 KIR genotyping was performed using PCR with sequence-specific primers.27 HLA typing was combined with KIR typing to stratify patients according to predicted KIR-ligand interactions and binding affinities. The selected KIR-HLA pairs were KIR2DL1/2DS1-HLA-C2, 2DL2/2DS2-HLA-C1, 3DL1/3DS1-HLA-Bw4, and 3DL2-HLA-A3 and -A11.
Statistical analysis
Categorical variables were compared by Pearson’s chi-squared test or Fisher’s exact test, as appropriate, and continuous variables were compared using the Mann-Whitney U test. Optimal cut-off values were determined by the Youden index. Clinical outcomes as of December 2018 were recorded as liver-related death. Genetic power was calculated using the EZR program of R commander software.28 Logistic regression analysis was performed to evaluate associations between HLA-DRB1*04:05 and KIR3DL1/HLA-B Bw4-80Ile in patients with AIH compared to healthy individuals. The Kaplan-Meier method and log-rank test were used to estimate progression to liver decompensation and survival rates of patients. Multivariate analysis was performed using the Cox proportional hazards model with stepwise method to identify independent factors associated with progression to liver decompensation and liver-related death. Variables with a p ≪0.1 in univariate analysis were included in this step. A p value of ≪0.05 was considered statistically significant. Statistical analyses were performed using IBM SPSS Statistics version 24 software (IBM, Chicago, IL).
Results
Baseline clinical characteristics of patients
The clinical profile of the experimental patient cohort is shown in Table 1. Median age was 59 years and 88% were female. Median follow-up was 11.1 years (interquartile range: 6.4–17.3 years). According to the IAIHG revised scoring system, 135 and 19 patients fulfilled the criteria for definite and probable AIH, respectively. No patient was positive for anti-SLA in our cohort. At diagnosis, liver biopsy was performed in 152 of 154 patients. One hundred and thirty-eight patients (90%) had chronic hepatitis and 16 patients (10%) had cirrhosis; 14 were diagnosed by liver histology and 2 were identified by liver imaging. Prednisolone was administered to all patients and 97% achieved a complete biochemical response. Prednisolone therapy was reinstituted for 64 patients (42%) who relapsed.
Table 1.
Demographic and clinical characteristics of 154 patients with type 1 AIH.
| Characteristic | Total (n = 154) |
|---|---|
| Age at diagnosis (years) | 59 (48-66) |
| Female, n (%) | 135 (88) |
| Follow-up (years) | 11.1 (6.4-17.3) |
| AIH score | 18 (17-19) |
| Definite AIH, n (%) | 135 (88) |
| AST (U/L) | 409 (148-818) |
| ALT (U/L) | 405 (161-948) |
| Total bilirubin (mg/dl) | 1.7 (0.9-6.5) |
| IgG (mg/dl) | 2,714 (2,051-3,497) |
| Platelets (×104/μl) | 17.4 (15.7-18.7) |
| Prothrombin % | 78.7 (60.0-96.6) |
| ANA, n (%) | 150 (97) |
| SMA, n (%) (n = 122) | 66 (54) |
| SLA, n (%) | 0 (0) |
| Cirrhosis, n (%) | 16 (10) |
| Complete remission, n (%) | 149 (97) |
| Relapse, n (%) | 64 (42) |
Values are expressed as the median (interquartile range) unless otherwise noted. AIH, autoimmune hepatitis; ALT, alanine aminotransferase; ANA, anti-nuclear antibody; AST, aspartate aminotransferase; SLA, anti-soluble liver antigen; SMA, anti-smooth muscle antibody.
Distribution of HLA, KIR, and KIR/HLA in patients with type 1 AIH and controls
We determined the KIR ligands (HLA-A3 and -A11, HLA-Bw4, HLA-C1, and HLA-C2) of 154 patients with type 1 AIH and 201 controls (Table S2). The frequency of HLA-B Bw4 was identical between AIH and controls (77%). The frequency of HLA-B Bw4-80Ile was significantly higher in patients with AIH (69%) than in controls (55%) (odds ratio [OR] 1.83; 95% CI 1.18–2.84; p = 0.0070), while that of HLA-B Bw4-80Thr was lower (16% vs. 28%; OR 0.47; 95% CI 0.27–0.79; p = 0.0045). The positivity rate of HLA-C2 was significantly less prevalent in patients with AIH (12%) than in controls (24%) (OR 0.41; 95% CI 0.23–0.74; p = 0.0025). There were no significant differences between the groups for the frequencies of HLA-A3 or -A11 or HLA-C1. Further analysis of 14 KIR genes in patients with AIH and controls showed the frequencies of all KIR genes to be comparable (Table S2).
Since KIRs interact with specific HLA class I molecules, we examined KIR/KIR-ligand combinations in AIH and control groups to investigate susceptibility or resistance to the disease (Table 2). The frequency of KIR3DL1/HLA-B Bw4-80Ile in patients with AIH was 64% and significantly higher than the 50% observed in healthy individuals (OR 1.82; 95% CI 1.18–2.80; p = 0.0062). In contrast, protective effects were seen for KIR3DL1/HLA-B Bw4-80Thr (15% vs. 26%, OR 0.49; 95% CI 0.28–0.84; p = 0.0092). Patients with AIH had a significantly lower frequency of KIR2DL1/HLA-C2 compared with controls (12% vs. 24%, OR 0.41; 95% CI 0.23–0.74; p = 0.0025). There were no remarkable differences in the frequencies of other KIR/HLA pairs between the groups.
Table 2.
KIR/HLA frequencies in type 1 AIH and healthy individuals.
| AIH (n = 154) |
Healthy individuals (n = 201) |
OR (95% CI) | p value | |
|---|---|---|---|---|
| KIR2DL1/HLA-C2 | 18 (12%) | 49 (24%) | 0.41 (0.23-0.74) | 0.0025 |
| KIR2DL2/HLA-C1 | 24 (16%) | 28 (14%) | 0.662 | |
| KIR2DS1/HLA-C2 | 7 (5%) | 19 (9%) | 0.079 | |
| KIR2DS2/HLA-C1 | 26 (17%) | 32 (16%) | 0.808 | |
| KIR3DL1/HLA-B Bw4 | 111 (72%) | 146 (73%) | 0.907 | |
| KIR3DL1/HLA-B Bw4-80Ile | 99 (64%) | 100 (50%) | 1.82 (1.18-2.80) | 0.0062 |
| KIR3DL1/HLA-B Bw4-80Thr | 23 (15%) | 53 (26%) | 0.49 (0.28-0.84) | 0.0092 |
| KIR3DS1/HLA-Bw4 | 50 (33%) | 70 (35%) | 0.642 | |
| KIR3DL2/HLA-A3, -A11 | 38 (25%) | 48 (24%) | 0.862 |
AIH, autoimmune hepatitis; HLA, human leukocyte antigen; KIR, killer cell immunoglobulin-like receptor; OR, odds ratio.
The frequency of the HLA-DRB1*04:05-DQB1:04:01 haplotype was significantly higher in patients with AIH (69%) than in healthy individuals (28%) (OR 5.62; 95% CI 3.52–8.97; p ≪0.001) as in previous studies (Table S2).29 Next, we evaluated this haplotype and the KIR3DL1/HLA-B Bw4-80Ile found in association with AIH for independence by logistic regression analysis. Both the HLA-DRB1*04:05-DQB1:04:01 haplotype (OR 5.67; 95% CI 3.53–9.11; p ≪0.001) and KIR3DL1/HLA-B Bw4-80Ile (OR 1.86; 95% CI 1.17–2.98; p = 0.009) were independent susceptibility genes related to AIH. AIH susceptibility correlated positively with the combination of the HLA-DRB1*04:05-DQB1:04:01 haplotype and KIR3DL1/HLA-B Bw4-80Ile (59 of 154 [38%] vs. 20 of 201 [10%]; OR 5.32; 95% CI 3.20–9.89; p ≪0.001). We therefore tested for synergic effects between the genetic factors as reported previously.30 Based on logistic regression analysis, this method evaluated whether the observed ORs of the 2 independent factors were greater combined than separately. No synergy was observed between the 2 factors in the AIH group (synergy factor = 1.10; 95% CI 0.42–2.85; Psynergy = 0.852), which confirmed their independence from each other.
Prediction and risk factors associated with liver decompensation and liver-related death
As shown in Table 3, 19 patients experienced liver decompensation (7 with ascites, 6 with hepatocellular carcinoma, 3 with hepatic encephalopathy, and 3 with bleeding esophageal varices) during a median follow-up period of 10.5 (interquartile range: 6.4–16.7) years. Serum ALT levels (222 vs. 428 U/L, p = 0.021), platelet count (15.5 vs. 17.4x104/μl, p = 0.002), and prothrombin time (PT) % (46.9 vs. 83.8%, p ≪0.001) were all significantly lower in patients who developed liver decompensation than in those who did not. The presence of cirrhosis at diagnosis was also significantly associated with future liver decompensation (47 vs. 5%, p ≪0.001).
Table 3.
Demographic and clinical characteristics of patients with type 1 AIH.
| Characteristic | Liver decompensation (n = 19) |
Liver compensation (n = 135) |
p value | Death (n = 13) |
Survival (n = 141) |
p value |
|---|---|---|---|---|---|---|
| Age (years) | 63 (54-72) | 59 (48-66) | 0.241 | 63 (55-70) | 59 (48-66) | 0.198 |
| Female, n (%) | 18 (95) | 117 (87) | 0.529 | 13 (100) | 122 (87) | 0.331 |
| Follow-up (years) | 6.3 (0.9-16.3) | 10.5 (6.6-16.8) | 0.146 | 19.1 (4.7-25.0) | 10.5 (6.5-16.8) | 0.326 |
| AIH score | 18 (17-19) | 18 (17-19) | 0.686 | 19 (17-19) | 18 (17-19) | 0.353 |
| Definite AIH, n (%) | 17 (90) | 118 (87) | 0.908 | 11 (85) | 124 (88) | 0.927 |
| AST (U/L) | 376 (78-594) | 410 (149-835) | 0.235 | 428 (65-595) | 408 (149-834) | 0.387 |
| ALT (U/L) | 222 (95-591) | 428 (177-997) | 0.021 | 222 (41-528) | 423 (174-983) | 0.049 |
| Total bilirubin (mg/dl) | 3.5 (1.4-10.3) | 1.6 (0.9-6.5) | 0.054 | 3.5 (1.3-10.9) | 1.6 (0.9-6.4) | 0.194 |
| IgG (mg/dl) | 2,981 (2,040-4,306) | 2,692 (2,053-3,464) | 0.401 | 2,981 (1,950-4,121) | 2,698 (2,060-3,465) | 0.609 |
| Platelets (×104/μl) | 15.5 (10.6-17.5) | 17.4 (16.3-20.3) | 0.002 | 15.5 (11.0-17.5) | 17.5 (16.3-19.9) | 0.020 |
| Prothrombin % | 46.9 (41.0-75.2) | 83.8 (67.3-98.0) | ≪0.001 | 46.0 (36.5-75.0) | 80.6 (65.0-98.0) | 0.001 |
| ANA, n (%) | 18 (95) | 132 (98) | 0.992 | 13 (100) | 137 (97) | 0.767 |
| SMA, n (%) | 8/12 (67) | 58/110 (53) | 0.358 | 6/9 (67) | 60/113 (53) | 0.661 |
| SLA, n (%) | 0 (0) | 0 (0) | - | 0 (0) | 0 (0) | – |
| Cirrhosis, n (%) | 9 (47) | 7 (5) | ≪0.001 | 6 (46) | 10 (7) | ≪0.001 |
| Complete remission, n (%) | 18 (95) | 131 (97) | 0.872 | 12 (92) | 137 (97) | 0.899 |
| Relapse, n (%) | 11 (58) | 53 (39) | 0.123 | 9 (69) | 55 (39) | 0.068 |
Values are expressed as the median (interquartile range) unless otherwise noted. AIH, autoimmune hepatitis; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ANA, anti-nuclear antibody; SLA, anti-soluble liver antigen; SMA, anti-smooth muscle antibody.
When HLA, KIR, and KIR/HLA were compared between patients with and without liver decompensation, the frequencies of HLA-B Bw4 (53 vs. 80%; OR 0.28; 95% CI 0.10–0.74; p = 0.008) and HLA-B Bw4-80Ile (42 vs. 73%; OR 0.25; 95% CI 0.08–0.80; p = 0.007) were significantly lower in patients with liver decompensation (Table S3). Although no KIRs were notably associated with liver decompensation development (Table S3), KIR3DL1/HLA-B Bw4 (47 vs. 76%; OR 0.29; 95% CI 0.11–0.78; p = 0.010) and KIR3DL1/HLA-B Bw4-80Ile (37 vs. 68%; OR 0.27; 95% CI 0.10–0.74; p = 0.008) were significantly less frequent in patients who experienced decompensation (Table 4).
Table 4.
KIR/HLA frequencies in patients with type 1 AIH with or without liver decompensation.
| Liver decompensation (n = 19) |
Liver compensation (n = 135) |
OR (95% CI) | p value | |
|---|---|---|---|---|
| KIR2DL1/HLA-C2 | 3 (15%) | 15 (11%) | 0.831* | |
| KIR2DL2/HLA-C1 | 1 (5%) | 23 (17%) | 0.324* | |
| KIR2DS1/HLA-C2 | 2 (11%) | 5 (4%) | 0.454* | |
| KIR2DS2/HLA-C1 | 1 (5%) | 25 (19%) | 0.264* | |
| KIR3DL1/HLA-B Bw4 | 9 (47%) | 102 (76%) | 0.29 (0.11–0.78) | 0.010 |
| KIR3DL1/HLA-B Bw4-80Ile | 7 (37%) | 92 (68%) | 0.27 (0.10–0.74) | 0.008 |
| KIR3DL1/HLA-B Bw4-80Thr | 2 (11%) | 21 (16%) | 0.816* | |
| KIR3DS1/HLA-B Bw4 | 5 (26%) | 45 (33%) | 0.726* | |
| KIR3DL2/HLA-A3, -A11 | 7 (37%) | 31 (23%) | 0.303 |
Fisher’s exact test. AIH, autoimmune hepatitis; HLA, human leukocyte antigen; KIR, killer cell immunoglobulin-like receptor; OR, odds ratio.
Among the 154 patients with AIH, 13 (8%) succumbed to liver-related death (6 from advanced hepatocellular carcinoma and 7 from liver failure) during follow-up. None had received liver transplantation. Those patients had significantly lower median ALT levels (222 vs. 423 U/L, p = 0.049), platelet count (15.5 vs. 17.5 ×104/μl, p = 0.020), and PT % (46.0 vs. 80.6%, p = 0.001) compared with survivors. Moreover, patients with a poor outcome more frequently had cirrhosis than those with a favorable outcome (46 vs. 7%, p ≪0.001) (Table 3).
The presence of HLA-B Bw4 (79 vs. 46%; OR 0.22; 95% CI 0.07–0.71; p = 0.007) and Bw4-80Ile (72 vs. 39%; OR 0.25; 95% CI 0.08–0.80; p = 0.031) were significantly associated with a favorable outcome (Table S4), with no remarkable associations seen for HLA-A3 or -A11, HLA-C1, or HLA-C2. The frequencies of KIR genes did not differ noticeably between survivors and patients with liver-related death (Table S4). Among KIR/HLA combinations, KIR3DL1/HLA-B Bw4 and KIR3DL1/HLA-B Bw4-80Ile were significantly associated with survival versus liver-related death (75 vs. 39%; OR 0.21; 95% CI 0.06–0.67; p = 0.012 and 67 vs. 31%; OR 0.22; 95% CI 0.06–0.74; p = 0.020, respectively) (Table 5).
Table 5.
KIR/HLA frequencies in patients with type 1 AIH according to survival and liver-related death.
| Death (n = 13) |
Survival (n = 141) |
OR (95% CI) | p value | |
|---|---|---|---|---|
| KIR2DL1/HLA-C2 | 0 (0%) | 18 (13%) | 0.358 | |
| KIR2DL2/HLA-C1 | 1 (8%) | 23 (16%) | 0.674 | |
| KIR2DS1/HLA-C2 | 0 (0%) | 7 (5%) | 0.899 | |
| KIR2DS2/HLA-C1 | 1 (8%) | 25 (18%) | 0.591 | |
| KIR3DL1/HLA-B Bw4 | 5 (39%) | 106 (75%) | 0.21 (0.06-0.67) | 0.012 |
| KIR3DL1/HLA-B Bw4-80Ile | 4 (31%) | 95 (67%) | 0.22 (0.06-0.74) | 0.020 |
| KIR3DL1/HLA-B Bw4-80Thr | 1 (8%) | 22 (16%) | 0.720 | |
| KIR3DS1/HLA-B Bw4 | 3 (23%) | 47 (33%) | 0.655 | |
| KIR3DL2/HLA-A3, -A11 | 4 (31%) | 34 (24%) | 0.844 |
AIH, autoimmune hepatitis; HLA, human leukocyte antigen; KIR, killer cell immunoglobulin-like receptor; OR, odds ratio.
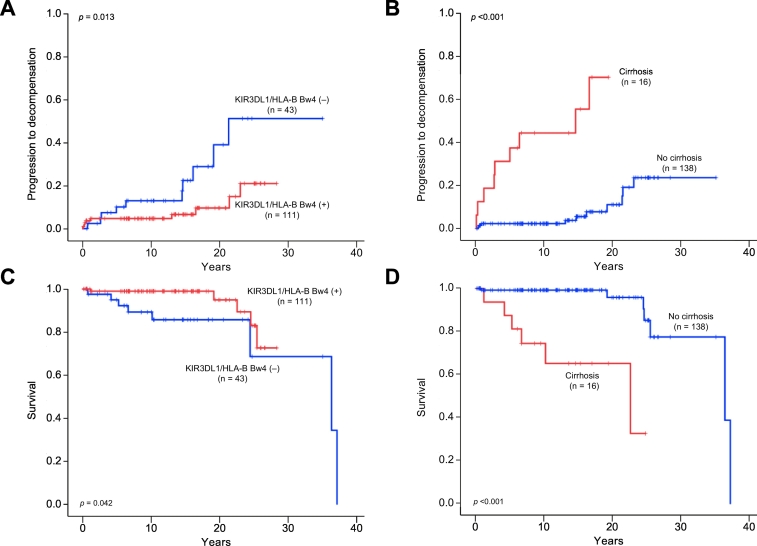
According to Cox proportional hazards regression analysis, liver decompensation and liver-related death had significant associations with KIR3DL1/HLA-B Bw4 (hazard ratio [HR] 0.37; 95% CI 0.14–0.94; p = 0.037 and HR 0.26; 95% CI 0.07–0.93; p = 0.038, respectively) and histological evidence of cirrhosis at initial diagnosis (HR 9.00; 95% CI 3.11–26.05; p ≪0.001 and HR 17.87; 95% CI 4.61–69.18; p ≪0.001, respectively) (Table 6). A PT % of ≫47.5% was a protective factor against liver decompensation (HR 0.21; 95% CI 0.08–0.55; p = 0.002), but not against liver-related death.
Table 6.
Factors associated with liver decompensation and liver-related death in AIH.
| Factor | HR | 95% CI | p value |
|---|---|---|---|
| Association with liver decompensation | |||
| KIR3DL1/HLA-B Bw4 | |||
| Positive | 0.37 | 0.14-0.94 | 0.037 |
| Negative | 1 | ||
| Cirrhosis | |||
| Yes | 9.00 | 3.11-26.05 | ≪0.001 |
| No | 1 | ||
| Prothrombin % | |||
| ≫47.5 | 0.21 | 0.08-0.55 | 0.002 |
| ≤47.5 | 1 | ||
| Association with liver-related death | |||
| KIR3DL1/HLA-B Bw4 | |||
| Positive | 0.26 | 0.07-0.93 | 0.038 |
| Negative | 1 | ||
| Cirrhosis | |||
| Yes | 17.87 | 4.61-69.18 | ≪0.001 |
| No | 1 | ||
Cox proportional hazard model. AIH, autoimmune hepatitis; HLA, human leukocyte antigen; HR, hazard ratio; KIR, killer cell immunoglobulin-like receptor.
Cumulative incidence of liver decompensation and liver-related death by risk factor
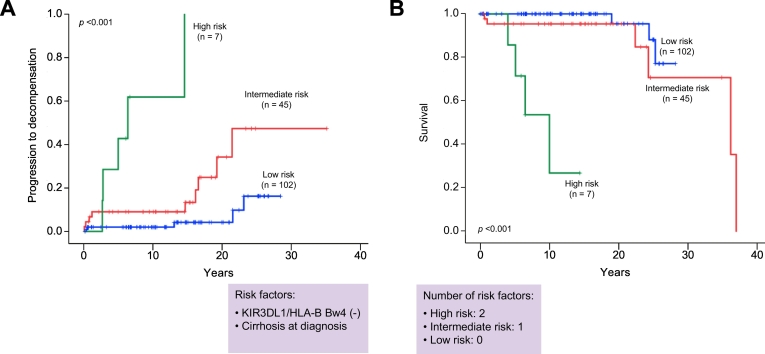
Kaplan-Meier survival estimates showed that patients with KIR3DL1/HLA-B Bw4 had a significantly lower risk of liver decompensation and liver-related death (log-rank test; p = 0.013 and p = 0.042, respectively) (Fig. 1A and 1C). In Kaplan-Meier plots for patients with or without cirrhosis, the cumulative liver decompensation and survival rates between the groups were significantly different (log-rank test; p ≪0.001 and p ≪0.001, respectively) (Fig. 1B and 1D). We further divided the patients into 3 groups according to the number of risk factors identified by multivariate analysis (KIR3DL1/HLA-B Bw4-negative and cirrhosis at diagnosis). Patients with both risk factors were assigned to the high-risk group, those with 1 factor to the intermediate-risk group, and those with no factors to the low-risk group. Kaplan-Meier testing showed that the cumulative incidences of liver decompensation and liver-related death were highest in the high-risk group, followed next by the intermediate- and low-risk groups (log-rank test; p ≪0.001 and p ≪0.001, respectively) (Fig. 2A and 2B).
Fig. 1.
Cumulative progression to liver decompensation and survival rates of AIH patients were analyzed using the Kaplan-Meier method. Progression to liver decompensation was significantly lower in (A) patients with KIR3DL1/HLA-B Bw4 (p = 0.043) and higher in (B) those with cirrhosis at diagnosis (p ≪0.001). Survival was significantly higher in (C) patients with KIR3DL1/HLA-B Bw4 (p = 0.042) and lower in (D) those with cirrhosis at diagnosis (p ≪0.001). p values were calculated by the log-rank test. AIH, autoimmune hepatitis.
Fig. 2.
Cumulative progression to liver decompensation and survival according to the number of risk factors of KIR3DL1/HLA-B Bw4 (-) and cirrhosis at diagnosis. Kaplan-Meier testing showed that the cumulative incidences of (A) liver decompensation and (B) liver-related death were highest in the high-risk group, followed next by the intermediate- and low-risk groups (log-rank test; p ≪0.001 and p ≪0.001, respectively). Green, red, and blue lines indicate high-risk patients (2 risk factors), intermediate-risk patients (1 risk factor), and low-risk patients (no risk factors), respectively. p values were calculated by the log-rank test.
Discussion
The present study examined HLA class I and II alleles, 14 KIR genes, and KIR/HLA combinations in type 1 AIH in a Japanese population. The KIR gene family is highly polymorphic and not well captured by standard genome-wide association study approaches. The key findings were: i) KIR3DL1/HLA-B Bw4-80Ile was significantly associated with the disease and was an independent susceptibility gene along with the HLA-DRB1*04:05-DQB1*04:01 haplotype; ii) patients with AIH had significantly lower frequencies of KIR2DL1/HLA-C2 and KIR3DL1/HLA-B Bw4-80Thr, suggesting protection by those KIR-HLA pairs against the disease; iii) KIR3DL1/HLA-B Bw4 and KIR3DL1/HLA-B Bw4-80Ile were protective against liver decompensation and liver-related death; and iv) the absence of KIR3DL1/HLA-B Bw4 and the presence of cirrhosis at diagnosis were strongly related to AIH progression.
This study revealed KIR3DL1/HLA-B Bw4-80Ile to be significantly increased in AIH in addition to the HLA-DRB1*04:05-DQB1*04:01 haplotype, a confirmed susceptibility factor in Japan.3,29 This haplotype has also been associated with primary biliary cholangitis in the Japanese.24 Although both genetic factors correlated positively with AIH, we did not observe any synergistic properties. The synergy factor calculations in the present study were designed to be robust for small sample sizes, even when individual cells were zero. The results corroborated the findings that the combination of KIR3DL1/HLA-B Bw4-80Ile and HLA-DRB1*04:05-DQB1:04:01 haplotypes had no significant advantage over each factor in isolation towards AIH susceptibility, confirming them both as independent disease risk factors.
All HLA-B molecules express 1 of 2 mutually exclusive serological epitopes (Bw4 or Bw6) that are encoded by 5 variable amino acids spanning positions 77–83. Bw6 is exclusive to HLA-B molecules and is present in roughly two-thirds of molecules, while Bw4 is present in the remaining third of HLA-B molecules, as well as in several HLA-A molecules. In this study, Bw4 motifs in HLA-B and HLA-A alleles were further analyzed.31,32 Receptor-ligand binding and lysis inhibition assays have shown that HLA-B molecules containing Bw4-80Ile may be more effective ligands for KIR3DL1 than those containing Bw4-80Thr.13,33 This difference might have accounted for the susceptibility observed for KIR3DL1/HLA-B Bw4-80Ile and protection for KIR3DL1/HLA-B Bw4-80Thr in AIH. At present, we can only describe the observation of an inverse effect on AIH development without a sound scientific basis for its occurrence.
Importantly, the present study revealed that the KIR2DL1/HLA-C2 combination, which also has the strongest inhibitory receptor-ligand interaction, was protective against AIH. In Japan, no correlations have been found between KIR2DL1/HLA-C2 and autoimmune diseases. Only a prior study from our group has detected an association of KIR2DL1/HLA-C2 with ulcerative colitis.34 Concurrent AIH and inflammatory bowel disease might represent unique entities from either process alone.35 Several reports have described that some patients with AIH had ulcerative colitis in addition to features of primary sclerosing cholangitis and younger age of onset.36,37 In our cohort, however, no case exhibited concurrent inflammatory bowel disease, primary sclerosing cholangitis, or young age of onset, although we could not exclude the possibility that HLA-C2 alone might play a prominent role in AIH resistance since all patients and controls possessed KIR2DL1. The decreased frequency of the strongly inhibitory KIR2DL1/HLA-C2 pair might have been difficult to overcome by simultaneous activation signals to promote autoimmunity in patients with AIH. Previous investigations have revealed that KIR allotypes modulate major NK cell effector function38 and KIR3DL1 allotypes modify HLA-B*57 protection against human immunodeficiency virus-1.39 Thus, further studies are required on KIR allotypes to clarify their relationship with AIH.
Interestingly, we observed novel protective effects for KIR3DL1/HLA-B Bw4 and KIR3DL1/HLA-B Bw4-80Ile against progression to liver decompensation and liver-related death in AIH, with KIR3DL1/HLA-B Bw4 remaining an independent predictor of disease progression in multivariate analysis. Several reports have identified that genetic features, in particular HLA DRB1 alleles, influence disease outcome in AIH.3,[40], [41], [42] However, none have demonstrated that KIR/HLA combinations influence disease progression and mortality in AIH despite significant associations between KIR/HLA pairs and hepatocellular carcinoma in hepatitis C virus infection.27 The precise reasons for this observation are unknown, but the association of this KIR/HLA pair and protection against disease progression is striking.
Kirstein et al. reported poor survival in patients with AIH who were anti-SLA positive.42 Although none of our patients were positive for anti-SLA antibodies, previous data have suggested an association between SLA antibodies and HLA class II DRB1*03:01 in Caucasians.42 HLA-DRB1*03:01 was absent in our cohort as its allele frequency is less than 0.15% in the Japanese population.
Lastly, we noticed that patients with cirrhosis at diagnosis had significantly increased risks of liver-related death and liver decompensation in Cox regression and Kaplan-Meier analyses, which was in agreement with studies in Germany and the Netherlands.42,43. No significant associations were seen between the frequencies of cirrhosis and KIR3DL1/HLA-B Bw4 by the chi-square test. When the factors of cirrhosis at diagnosis and KIR3DL1/HLA-B Bw4 negativity were combined, patients with both risk factors tended to exhibit a more rapid progression to liver decompensation and liver-related death. In contrast, patients without either risk factor seldom showed liver failure or liver-related death, even at 20 years after diagnosis.
There were several limitations to this study. It was retrospective in nature and contained a small sample size despite its long median follow-up period of 11.1 years. As the numbers of patients who developed liver-related death and liver decompensation were small, further longitudinal multicentric studies including a larger number of cases and controls are required. However, power calculations based on the study of 154 patients with AIH and 201 controls, and an OR of 1.82 at KIR3DL1/HLA-B Bw4, demonstrated sufficient power (0.814) at the 0.05 level of significance.
In conclusion, KIR3DL1/HLA-B Bw4-80Ile, KIR3DL1/HLA-B Bw4-80Thr, and KIR2DL1/HLA-C2 were associated with either AIH susceptibility or protection in a Japanese population. KIR3DL1/HLA-B Bw4 is a novel KIR/HLA pair related to a favorable clinical outcome, while histological cirrhosis at the initial diagnosis is a risk factor for poor prognosis. Thus, frequent and careful surveillance is advised for KIR3DL1/HLA-B Bw4-negative patients with cirrhosis. It should be noted, however, that genetic analysis alone is insufficient to pinpoint the contributions of predisposing KIR-HLA factors; functional clinical and basic studies are also needed to identify the mechanisms by which NK cells protect or predispose individuals to AIH pathogenesis and progression.
Abbreviations
ALT, alanine aminotransferase; ANA, anti-nuclear antibody; AST, aspartate aminotransferase; HLA, human leukocyte antigen; HR, hazard ratio; KIR, killer cell immunoglobulin-like receptor; NK, natural killer; OR, odds ratio; PT, prothrombin time; SLA, soluble liver antigen; SMA, anti-smooth muscle antibody.
Financial support
This study was funded by a JSPS grant-in-aid (grant number; 17K09416 to TU).
Conflict of interest
Gary L. Norman is an employee of Inova Diagnostics, Inc. The remaining authors declare no conflicts of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
TU and MO contributed to the conception and design of the study. TU, SJ, KY, and ET contributed to acquisition of the data. HS contributed to the analysis of the data. GLN contributed reagents/materials/analysis tools. TU, SJ, HS, GLN, and MO contributed to interpretation of the data. TU, SJ, and MO contributed to the drafting of the article. All authors contributed to critical revision and had final approval of the version of the manuscript to be published.
Acknowledgments
The authors thank Trevor Ralph for his editorial assistance.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jhepr.2019.09.003.
Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
References
- 1.Heneghan MA, Yeoman AD, Verma S, Smith AD, Longhi MS. Autoimmune hepatitis. Lancet. 2013;382:1433–1444. doi: 10.1016/S0140-6736(12)62163-1. [DOI] [PubMed] [Google Scholar]
- 2.Bossen L, Gerussi A, Lygoura V, Mells GF, Carbone M, Invernizzi P. Support of precision medicine through risk-stratification in autoimmune liver diseases - histology, scoring systems, and non-invasive markers. Autoimmun Rev. 2018;17:854–865. doi: 10.1016/j.autrev.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Umemura T, Katsuyama Y, Yoshizawa K, Kimura T, Joshita S, Komatsu M. Human leukocyte antigen class II haplotypes affect clinical characteristics and progression of type 1 autoimmune hepatitis in Japan. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strettell MD, Donaldson PT, Thomson LJ, Santrach PJ, Moore SB, Czaja AJ. Allelic basis for HLA-encoded susceptibility to type 1 autoimmune hepatitis. Gastroenterology. 1997;112:2028–2035. doi: 10.1053/gast.1997.v112.pm9178696. [DOI] [PubMed] [Google Scholar]
- 5.Fainboim L, Marcos Y, Pando M, Capucchio M, Reyes GB, Galoppo C. Chronic active autoimmune hepatitis in children. Strong association with a particular HLA-DR6 (DRB1*1301) haplotype. Hum Immunol. 1994;41:146–150. doi: 10.1016/0198-8859(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 6.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 7.Jeffery HC, Braitch MK, Bagnall C, Hodson J, Jeffery LE, Wawman RE. Changes in natural killer cells and exhausted memory regulatory T Cells with corticosteroid therapy in acute autoimmune hepatitis. Hepatol Commun. 2018;2:421–436. doi: 10.1002/hep4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mele D, Bossi G, Maggiore G, Oliviero B, Mantovani S, Bonelli B. Altered natural killer cell cytokine profile in type 2 autoimmune hepatitis. Clin Immunol. 2018;188:31–37. doi: 10.1016/j.clim.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Xiao F, Ai G, Yan W, Wan X, Luo X, Ning Q. Intrahepatic recruitment of cytotoxic NK cells contributes to autoimmune hepatitis progression. Cell Immunol. 2018;327:13–20. doi: 10.1016/j.cellimm.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15:243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 11.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 12.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Döhring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- 15.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 16.Littera R, Chessa L, Onali S, Figorilli F, Lai S, Secci L. Exploring the Role of Killer Cell Immunoglobulin-Like Receptors and Their HLA Class I Ligands in Autoimmune Hepatitis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podhorzer A, Paladino N, Cuarterolo ML, Fainboim HA, Paz S, Theiler G. The early onset of type 1 autoimmune hepatitis has a strong genetic influence: role of HLA and KIR genes. Genes Immun. 2016;17:187–192. doi: 10.1038/gene.2016.7. [DOI] [PubMed] [Google Scholar]
- 18.Saito S, Ota S, Yamada E, Inoko H, Ota M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens. 2000;56:522–529. doi: 10.1034/j.1399-0039.2000.560606.x. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez F, Berg PA, Bianchi FB, Bianchi L, Burroughs AK, Cancado EL. International Autoimmune Hepatitis Group Report: review of criteria for diagnosis of autoimmune hepatitis. J Hepatol. 1999;31:929–938. doi: 10.1016/s0168-8278(99)80297-9. [DOI] [PubMed] [Google Scholar]
- 20.Umemura T, Zen Y, Hamano H, Kawa S, Nakanuma Y, Kiyosawa K. Immunoglobin G4-hepatopathy: association of immunoglobin G4-bearing plasma cells in liver with autoimmune pancreatitis. Hepatology. 2007;46:463–471. doi: 10.1002/hep.21700. [DOI] [PubMed] [Google Scholar]
- 21.Yoshizawa K, Matsumoto A, Ichijo T, Umemura T, Joshita S, Komatsu M. Long-term outcome of Japanese patients with type 1 autoimmune hepatitis. Hepatology. 2012;56:668–676. doi: 10.1002/hep.25658. [DOI] [PubMed] [Google Scholar]
- 22.European Association for the Study of the L EASL Clinical Practice Guidelines: Autoimmune hepatitis. J Hepatol. 2015;63:971–1004. doi: 10.1016/j.jhep.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 23.van Gerven NM, Verwer BJ, Witte BI, van Hoek B, Coenraad MJ, van Erpecum KJ. Relapse is almost universal after withdrawal of immunosuppressive medication in patients with autoimmune hepatitis in remission. J Hepatol. 2013;58:141–147. doi: 10.1016/j.jhep.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Umemura T, Joshita S, Ichijo T, Yoshizawa K, Katsuyama Y, Tanaka E. Human leukocyte antigen class II molecules confer both susceptibility and progression in Japanese patients with primary biliary cirrhosis. Hepatology. 2012;55:506–511. doi: 10.1002/hep.24705. [DOI] [PubMed] [Google Scholar]
- 25.Muller CA, Engler-Blum G, Gekeler V, Steiert I, Weiss E, Schmidt H. Genetic and serological heterogeneity of the supertypic HLA-B locus specificities Bw4 and Bw6. Immunogenetics. 1989;30:200–207. doi: 10.1007/BF02421207. [DOI] [PubMed] [Google Scholar]
- 26.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 27.Saito H, Umemura T, Joshita S, Yamazaki T, Fujimori N, Kimura T. KIR2DL2 combined with HLA-C1 confers risk of hepatitis C virus-related hepatocellular carcinoma in younger patients. Oncotarget. 2018;9:19650–19661. doi: 10.18632/oncotarget.24752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka S, Furukawa H, Yasunami M, Kawasaki A, Nakamura H, Nakamura M. HLA-DRB1 and DQB1 alleles in Japanese type 1 autoimmune hepatitis: The predisposing role of the DR4/DR8 heterozygous genotype. PLoS One. 2017;12 doi: 10.1371/journal.pone.0187325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortina-Borja M, Smith AD, Combarros O, Lehmann DJ. The synergy factor: a statistic to measure interactions in complex diseases. BMC Res Notes. 2009;2:105. doi: 10.1186/1756-0500-2-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foley BA, De Santis D, Van Beelen E, Lathbury LJ, Christiansen FT, Witt CS. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2008;112:435–443. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- 32.Thons C, Senff T, Hydes TJ, Manser AR, Heinemann FM, Heinold A. HLA-Bw4 80(T) and multiple HLA-Bw4 copies combined with KIR3DL1 associate with spontaneous clearance of HCV infection in people who inject drugs. J Hepatol. 2017;67:462–470. doi: 10.1016/j.jhep.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 33.Rojo S, Wagtmann N, Long EO. Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol. 1997;27:568–571. doi: 10.1002/eji.1830270231. [DOI] [PubMed] [Google Scholar]
- 34.Saito H, Hirayama A, Umemura T, Joshita S, Mukawa K, Suga T. Association between KIR-HLA combination and ulcerative colitis and Crohn's disease in a Japanese population. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeFilippis EM, Kumar S. Clinical Presentation and Outcomes of Autoimmune Hepatitis in Inflammatory Bowel Disease. Dig Dis Sci. 2015;60:2873–2880. doi: 10.1007/s10620-015-3699-4. [DOI] [PubMed] [Google Scholar]
- 36.Gregorio GV, Portmann B, Karani J, Harrison P, Donaldson PT, Vergani D. Autoimmune hepatitis/sclerosing cholangitis overlap syndrome in childhood: a 16-year prospective study. Hepatology. 2001;33:544–553. doi: 10.1053/jhep.2001.22131. [DOI] [PubMed] [Google Scholar]
- 37.Deneau M, Jensen MK, Holmen J, Williams MS, Book LS, Guthery SL. Primary sclerosing cholangitis, autoimmune hepatitis, and overlap in Utah children: epidemiology and natural history. Hepatology. 2013;58:1392–1400. doi: 10.1002/hep.26454. [DOI] [PubMed] [Google Scholar]
- 38.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin MP, Naranbhai V, Shea PR, Qi Y, Ramsuran V, Vince N. Killer cell immunoglobulin-like receptor 3DL1 variation modifies HLA-B*57 protection against HIV-1. J Clin Invest. 2018;128:1903–1912. doi: 10.1172/JCI98463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czaja AJ, Strettell MD, Thomson LJ, Santrach PJ, Moore SB, Donaldson PT. Associations between alleles of the major histocompatibility complex and type 1 autoimmune hepatitis. Hepatology. 1997;25:317–323. doi: 10.1002/hep.510250211. [DOI] [PubMed] [Google Scholar]
- 41.Muratori P, Granito A, Quarneti C, Ferri S, Menichella R, Cassani F. Autoimmune hepatitis in Italy: the Bologna experience. J Hepatol. 2009;50:1210–1218. doi: 10.1016/j.jhep.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Kirstein MM, Metzler F, Geiger E, Heinrich E, Hallensleben M, Manns MP. Prediction of short- and long-term outcome in patients with autoimmune hepatitis. Hepatology. 2015;62:1524–1535. doi: 10.1002/hep.27983. [DOI] [PubMed] [Google Scholar]
- 43.van den Brand FF, van der Veen KS, de Boer YS, van Gerven NM, Lissenberg-Witte BI, Beuers U. Increased Mortality Among Patients With vs Without Cirrhosis and Autoimmune Hepatitis. Clin Gastroenterol Hepatol. 2019;17:940–947. doi: 10.1016/j.cgh.2018.09.046. e942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3