Abstract
Biological reconstruction of allografts and recycled autografts have been widely implemented in high-grade osteogenic sarcoma. For treating tumor-bearing autografts, extracorporeal irradiation (ECIR) and liquid nitrogen (LN) freezing techniques are being used worldwide as a gold standard treatment procedure. Both the methods aim to eradicate the tumor cells from the local recurrence and restore the limb function. Therefore, it is essential and crucial to find, and compare the alterations at molecular and physiological levels of the treated and untreated OGS recycled autografts to obtain valuable clinical information for better clinical practice. Thus, we aimed to investigate the significantly expressed altered proteins from ECIR-and cryotherapy/freezing- treated OGS (n = 12) were compared to untreated OGS (n = 12) samples using LC-ESI-MS/MS analysis, and the selected proteins from this protein panel were verified using immunoblot analysis. From our comparative proteomic analysis identified a total of 131 differentially expressed proteins (DEPs) from OGS. Among these, 91 proteins were up-regulated (2.5 to 3.5-folds), and 40 proteins were down-regulated (0.2 to 0.5 folds) (p < 0.01 and 0.05). The functional enrichment analysis revealed that the identified DEPs have belonged to more than 10 different protein categories include cytoskeletal, extracellular matrix, immune, enzyme modulators, and cell signaling molecules. Among these, we have confirmed two potential candidates’ expressions levels such as Fibronectin and Protein S100 A4 using western blot analysis. Our proteomic study revealed that LN-freezing and ECIR treatments are effectively eradicating tumor cells, and reducing the higher expressions of DEPs at molecular levels which may help in restoring the limb functions of OGS autografts effectively. To the best of our knowledge, this is the first proteomic study that compared proteomic profiles among freezing, ECIR treated with untreated OGS in recycled autografts. Moreover, the verified proteins could be used as prognostic or diagnostic markers that reveal valuable scientific information which may open various therapeutic avenues in clinical practice to improve patient outcomes.
Subject terms: Proteome, Proteomic analysis, Bone cancer, Predictive markers, Bone cancer
Introduction
High-grade osteogenic sarcomas (OGS) are the most common primary malignant bone sarcomas that distress the bone and forms a matrix and osteoid around the knees1–3. It accounts one to three per million each year worldwide and has a high rate of incidence in children and adults3,4. Currently, the standard treatment procedures applicable for patients are neoadjuvant chemotherapy drugs combined with surgery, precision diagnostic instruments, and limb salvage operations5. At present, there are three reconstructive procedures available after resection of tumors that are affected with major joints, include tumor prosthesis, an osteoarticular allograft, and a composite biological reconstruction. Among these three options, biological reconstruction of allograft and autografts (recycled from the resected autogenic bone segment) technique has been widely implemented and become a gold standard procedure for patients with sarcomas6. In order to eliminate the residual tumor cells from recycled autografts extracorporeal irradiation (ECIR) and cryotherapy/liquid nitrogen (LN)freezing are the two commonly used treatment methods employed in the biological reconstruction7,8. This technique can improve the regeneration of the bone, help to attain union and subsequent remodeling, and especially it restores limb function by supplying blood, osteogenic cells and proteins to the graft interface.
There is an abundant amount of proteins in the human body play a prominent role in numerous biological and physiological processes. Especially every single protein has a unique function and play a crucial role in organs growth, development, metabolic regulation, disease progression, and pathophysiology. Thus, the altered levels of these proteins are extremely useful in the classification of cells and tissues in disease states9. Moreover, Proteomics is a composition of global proteins and their isoforms that helps to understand the different biological mechanisms of cells and organisms10. It is an emerging field of science that reveals numerous scientific and pathological information about any clinical specimen’s disease condition and treatment effects. The identified significantly expressed proteins could serve as therapeutic and diagnostic markers for cancers. By using the advanced proteomic technologies, we can identify the differentially expressed proteins (DEPs), and their functions, interactions, and structural changes in any clinical specimen10. On top of this, there are no reports available to this date related to the changes in protein expressions after ECIR and cryotherapy/LN-freezing treatments. In order to identify the molecular and proteomic changes after these treatments in recycled autografts of OGS helps to distinguish the status of the disease, and the effect of the treatments. In addition to this, a biomarker plays a significant role in monitoring the disease and provides valuable clinical information regarding the treatment concerning the tumor development, and its progression at the physiological and biological state
There is some evidence demonstrated about the effective irradiation dosage and the levels of protein change among the tumor samples11. In addition to this, our recent study has successfully evaluated the preservation of bone morphogenetic protein activity with ECIR and LN-freezing in the tumor-bearing recycled autografts for biological reconstruction12. But there is no complete protein profile report on alterations of proteins in recycled autografts especially after treatment with LN-freezing and ECIR. Therefore, we aimed to screen the DEPs (p < 0.05) from treated recycled OGS autografts compared with untreated OGS using high-resolution electron spray ionization liquid chromatography (LC-ESI-MS/MS) and tandem mass spectrometry analysis. The identified DEPs from OGS untreated samples help us to understand the tumor microenvironment, recurrence, metastasis, and prognosis of OGS. On the other hand, the altered protein expressions from OGS treated autografts with cryotherapy/LN-freezing and ECIR will provide crucial information about how both the treatments are effectively playing a key role in treating the autografts by eradicating the tumor cells and restores limb function by preserving its essential proteins. Since recycled autografts are widely being used in the biological reconstruction, this comparative proteomic study will provide numerous clues at molecular, and physiological levels such as bone healing, repair, remodeling, and pathophysiology of OGS tumor. Therefore, the identified protein profiles from our study will open new therapeutic avenues to improve patients’ outcomes in clinical practice.
Results
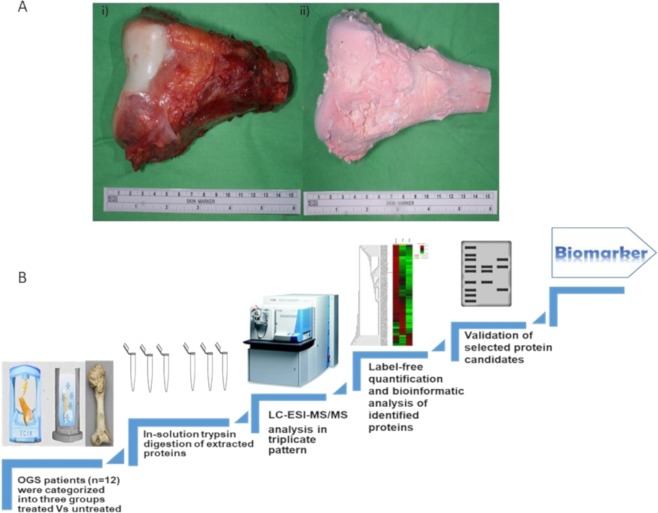
Proteins that are expressed significantly after any clinical treatment in human biological fluids have tremendous essential scientific information that helps to reveal potential diagnostic and prognostic features. Biological reconstruction of autografts and allografts is a gold standard procedure using worldwide that helps to fully restores the functions of affected OGS limbs. Cryotherapy/LN-freezing and ECIR are effectively eradicating the tumor cells from OGS bones and the rate of recurrence is very less. Therefore, to elucidate the biological and physiological impact of these methodologies on recycled autografts it is necessary to investigate the proteomic changes to understand the molecular alterations before and after the treatments. Therefore, a total of 36 bone tissue specimens from 12 OGS patients were characterized into three different groups, two groups were subjected as OGS treated such as with cryotherapy/freezing (n = 12) for 15 mins, and the other is ECIR treated at 15,000 gamma irradiations (n = 12), and the third is an untreated group (n = 12) which is a negative control. The complete workflow of our study along with freezing treated and ECIR treated OGS bone illustrations were shown in Fig. 1A,B.
Figure 1.
(A) Osteosarcoma resected autografts for treatment (i) OGS bone treated using irradiation and (ii) freezing. (B) The complete protein profiling workflow of comparative proteomic analysis between treated and untreated samples of osteosarcoma.
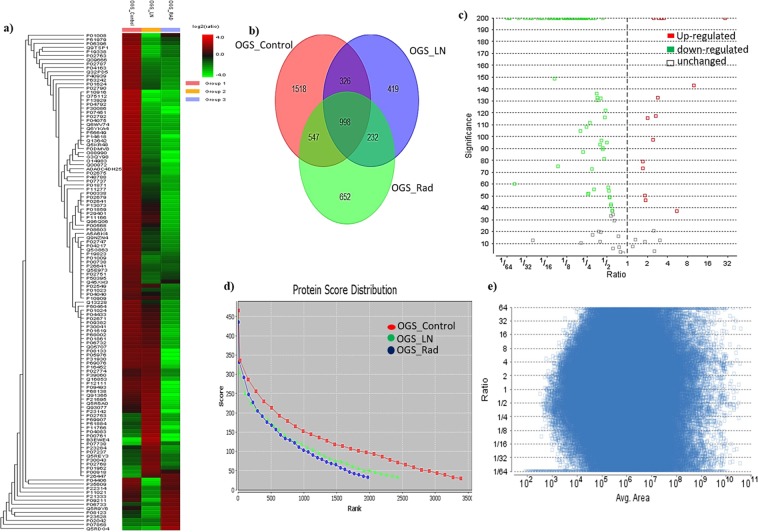
Protein samples were extracted from both treated and untreated OGS groups were subjected to triplicate LC-ESI-MS/MS analysis. Our comparative proteomic evaluations identified a total of 1518 proteins from untreated OGS, and 419 proteins from freezing treated, and 652 proteins from ECIR treated samples. In a heatmap generated by PEAKS Q software version X shows the differences in protein abundances of the identified proteins and their fold changes in Fig. 2a. The commonly identified proteins from three-groups are 998, and the commonly identified proteins among OGS control and freezing treated were 326. On the other hand, between OGS control and ECIR treated equally identified proteins were 527. When compared to OGS freezing and ECIR groups 232 proteins were identified (Fig. 2b). In order to obtain the highly significant differentially expressed proteins from our study, we filtered these proteins using the false discovery rate (FDR) of <0.1%, and the highest protein scores of >70 with a significance score of <20, and at least 2 up to ten unique peptides should be identified (Fig. 2c & Supplementary Table 1). Therefore, from our analysis, we have successfully identified a total of 131 proteins were significantly expressed in OGS treated vs untreated. About 99% of identified proteins from the three groups were quantified and the ratios were measured between triplicate individual samples and the results were highly correlated (r = 0.91, Supplementary Table 2). Among 131 proteins, 90 were up-regulated with >3.5–1.5-fold (p < 0.05 or 0.01) in OGS untreated/treated samples. The complete list of significantly identified up-regulated proteins from OGS control compared to the treatment groups was shown in Table 1. On the other hand, the complete list of 40 proteins which are down-regulated with <0.2–0.5 folds (p < 0.01 or 0.05) were presented in Tables 2 and 3. All the identified proteins and their abundances were compared and quantified using one-way ANOVA and the statistical significances were measured as 0.01 to 0.05.
Figure 2.
Identification of differentially expressed proteins from high-grade osteosarcoma compared after freezing and irradiation using liquid chromatography and tandem mass spectrometry label-free quantification (a) Heat map of OGS comparative proteomic profile among treated and untreated OGS was generated using PEAKS X software. (b) Venn diagram showing the differentially expressed proteins from comparative proteomic analysis of OGS control (untreated), and treated with freezing and ECIR. (c) Protein volcano plot illustration of significantly identified proteins red dots represents the up-regulated proteins green dots represents the down-regulated and the no colored box represents the unchanged proteins of the comparative analysis. (d) The protein score distribution among the three groups. (e) The distribution of feature vector ratio by intensity quantified using area under the curve by label-free quantification.
Table 1.
List of Up-regulated proteins in OGS untreated/control compared to OGS treated with LN-freezing and ECIR groups.
| S.NO | Description | Accession | Avg. Mass | FDR | Coverage (%) | #Peptides | #Unique | OGS Control Area |
OGS LN Area |
OGS RAD Area |
P-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-regulated proteins in OGS untreated/control compared to the treatment groups | |||||||||||
| 1 | Serotransferrin | P02787 | 77064 | <0.02 | 36 | 24 | 2 | 5.94E + 08 | 1.86E + 08 | 1.66E + 08 | 0.03 |
| 2 | Collagen alpha-1(XIV) chain | Q05707 | 193513 | <0.01 | 7 | 10 | 6 | 5.87E + 07 | 5.72E + 07 | 1.44E + 07 | 0.03 |
| 3 | Spectrin beta chain | P11277 | 246466 | <0.02 | 1 | 2 | 1 | 8.57E + 06 | 4.67E + 06 | 1.78E + 06 | 0.0008 |
| 4 | Alpha-actinin-2 | P35609 | 103854 | <0.01 | 4 | 4 | 1 | 6.28E + 06 | 7.35E + 05 | 5.40E + 06 | 0.0003 |
| 5 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | O14983 | 110252 | <0.02 | 2 | 2 | 1 | 1.08E + 08 | 4.02E + 07 | 8.01E + 05 | 0.00213 |
| 6 | Fibrinogen beta chain | P02675 | 55928 | <0.01 | 31 | 11 | 6 | 4.34E + 08 | 1.99E + 08 | 4.75E + 07 | 0.021 |
| 7 | Alpha-actinin-3 | O88990 | 103043 | <0.02 | 5 | 5 | 1 | 1.64E + 07 | 4.81E + 06 | 1.67E + 05 | 0.00012 |
| 8 | Creatine kinase M-type | P06732 | 43101 | <0.01 | 21 | 7 | 3 | 2.07E + 09 | 1.71E + 09 | 3.79E + 08 | 0.00014 |
| 9 | Fibrinogen alpha chain | P02671 | 94973 | <0.02 | 10 | 8 | 8 | 4.76E + 08 | 3.76E + 08 | 1.11E + 08 | 0.00052 |
| 10 | Apolipoprotein A-I | P02647 | 30778 | <0.01 | 56 | 14 | 4 | 7.88E + 08 | 2.59E + 08 | 3.75E + 07 | 0.000542 |
| 11 | Annexin A6 | P08133 | 75873 | <0.02 | 4 | 2 | 1 | 3.01E + 07 | 2.95E + 07 | 5.11E + 05 | 0.002 |
| 12 | Phosphoglycerate kinase 1 | P00558 | 44615 | <0.01 | 35 | 13 | 3 | 4.56E + 07 | 2.35E + 07 | 1.05E + 07 | 0.002 |
| 13 | Myosin-binding protein C | Q00872 | 128294 | <0.02 | 3 | 3 | 3 | 3.36E + 07 | 1.27E + 07 | 3.03E + 06 | 0.001 |
| 14 | Glyceraldehyde-3-phosphate dehydrogenase | P04406 | 36053 | <0.01 | 43 | 13 | 1 | 6.27E + 06 | 4.22E + 05 | 4.96E + 06 | 0.00000003 |
| 15 | Beta-enolase | P13929 | 46987 | <0.02 | 30 | 9 | 2 | 1.28E + 08 | 7.23E + 06 | 1.33E + 07 | 0.005 |
| 16 | Fructose-bisphosphate aldolase A | P04075 | 39420 | <0.01 | 44 | 13 | 2 | 2.06E + 08 | 3.48E + 07 | 2.37E + 07 | 0.005 |
| 17 | Complement factor H | P08603 | 139096 | <0.02 | 3 | 4 | 3 | 1.03E + 08 | 5.96E + 07 | 2.02E + 07 | 0.00054 |
| 18 | Heat shock 70 kDa protein 1 A | P0DMV8 | 70052 | <0.01 | 12 | 5 | 1 | 1.82E + 07 | 4.85E + 06 | 2.15E + 06 | 0.000569 |
| 19 | Gelsolin | P06396 | 85697 | <0.02 | 10 | 5 | 1 | 6.40E + 06 | 1.36E + 06 | 2.92E + 06 | 0.000425 |
| 20 | Tropomyosin beta chain | P07951 | 32851 | <0.01 | 23 | 7 | 1 | 1.78E + 07 | 4.81E + 06 | 1.44E + 06 | 0.00041 |
| 21 | Trifunctional enzyme subunit alpha mitochondrial | P40939 | 83000 | <0.02 | 4 | 2 | 2 | 1.04E + 07 | 4.38E + 06 | 3.19E + 06 | 0.001 |
| 22 | Fibrinogen gamma chain | P02679 | 51512 | <0.01 | 35 | 12 | 10 | 3.06E + 08 | 2.19E + 08 | 5.18E + 07 | 0.001 |
| 23 | L-lactate dehydrogenase A chain | P00338 | 36689 | <0.02 | 14 | 5 | 1 | 1.43E + 08 | 7.39E + 07 | 1.87E + 07 | 0.001 |
| 24 | Transketolase | P29401 | 67878 | <0.01 | 10 | 6 | 1 | 4.78E + 07 | 2.89E + 07 | 6.49E + 06 | 0.001 |
| 25 | Carbonic anhydrase 3 | P07451 | 29557 | <0.02 | 13 | 2 | 1 | 2.06E + 08 | 2.71E + 07 | 6.53E + 05 | 0.001 |
| 26 | Dihydropyrimidinase-related protein 2 | Q16555 | 62294 | <0.01 | 11 | 6 | 1 | 2.73E + 06 | 8.99E + 05 | 4.43E + 06 | 0.001 |
| 27 | Peroxiredoxin-6 | P30041 | 25035 | <0.02 | 22 | 4 | 3 | 1.19E + 08 | 8.58E + 07 | 2.08E + 07 | 0.001 |
| 28 | Perilipin-4 | Q96Q06 | 134431 | <0.01 | 2 | 2 | 2 | 1.72E + 07 | 1.18E + 07 | 2.83E + 06 | 0.001 |
| 29 | Four and a half LIM domains protein 1 | Q13642 | 36263 | <0.02 | 13 | 4 | 3 | 1.19E + 08 | 3.90E + 07 | 1.31E + 07 | 0.001 |
| 30 | Adenylate kinase isoenzyme 1 | P00568 | 21635 | <0.01 | 28 | 3 | 3 | 6.11E + 07 | 3.58E + 07 | 1.00E + 07 | 0.001 |
| 31 | Immunoglobulin heavy constant gamma 2 | P01859 | 35901 | <0.02 | 22 | 8 | 2 | 3.16E + 08 | 1.53E + 08 | 3.00E + 07 | 0.001 |
| 32 | Immunoglobulin heavy constant mu | P01871 | 49440 | <0.01 | 10 | 4 | 1 | 2.97E + 07 | 1.47E + 07 | 5.54E + 06 | 0.001 |
| 33 | Troponin I | P48788 | 21339 | <0.02 | 15 | 3 | 3 | 6.11E + 07 | 2.60E + 07 | 1.02E + 07 | 0.001 |
| 34 | Immunoglobulin heavy constant gamma 4 | P01861 | 35941 | <0.01 | 43 | 11 | 3 | 3.93E + 08 | 3.31E + 08 | 8.71E + 07 | 0.001 |
| 35 | Nucleolin | P19338 | 76615 | <0.02 | 1 | 1 | 1 | 2.07E + 07 | 2.28E + 06 | 6.46E + 06 | 0.001 |
| 36 | Histone H3 | P69076 | 15402 | <0.01 | 14 | 3 | 1 | 2.09E + 07 | 2.01E + 07 | 1.69E + 06 | 0.001 |
| 37 | Myosin light chain 1/3 skeletal muscle isoform | P05976 | 21145 | <0.02 | 11 | 2 | 1 | 2.55E + 07 | 2.48E + 07 | 7.31E + 05 | 0.001 |
| 38 | Ferritin light chain | P02792 | 20020 | <0.01 | 23 | 3 | 3 | 4.67E + 09 | 7.61E + 08 | 5.25E + 08 | 0.001 |
| 39 | Histone H4 | Q6WV74 | 11395 | <0.02 | 50 | 7 | 1 | 8.83E + 07 | 1.65E + 07 | 1.30E + 07 | 0.001 |
| 40 | Heat shock protein beta-1 | P04792 | 22783 | <0.01 | 33 | 4 | 2 | 2.33E + 08 | 2.29E + 07 | 1.69E + 07 | 0.001 |
| 41 | Cofilin-1 | P23528 | 18502 | <0.02 | 23 | 2 | 2 | 1.58E + 07 | 6.81E + 06 | 2.91E + 07 | 0.001 |
| 42 | Troponin T fast skeletal muscle | P02641 | 33034 | <0.01 | 3 | 1 | 1 | 2.29E + 07 | 1.26E + 07 | 2.53E + 06 | 0.001 |
| 43 | Heterogeneous nuclear ribonucleoprotein K | P61978 | 50976 | <0.02 | 3 | 1 | 1 | 1.23E + 07 | 2.52E + 06 | 6.43E + 06 | 0.001 |
| 44 | LIM domain-binding protein 3 | O75112 | 77135 | <0.01 | 1 | 1 | 1 | 9.52E + 06 | 4.70E + 05 | 6.32E + 05 | 0.001 |
| 45 | Phosphatidylethanolamine-binding protein 1 | P30086 | 21057 | <0.02 | 12 | 1 | 1 | 5.35E + 07 | 5.78E + 06 | 6.36E + 05 | 0.001 |
| 46 | High mobility group protein B1 | P09429 | 24894 | <0.01 | 7 | 1 | 1 | 8.70E + 06 | 1.50E + 06 | 1.46E + 06 | 0.001 |
| 47 | Methanethiol oxidase | Q13228 | 52391 | <0.02 | 3 | 1 | 1 | 1.15E + 07 | 6.95E + 06 | 2.75E + 06 | 0.001 |
| 48 | Erythrocyte membrane protein band 4.2 | P16452 | 77009 | <0.01 | 2 | 1 | 1 | 1.41E + 07 | 1.35E + 07 | 2.15E + 06 | 0.001 |
| 49 | 40 S ribosomal protein S19 | P39019 | 16060 | <0.02 | 14 | 2 | 2 | 9.14E + 06 | 3.59E + 06 | 2.60E + 06 | 0.001 |
| 50 | Antithrombin-III | P01008 | 52602 | <0.01 | 2 | 1 | 1 | 4.62E + 06 | 3.01E + 05 | 2.62E + 06 | 0.001 |
| 51 | Alpha-1-acid glycoprotein 1 | P02763 | 23512 | <0.02 | 7 | 1 | 1 | 8.83E + 08 | 1.21E + 08 | 3.00E + 08 | 0.001 |
| 52 | Solute carrier family 2 facilitated glucose transporter member 1 | P11166 | 54084 | <0.01 | 5 | 2 | 1 | 3.45E + 06 | 2.29E + 06 | 5.16E + 05 | 0.001 |
| 53 | Voltage-dependent anion-selective channel protein 2 | P45880 | 31567 | <0.02 | 6 | 2 | 2 | 1.90E + 07 | 1.59E + 07 | 2.64E + 06 | 0.001 |
| 54 | Eukaryotic translation initiation factor 5A-1 | P63241 | 16832 | <0.01 | 8 | 1 | 1 | 1.55E + 07 | 6.71E + 06 | 3.84E + 06 | 0.001 |
| 55 | Protein S100-A10 | P60903 | 11203 | <0.02 | 18 | 1 | 1 | 2.67E + 07 | 9.67E + 06 | 7.50E + 06 | 0.001 |
| 56 | Cytochrome c oxidase subunit 4 isoform 1 | P13073 | 19577 | <0.01 | 7 | 1 | 1 | 2.20E + 07 | 1.27E + 07 | 2.79E + 06 | 0.001 |
| 57 | Immunoglobulin kappa variable 3–11 | P04433 | 12575 | <0.02 | 8 | 1 | 1 | 2.36E + 07 | 1.71E + 07 | 5.73E + 06 | 0.01 |
| 58 | Immunoglobulin kappa variable 3–20 | P01619 | 12557 | <0.01 | 8 | 1 | 1 | 4.11E + 07 | 3.25E + 07 | 5.51E + 06 | 0.001 |
| 59 | Immunoglobulin kappa variable 3–15 | P01624 | 12496 | <0.02 | 8 | 1 | 1 | 5.88E + 07 | 2.81E + 07 | 1.40E + 07 | 0.001 |
| 60 | Immunoglobulin kappa variable 3D-20 | A0A0C4DH25 | 12515 | <0.01 | 8 | 1 | 1 | 2.55E + 06 | 1.15E + 06 | 2.07E + 05 | 0.001 |
| 61 | Neutrophil defensin 3 | P59666 | 10245 | <0.02 | 10 | 1 | 1 | 1.84E + 07 | 1.16E + 07 | 6.38E + 06 | 0.001 |
| 62 | Alpha-1-antitrypsin | P01009 | 46737 | <0.01 | 17 | 9 | 6 | 1.57E + 09 | 6.85E + 08 | 7.21E + 08 | 0.001 |
| 63 | Complement c1 q | P02747 | 25774 | <0.02 | 5 | 1 | 1 | 5.86E + 06 | 3.29E + 06 | 1.62E + 06 | 0.001 |
| 64 | Galectin-1 | P09382 | 14716 | <0.01 | 33 | 6 | 6 | 2.20E + 08 | 1.59E + 08 | 5.28E + 07 | 0.001 |
| 65 | Profilin-1 | P07737 | 15054 | <0.02 | 26 | 4 | 1 | 9.53E + 07 | 3.81E + 07 | 1.89E + 07 | 0.001 |
| 66 | Ubiquitin-like modifier-activating enzyme 1 | P22314 | 117849 | <0.01 | 2 | 1 | 1 | 8.17E + 06 | 3.38E + 06 | 7.22E + 06 | 0.001 |
| 67 | Haptoglobin | P00738 | 45205 | <0.02 | 26 | 9 | 3 | 2.68E + 07 | 1.15E + 07 | 1.45E + 07 | 0.00005 |
| 68 | Inter-alpha-trypsin inhibitor heavy chain H2 | P19823 | 106463 | <0.01 | 3 | 2 | 1 | 7.48E + 06 | 3.67E + 06 | 3.22E + 06 | 0.00001 |
| 69 | Hemopexin | P02790 | 51676 | <0.02 | 8 | 2 | 2 | 3.22E + 07 | 1.75E + 07 | 7.03E + 06 | 0.00008 |
| 70 | Spectrin alpha chain erythrocytic 1 | P02549 | 280013 | <0.01 | 2 | 6 | 5 | 1.96E + 07 | 1.10E + 07 | 6.10E + 06 | 0.001 |
| 71 | Serpin H1 | P50454 | 46441 | <0.02 | 8 | 2 | 1 | 4.41E + 06 | 2.93E + 06 | 1.30E + 06 | 0.0041 |
| 72 | Pyruvate kinase PKM | P14618 | 57937 | <0.01 | 28 | 10 | 1 | 1.90E + 06 | 4.08E + 05 | 3.86E + 04 | 0.00001 |
| 73 | Alpha-2-macroglobulin | P01023 | 163290 | <0.02 | 7 | 8 | 2 | 4.89E + 07 | 3.27E + 07 | 2.53E + 07 | 0.0051 |
| 74 | Alpha-1B-glycoprotein | P04217 | 54254 | <0.01 | 9 | 4 | 4 | 5.42E + 07 | 3.08E + 07 | 1.68E + 07 | 0.001 |
| 75 | Elongation factor 1-gamma | P26641 | 50119 | <0.02 | 2 | 1 | 1 | 3.94E + 07 | 2.17E + 07 | 2.07E + 07 | 0.001 |
| 76 | EH domain-containing protein 2 | Q9NZN4 | 61162 | <0.01 | 6 | 4 | 2 | 2.17E + 07 | 1.18E + 07 | 5.73E + 06 | 0.001 |
| 77 | Complement C3 | P01024 | 187147 | <0.02 | 12 | 19 | 19 | 4.68E + 08 | 3.75E + 08 | 1.59E + 08 | 0.001 |
| 78 | Rab GDP dissociation inhibitor beta | P50395 | 50663 | <0.01 | 3 | 1 | 1 | 1.69E + 07 | 9.20E + 06 | 1.17E + 07 | 0.001 |
| 79 | Hemoglobin subunit epsilon | Q45XH3 | 16156 | <0.02 | 22 | 3 | 1 | 6.31E + 09 | 3.45E + 09 | 4.93E + 09 | 0.00001 |
| 80 | Fibronectin | P02751 | 262622 | <0.01 | 2 | 2 | 1 | 1.58E + 06 | 9.03E + 05 | 1.06E + 06 | 0.001 |
| 81 | Catalase | P04040 | 59756 | <0.02 | 14 | 5 | 2 | 4.19E + 06 | 7.00E + 06 | 2.45E + 06 | 0.001 |
| 82 | Clusterin | P10909 | 52495 | <0.01 | 6 | 2 | 2 | 2.51E + 06 | 2.22E + 06 | 1.49E + 06 | 0.001 |
| 83 | 60 S ribosomal protein L18 | Q07020 | 21634 | <0.02 | 5 | 1 | 1 | 5.08E + 06 | 3.15E + 06 | 3.04E + 06 | 0.001 |
| 84 | Transthyretin | P02766 | 15887 | <0.01 | 9 | 1 | 1 | 1.52E + 07 | 1.20E + 07 | 9.30E + 06 | 0.001 |
| 85 | Trifunctional enzyme subunit beta | P55084 | 51294 | <0.02 | 4 | 2 | 1 | 3.46E + 06 | 2.13E + 06 | 1.96E + 06 | 0.001 |
| 86 | 40 S ribosomal protein S25 | P62851 | 13742 | <0.01 | 7 | 1 | 1 | 4.78E + 06 | 3.05E + 06 | 3.82E + 06 | 0.001 |
| 87 | Band 3 anion transport protein | P02730 | 101792 | <0.02 | 13 | 7 | 6 | 2.06E + 08 | 1.21E + 08 | 1.07E + 08 | 0.001 |
| 88 | Eukaryotic translation initiation factor 3 subunit I | Q13347 | 36502 | <0.01 | 3 | 1 | 1 | 6.44E + 05 | 1.95E + 05 | 3.58E + 05 | 0.001 |
| 89 | Carbonic anhydrase 1 | P00915 | 28870 | <0.02 | 21 | 5 | 4 | 2.79E + 08 | 2.12E + 08 | 2.24E + 08 | 0.001 |
| 90 | Peroxiredoxin-2 | P32119 | 21892 | <0.01 | 40 | 9 | 2 | 2.99E + 07 | 6.00E + 06 | 1.78E + 07 | 0.001 |
LN: Liquid Nitrogen; OGS: Osteosarcoma RAD: Irradiation/ECIR, FDR: False Discovery Rate.
Table 2.
List of down-regulated proteins in OGS untreated/control compared to OGS treated with LN-freezing and ECIR groups.
| S.NO | Description | Accession | Avg. Mass | FDR | Coverage (%) | #Peptides | #Unique | OGS Control Area |
OGS LN Area |
OGS RAD Area |
P-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Down-regulated proteins in OGS untreated/control compared to treated groups | |||||||||||
| 1 | Collagen alpha-3(VI) chain | P12111 | 343668 | <0.01 | 2 | 6 | 6 | 2.44E + 08 | 2.72E + 08 | 1.28E + 07 | 0.001 |
| 2 | Actin alpha skeletal | P68133 | 42051 | <0.01 | 43 | 17 | 1 | 2.83E + 07 | 3.35E + 07 | 2.21E + 06 | 0.00021 |
| 3 | Annexin A2 | P07355 | 38604 | <0.01 | 28 | 8 | 1 | 2.33E + 07 | 3.73E + 07 | 3.03E + 06 | 0.00012 |
| 4 | Tropomyosin alpha-1 chain | P09493 | 32709 | <0.01 | 23 | 7 | 1 | 3.36E + 07 | 3.85E + 07 | 1.76E + 06 | 0.00025 |
| 5 | Hemoglobin subunit alpha | P69905 | 15258 | <0.01 | 70 | 15 | 2 | 8.97E + 08 | 2.34E + 09 | 4.89E + 08 | 0.0025 |
| 6 | Annexin A1 | P04083 | 38714 | <0.01 | 15 | 4 | 1 | 1.32E + 07 | 3.86E + 07 | 5.46E + 06 | 0.001 |
| 7 | Carbonic anhydrase 2 | P00918 | 29246 | <0.01 | 5 | 1 | 1 | 2.86E + 06 | 9.52E + 06 | 6.45E + 06 | 0.001 |
| 8 | Membrane primary amine oxidase | Q16853 | 84622 | <0.01 | 8 | 6 | 1 | 1.80E + 07 | 2.08E + 07 | 4.50E + 06 | 0.001 |
| 9 | Glycerol-3-phosphate dehydrogenase | P21695 | 37568 | <0.01 | 11 | 4 | 1 | 1.70E + 07 | 2.62E + 07 | 1.56E + 06 | 0.001 |
| 10 | Lumican | P51884 | 38429 | <0.01 | 26 | 9 | 9 | 3.52E + 08 | 6.69E + 08 | 7.48E + 07 | 0.001 |
| 11 | Fibulin-1 | P23142 | 77214 | <0.01 | 3 | 2 | 1 | 1.72E + 06 | 3.25E + 06 | 5.65E + 04 | 0.001 |
| 12 | Immunoglobulin heavy constant gamma 3 | P01860 | 41287 | <0.01 | 29 | 11 | 2 | 4.41E + 06 | 5.82E + 06 | 5.66E + 06 | 0.001 |
| 13 | 40 S ribosomal protein S11 | P62280 | 18431 | <0.01 | 8 | 1 | 1 | 1.90E + 06 | 2.73E + 06 | 7.27E + 04 | 0.001 |
| 14 | Cytochrome b-c1 complex subunit 1 | P31930 | 52646 | <0.01 | 5 | 2 | 1 | 5.74E + 06 | 5.72E + 06 | 2.17E + 05 | 0.001 |
| 15 | Histone H2A | Q93077 | 14105 | <0.01 | 38 | 5 | 1 | 7.21E + 06 | 1.22E + 07 | 1.57E + 06 | 0.001 |
| 17 | Bisphosphoglycerate mutase | P07738 | 30005 | <0.01 | 5 | 1 | 1 | 1.95E + 05 | 1.93E + 06 | 6.50E + 05 | 0.001 |
| 18 | Adipocyte plasma membrane-associated protein | Q9HDC9 | 46480 | <0.01 | 6 | 1 | 1 | 3.17E + 06 | 7.86E + 06 | 2.85E + 06 | 0.001 |
| 19 | Protein disulfide-isomerase | P07237 | 57116 | <0.01 | 4 | 2 | 1 | 2.37E + 06 | 4.49E + 06 | 1.30E + 06 | 0.007 |
| 20 | Protein S100-A4 | P26447 | 11729 | <0.01 | 9 | 1 | 1 | 4.77E + 05 | 1.56E + 06 | 1.69E + 06 | 0.001 |
| 21 | Centrosomal protein of 162 kDa (Fragment) | Q91365 | 143966 | <0.01 | 1 | 1 | 1 | 4.47E + 05 | 6.77E + 05 | 1.06E + 04 | 0.001 |
| 22 | Retinol-binding protein 4 | P02753 | 23010 | <0.01 | 5 | 1 | 1 | 1.12E + 06 | 2.71E + 06 | 5.42E + 05 | 0.001 |
| 23 | Serum albumin | P02768 | 69367 | <0.01 | 80 | 70 | 11 | 7.61E + 09 | 6.03E + 09 | 2.39E + 09 | 0.001 |
| 24 | Hemoglobin subunit delta | P02042 | 16055 | <0.01 | 95 | 14 | 2 | 2.42E + 08 | 2.52E + 08 | 6.44E + 08 | 0.001 |
| 25 | Collagen alpha-1(XVIII) chain | P39060 | 178187 | <0.01 | 1 | 1 | 1 | 2.23E + 06 | 2.73E + 06 | 1.03E + 06 | 0.001 |
| 26 | Vitamin D-binding protein | P02774 | 52918 | <0.01 | 6 | 3 | 3 | 1.48E + 07 | 1.75E + 07 | 7.32E + 06 | 0.0001 |
| 27 | Peptidyl-prolyl cis-trans isomerase B | P23284 | 23743 | <0.01 | 8 | 2 | 2 | 6.12E + 06 | 1.03E + 07 | 2.41E + 06 | 0.00001 |
| 28 | Alcohol dehydrogenase class-3 | P11766 | 39724 | <0.01 | 3 | 1 | 1 | 1.15E + 06 | 2.91E + 06 | 1.06E + 05 | 0.001 |
| 29 | Thioredoxin-dependent peroxide reductase | P30048 | 27693 | <0.01 | 4 | 1 | 1 | 3.14E + 06 | 5.38E + 06 | 2.52E + 06 | 0.00051 |
| 30 | Flavin reductase (NADPH) | P30043 | 22119 | <0.01 | 11 | 2 | 1 | 2.63E + 06 | 4.85E + 06 | 2.26E + 06 | 0.00001 |
LN: Liquid Nitrogen; OGS: Osteosarcoma RAD: Irradiation/ECIR, FDR: False Discovery Rate.
Table 3.
List of down-regulated proteins after the treatment of OGS irradiation compared to the control.
| S.NO | Description | Accession | Avg. Mass | Significance | Coverage (%) | #Peptides | #Unique | OGS Control Area |
OGSLN Area | OGSRAD Area | P-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Down-regulated proteins after Irradiation treatment | |||||||||||
| 1 | Endoplasmic reticulum chaperone BiP | P11021 | 72333 | 132 | 10 | 5 | 2 | 1.48E + 07 | 5.97E + 06 | 1.69E + 07 | 0.001 |
| 2 | Cathepsin B | P07858 | 37822 | 132.88 | 5 | 1 | 1 | 5.58E + 06 | 5.05E + 06 | 1.59E + 07 | 0.001 |
| 3 | Glutathione S-transferase P | P09211 | 23356 | 200 | 8 | 1 | 1 | 2.05E + 07 | 3.86E + 06 | 2.77E + 07 | 0.001 |
| 4 | Collagen alpha-2(I) chain | P08123 | 129314 | 130.34 | 2 | 1 | 1 | 1.73E + 06 | 6.16E + 05 | 3.11E + 06 | 0.001 |
| 5 | Filamin-A | P21333 | 280737 | 121.83 | 3 | 6 | 4 | 1.07E + 07 | 5.24E + 06 | 1.29E + 07 | 0.001 |
| 6 | Alpha-enolase | P06733 | 47169 | 79.24 | 29 | 9 | 2 | 1.67E + 07 | 1.20E + 07 | 2.87E + 07 | 0.001 |
| 7 | Transaldolase | P37837 | 37540 | 11.26 | 9 | 4 | 2 | 5.26E + 06 | 4.11E + 06 | 9.62E + 06 | 0.001 |
| 8 | Immunoglobulin gamma-1 heavy chain | P0DOX5 | 49329 | 11.21 | 27 | 11 | 2 | 5.13E + 07 | 2.01E + 07 | 5.31E + 07 | 0.001 |
| 9 | Protein disulfide-isomerase A3 | P30101 | 56782 | 36.97 | 5 | 2 | 1 | 1.77E + 07 | 1.06E + 07 | 9.84E + 07 | 0.001 |
| 10 | Alpha-1-acid glycoprotein 2 | P19652 | 23603 | 12.51 | 5 | 1 | 1 | 8.34E + 05 | 6.97E + 05 | 2.58E + 06 | 0.001 |
LN: Liquid Nitrogen; OGS: Osteosarcoma RAD: Irradiation/ECIR, FDR: False Discovery Rate.
Protein profile of the differentially expressed proteins
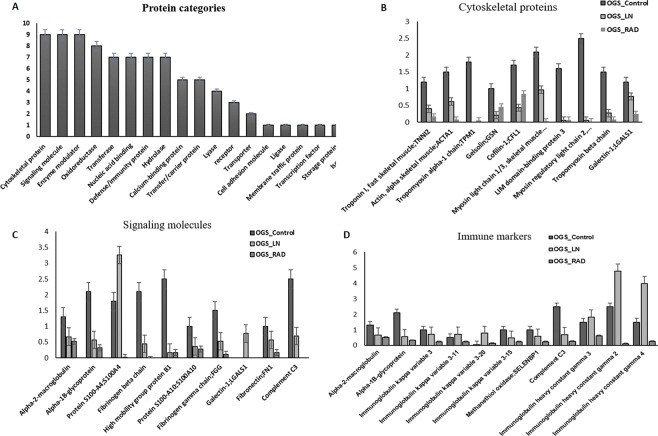
The functional enrichment analysis categorized the identified significantly altered proteins from this study into more than 10 different classes (Fig. 3A). The top 7 categories which hit the highest number of proteins from our study are cytoskeletal proteins, calcium-binding group, signaling molecules, extracellular matrix proteins (ECM) proteins, immunity or defense proteins, enzyme modulators, transfer, and carrier proteins, etc. We have identified more than 10 cytoskeletal proteins such as Troponin I (TNNI), Alpha-Actin (ACTN1), Tropomyosin alpha-1(TPM1), Gelsolin (GSN) Myosin light and heavy chain (MYL) Cofilin (CFL1), Lumican (LUM), Fibulin-1(FBLN-1), and Galectin (GAL) showed higher expressions levels in OGS control groups. After treatment with LN-freezing/cryotherapy and ECIR treatment, all cytoskeletal protein expressions were reduced from our evaluation except cofilin (Fig. 3B). The expressions of the signaling proteins like Alpha-2-macroglobulin (A2M), Alpha-1B-glycoprotein (A1BG), Protein S100A4, Fibrinogen beta and alpha chains (FGA & FBA) High-mobility group box proteins B1(HMGB1), Galectin (GAL), Fibronectin (FN) and complement C3 proteins from OGS untreated groups showed relatively higher expressions with more than 1-fold (Fig. 3C), and after the treatment with ECIR, and freezing these proteins levels were reduced signifying a numerous clue for further studies.
Figure 3.
Differential expressions of the identified proteins between treated (OGS n = 12) and untreated (n = 12) D) were categorized based on the functional enrichment analysis. Sample groups statistics (mean ± s.d) obtained for (A). Various categories of proteins from this study. (B) Altered expressions of cytoskeletal proteins. (C) Differential expressions of signaling molecules. (D) Immune markers expressions in OGS (all categories were quantified among three groups of OGS samples p < 0.01, p < 0.05 one-way ANOVA, Mann-Whitney U-test, triplicate analysis).
Over 10 immune/defense proteins (Fig. 3D) have been identified from our analysis, which plays an important role in inflammation, regulation of the immune system and immunotherapy. Most of the proteins associated with the immune system were dysregulated in untreated OGS groups. Surprisingly, after freezing treatment, the heavy and light chain expressions of immunoglobulins were increased. On the other hand, after irradiation treatment, most of the defense proteins were drastically reduced. Additionally, most of the enzyme modulators showed increased expressions with 1.5 to 2.5 folds in OGS control than the treatment group. Other promising results were ECM and calcium-binding proteins which are identified with 2.5 to 3.0 folds elevated in untreated groups and reduced expressions (0.5 to 1.0 folds) were observed in treatment groups. Moreover, from the literature, we came to understand that these proteins play a prominent role in OGS pathogenesis, cell cycle regulation, injury, tissue remodeling, and healing process. This led us to the next confirmation analysis to determine the expression levels of ECM proteins in OGS.
Gene ontology enrichment analysis
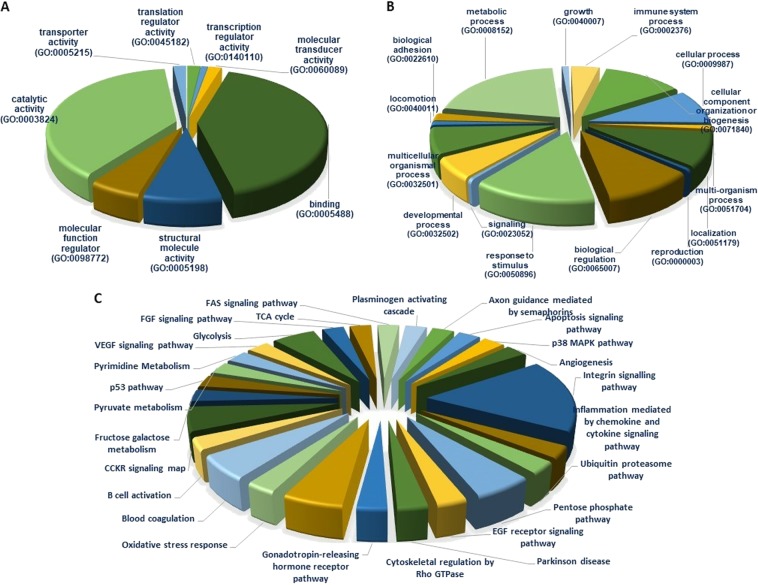
The functional enrichment analysis of Gene Ontology (GO), Protein Analysis Through Evolutionary Relationships (PANTHER), and Database Annotation Visualization, and Integrated Discovery (DAVID) were performed with the total proteins identified from OGS revealed that the identified proteins were involved in various crucial biological and molecular functions such as biological regulation (22%), metabolic process (36.4%), developmental process (10%), cellular component organization (20.9%) and multicellular organismal process (12.7%). The identified proteins intricate in significant molecular functions such as catalytic activity (38%), binding (41%), molecular function regulation (5.50%), transcription regulation activity (1.8%), etc. The top ten significantly enriched GO biological process and the identified proteins involvement in various molecular functions were shown in Fig. 4A,B.
Figure 4.
Functional enrichment analysis of identified proteins from comparative proteomic analysis of OGS. using DAVID, KEGG and PANTHER. (A) Biological function. (B) Molecular functions. (C) Identified proteins involved in various important pathways
Pathway Analysis
The identified DEPs from our study are involved in several significant pathways which play an important role in tumor growth and progression was revealed by Ingenuity Pathway Analysis (IPA) evaluations. Majority of the screened proteins from our study were involved in integrin signaling (17%), inflammation-mediated chemokine and cytokine signaling (3%), and cytoskeletal regulation (8%), etc. Next majority of the significant proteins involved in apoptosis signaling (3%), EGF receptor signaling (3%), VEGF signaling (3%), and p53 pathway (2.5%). Additionally, the greatest number of key proteins and enzyme modulators were taken part in glycolysis pathways (8%) as illustrated in Fig. 4C.
Protein-protein interaction (PPI) networks
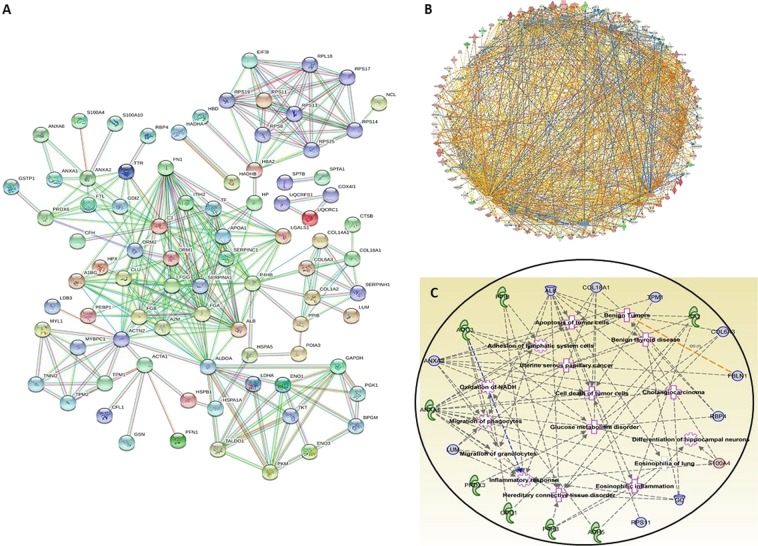
PPI network analysis was employed by STRING and IPA showed a tight and strong interaction network of all the identified proteins at the highest confidence score of 0.9 was displayed in Fig. 5A,B. Additionally, the IPA analysis also revealed that the identified proteins involved in several carcinomas were demonstrated in Fig. 5C.
Figure 5.
Protein-Protein Interaction (PPI) analysis of the identified proteins from comparative proteomic analysis of OGS using IPA and String networks. (A) PPI illustration of tight interaction of altered proteins. (B) PPI network showing all the proteins are tightly networked at highest confidence of 0.900 at STRING network analysis.
Verification of ECM proteins using western blotting
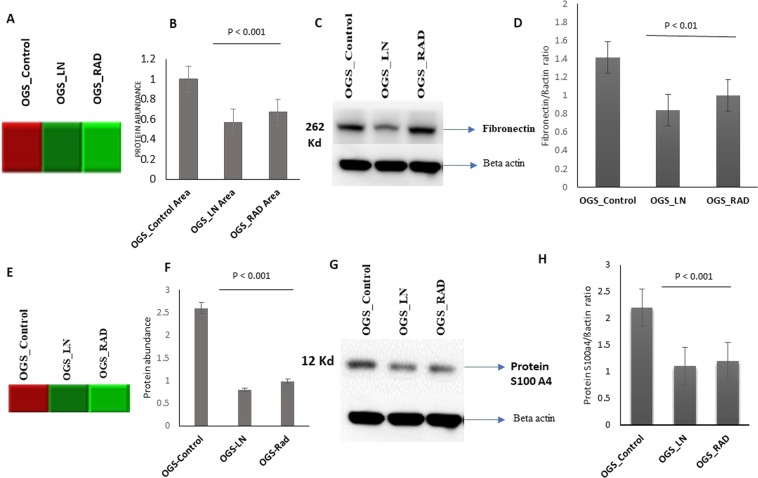
ECM proteins significantly contributed to cancer progression (Lu et al., 2012). From our proteomic evaluations, ECM’s were significantly down-regulated after freezing and irradiation treatments of OGS. Therefore, from our protein panel, we focused on the expressions of FN and Protein S100A4 which were up-regulated with more than two folds in OGS untreated groups. After the treatment with freezing and irradiation, the expression levels of these proteins were predominantly reduced. Therefore, we have determined to validate and compare the expressions of FN and Protein S100A4 proteins using another set of OGS (n = 6) untreated and treated samples (n = 6) using western blot analysis. Our verification study confirmed the consistent expression patterns FN and Protein S100A4 were significantly correlated with the mass spectrometric analysis (p < 0.01) (Fig. 6A,B).
Figure 6.
Validation of Fibronectin and Protein S100 A4 expression levels using immunoblot analysis. (A) Heat map showing the FN levels evaluated by mass spectrometry. (B) Quantification of FN levels from mass spectrometry were showed in a bar chart. (C) Immunoblot analysis of validating the FN levels in OGS freezing treated samples compared to OGS untreated and the data was normalized using beta actin. Full length immunoblots were showed in Fig. S1. (D) Bar chart showing the validation results of FN. (E) Heat map showing Protein S100 A4 levels evaluated by mass spectrometry. (F) Quantification of protein S100A4 levels from mass spectrometry were showed in a bar chart. (G) Immunoblot analysis validating Protein S100 A4 levels in OGS treated samples, data was normalized using beta actin. Full-length immunoblots were shown in Fig. S2. (H) Bar chart showing the quantification values of validation analysis.
Discussion
High-grade OGS is a malignant mesenchymal tumor which produces a matrix or osteoid around the joint either in the distal femur or proximal tibia. It is the most common primary malignant bone sarcoma in adolescents and children4. Currently, neoadjuvant chemotherapy drugs in combination with limb salvage surgery is a gold standard treatment option to diagnose OGS. Our recent evidence by Wu et al., 2018 concerning biological reconstruction of allografts and autografts have been widely implemented and ECIR and LN-freezing/cryotherapy are being successfully used for treating autografts to eradicate tumor cells from local recurrence13. Freezing and irradiation are the most widely used therapeutic modalities to treat autografts14,15. Nevertheless, there is a tremendous amount of clinical progress has been made so far in terms of diagnosis, therapeutics and understanding of OGS still there are so many aspects remain unclear, especially concerning the diagnosis, prognosis, tumor development, metastasis, and invasion.
Moreover, the critical challenge in diagnosing OGS is the lack of validated prognostic and diagnostic markers. We hypothesized that freezing and ECIR treatment applications would result in huge changes in the metabolome and transcriptome of bone tissues in the biological reconstruction. In addition to this, there is no evidence available yet regarding the proteomic changes after using both the treatment options on OGS autografts. Therefore, it is necessary to investigate the protein expression changes to understand the molecular and biological levels after using these treatments on OGS. The altered expression levels of proteins will provide numerous keys to open various therapeutic and diagnostic options to treat OGS and improve the current situation efficiently. Based on our vigorous literature search we did not find any proteomics study which has looked at the whole proteome changes on OGS freezing, and irradiation treated samples. Therefore, we aimed to distinguish the DEPs from cryotherapy and ECIR treated autografts compared to untreated samples of OGS (control). From our triplicate proteomic analysis, we have successfully identified a total of 131 proteins with highly significant differences among the three groups. A total of 131 proteins including 90 were up-regulated and 40 were down-regulated with a high score of more than 90 with >2 unique peptides with an FDR of <0.1%. All proteins were consistently identified in triplicate mass spectrometric analysis with the highest accuracy. Based on the literature review, the majority of the up-regulated proteins from our study make a substantial contribution to various crucial pathways which are directly related to the development of tumor growth.
Cytoskeletal proteins in osteosarcoma
Our proteomic evaluations identified more than 10 altered cytoskeletal proteins showed higher expressions in OGS untreated groups that are down-regulated after freezing and ECIR treatments. TNN1 and TPM1 are actin-binding proteins were elevated in our study are involved in the cytoskeleton’s contractile striated and smooth muscle system16. The up-regulated expressions of these proteins are a sign of tumor development and metastasis in OGS. After LN-freezing/cryotherapy, and ECIR treatment these proteins were down-regulated in treated groups. An ezrin family protein MYL that connects major cytoskeletal structures of the plasma membrane showed higher expressions in OGS untreated/control group. Previously, the up-regulated expressions of this protein have been reported to be closely related to Enneking classification and lung metastasis17. After ECIR and freezing treatments the higher expressions of MYL were efficiently reduced.
Another actin superfamily ubiquitously expressed member GSN was up-regulated in OGS control. Ma et al. group recently reported that the higher GSN expressions are correlated with tumor growth and poor prognosis18. Moreover, GSN also supports the growth and metastasis of tumor cells19. Interestingly, GSN expression levels were significantly decreased after freezing and irradiation. From earlier evidence reduced expressions of GSN found to reduce human breast, gastric, non-small cell lung cancers20–23. Likewise, lower levels of GSN inhibited cell growth, invasion, cell cycle arrest and also played a potential role in OGS as an oncogene22. GN is a multifunctional beta-galactoside-binding protein that has been playing a crucial role in carcinogenesis24, and has shown up-regulated expressions in untreated groups were showed contrary results in the treated category. Recently, Zhou et al. reported that over expressions of GN correlated with Enneking stage of cancer and metastasis occurrence in OGS25. Furthermore, over expressions of fibulin have been observed in untreated OGS groups, which is positively correlated with the development of OGS, and its progression, and poor prognosis26. Interestingly, after freezing and irradiation fibulin levels have been reduced.
A small proteoglycan-rich in leucine LUM is elevated in OGS untreated groups. It has been demonstrated to contribute to numerous biological and physiological processes. Interestingly the levels have been reduced after the treatment with freezing and irradiation. LUM expressions have been suggested to be positively correlated with the differentiation and negatively associated with OGS progression27. Additionally, over expressions of Profilin and Cofilin were also identified in OGS control which can inhibit actin the polymerization28,29. It plays a key role in the dynamic change in actin filaments structure and is associated with proliferation, invasion, and metastasis of tumor cells. Our proteomic analysis suggests that both the treatments act effectively on the cytoskeletal proteome level to suppress the higher protein expressions in OGS.
Signaling proteins expression in OGS
Signaling proteins play a key role in the diagnosis and prognosis of cancer. In our OGS comparative proteomic study, we identified more than 10 significant molecular proteins. All of them showed differential expressions between samples of untreated and treated. Most importantly the calcium-binding Protein S100 A4 and A10 showed higher levels of expression in OGS control were reported earlier in patients with metastasis and poor prognosis30,31. Interestingly, after freezing and irradiation treatment, both the proteins were reduced. An ECM family HMGB1 that functions as a signaling molecule and take part in inflammation and carcinogenic ability32 has been identified with relatively higher expressions in OGS control compared to the treatment groups. Previous studies reported a poor recurrence and free survival rate associated with the higher expressions in OGS. Another recent study also identified similar results with us demonstrated that the higher expressions of HMGB1 related to cancer development, tumor progression, and metastasis of lymph nodes33. On the other hand, we observed reduced levels of HMGB1 in treatment groups revealing that both the treatments successfully reduced the over expressions.
Immune markers
In response to any disease, the immune system, and its regulation play an outstanding role, especially for malignant tumors, it is a major determinant for the ultimate prognosis. It is evident that the huge cell diversity of the immune niche regulates the OGS microenvironment34,35. It has been stated recently that altered IGg’s expressions could promote tumor cell growth36. From our analysis, we have identified differential expressions of several IGg’s such as Immunoglobulin kappa variable 3 chains from 3–11, 3–15, 3–20 and Immunoglobulin constant gamma 2, 3 and 4 were significantly down-regulated in OGS control groups were remarkably showed increased expressions after LN-freezing/cryotherapy treatment. A recent study by Kato et al. in-kidney cancer patients demonstrated that cryoablation could induce strong immune reactions in tumors with an oligoclonal expansion of antitumor T cells, which circulate systemically37. Earlier investigations on cryoablation supports our results that freezing can improve and regulate the immune system process and antitumor response to fight against cancer. By contrast, ECIR treatment drastically reduced IGg’s expressions. Besides, we also evaluated higher expressions of complement C3 and complement factor H in OGS untreated samples, which were reduced remarkably in treatment groups. Since complement is not only an effector of innate immunity but also a contributor to inflammation, hemostasis, adaptive immune response and regulation of the immune system process38. Recent findings demonstrated that activation of complement has traditionally been considered as a part of the body’s immunosurveillance against cancer39. In addition, elevated expressions of A2M, A1BG, and Methanethiol oxidase have also been identified in OGS control samples which were reduced after treatment indicating that freezing can be an effective immunotherapy target for treating OGS.
Catalytic proteins
A catalytic member Protein disulfide isomerase-1 (PD1) identified with higher expressions in untreated samples of OGS. Based on the literature, PD1 has been abruptly identified in many tissues and expressed during endoplasmic reticulum stress. Xu et al. team demonstrated higher levels of PD1 associated with numerous types of cancer cells including kidney, lungs, brain, ovarian and prostate cancers40. In our study, PD1 showed reduced after freezing and irradiation treatment suggesting the fact that both treatments are efficiently reducing the elevated levels of proteins. Recent evidence described that lower levels of PD1 expression could increase the survival rate in breast cancer patients41. In addition to this, we have also identified several transcriptions and translation factors such as elongation factor gamma, eukaryotic translation initiation factor 3 subunit, carbonic anhydrase 1, band 3 anion translation initiation factor 3 subunit 1, trifunctional enzyme subunit beta, carbonic anhydrase, cathepsin B, EH domain-containing protein 2, Catalase, Cytochrome c oxidase subunit 4 isoform 1 and so on were differentially expressed in our proteomic study. Furthermore, we have also screened some key glycolysis-related proteins such as glyceraldehyde 2-phosphate dehydrogenase, fructose-bisphosphate aldolase, phosphoglycerate kinase, transketolase, and L-lactate dehydrogenase were up-regulated in untreated groups of OGS were reduced their expressions after freezing and irradiation treatments. Therefore, LN-freezing/cryotherapy and ECIR competently reducing the over expressions of critical proteins and preserve them for biological autografting.
Extra cellular matrix (ECM) proteins
The ECM proteins play a serious role in the development of cancer by regulating the dynamic behaviors of endothelial cells through different receptors of cell adhesion in cytoskeletal organization, remodeling, and tumor angiogenesis42. Thus, ECM proteins are promising therapeutic targets for tumors. From our OGS proteomic study, we have focused on a glycoprotein FN from ECM family which showed up-regulated expressions in OGS control samples and reduced after freezing and irradiation treatments. FN has played a prominent role in cell adhesion, differentiation, cell-matrix regulation, and tumor development43,44. We, therefore, choose this multifunctional protein for our validation studies and successfully confirmed its levels of expression using immunoblot analysis. FN levels were tremendously reduced after freezing and ECIR, and in fact, freezing showed more prominent results than ECIR. Thus, FN could be a valuable marker for the prognosis and diagnosis of OGS treatment. We have also validated Protein S100 A4, another important protein of interest from our study. This calcium-binding protein promotes metastasis and has been associated with patient’s outcome in various tumor types. Earlier studies reported the overexpression of Protein S100A4 is associated with tumorigenesis, poor prognosis, prediction of metastasis potency and has been stated as a prognostic marker for OGS45,46. From our investigations, we observed a significant amount of reduction in protein S100A4 levels after treated with freezing and irradiation. Besides, it can be a valuable marker to predict metastasis, tumor prognosis and development.
In summary, our comparative proteomic study successfully identified more than 10 different protein categories that are significantly altered expressions were identified after freezing and irradiation treatments compared to untreated OGS group. From our evaluations, both the treatments have effectively reduced the expression levels of highly regulating proteins that directly related to tumor development, recurrence, and metastasis. Besides the majority of the identified proteins from our study are associated with various biological, physiological and molecular functions of OGS. Especially most of the protein expressions have been reduced to the required levels in freezing treatment. On the other hand, in the ECIR treatment, some category of proteins such as immune markers, signaling molecules, calcium-binding protein expressions have been drastically reduced (−0.03 to −0.004) or diminished. Exposure to irradiation may cause cell damage that leads to protein degradation could be one of the reasons behind extreme changes in protein expressions with ECIR treatment. At the same time, LN-freezing showed better results than irradiation. However, we would like to emphasize that both the treatment options are successfully reducing overly expressing proteins which helps for the proper functioning of recycled autografts in the biological reconstruction. Abnormal protein levels can increase tumor growth and metastasis. Therefore, to gain longevity and proper function of the biological autografts, it is important to attain the normal levels of important proteins.
Conclusion
From our comparative proteomic study among OGS treated with cryotherapy/LN-freezing, ECIR and untreated were successfully identified a set of potential protein markers and their tremendous changes. The identified significantly expressed proteins from OGS non-treated groups play a crucial role in the tumor development, recurrence, metastasis and bone matrix formation which provides numerous clues for diagnosis and OGS management. On the other hand, DEPs from treated OGS groups play an important role in various crucial pathways which are directly related to tumor progression, metastasis, and OGS pathophysiology. We believe this is the first work that shows altered expressions of important protein profiles after freezing and ECIR treatment in recycled autografts. This study sheds new light on the role of freezing and ECIR treatments in biological reconstruction. Most importantly, the identified proteomic patterns and the verified protein candidates from our study help us to understand osteosarcoma in biological, physiological and molecular levels that could open various diagnostic avenues in therapeutics.
Materials and Methods
Patients and clinical information
This study included a total of 36 high-grade OGS bone tissue samples from 12 patients (male/female; 10/2; age ranging from 23–65 years) were collected from Taipei Veterans General Hospital (VGH-TPE). The collected OGS samples were categorized into three different groups such as LN-freezing, ECIR treated, and untreated OGS groups for comparative proteomic analysis. The tumor bone samples were collected from all the patients during the surgery. The demographic and clinical features of the obtained samples were shown in Table 4. All samples were freshly collected from the operation theatre after the surgery before chemotherapy, radiation and immunosuppressive medication or any treatment. The collected samples were stored at −80 °C for further analysis. Diagnostic criteria of all the collected OGS patient samples was confirmed by a certified surgeon as well as a pathologist by the tissue biopsy examinations. This study and all the materials and methodology was approved by the institutional review board47 of VGH-TPE, Taiwan (IRB Approval No.2019-02-021 A), and informed consent was obtained from all the patients. This study confirmed and conducted all the experiments according to the guidelines and regulations of IRB.
Table 4.
Demographic and tumor characteristics of OGS Patients.
| Patient number | Gender | Age | OGS Tumor location | Tumor Length | Status |
|---|---|---|---|---|---|
| 1 | M | 49 | Distal tibia | 13.7 | AWD |
| 2 | M | 45 | Distal femur | 4.3 | NED |
| 3 | M | 23 | Distal tibia | 9.3 | AWD |
| 4 | M | 36 | Proximal tibia | 6.2 | AWD |
| 5 | M | 53 | Distal femur | 8.5 | AWD |
| 6 | M | 55 | Proximal femur | 13.7 | AWD |
| 7 | M | 65 | Distal tibia | 15 | AWD |
| 8 | F | 45 | Proximal tibia | 4.1 | NED |
| 9 | F | 24 | Distal femur | 9.1 | NED |
| 10 | M | 36 | Proximal tibia | 9.8 | NED |
| 11 | M | 56 | Proximal femur | 14.4, 3.1 | AWD |
| 12 | M | 65 | Distal tibia | 13.3 | NED |
AWD: Alive with disease NED: No evidence of disease.
Protein extraction from OGS samples
Cryotherapy treated samples preparation
A total of 12 OGS patients bone tissues were collected, and subjected to Cryotherapy/LN-freezing treatment for 15 minutes under complete sterilization conditions14,48. All the freeze tissues were kept at room temperature and thawed for 20–25 mins before protein extraction. To extract the total protein all the samples were subjected to pulverized with a mortar and pestle using liquid nitrogen49. Then, the samples were transferred to new Eppendorf tubes and kept on ice until ready for extraction. Later, RIPA lysis buffer (50 mM Tris-HCl pH7.2, 150 Mm NaCl,1% NP40, 0.1% SDS, 0.5% DOC, 1 mM PMSF, 25 mM MgCl2) (sigma; R0278) supplemented with a phosphatase inhibitor cocktail (Thermo; 78420) was used to extract the protein from the samples and centrifuged at 13000 g for 15 min. Then, separated the supernatant to new tubes, and the extracted purified protein from all the freezing treated OGS were subjected to total protein concentration determination assays such as BCA and Bradford (Bio-Rad Laboratories, Hercules, CA)50.
Extra corporeal irradiation (ECIR) treated samples preparation
A total of 12 OGS samples were subjected to ECIR treatment at 15,000 gamma irradiations51. After irradiation the samples were subjected to protein extraction using RIPA lysis buffer (50 mM Tris-HCl pH7.2, 150 Mm NaCl,1% NP40, 0.1% SDS, 0.5% DOC, 1 mM PMSF, 25 mM MgCl2 (sigma; R0278)) supplemented with a phosphatase inhibitor cocktail (Thermo; 78420). Later, all the ECIR treated samples were centrifuged at 13000 g for 15 min. Then, separated the supernatant to a new tube and the extracted purified protein concentration was determined using BCA and Bradford assay (Bio-Rad Laboratories, Hercules, CA)50,52. On the other hand, the untreated OGS samples protein was extracted by using the same methodology as above without any prior treatment was employed and protein concentration was determined.
Protein precipitation and in-solution digestion
We tried to analyze the autogenous OGS host bone grafts for proteomic analysis using (LC ESI-MS/MS analysis using the same methodology as our previous studies54. The extracted protein samples from treated and untreated OGS autografts were precipitated with a fourfold volume of 100% ice-cold acetone and incubated overnight at −20 °C. The precipitated samples were centrifuged at 14000 X g for 10 min, and the pellets were dissolved in 100 µl of 25 mM NH4HCO3 with 6.5 M urea (0. 1–1 µg/µl) followed by an in-solution digestion procedure illustrated by earlier groups53. All the protein samples were reduced at 37 °C by 100 mM DTT (Dithiothreitol)53 for 30–40 min, later alkylated with 200 mM IAA (Iodoacetamide) in the dark at room temperature for 25–35 min, respectively. The proteins were digested overnight (16–18 hours) with sequencing grade trypsin (Promega, Madison, WI, USA; V5111) in 50:1 ratio at 37 °C. The reaction was quenched by adding 2 µl of 50% formic acid (FA) mixed briefly and incubated for 10 minutes. The digest was briefly vortexed and centrifuged then the supernatant containing peptides were collected. The final solution was lyophilized and desalted using C18 zip-tip procedure54.
Nano UPLC and mass spectrometry conditions
A slightly modified mass spectrometry conditions from our previously described method by Madda R et al.55 were employed successfully in this proteomic study. At 10000 full-width half maximum (FWHM) resolution an interface of ESI-Q-TOF MS/MS was reached as we performed in our earlier studies55. An external standard of lock mass BSA was constantly infused using the Nano-ACQUITY auxiliary pump at an interval of 20 secs (lock spray frequency) for calibrating the instrument at a flow rate of 0.25 µl/min. To obtain the accuracy precursor mass error was chosen as <2 ppm and the lock mass data were averaged. By using the positive V mode all the attained peptide spectra were eluted with a scan mass range of 50–200 m/z at a scan time of 1 sec. The digested 400 ng peptides were reconstituted in 3% ACN (Acetonitrile) and 0.1% FA (Formic Acid), then injected into an online nano-ACQUITY, UPLC coupled Q-TOF, Synapt-HDMS mass spectrometer (Waters Corporation, Milford, MA, USA). Next, the peptides were separated using a C18 reverse-phase column (1.7 µm × 75 µm × 250 mm) (Waters Corporation, Milford, MA, USA). A binary solvent system consisted of 99.9% water and 0.1% FA (mobile phase A) and 99.9% ACN and 0.1% FA (mobile phase B). The peptides were initially pre-concentrated and desalted online at a flow rate of 5 µl/min using a 5 µm symmetry C18 trapping column (internal diameter 180 mm, length 20 mm) (Waters Corporation, Milford, MA, USA) with 0.1% FA. After each injection, the peptides were eluted into the Nano-LockSpray ion source at a flow rate of 300 n/L and a gradient of 2% to 40% for 120 min. Later, the column was washed and equilibrated. The digested OGS treated with freezing, ECIR and untreated/control samples were run in triplicates and the data were analyzed by ProteinLynx Global Server 4.2 software (PLGS: Waters Corporation, Milford, MA, USA)56. Each sample was injected three times to obtain technical triplicates.
Protein quantification
We tried to analyze the autogenous OGS host bone grafts for proteomic analysis using high- resolution electron spray ionization liquid chromatography and tandem mass spectrometry (LC-ESI-MS/MS) analysis. The identified proteins from the LC-ESI-MS/MS analysis were quantified using label-free quantification by PEAKS Studio X (Bioinformatics Solutions Inc. Waterloo, ON, USA)47,57. Analyzed triplicate independent samples were compared among the treated and untreated groups of OGS. All the obtained raw data files from the mass spectrometry analysis were imported from the machine and uploaded to the PEAKS software program47 for quantification and interpretation of the spectra, and alignment of the total ion chromatograms along with the retention times were performed. A specific retention time of 600 to 10,500 seconds was specified. The protein identification from the raw data was performed same as we described in our earlier study55, an Uniprot’s reference database of Homo sapiens (release 03_2014)58 contained 20,272 entries were added and combined with a decoy database (the sequences were reversed) was used. The following parameters were specified for label-free quantification: digested by trypsin, with two missed cleavages; precursor mass tolerance was 10 ppm; fragment mass tolerance: 0.7 Da, minimum charge: 2, maximum charge: 3, carbamidomethylation, oxidation (M), and deamidated (N and Q) were specified as fixed and variable modifications. For determining the false-positive identification rate, the estimated spectra were used against the decoy database. The confident protein identifications and quantifications were estimated using a false discovery rate (FDR) of <1%, containing the peptide score of −10 log p ≥ 20 was employed. To determine the relative protein and peptide abundance in the tested samples, peptide feature-based quantification was performed59. For the accurate identification of peptide intensity differences among two samples the peptide signal intensity is directly proportional to the abundance of the peptides in the sample; hence the estimated peptide features were matched perfectly and the differences in peptide intensity between two samples were quantified effectively. Then, the area under the curve (AUC) of the extracted ion chromatograms (XICs) were measured and compared among the three analyzed runs. To determine the total cumulative peak area of the identified proteins, only unique peptides that are specifically assigned to the particular proteins were designated. FDR was calculated based on the target/decoy database as mentioned in the earlier studies60, and the >1% FDR peptides were chosen as true positive hits (considering the chance of getting one false positive in 20 observations). With this active feature-based quantitative approach61 the identified peptides with p-values < 0.05 and 0.01 that were identified in at least three observations from the OGS treated and untreated were compared and measured.
For identifying the significantly differential protein expressions among the tested groups’ one-way analysis of variance (ANOVA)62 was performed. The quantified spectral datasets were normalized with their spectral abundance factor values (triplicate experiments were averaged) that were used to generate a heat map showing the differentially expressed proteins between three groups. To minimize false positives, we excluded the proteins with an individual false detection rate p > 0.05 from further analysis. The quantified proteins with an XIC value lower than 100,000 were observed as absent (noise) and identified in only one of the three technical replicates were also excluded in this study. Each sample technical replicate XIC values were averaged and quantified for each OGS untreated and treated samples group, and the ratios of OGS-untreated/OGS-treated with cryotherapy, and ECIR were employed to identify the differentially expressed proteins as down-regulated proteins with <0.3–0.5 folds. Upregulated proteins were denoted with OGS-untreated/OGS-treated with a fold change of <1.5 to 2.
Protein identification
The Mascot software program (Matrix Science version 2.2, http://www.matrixscience.com) search engine along with the UniProtKB database (UniProt release 2015-10) and National Center for Biotechnology non-redundant (NCBInr) was used for the protein identification of the analyzed samples. To distinguish the altered proteins the following parameters have been specified: trypsin was specified for the enzymatic digestion with two missed cleavages, the protein modification changes observation was employed by specifying carbamidomethyl as a constant modification, and oxidation (M) as variable modification. The peptide mass tolerance of 50 ppm, and 0.1 Da MS/MS tolerance with an FDR of <1% were used for the accurate protein observation. Based on the specified parameters the proteins that are consistently identified from all the three technical replicates or at least two of the three analyses were selected for further evaluations. The theoretical molecular mass (MW) and isoelectric point (pI) of the identified proteins from this study were determined using the Mascot database.
Bioinformatics analysis
To understand the identified proteins involvement in various biological processes (BPs) and their molecular functions (MFs), along with the protein categories and cellular components (CCs) an international standardized gene function classification system of gene-ontology(GO) (http://www.geneontology.org/), and the DAVID (http://david.abcc.ncifcrf.gov/) (Database Annotation Visualization, and Integrated Discovery) database for functional analysis were performed63–66. For evaluate the protein-protein interactions among the identified proteins from OGS untreated Vs. treated we further analyzed our results using protein-protein interaction (PPI) networks with STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, Version 9.1) networks (website: http://string-db.org/) and specified high score of 0.09 along with the default parameters for the significant results.
Statistical analysis
To differentiate the changes in the protein profiles from the untreated Vs. treated OGS patients with LN-freezing/cryotherapy and ECIR, each patient sample was analyzed in a triplicate, and the variations in the percentage of volume and relative intensity were confirmed by statistical analysis. The differential expressions of the proteins quantified using spectral counting assessments for the LC-ESI-MS/MS data evaluations. Each sample was evaluated in three technical replicates and the average of the obtained abundance spectra was calculated. The data are expressed as mean ± standard deviation (SD) was determined using analysis of variance (ANOVA)62 assessment, and Mann-Whitney U-test was performed by SPSS statistical package (SPSS19, SPSS Ltd., Woking, Surrey, UK) for Windows. A probability value < 0.05 was considered as statistically significant and <0.01 was considered as highly significant.
Functional annotation of protein cohort
The identified DEPs from OGS treated and untreated samples were characterized using sub-cellular localization (SC), molecular function (MF), biological process (BP) and pathway analysis were performed using GO, PANTHER version 7.1, DAVID functional enrichment, and Inequity pathway (IPA) was used65,67. From our analysis we have gained a better understanding and biological context of the identified proteins and their involvement with the disease and its pathobiology of involvement of various physiological pathways.
Western blot analysis
Validation of the selected proteins was carried using western blotting analysis in another set of OGS bone tissue samples (n = 6) as described in earlier studies68,69. Proteins were separated by SDS-PAGE on to an electro transferred PVDF membrane (Millipore Corporation, Bedford, MA, USA) at 100 V for 60 min. In a TTBS solution [0.2 M TRIS-HCl (pH 7.6), 1.37 M NaCl, 0.1%Tween-20]70, the transferred protein membranes were immersed in 5% non-fat milk for 1 hr at room temperature. The proteins were incubated with primary antibodies, Fibronectin (FN) rabbit monoclonal antibody71 (catalog no. ab2413, 1:1000 dilution), protein S100a4 rabbit mAb (catalog ab124805, :10000 dilution), beta-actin rabbit mAb (catalog no. ab8227, 1:1000 dilution) at 4 °C overnight. All the antibodies were purchased from Abcam (www.abcam.com) (Cambridge, United Kingdom). Then the membranes were washed and incubated in 5% non-fat milk in a TTBS solution for 3 h at room temperature and subjected to three 5 min rinses in a TTBS solution. Later membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit antibody (Zhongshan Golden Bridge Biotechnology Co., Ltd, Beijing, China; catalog no. 7074) for 1 h at room temperature, and subjected to three 5 min rinses in a TTBS solution. The blot was developed with a Super ECL Plus kit (Applygen, Beijing, China), and the signal was exposed with X-ray film. The images were scanned, and the intensity of each band was captured using an Image Master 2D Platinum version 5.0 (GE Healthcare Amersham Bioscience). The intensity of each band was standardized as a percentage of the total intensity and the results were referred to as a relative volume that represents the relative expression abundance of the gene in the samples tested. The relative expression abundance was used to evaluate protein expression stability. Western blotting and quantification analysis were performed in at least three biological replications.
Ethics approval
This study was approved from the institutional review board of VGH-TPE, Taiwan (IRB Approval no.2019-02-021 A). And an informed consent was obtained from all the patients.
Supplementary information
Acknowledgements
We would like to thank Taipei’s Veterans General Hospital (VGH-TPE) for providing Osteosarcoma tumor patient’s samples for our proteomic analysis. Mass spectrometry instrumentation resources from the instrumentation center, National Taiwan University (NTU) Taipei. The funding for this study (OGS-Proteomics) was provided by the Ministry of Science and Technology (MOST- 108-2314-B-0101-041), Taiwan, ROC. There is no non-funding materials to declare in this study.
Author contributions
R.M. designed, conducted the experiments, analyzed and interrupted the entire data of this study, and wrote the manuscript; J.Y.W. and C.M.C. involved in the data analysis discussions, and edited manuscript; K.Y.C., H.Y.W. provided analytical tools; Y.Y.M. sample procurement, W.M.C. interpreted results and edited manuscript; P.K.W. interpreted results, raised funding, designed study, interpreted results and edited manuscript.
Data availability
All the triplicated datasets generated during and/or analyzed during the current study are available from the corresponding author upon request. However, the required data which was analyzed in this study are included in Supplementary Information Files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56024-7.
References
- 1.Pfaller MA, Krogstad DJ, Granich GG, Murray PR. Laboratory evaluation of five assay methods for vancomycin: bioassay, high-pressure liquid chromatography, fluorescence polarization immunoassay, radioimmunoassay, and fluorescence immunoassay. J Clin Microbiol. 1984;20:311–316. doi: 10.1128/JCM.20.3.311-316.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajpai J, et al. Outcomes in Treatment-Naive Patients With Metastatic Extremity Osteosarcoma Treated With OGS-12, a Novel Non-High-Dose Methotrexate-Based, Dose-Dense Combination Chemotherapy, in a Tertiary Care Cancer Center. J Glob Oncol. 2018;4:1–10. doi: 10.1200/JGO.17.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friebele JC, Peck J, Pan X, Abdel-Rasoul M, Mayerson JL. Osteosarcoma: A Meta-Analysis and Review of the Literature. Am J Orthop (Belle Mead NJ) 2015;44:547–553. [PubMed] [Google Scholar]
- 4.Misaghi A, Goldin A, Awad M, Kulidjian AA. Osteosarcoma: a comprehensive review. SICOT J. 2018;4:12. doi: 10.1051/sicotj/2017028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan G, Chen J, Wu D, Gao C. Neoadjuvant chemotherapy combined with limb salvage surgery in patients with limb osteosarcoma of Enneking stage II: a retrospective study. Onco Targets Ther. 2017;10:2745–2750. doi: 10.2147/OTT.S136621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed AR, Manabe J, Kawaguchi N, Matsumoto S, Matsushita Y. Radiographic analysis of pasteurized autologous bone graft. Skeletal Radiol. 2003;32:454–461. doi: 10.1007/s00256-003-0657-6. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi T, et al. Successful joint preservation of distal radius osteosarcoma by en bloc tumor excision and reconstruction using a tumor bearing frozen autograft: a case report. BMC Surg. 2018;18:12. doi: 10.1186/s12893-018-0346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi Y, et al. Radiation enhanced the local and distant anti-tumor efficacy in dual immune checkpoint blockade therapy in osteosarcoma. PLoS One. 2017;12:e0189697. doi: 10.1371/journal.pone.0189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Dang TA, Man TK. Plasma proteomic profiling of pediatric osteosarcoma. Methods Mol Biol. 2012;818:81–96. doi: 10.1007/978-1-61779-418-6_6. [DOI] [PubMed] [Google Scholar]
- 10.Cho WC. Contribution of oncoproteomics to cancer biomarker discovery. Mol Cancer. 2007;6:25. doi: 10.1186/1476-4598-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 12.Chen CM, et al. Bone morphogenetic protein activity preservation with extracorporeal irradiation- and liquid nitrogen freezing-treated recycled autografts for biological reconstruction in malignant bone tumor. Cryobiology. 2019;89:82–89. doi: 10.1016/j.cryobiol.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Chen WM, et al. High-grade osteosarcoma treated with hemicortical resection and biological reconstruction. J Surg Oncol. 2012;105:825–829. doi: 10.1002/jso.23005. [DOI] [PubMed] [Google Scholar]
- 14.Gage AA, Baust JM, Baust JG. Experimental cryosurgery investigations in vivo. Cryobiology. 2009;59:229–243. doi: 10.1016/j.cryobiol.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchiya H, Abdel-Wanis ME, Tomita K. Biological reconstruction after excision of juxta-articular osteosarcoma around the knee: a new classification system. Anticancer Res. 2006;26:447–453. [PubMed] [Google Scholar]
- 16.Dutour A, et al. Is troponin I gene therapy effective for osteosarcoma treatment? Study on a human-like orthotopic rat model. Anticancer Res. 2004;24:3977–3982. [PubMed] [Google Scholar]
- 17.Zhou W, et al. The expression of MYH9 in osteosarcoma and its effect on the migration and invasion abilities of tumor cell. Asian Pac J Trop Med. 2016;9:597–600. doi: 10.1016/j.apjtm.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Ma X, et al. Gelsolin promotes cell growth and invasion through the upregulation of p-AKT and p-P38 pathway in osteosarcoma. Tumour Biol. 2016;37:7165–7174. doi: 10.1007/s13277-015-4565-x. [DOI] [PubMed] [Google Scholar]
- 19.Deng R, et al. Gelsolin regulates proliferation, apoptosis, migration and invasion in human oral carcinoma cells. Oncol Lett. 2015;9:2129–2134. doi: 10.3892/ol.2015.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniuchi K, et al. Podocalyxin-like protein, linked to poor prognosis of pancreatic cancers, promotes cell invasion by binding to gelsolin. Cancer Sci. 2016;107:1430–1442. doi: 10.1111/cas.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Plasma Gelsolin Promotes Proliferation of Mesangial Cell in IgA Nephropathy. Cell Physiol Biochem. 2016;40:1473–1486. doi: 10.1159/000453199. [DOI] [PubMed] [Google Scholar]
- 22.Klampfer L, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. Oncogenic Ras promotes butyrate-induced apoptosis through inhibition of gelsolin expression. J Biol Chem. 2004;279:36680–36688. doi: 10.1074/jbc.M405197200. [DOI] [PubMed] [Google Scholar]
- 23.Boni F, et al. Gelsolin pathogenic Gly167Arg mutation promotes domain-swap dimerization of the protein. Hum Mol Genet. 2018;27:53–65. doi: 10.1093/hmg/ddx383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esmailiejah, A. A. et al. RETRACTED ARTICLE: Analysis of serum levels and tissue expression of galectin-1 and galectin-3 as noninvasive biomarkers in osteosarcoma patients. Tumour Biol, 10.1007/s13277-015-4194-4 (2015). [DOI] [PubMed]
- 25.Zhou X, et al. Expression and clinical significance of galectin-3 in osteosarcoma. Gene. 2014;546:403–407. doi: 10.1016/j.gene.2014.04.066. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D, et al. Fibulin-4 promotes osteosarcoma invasion and metastasis by inducing epithelial to mesenchymal transition via the PI3K/Akt/mTOR pathway. Int J Oncol. 2017;50:1513–1530. doi: 10.3892/ijo.2017.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikitovic D, et al. Lumican regulates osteosarcoma cell adhesion by modulating TGFbeta2 activity. Int J Biochem Cell Biol. 2011;43:928–935. doi: 10.1016/j.biocel.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Coumans JVF, Davey RJ, Moens PDJ. Cofilin and profilin: partners in cancer aggressiveness. Biophys Rev. 2018;10:1323–1335. doi: 10.1007/s12551-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaiyawat P, et al. Protein profiling of osteosarcoma tissue and soft callus unveils activation of the unfolded protein response pathway. Int J Oncol. 2019;54:1704–1718. doi: 10.3892/ijo.2019.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, Zheng HY, Liu ZL, Zhang L. Prognostic significance of relaxin-2 and S100A4 expression in osteosarcoma. Eur Rev Med Pharmacol Sci. 2014;18:2828–2834. [PubMed] [Google Scholar]
- 31.Gebhard C, et al. Comparative proteome analysis of monolayer and spheroid culture of canine osteosarcoma cells. J Proteomics. 2018;177:124–136. doi: 10.1016/j.jprot.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 32.He J, Zhang P, Li Q, Zhou D, Liu P. Expression of high mobility group box 1 protein predicts a poorer prognosis for patients with osteosarcoma. Oncol Lett. 2016;11:293–298. doi: 10.3892/ol.2015.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Yang L. The function and mechanism of HMGB1 in lung cancer and its potential therapeutic implications. Oncol Lett. 2018;15:6799–6805. doi: 10.3892/ol.2018.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endo-Munoz L, et al. Progression of Osteosarcoma from a Non-Metastatic to a Metastatic Phenotype Is Causally Associated with Activation of an Autocrine and Paracrine uPA Axis. PLoS One. 2015;10:e0133592. doi: 10.1371/journal.pone.0133592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Théoleyre S, et al. Phenotypic and functional analysis of lymphocytes infiltrating osteolytic tumors: use as a possible therapeutic approach of osteosarcoma. BMC Cancer. 2005;5:123. doi: 10.1186/1471-2407-5-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao Q, et al. Aberrant high expression of immunoglobulin G in epithelial stem/progenitor-like cells contributes to tumor initiation and metastasis. Oncotarget. 2015;6:40081–40094. doi: 10.18632/oncotarget.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato T, et al. Characterization of the cryoablation-induced immune response in kidney cancer patients. Oncoimmunology. 2017;6:e1326441. doi: 10.1080/2162402X.2017.1326441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min X, et al. Expression and regulation of complement receptors by human natural killer cells. Immunobiology. 2014;219:671–679. doi: 10.1016/j.imbio.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Pio R, Corrales L, Lambris JD. The role of complement in tumor growth. Adv Exp Med Biol. 2014;772:229–262. doi: 10.1007/978-1-4614-5915-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu S, Sankar S, Neamati N. Protein disulfide isomerase: a promising target for cancer therapy. Drug discovery today. 2014;19:222–240. doi: 10.1016/j.drudis.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Thongwatchara P, et al. Differential protein expression in primary breast cancer and matched axillary node metastasis. Oncol Rep. 2011;26:185–191. doi: 10.3892/or.2011.1266. [DOI] [PubMed] [Google Scholar]
- 42.Walker, C., Mojares, E. & Del Rio Hernandez, A. Role of Extracellular Matrix in Development and Cancer Progression. Int J Mol Sci19, 10.3390/ijms19103028 (2018). [DOI] [PMC free article] [PubMed]
- 43.Wang JP, Hielscher A. Fibronectin: How Its Aberrant Expression in Tumors May Improve Therapeutic Targeting. J Cancer. 2017;8:674–682. doi: 10.7150/jca.16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kun-Peng Z, Chun-Lin Z, Xiao-Long M, Lei Z. Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J Cell Physiol. 2019;234:6927–6939. doi: 10.1002/jcp.27435. [DOI] [PubMed] [Google Scholar]
- 45.Ochiya T, Takenaga K, Endo H. Silencing of S100A4, a metastasis-associated protein, in endothelial cells inhibits tumor angiogenesis and growth. Angiogenesis. 2014;17:17–26. doi: 10.1007/s10456-013-9372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao CM, Yang FX, Wang PL, Yang QX, Sun XR. Clinicopathologic significance of S100A4 expression in osteosarcoma. Eur Rev Med Pharmacol Sci. 2014;18:833–839. [PubMed] [Google Scholar]
- 47.Ma B, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 48.Kunz P, Bernd L. Methods of biological reconstruction for bone sarcoma: indications and limits. Recent Results Cancer Res. 2009;179:113–140. doi: 10.1007/978-3-540-77960-5_9. [DOI] [PubMed] [Google Scholar]
- 49.Acquadro A, et al. Proteomics in globe artichoke: protein extraction and sample complexity reduction by PEG fractionation. Electrophoresis. 2009;30:1594–1602. doi: 10.1002/elps.200800549. [DOI] [PubMed] [Google Scholar]
- 50.Walker JM. The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol Biol. 1994;32:5–8. doi: 10.1385/0-89603-268-X:5. [DOI] [PubMed] [Google Scholar]
- 51.Pruksakorn D, et al. Surgical outcomes of extracorporeal irradiation and re-implantation in extremities for high grade osteosarcoma: A retrospective cohort study and a systematic review of the literature. J Bone Oncol. 2019;14:100210. doi: 10.1016/j.jbo.2018.100210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Sun H, Zhang J, Brasier AR, Zhao Y. Quantitative Assessment of the Effects of Trypsin Digestion Methods on Affinity Purification-Mass Spectrometry-based Protein-Protein Interaction Analysis. J Proteome Res. 2017;16:3068–3082. doi: 10.1021/acs.jproteome.7b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erve JC, Demaio W, Talaat RE. Rapid metabolite identification with sub parts-per-million mass accuracy from biological matrices by direct infusion nanoelectrospray ionization after clean-up on a ZipTip and LTQ/Orbitrap mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:3015–3026. doi: 10.1002/rcm.3702. [DOI] [PubMed] [Google Scholar]
- 55.Madda, R., Lin, S. C., Sun, W. H. & Huang, S. L. Plasma proteomic analysis of systemic lupus erythematosus patients using liquid chromatography/tandem mass spectrometry with label-free quantification. Peerj6, 10.7717/peerj.4730 (2018). [DOI] [PMC free article] [PubMed]
- 56.Bond NJ, Shliaha PV, Lilley KS, Gatto L. Improving Qualitative and Quantitative Performance for MSE-based Label-free Proteomics. Journal of Proteome Research. 2013;12:2340–2353. doi: 10.1021/pr300776t. [DOI] [PubMed] [Google Scholar]
- 57.Tran NH, et al. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nature methods. 2019;16:63–66. doi: 10.1038/s41592-018-0260-3. [DOI] [PubMed] [Google Scholar]
- 58.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang T, et al. Comparison of liquid chromatography-tandem mass spectrometry-based targeted proteomics and conventional analytical methods for the determination of P-glycoprotein in human breast cancer cells. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;936:18–24. doi: 10.1016/j.jchromb.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 60.Aggarwal S, Yadav AK. False Discovery Rate Estimation in Proteomics. Methods Mol Biol. 2016;1362:119–128. doi: 10.1007/978-1-4939-3106-4_7. [DOI] [PubMed] [Google Scholar]
- 61.Zhong Z, Mao S, Lin H, Lin JM, Lin J. Comparative proteomics of cancer stem cells in osteosarcoma using ultra-high-performance liquid chromatography and Orbitrap Fusion mass spectrometer. Talanta. 2018;178:362–368. doi: 10.1016/j.talanta.2017.09.053. [DOI] [PubMed] [Google Scholar]
- 62.Nyaga VN, Aerts M, Arbyn M. ANOVA model for network meta-analysis of diagnostic test accuracy data. Stat Methods Med Res. 2018;27:1766–1784. doi: 10.1177/0962280216669182. [DOI] [PubMed] [Google Scholar]
- 63.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 64.Ren YM, et al. Exploring the key genes and pathways of side population cells in human osteosarcoma using gene expression array analysis. J Orthop Surg Res. 2018;13:153. doi: 10.1186/s13018-018-0860-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou X, Su Z. EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics. 2007;8:246. doi: 10.1186/1471-2164-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blancher C, Jones A. SDS -PAGE and Western Blotting Techniques. Methods Mol Med. 2001;57:145–162. doi: 10.1385/1-59259-136-1:145. [DOI] [PubMed] [Google Scholar]
- 69.Li X, et al. Identification and validation of rice reference proteins for western blotting. Journal of Experimental Botany. 2011;62:4763–4772. doi: 10.1093/jxb/err084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.TBST for Western Blotting. Cold Spring Harbor Protocols2013, pdb.rec074104, 10.1101/pdb.rec074104 (2013).
- 71.Miyaji Y, et al. Use of an intravenous microdose of 14C-labeled drug and accelerator mass spectrometry to measure absolute oral bioavailability in dogs; cross-comparison of assay methods by accelerator mass spectrometry and liquid chromatography-tandem mass spectrometry. Drug Metab Pharmacokinet. 2009;24:130–138. doi: 10.2133/dmpk.24.130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the triplicated datasets generated during and/or analyzed during the current study are available from the corresponding author upon request. However, the required data which was analyzed in this study are included in Supplementary Information Files.