Abstract
The mite Varroa destructor is a serious threat to honeybee populations. Selective breeding for Varroa mite tolerance could be accelerated by biomarkers within individual bees that could be applied to evaluate a colony phenotype. Previously, we demonstrated differences in kinase-mediated signaling between bees from colonies of extreme phenotypes of mite susceptibility. We expand these findings by defining a panel of 19 phosphorylation events that differ significantly between individual pupae from multiple colonies with distinct Varroa mite tolerant phenotypes. The predictive capacity of these biomarkers was evaluated by analyzing uninfested pupae from eight colonies representing a spectrum of mite tolerance. The pool of biomarkers effectively discriminated individual pupae on the basis of colony susceptibility to mite infestation. Kinome analysis of uninfested pupae from mite tolerant colonies highlighted an increased innate immune response capacity. The implication that differences in innate immunity contribute to mite susceptibility is supported by the observation that induction of innate immune signaling responses to infestation is compromised in pupae of the susceptible colonies. Collectively, biomarkers within individual pupae that are predictive of the susceptibility of colonies to mite infestation could provide a molecular tool for selective breeding of tolerant colonies.
Subject terms: Biochemistry, Biomarkers
Introduction
Western honeybee (Apis mellifera L.) populations are showing increased annual losses worldwide1–4. This trend is cause for considerable concern as many food crops depend on honeybees for pollination5. A number of causative, or contributing, factors to the declining health of honeybee colonies have been proposed including: Varroa destructor (Anderson and Trueman)6 parasitism and associated viral pathogens7,8, increased use of pesticides, lack of bee genetic diversity, and other factors9. Of these, the ectoparasitic mite Varroa destructor is typically considered the most significant threat to honeybee health7,8,10. In large part this is because these mites serve as a vector for a number of viral pathogens including: Deformed wing virus7,11, Israeli acute paralysis virus12, Acute bee paralysis virus, and Kashmir bee virus13. Varroa also compromises bee health by removing hemolymph14 and feeding on bees’ fat body tissue15.
In response to the threats imposed by Varroa mites, many producers have incorporated miticides into their management practices. Unfortunately, these treatments can have detrimental effects on honeybee health and introduce dangerous residues into the wax16. Further, as mites develop resistance to these treatments, the use of miticides is unlikely to represent a viable long-term solution16. An alternative approach is to focus on selective breeding of honeybees with an enhanced capacity to resist and/or tolerate Varroa mite infestation and associated viruses.
The feasibility of genetic selection for Varroa mite resistance is supported by the historic example of Asian honeybees (Apis ceranae) who, in the face of evolutionary pressures imposed by the parasite, developed protective mechanisms, including behavioral characteristics (such as grooming and hygienic traits) and immune adaptations17,18. Western honeybees, whose exposure to this parasite extends only about fifty years, have yet to naturally develop resistance mechanisms and remain susceptible to infestation17. Although a number of breeding programs have identified colonies with some resistance to mites, these colonies are lacking in other economic traits, such as honey production19. Accelerated breeding of resistance traits within Western honeybees is complicated by our limited understanding of the mechanisms mediating susceptibility to mite parasitism, a shortage of biomarkers to identify resistant progeny, and instability of resistant phenotypes within a colony. Nevertheless, through genetic selection, a number of research teams have established colonies with an increased Varroa tolerant phenotype20–22. Varroa sensitive hygiene (VSH) lines are the best-defined genetic stock able to suppress mite infestation23,24. The susceptible and tolerant honeybee colony phenotypes of the current investigation were developed and characterized by the Saskatraz natural selection project (http://www.saskatraz.com) with the goal of increasing the frequency of the alleles associated with economic traits and eventual distribution to commercial beekeepers. This is accomplished through recurrent natural selection of survivor colonies, in the absence of synthetic miticides. The project selects for quantitative traits, including honey production, wintering ability, mite tolerance, and general colony health25,26.
A number of molecular approaches, including at the levels of the transcriptome26–29 and proteome30,31, have been applied to honeybees in efforts to understand the cellular mechanisms of Varroa resistance as well as to identify biomarkers of these traits. This is a daunting task due to the potential complexities of the molecular mechanisms underlying these phenotypes, coupled with the challenges of deciphering biology within a mixed genetic population. This last point is particularly true of bees where multiple mating events by individual queen bees, high recombination rates, caste-specific influences on signaling, and supersedure events can increase the variability of individual bees within a colony. In addition, there are inherent limitations to the extent to which patterns of gene and/or protein expression can accurately predict or rationalize complex phenotypes due to a variety of post-transcriptional and post-translation regulatory events.
As protein phosphorylation often serves as a central mechanism for direct regulation of cellular processes, characterization of cellular responses at the level of kinase-mediated phosphorylation (kinome analysis) has the potential to offer unobstructed insight into complex biology32. That phosphorylation events can be represented by short peptides, enabling the creation of arrays for high throughput analysis of global cellular kinase activity, in particular in a manner that can be customized for species of interest33. Kinome analysis through species-specific peptide arrays has proven to be a powerful approach for deciphering biology, in particular in the context of host-pathogen interactions within outbred populations including cattle34–36, pigs37, and chickens38. Our previous efforts to develop a honeybee-specific kinome array (when applied to bees with a well-defined phenotype) provided evidence for signaling differences between individual bees, representing a variety of developmental stages, selected from colonies representing extreme phenotypes of Varroa mite tolerance and susceptibility25.
While providing evidence for the potential utility of this technology to identify phosphorylation biomarkers that inform breeding programs, this initial study was limited in that the bees considered represented only a single colony of each phenotype of mite susceptibility. Practical application of the technology would require the capacity to discriminate among bees representing colonies of a spectrum of Varroa mite tolerant phenotypes as well as greater consideration of the variability that exists within individuals of the same colony. In our initial kinome investigation, variability among individual kinome profiles was observed, even within bees of the same colony and developmental stage25. This variability can reflect the previously discussed unique aspects of bee biology (multiple mating events by individual queen bees, high recombination rates, caste-specific influences on signaling, and supersedure events), individual responses to environmental stimuli, and inherent variability of signaling within individuals’ “kinotypes”39. Despite this internal variability, distinct kinome patterns associated with colony phenotypes were observed, motivating further efforts to confirm the effectiveness of the adopted approach.
In the current investigation, we expand on the findings of our previous investigation of the utility for kinome analysis to reveal biomarkers and mechanisms of Varroa mite tolerance in honeybees by identifying a panel of 19 phosphorylation events with significant (p < 0.01) differences in phosphorylation when comparing individual pupae (n = 5/phenotype) selected from two colonies of each high and low susceptibility to the parasite. Bees evaluated were at the dark-eyed (white-bodied) pupae stage of development to minimize signaling differences between individuals of the same colony that could result from caste specializations or environmental influences. The utility of these biomarkers was validated by analyzing individual pupae (n = 18) from multiple colonies (n = 8) representing a spectrum of mite susceptibilities. The biomarkers effectively discriminated among individuals on the basis of colony phenotype and survival time. Further, analysis of the kinome profiles indicated that differential susceptibilities to the parasite were associated with innate immune capabilities even in the absence of infestation. Specifically, relative to pupae from Varroa mite tolerant colonies, Toll-like receptor (TLR) signaling40–42 – a central pathway of the innate immune response – was down-regulated in pupae from mite susceptible colonies. The hypothesis that differential susceptibilities to the parasite were associated with innate immune capabilities is supported by the observation that mite infestation induced elevated innate immune signaling responses in pupae from the tolerant, but not susceptible, colonies. These results indicate that innate immune capabilities contribute to mite resistance, either directly or possibly indirectly by influencing vulnerability to secondary infections by mite-associated viruses. Collectively, in addition to offering insight into the cellular processes underlying Varroa mite tolerance, this investigation enabled identification of a group of biomarkers that could be applied at the level of individual pupae to predict phenotypes at a population (colony) level.
Results
Characterization of colony phenotypes
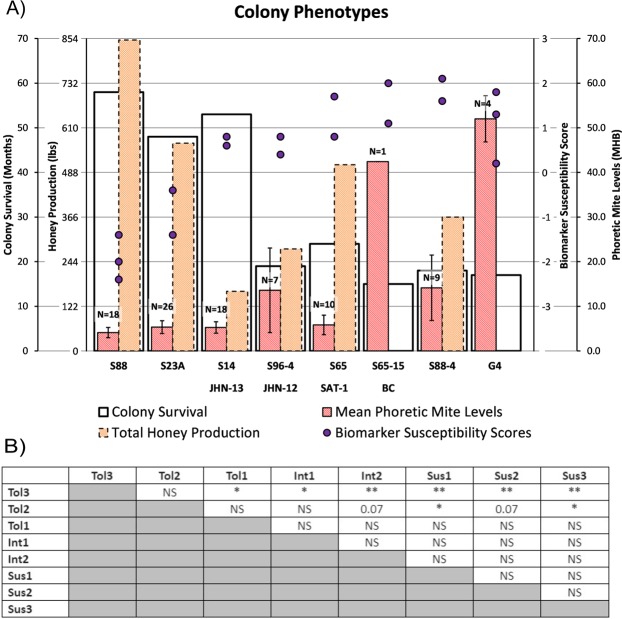
Eight colony phenotypes were analyzed in this investigation [Table 1]. Three colonies (S88, S23A, and S14 JHN-13) which showed the longest survival times and maintained lower Varroa mite populations over their lifetime were classified as tolerant. Three colonies (S65–15 BC, S88-4, and G4) which showed shorter survival times with mite levels showing sudden dramatic increases in population growth were classified as susceptible. Two colonies (S96-4 JHN-12 and S65 SAT-1) which demonstrated intermediate levels of Varroa infestation with shorter survival times than the tolerant colonies, but maintained lower mite levels than the susceptible colonies, were classified as intermediate phenotype [Table 1]. Total honey production of each colony over its survival time, as an indicator of colony strength and productivity, is also presented [Table 1].
Table 1.
Varroa mite Burden and Colony Survival Time.
| Phenotype | Tolerant | Intermediate | Susceptible | |||||
|---|---|---|---|---|---|---|---|---|
| Colony ID | S88 | S23A | S14 JHN-13 | S96-4 JHN-12 | S65 SAT-1 | S65-15 BC | S88-4 | G4 |
| Sampling Date | 09-2010 | 08-2011 | 07- 2015 | 08- 2013 | 09- 2011 | 08-2016 | 09-2011 | 09-2010 |
| Mite Infestation at Sampling Date (MHB) | 5.50 | 0.00 | 1.69 | 9.03 | 16.50 | 42.42 | 50.00 | 67.00 |
| Mean Mite Infestation (MHB) | 4.11 | 5.33 | 5.26 | 13.58 | 5.82 | N/A | 14.15 | 52.00 |
| SE ± | 1.2 | 1.4 | 1.3 | 9.5 | 2.2 | N/A | 7.3 | 5.2 |
| # Samples | 18 | 26 | 18 | 7 | 10 | 1 | 9 | 4 |
| Total Honey Production (lbs.) | 850 | 568 | 163 | 279 | 509 | 0 | 366 | 0 |
| Colony Survival (Months) | 58 | 48 | 53 | 19 | 24 | 15 | 18 | 17 |
Biomarkers of varroa mite susceptibility
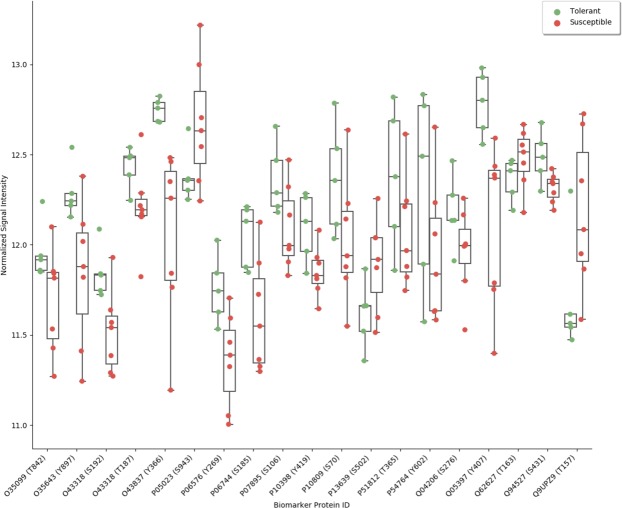
To identify biomarkers associated with Varroa mite susceptibility, kinome datasets corresponding to uninfested pupae from colonies representing the extremes of mite susceptibility and tolerance were analyzed. Nineteen peptides had a significant difference (p < 0.01) in levels of phosphorylation when comparing pupae from colonies of the tolerant and susceptible phenotypes [Table 2]. These differentially phosphorylated peptides can be grouped according to biological processes (innate immunity, stress responses, and metabolism) implicated in Varroa mite susceptibility40–42. With a coefficient of variance of less than one percent, the normalized data for each biomarker peptide was highly consistent across the replicate pupae of the two representative colonies of each phenotype. For many of the biomarker peptides the range of intensities associated with individuals of the susceptible phenotype partially overlap with intensities of phosphorylation of that peptide by individuals of the tolerant phenotype. This likely reflects the complexity of the phenotype as well as diversity of signaling in individual pupae within a colony [Fig. 1].
Table 2.
Biomarker Peptides: Differently Phosphorylated Peptides Between Pupae Collected from Varroa Susceptible and Tolerant Colonies.
| Protein | ID | Sequence | P | |
|---|---|---|---|---|
| Innate Immunity | TAK1 kinase | O43318 | YMTNNKGSAAWMAPE | 0.001 |
| TAK1 kinase | O43318 | CDLNTYMTNNKGSAA | 0.003 | |
| Mitogen-activated protein kinase kinase kinase_5 | O35099 | TETFTGTLQYMAPE | 0.009 | |
| Nuclear factor NF-kappa-B p110 subunit Rel-p110 | Q94527 | YIQLKRPSDGATSEP | 0.005 | |
|
Transcription_factor p65 Nuclear factor NF-kappa-B |
Q04206 | IQLKRPSDGALSEP | 0.005 | |
| Focal adhesion kinase 1 FADK1 | Q05397 | IVDEEGDYSTPATRD | 0.005 | |
| AP-1 complex subunit beta-1 | O35643 | VEGQDMLYQSLKLTN | 0.008 | |
| Metabolism | ATP synthase_subunit_beta | P06576 | TSKVALVYGQMNEPP | 0.004 |
| Na-K transporting ATPase subunit alpha1 | P05023 | ICKTRRNSLFRQGM | 0.009 | |
| Glucose-6-phosphate isomerase | P06744 | GPRVHFVSNIDGTHI | 0.005 | |
| Isocitrate_dehydrogenase subunit_beta, | O43837 | TKDLGGQSSTTEF | 0.006 | |
|
Stress Responses |
Ribosomal protein S6 kinase alpha | P51812 | DSEFTCKTPKDSPGV | 0.006 |
| Elongation factor 2 (EF-2) | P13639 | KVMKFSVSPVVRVAV | 0.007 | |
| 60_kDa_heat_shock_protein | P10809 | ILEQSWGSPKITKDG | 0.016 | |
| Superoxide dismutase | P07895 | SIFWCNLSPNGG | 0.008 | |
| Other | Ephrin type-A receptor 4 EPH-like kinase 8 (EK8) | P54764 | SYVDPHTYEDPNQAV | 0.006 |
| PRKC_apoptosis_WT1 regulator_protein__ | Q62627 | LREKRRSTGVVHLPS | 0.006 | |
| A-Raf Kinase | P10398 | QTAQGMDYLHAKNII | 0.010 | |
| Intestinal cell kinase (ICK) | Q9UPZ9 | CKIRSRPPYTDYVSTRW | 0.010 |
Figure 1.
Swarm Plot Illustrating Biomarker Application. For each of the nineteen biomarker peptides, the intensities of signal from pupae from colonies of known phenotype are represented by color-coded dots: Varroa mite tolerant (green), Varroa mite susceptible (red).
Application of kinome biomarkers to individual bees to predict colony phenotype
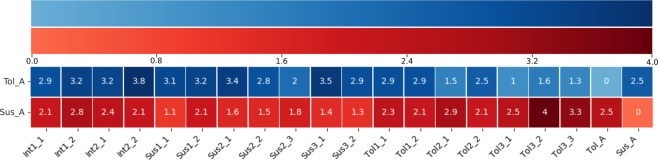
The predictive power of the biomarker peptides was evaluated using kinome data from individual, uninfested dark-eyed pupae (n = 18) selected from colonies with a spectrum of susceptibilities to Varroa mite infestation. The priority was to determine the effectiveness of biomarkers selected at the level of a colony phenotype in anticipating molecular differences within individual pupae. Kinome analysis was performed blinded to the phenotypes of the individual pupae. From the resulting kinome data, each pupa was assigned scores based on similarity to the mean of the pupae representing the tolerant and susceptible phenotypes based on levels of phosphorylation across the nineteen biomarker peptides [Fig. 2]. From this, a cumulative biomarker susceptibility score was calculated as the difference between the scores relative to the tolerant and susceptible phenotypes [Table 3]. In general, pupae of the same colony had consistent biomarker scores, supporting the feasibility of using this type of approach to guide breeding efforts at the level of a colony.
Figure 2.
Euclidian Distances of Individual Pupae measured by comparing the phosphorylation levels in each phenotype to the mean phosphorylation levels of the pupae representing the tolerant (Tol_A) and susceptible phenotypes (Sus_A). Higher values represent increasing distance in Euclidian space and decreasing similarity, while low values indicate high similarity and a value of zero represents identical vectors or self-comparison.
Table 3.
Variance in Phosphorylation Levels of Biomarker Peptides Across Individual Pupae of Colonies of Different Mite Susceptibility Phenotypes.
| Colony | Pupae | Distance from mean tolerant phenotypeX | Distance from mean susceptible phenotypeY | Biomarker Score (x-y) |
|---|---|---|---|---|
| G4 | Sus2_1 | 3.4 | 1.6 | 1.8 |
| Sus2_2 | 2.8 | 1.5 | 1.3 | |
| Sus2_3 | 2 | 1.8 | 0.2 | |
| S88-4 | Sus3_1 | 3.5 | 1.4 | 2.1 |
| Sus3_2 | 2.9 | 1.3 | 1.6 | |
| S65-15 BC | Sus1_1 | 3.1 | 1.1 | 2.0 |
| Sus1_2 | 3.2 | 2.1 | 1.1 | |
| S96-4 JHN-12 | Int1_1 | 2.9 | 2.1 | 0.8 |
| Int1_2 | 3.2 | 2.8 | 0.4 | |
| S65 SAT-1 | Int2_1 | 3.2 | 2.4 | 0.8 |
| Int2_2 | 3.8 | 2.1 | 1.7 | |
| S14 JHN-13 | Tol1_1 | 2.9 | 2.3 | 0.6 |
| Tol1_2 | 2.9 | 2.1 | 0.8 | |
| S23A | Tol2_1 | 1.5 | 2.9 | −1.4 |
| Tol2_2 | 2.5 | 2.1 | −0.4 | |
| S88 | Tol3_1 | 1.0 | 2.5 | −1.4 |
| Tol3_2 | 1.6 | 4.0 | −2.4 | |
| Tol3_3 | 1.3 | 3.3 | −2.0 |
When the calculated kinome biomarker scores are compared to the survival times and mite loads of the eight colonies under consideration there is an apparent trend with the observed phenotype, in particular for pupae from colonies of the tolerant phenotype [Fig. 3A]. This trend was confirmed through pairwise analysis of the biomarker scores of the various colonies. In particular, the S88 tolerant colony (Tol3) was significantly different (p < 0.01) from all three susceptible colonies [Fig. 3B]. The biomarker scores of the S88 colony were also significantly different from those of both colonies of intermediate phenotype; p < 0.01 for S96-4 JHN-12 (Int1) and p < 0.05 S65 SAT-1 (Int2). Pupae from the S23A colony (Tol2) had biomarker scores significantly different (p < 0.05) from two of the three susceptible colonies; S65-15BC (Sus1) and S88-4 (Sus3). The biomarker scores of pupae from the tolerant phenotype S14 JHN-13 (Tol1) were not significantly different from those of any of the susceptible colonies but were statistically different (p < 0.05) from those of Tol3 [Fig. 3B]. Collectively indicating that the biomarkers are most effective at distinguishing pupae of the tolerant colonies from those of intermediate and susceptible phenotypes.
Figure 3.
Mite loads, Colony Survival, Honey Production, Varroa mite Susceptibility Scores. (A) The survival time, mean phoretic mite infestation, honey production, and biomarker susceptibility scores for 8 colony phenotypes are shown here. Error bars are shown as ± SE of the mean phoretic mite level where N is the number of samples tested to calculate the mean [Table 1]; where S65-15 BC is represented only by a single sample. The purple dots represent the biomarker susceptibility scores calculated from the kinome array (n = 299 peptides) analyses of dark eyed pupae. Each dot represents a score calculated from one pupa. (B) Significance of difference between biomarker scores 28 possible pairs of colonies. *p < 0.05, **p < 0.01.
Cellular mechanisms of varroa mite susceptibility
To investigate molecular mechanisms of Varroa mite tolerance, pathway over-representation analysis was performed on kinome datasets corresponding to uninfested bee pupae (n = 5/phenotype) from two colonies representing each phenotype of Varroa susceptibility25. A considerable number of peptides (58 of 299) were differentially phosphorylated (p < 0.05) when comparing pupae representing the two phenotypes. This suggests that these phenotypes reflect complex, multi-faceted molecular mechanisms and that these differences exist even in the absence of Varroa mite infestation. Consistent with this hypothesis, a large number of pathways were found to be differentially activated when comparing bees of the two different phenotypes [Table 4]. Notably, relative to bees from the tolerant colonies, there was a down-regulation of Toll-like receptor (TLR) signaling in pupae from susceptible colonies. The process of recurrent natural selection for Varroa tolerance may be resulting in the enrichment of genetic mechanisms involved in signaling innate immune responses, in response to pathogens associated with mite infestation. TLR signaling activates antimicrobial factors and other defensive mechanisms at the cellular level39,40. This provides for colonies expressing these traits a selective advantage with higher survival rate in presence of mite infestation and largest honey production.
Table 4.
Pathway Over-Representation Analysis of Uninfested Dark-Eyed Pupae from Varroa mite Susceptible versus Tolerant Colonies.
| Direction | Pathway | ID | Source | P |
|---|---|---|---|---|
| Up-Regulated | N-cadherin signaling events | 15853 | PID NCI | 0.01 |
| beta-catenin independent WNT signaling | 17081 | REACTOME | 0.02 | |
| RAC1 signaling pathway | 15344 | PID NCI | 0.02 | |
| Ca2 + pathway | 16886 | REACTOME | 0.03 | |
| Ctcf: first multivalent nuclear factor | 4040 | PID BIOCARTA | 0.03 | |
| S1P2 pathway | 15183 | PID NCI | 0.03 | |
| TGF-beta receptor signaling | 15571 | PID NCI | 0.04 | |
| ALK1 signaling events | 15251 | PID NCI | 0.04 | |
| Arf1 pathway | 15337 | PID NCI | 0.04 | |
| Glypican 1 network | 15119 | PID NCI | 0.04 | |
| Il12 and stat4 dependent signaling pathway | 4054 | PID BIOCARTA | 0.04 | |
| NRAGE signals death through JNK | 13200 | REACTOME | 0.04 | |
| Regulation of RAC1 activity | 14985 | PID NCI | 0.04 | |
| CDC42 signaling events | 15467 | PID NCI | 0.05 | |
| Down-Regulated | ERK1 activation | 13000 | REACTOME | 0.02 |
| Axon guidance | 17789 | REACTOME | 0.02 | |
| Toll Like Receptor 9 (TLR9) Cascade | 13042 | REACTOME | 0.04 | |
| EPH-ephrin mediated repulsion of cells | 19071 | REACTOME | 0.05 |
Global signaling responses to varroa mite challenge
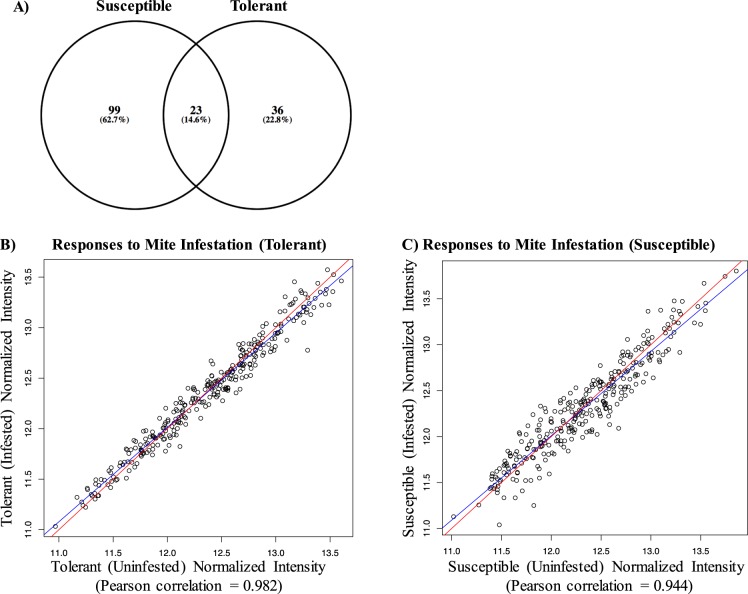
Kinome responses to Varroa mite infestation were evaluated in individual pupae representing susceptible and tolerant phenotypes (n = 5/phenotype) in the presence and absence of mites. In pupae from the tolerant colonies, 59 peptides underwent significant changes (p < 0.05) in their phosphorylation levels in response to mite infestation. In contrast, mite infestation of pupae from susceptible colonies resulted in significant (p < 0.05) changes in the phosphorylation status of 122 peptides, nearly half the array and double the number observed for the pupae from tolerant colonies [Fig. 4A]. Further, the majority of the peptides which underwent mite-induced differential phosphorylation were unique to susceptible pupae and only 23 differentially phosphorylated peptides were common to both phenotypes. Thus, signaling responses induced by the parasite were, to a large extent, distinct when comparing pupae from susceptible and tolerant colonies. This trend for greater kinomic changes within susceptible pupae in response to Varroa mite infestation is supported by scatterplots which illustrate a greater number, and magnitude, of changes within pupae of the susceptible phenotype [Fig. 4B,C].
Figure 4.
Kinome responses of pupae from colonies susceptible and tolerant to Varroa infestation. (A) Venn diagram shows the number of differently phosphorylated peptides in susceptible and tolerant pupae in presence of mite infestation. (B,C) Scatterplots of kinome peptides signal intensity of infested versus uninfested pupae from tolerant and susceptible colonies, respectively.
Pathway analysis of responses to varroa mite infestation
To gain insight into the cellular basis of the differential susceptibilities to Varroa mite infestation, pathway analysis was performed on the kinome data corresponding to responses to mite infestation in pupae of the tolerant [Table 5] and susceptible [Table 6] phenotypes. Consistent with the observation of a greater number of differentially phosphorylated peptides in susceptible pupae in response to infestation [Fig. 4A], more pathways were differentially activated within pupae from colonies of the susceptible versus the tolerant phenotype. Pupae of the tolerant phenotype displayed activation of pathways relating to innate immune responses, including activation of TLR signaling [Table 5]. In contrast, pupae of the susceptible phenotype displayed no evidence for innate immune activation in response to the parasite. Instead, the upregulated pathways were associated with activation of stress-associated responses [Table 6].
Table 5.
Pathway Over-Representation Analysis of Responses of Dark-Eyed Pupae from Varroa Tolerant Colonies to Mite Infestation.
| Direction | Pathway | ID | Source | P |
|---|---|---|---|---|
| Tolerant | ||||
| Up-Regulated | TNF receptor signaling pathway | 15154 | PID NCI | 0.01 |
| Adipocytokine signaling pathway | 590 | KEGG | 0.02 | |
| Protein processing in endoplasmic reticulum | 10363 | KEGG | 0.02 | |
| IL-1 signaling pathway | 16110 | INOH | 0.04 | |
| Toll-like receptor signaling pathway | 16121 | INOH | 0.05 | |
| Down- Regulated | None | None | None | None |
Table 6.
Pathway Over-Representation Analysis of Responses of Dark-Eyed Pupae from Varroa Susceptible Colonies to Mite Infestation.
| Direction | Pathway | ID | Source | P |
|---|---|---|---|---|
| Susceptible | ||||
| Up- Regulated | IRE1alpha activates chaperones | 13378 | REACTOME | 9.23E-04 |
| Unfolded Protein Response (UPR) | 16784 | REACTOME | 9.23E-04 | |
| XBP1(S) activates chaperone genes | 13377 | REACTOME | 0.002 | |
| Metabolism of proteins | 18366 | REACTOME | 0.003 | |
| Dual incision reaction in TC-NER | 13847 | REACTOME | 0.02 | |
| Formation of transcription-coupled NER (TC-NER) repair complex | 13844 | REACTOME | 0.02 | |
| Nucleotide Excision Repair | 19742 | REACTOME | 0.02 | |
| RNA Polymerase I Transcription Initiation | 13738 | REACTOME | 0.02 | |
| Sonic hedgehog receptor ptc1 regulates cell cycle | 3999 | PID BIOCARTA | 0.02 | |
| Transcription-coupled NER (TC-NER) | 13848 | REACTOME | 0.02 | |
| DNA Repair | 19821 | REACTOME | 0.04 | |
| RNA Polymerase I Transcription | 16984 | REACTOME | 0.04 | |
| Regulation of ck1/cdk5 by glutamate receptors | 4104 | PID BIOCARTA | 0.04 | |
| TAK1 mediates p38 MAPK activation | 13024 | REACTOME | 0.04 | |
| Down- Regulated | ALK1 signaling events | 15251 | PID NCI | 0.01 |
| Downregulation of TGF-beta receptor signaling | 13267 | REACTOME | 0.01 | |
| Glypican 1 network | 15119 | PID NCI | 0.01 | |
| Citric acid cycle/respiratory electron transport | 17057 | REACTOME | 0.01 | |
| Signaling by NOTCH1 | 19096 | REACTOME | 0.03 | |
| Cytokine-cytokine receptor interaction | 515 | KEGG | 0.03 | |
Discussion
Previously, through the development and application of a honeybee specific peptide array for kinome analysis, our group provided evidence of differences in kinase-mediated signaling between bees from individual colonies of extreme Varroa mite susceptibility phenotypes. These differences were present at a number of developmental stages25. These efforts provided insight into the extent, and nature, of the signaling responses within each phenotype to mite infestation25. While providing proof-of-principle support for the utility of kinome analysis for identifying phosphorylation-associated biomarkers of mite tolerant phenotypes, the restricted scope of phenotypic diversity considered within this study limited the extent to which these findings can be translated in real-world scenarios. The current work builds on those efforts by investigating pupae representing multiple colonies of each mite susceptibility phenotype to identify biomarkers, as well as to determine the molecular mechanisms, of these traits. Analysis of multiple colonies of each phenotype enables identification of more robust biomarkers and reliable mechanistic explanations of these phenotypes; subsequent evaluation of these biomarkers within colonies of a spectrum of Varroa mite susceptibilities represents a more realistic scenario for biomarker application.
A number of investigations have shown honeybees have innate immune responses when challenged with pathogens25,40–42. This response could be a contributing factor to Varroa mite tolerant phenotypes. Kinome analysis indicates that differences in innate immune capabilities exist between the pupae of tolerant and susceptible phenotypes, both in the presence and absence of infestation. Specifically, that there was a down-regulation of Toll-like receptor (TLR) signaling in pupae from susceptible colonies. As TLR signaling is a central pathway in innate immune responses, the reduced TLR signaling in susceptible bees is consistent with increased susceptibility to mites as well as their associated viral pathogens.
In our previous kinome investigation, difference in innate immunity between the mite susceptible and tolerant phenotypes were only observed in response to mite infestation, which was interpreted as evidence that the molecular mechanism of susceptibility involved mite mediated suppression of innate immune responses rather than differences in innate immune capabilities between the phenotypes in the absence of infestation. In the current investigation, there is still evidence for suppression of innate immune responses in the susceptible bees by Varroa mites; however, the differences in innate immunity also appear to precede infestation. Notably, the initial investigation classified the differential responses at the level of gene ontology analysis (which provides a more general overview of biological processes) and considered bees from only a single colony of each phenotype. In contrast, the current investigation utilizes pathway over-representation analysis (which offers more specific insight into biological responses) and considers bees from multiple colonies of each phenotype, which collectively should offer more reliable and detailed insight into the biological differences between the phenotypes.
A panel of nineteen peptides showing different phosphorylation levels between uninfested pupae collected from mite susceptible and tolerant colonies were identified. The biological roles of the phosphorylation events represented by these biomarker peptides implicated differences in innate immunity, metabolism, and stress tolerance between the two phenotypes. For many of the individual biomarker peptides there is incomplete separation of pupae based on phenotype, which likely reflects the complexity of the phenotype as well as diversity of signaling in individual pupae. However, as a collective unit, these peptides are effective in predicting colony phenotype, in particular of identifying pupae of tolerant colonies. This emphasized the necessity to consider multiple peptides to effectively discriminate between tolerant and susceptible pupae. As would be expected, the biomarker peptides were less effective in discriminating colonies with corresponding reduction in colony survival time from intermediate phenotypes.
In the current investigation, emphasis was placed on the identification of biomarkers that discriminate individuals from colonies of the extreme phenotypes. This reflects the priority of bee breeding efforts in selecting for, or against, colonies with mite tolerance or susceptibility. From a practical perspective, identifying pupae representing either tolerant or susceptible phenotypes would be an effective approach for guiding breeding efforts. Importantly, these potential biomarkers were detected in the absence of mite infestation, indicating that these differences reflect innate (naturally selected) traits rather than differences in responses to infestation. The colonies that show these innate traits were previously subjected to natural selection in the presence of Varroa without miticide treatments (www.saskatraz.com). For bee breeding efforts, it would be important to have selectable markers that do not depend on infestation. Having established those biomarkers, future investigations will consider greater representation of colonies of intermediate phenotype to more definitively establish the accuracy of this approach.
The biomarker scores were generally consistent for pupae of the same colony, supporting the feasibility of using this type of approach to guide breeding efforts at the level of a colony. A degree of biological variability of individual pupae of a given colony is anticipated due to multiple mating events of individual queen bees, high recombination rates, and supersedure events. From a practical perspective, this would require that multiple pupae from a particular colony are analyzed to make a reliable determination of colony phenotype. Future investigations with greater numbers of pupae from individual colonies should be completed to determine the minimum number of individual pupae, and specific stage of development, which provides the most reliable prediction of colony phenotype. Such efforts will need to pay particular consideration to the priorities of application and the consequences associated with false positives versus false negatives. In the interest of developing a cost-effective screening tool, such investigations may also consider whether it is possible to obtain reliable colony phenotype predictions by performing kinome analysis on a pooled sample of 30 or more pupae from an individual colony.
Collectively, while the current investigation provides strong proof-of-principle support for the utility of kinome analysis for identifying phosphorylation-associated biomarker of the Varroa mite tolerance phenotype, further investigations which consider a greater number of bees from each colony, as well as a greater number of phenotypically distinct colonies, will ultimately determine the accuracy, and value, of these biomarkers. Alternatively, kinome analysis could be used to identify biological events which, from cost and practicality perspectives, are more amenable for use by bee breeders. For example, a previous kinome investigation of stress responses of livestock identified differences in signaling associated with carbohydrate metabolisms between high and low stress responding animals. This led to the demonstration that serum glucose levels, which are easy and inexpensive to monitor, had a greater predictive value of stress-associated behaviors than traditional stress biomarkers43,44.
In conclusion, the process of recurrent natural selection is resulting in phenotypes that can survive longer in the presence of Varroa infestations. This is due to a number of behavioural traits such as grooming and hygienic behaviour (VSH). However, in this work we have also shown some evidence that tolerant colonies are showing increased innate immune capabilities that would increase the ability of these colonies to tolerate the pathogens associated with Varroa infestation. In future kinome analyses it would be beneficial to look at other stocks of honeybees showing Varroa resistance for evidence of innate immune responses.
Materials and Methods
Colony phenotype selection
A detailed description of the honeybee breeding and selection program used to construct and identify the Varroa mite susceptible and tolerant phenotypes can be accessed at http://www.saskatraz.com and in Robertson, 201425. The Saskatraz natural selection apiaries were initially established with a diverse group of colonies represented by selections from a number of different races. Canadian (carnica and ligustica), Russian (caucasica, ligustica, carnica), and German (carnica) as well as hybrids such as Buckfast. The Saskatraz bees result from natural selection of a mixture of these races selected for honey production, winter survival, and mite tolerance in Saskatchewan, CA (52.1332° N, 106.6700° W). The natural selection apiaries were isolated for close population mating and ranged in size 32–40 colonies.
In this investigation, we studied eight colonies classified as either tolerant, susceptible, or intermediate phenotypes with respect to Varroa mite burdens and colony survival times [Table 1; Fig. 2]. This represented three colonies of tolerant (S88, S23A, and S14 JHN-13) and susceptible (S65-15 BC, S88-4, and G4) phenotypes, as well as two colonies deemed as an intermediate (S96-4 JHN-12 and S65 SAT-1) phenotype. Colonies were selected between 2010 and 2016. The length of survival under natural selection conditions defines the susceptibility or tolerance to Varroa infestation. S65-15 BC was identified as being very susceptible to mite population growth during 2016 with a survival time of 15 months [Table 1].
Data was collected periodically on the phoretic mite infestation from April – September/October each year at each of our natural selection apiaries. Varroa infestations on adult bees (phoretic phase) were determined by washing 200–300 bees from each colony in 100% methanol. Mite infestation levels are reported as mites per hundred bees (MHB). MHB = (# of Mites/# of Bees) * 100. For all natural selection apiaries honey production was measured by weighing all supers of honey harvested from each colony over the production season for each year that the colony was alive. Colonies with the defined phenotypes of susceptible, intermediate, and tolerant were subjected to brood analyses. Sealed brood frames were removed from the colonies and several stages of pupae (white, pink, dark-eyed, and dark-bodied) were randomly sampled from each frame from each colony in the month described [Table 1]. Clean stainless-steel forceps were used to open each brood cell and the pupae were removed and visually inspected for the presence or absence of Varroa. The pupae were immediately placed in liquid nitrogen after removal and stored at −80 °C until analysis. Varroa free pupae were defined as those that did not have a foundress, progeny, or any evidence of Varroa in the brood cell (i.e. Varroa feces).
All colonies except S65-15 BC were maintained in natural selection apiaries, where no miticide treatments were used. S65-15 BC was observed to be extremely susceptible to Varroa population growth in one of our commercial apiaries in August 2016. It was managed for honey production and treated for mites with Apistan (2 strips) on Sep 1, 2015 and Apivar (2 strips) and oxalic (3.2 grams per 100 ml in 50% w/v sucrose) on Apr 18, 2016. None of the other colonies in this apiary showed notable Varroa infestations. It is difficult to detect Varroa susceptible colonies because they die quickly.
Honey production
Honey production is measured by weighing all honey harvested from each colony over the honey production season for each year that the colony was alive.
Kinome Analysis
Peptide arrays for kinome analysis
The kinome peptide array customized for the honeybee phosphoproteome has been described23. The current investigation utilized the same array with no further modifications. Kinome analyses were performed on 36 individual dark-eyed pupae (18 mite-infested; 18 uninfested) collected from eight different colonies representing a phenotypic range of susceptibility to Varroa mite infestation. Prioritizing bees at the dark-eyed pupae stage of development minimizes potential signaling variability arising from different castes or environmental influences in adult bees. All samples were analyzed within the same kinome assay to minimize technical effects due to inter-assay variability. Individual frozen pupae were placed in a sealed plastic bag in the presence of 300 μl of lysis buffer25. The pupae were pulverized with a rubber mallet and the resulting suspension was centrifuged at 10,000 × g for 10 min. Supernatants were used for kinome analysis25.
Data analysis
The dataset for each array contains signal intensities associated with the nine technical replicates for each of the 299 peptides spotted on each array. Kinome data were processed through PIIKA 2, a pipeline for processing kinome array data45.
Identification of peptide biomarkers of varroa mite susceptibility
A one-sided paired student’s t-test of normal distribution was used to compare normalized signal intensity values for each of the 299 peptides of individual pupae (n = 5) representing the two high and low colony phenotypes of Varroa mite susceptibility. Specifically, mite tolerance was represented by pupae from the S88 (n = 3) and S23A (n = 2) colonies while mite susceptibility was represented by pupae from the G4 (n = 3) and S88-4 (n = 2) colonies in the absence of mite infestation. Peptides with significant (P-value < 0.01) differences in levels of phosphorylation between pupae of the two phenotypes were classified as potential biomarkers.
Application of kinome biomarkers to individual bees to predict colony phenotype
The predictive power of the biomarker peptides was evaluated using kinome data from individual, uninfested dark-eyed pupae (n = 18) selected from colonies with a spectrum of susceptibilities to Varroa mite infestation. This consideration included the pupae from the colonies used to identify the biomarker peptides as well as additional pupae from colonies representing a range of phenotypes, including those classified as tolerant (S88 (n = 3), S23A (n = 2), and S14 JHN13 (n = 2)), susceptible (G4 (n = 3), S88-4 (n = 2), and S65-15 BC (n = 2)) and intermediate (S96-4 JHN12 (n = 2) and S65 SAT-1 (n = 2)) phenotypes all in the absence of Varroa mite infestation. Each pupa was assigned scores based on similarity to the mean of the pupae representing the tolerant and susceptible phenotypes based on levels of phosphorylation across the nineteen biomarker peptides. The similarity between phenotypes’ phosphorylation pattern was assessed using pairwise distance from python package scipy (scipy.distance.spatial.pdist). For each phenotype, a vector was created consisting of the normalized values of 19 peptides. The Euclidian distance between each vector and a vector consisting of the mean of the same 19 peptides values in the most tolerant phenotypes (S88 and S23A) was measured, and the same was done with the most susceptible phenotypes (S88-4 and G4). Differences between the biomarker scores of the eight different colonies were analyzed using one-way ANOVA for multiple comparisons using the R package emmeans. Pairwise comparisons were assessed using the post-hoc Tukey HSD test.
Treatment-treatment variability analysis
For each peptide, a paired t-test was used to compare its normalized signal intensity values in the presence and absence of mite infestation. Peptides with significant (P-value < 0.10) changes in phosphorylation were identified. This level of significance was chosen to retain as much data as possible to facilitate subsequent pathway analysis35.
Pathway analysis
Pathway analysis was performed as described previously using the software InnateDB utilizing a hypergeometric algorithm with the Benjamini Hochberg correction method46.
Acknowledgements
This work was funded by Saskatchewan Agriculture (Agriculture Development Fund project number 20130110), Meadow Ridge Enterprises LTD, Saskatchewan Beekeepers’ Development Commission (SBDC) grants to Albert J. Robertson. Philip Griebel holds a Tier I Canada Research Chair funded by the Canadian Institutes of Health Research. We thank the Saskatraz research team members (Antonio Munoz Cerna, Neil Morrison, S Shaw, Yang Tan and Heloise Garez) for assistance with the bee breeding work, and Dr. Bob Danka (USDA) for critically reviewing the manuscript.
Author contributions
A.J.R. contributed to the experimental design and managed the bee breeding efforts, phenotypic characterizations of bees, sample collection, data analyses, presentation, and manuscript writing. E.S. performed all of the kinome analysis and was involved in data interpretation. M.M. was involved in the collection of phenotypic data of the bees. T.R. was involved in the management of bee colonies and collection of phenotypic data. C.D. was responsible for the calculation of biomarker scores. D.H. was responsible for identification of biomarker peptides. A.R. was responsible for gridding kinome arrays and normalization of data. C.R. was involved in the management of bee colonies, collection of phenotypic data, data presentation, and analyses. A.K. oversaw all bioinformatics work. P.G. was involved in experimental design, data interpretation and manuscript preparation. S.N. was involved in experimental design, data interpretation, and wrote the majority of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Albert J. Robertson, Email: a.j.robertson@saskatraz.com
Scott Napper, Email: scott.napper@usask.ca.
References
- 1.Lee KV, et al. A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie. 2015;46:292–305. doi: 10.1007/s13592-015-0356-z. [DOI] [Google Scholar]
- 2.Spleen AM, et al. A national survey of managed honey bee 2011–12 winter colony losses in the United States: results from the Bee Informed Partnership. J. Apic. Res. 2013;52:44–53. doi: 10.3896/IBRA.1.52.2.07. [DOI] [Google Scholar]
- 3.Kulhanek K, et al. A national survey of managed honey bee 2015–2016 annual colony losses in the USA. J. Api. Res. 2017;56:328–340. doi: 10.1080/00218839.2017.1344496. [DOI] [Google Scholar]
- 4.Steinhauer NA, et al. A national survey of managed honey bee 2012–2013 annual colony losses in the USA: results from the Bee Informed Partnership. J. Apic. Res. 2014;53:1–18. doi: 10.3896/IBRA.1.53.1.01. [DOI] [Google Scholar]
- 5.Greenleaf SS, Kremen C. Wild bees enhance honey bees’ pollination of hybrid sunflower. Proc. Natl. Acad. Sci. USA. 2006;103:13890–13895. doi: 10.1073/pnas.0600929103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson DL, Trueman JWH. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp. Appl. Acarol. 2000;24:165–189. doi: 10.1023/A:1006456720416. [DOI] [PubMed] [Google Scholar]
- 7.Martin SJ, et al. Global honey bee viral landscape altered by a parasitic mite. Science. 2012;336:1304–1306. doi: 10.1126/science.1220941. [DOI] [PubMed] [Google Scholar]
- 8.Nazzi F, et al. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 2012;8:e1002735. doi: 10.1371/journal.ppat.1002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullin CA, et al. High levels of miticides and agrochemicals in North American apiaries: implications for honey bee health. Plos One. 2010;5:e9754. doi: 10.1371/journal.pone.0009754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryabov Eugene V., Wood Graham R., Fannon Jessica M., Moore Jonathan D., Bull James C., Chandler Dave, Mead Andrew, Burroughs Nigel, Evans David J. A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees (Apis mellifera) Prevails after Varroa destructor-Mediated, or In Vitro, Transmission. PLoS Pathogens. 2014;10(6):e1004230. doi: 10.1371/journal.ppat.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin SJ, Ball BV, Carreck NL. The role of deformed wing virus in the initial collapse of varroa infested honey bee colonies in the UK. J. Apic. Res. 2013;52:251–258. doi: 10.3896/IBRA.1.52.5.12. [DOI] [Google Scholar]
- 12.Chen Yan Ping, Pettis Jeffery S., Corona Miguel, Chen Wei Ping, Li Cong Jun, Spivak Marla, Visscher P. Kirk, DeGrandi-Hoffman Gloria, Boncristiani Humberto, Zhao Yan, vanEngelsdorp Dennis, Delaplane Keith, Solter Leellen, Drummond Francis, Kramer Matthew, Lipkin W. Ian, Palacios Gustavo, Hamilton Michele C., Smith Barton, Huang Shao Kang, Zheng Huo Qing, Li Ji Lian, Zhang Xuan, Zhou Ai Fen, Wu Li You, Zhou Ji Zhong, Lee Myeong-L., Teixeira Erica W., Li Zhi Guo, Evans Jay D. Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health. PLoS Pathogens. 2014;10(7):e1004261. doi: 10.1371/journal.ppat.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Prisco G, et al. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011;92:151–155. doi: 10.1099/vir.0.023853-0. [DOI] [PubMed] [Google Scholar]
- 14.Annoscia D, et al. Haemolymph removal by Varroa mite destabilizes the dynamical interaction between immune effectors and virus in bees, as predicted by Volterra’s model. Proc. R. Soc. B. 2019;286:20190331. doi: 10.1098/rspb.2019.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsey Samuel D., Ochoa Ronald, Bauchan Gary, Gulbronson Connor, Mowery Joseph D., Cohen Allen, Lim David, Joklik Judith, Cicero Joseph M., Ellis James D., Hawthorne David, vanEngelsdorp Dennis. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proceedings of the National Academy of Sciences. 2019;116(5):1792–1801. doi: 10.1073/pnas.1818371116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lodesani M, Costa C. Limits of chemotherapy in beekeeping: development of resistance and the problem of residues. Bee World. 2005;86:102–109. doi: 10.1080/0005772X.2005.11417324. [DOI] [Google Scholar]
- 17.Sammataro D, Gerson U, Needham G. Parasitic mites of honey bees: life history, implications, and impact. Annu. Rev. Entomol. 2000;45:519–548. doi: 10.1146/annurev.ento.45.1.519. [DOI] [PubMed] [Google Scholar]
- 18.Rosenkranz P, Aumeier P, Ziegelmann B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2010;103:S96–S119. doi: 10.1016/j.jip.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Locke B. Natural Varroa mite-surviving Apis mellifera honeybee populations. Apidologie. 2016;47:467–482. doi: 10.1007/s13592-015-0412-8. [DOI] [Google Scholar]
- 20.Le Conte Y, et al. Honey bee colonies that have survived Varroa destructor. Apidologie. 2007;38:566–572. doi: 10.1051/apido:2007040. [DOI] [Google Scholar]
- 21.Seeley TD. Honey bees of the Arnot Forest: a population of feral colonies persisting with Varroa destructor in the northeastern United States. Apidologie. 2007;38:19–29. doi: 10.1051/apido:2006055. [DOI] [Google Scholar]
- 22.Danka RG, Harris JW, Dodds GE. Selection of VSH-derived “Pol-line” honey bees and evaluation of their Varroa-resistance characteristics. Apidologie. 2016;47:483–490. doi: 10.1007/s13592-015-0413-7. [DOI] [Google Scholar]
- 23.Harbo JR, Harris JW. Responses to Varroa by honey bees with different levels of varroa sensitive hygiene. J. Apic. Res. 2009;48:156–161. doi: 10.3896/IBRA.1.48.3.02. [DOI] [Google Scholar]
- 24.Tsuruda JM, Harris JW, Bourgeois L, Danka RG, Hunt GJ. High-resolution linkage analyses to identify genes that influence Varroa sensitive hygiene behavior in honey bees. PLoS ONE. 2012;7:e48276. doi: 10.1371/journal.pone.0048276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson, A. J. et al. Identification of developmentally-specific kinotypes and mechanisms of Varroa mite resistance through whole-organism, kinome analysis of honeybee. Front. Genetics. May 21, 5, 139, 10.3389/fgene.2014.00139 (2014) [DOI] [PMC free article] [PubMed]
- 26.S. Jiang S, et al. Differential Gene Expression of Two Extreme Honey bee (Apis mellifera) Colonies Showing Varroa Tolerance and Susceptibility. Insect Mol. Biol. 2016;25:272–282. doi: 10.1111/imb.12217. [DOI] [PubMed] [Google Scholar]
- 27.Navajas M, et al. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genomics. 2008;9:301. doi: 10.1186/1471-2164-9-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Conte Y, et al. Social immunity in honeybees (Apis mellifera): transcriptome analysis of varroa-hygienic behaviour. Insect Mol. Biol. 2011;20:399–408. doi: 10.1111/j.1365-2583.2011.01074.x. [DOI] [PubMed] [Google Scholar]
- 29.Brutscher, L. M., Daughenbaugh, K. F., & Flenniken, M. L. Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. Scientific Reports Jul 25, 7(1), 6448, 10.1038/s41598-017-06623-z (2017). [DOI] [PMC free article] [PubMed]
- 30.Parker R, et al. (2012). Correlation of proteome-wide changes with social immunity behaviors provides insight into resistance to the parasitic mite, Varroa destructor, in the honey bee (Apis mellifera) Genome Biol. 2012;13:R81. doi: 10.1186/gb-2012-13-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michaud S, et al. Response of the honey bee (Apis mellifera) proteome to Israeli acute paralysis virus (IAPV) infection. Can. J. Zoo. 2015;93:711–720. doi: 10.1139/cjz-2014-0181. [DOI] [Google Scholar]
- 32.Daigle, J. et al. Peptide arrays for kinome analysis of livestock species. Frontiers in Veterinary Science14, 10.3389/fvets.2014.00004 (2014). [DOI] [PMC free article] [PubMed]
- 33.Jalal S., Arsenault R., Potter A. A., Babiuk L. A., Griebel P. J., Napper S. Genome to Kinome: Species-Specific Peptide Arrays for Kinome Analysis. Science Signaling. 2009;2(54):pl1–pl1. doi: 10.1126/scisignal.254pl1. [DOI] [PubMed] [Google Scholar]
- 34.Arsenault RJ, et al. Mycobacterium avium subsp. paratuberculosis inhibits gamma interferon- induced signaling in bovine monocytes: insights into the cellular mechanisms of Johne’s disease. Infect. Immun. 2012;80:3039–3048. doi: 10.1128/IAI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arsenault RJ, et al. Altered toll-like receptor 9 signaling in Mycobacterium avium subsp. paratuberculosis-infected bovine monocytes reveals potential therapeutic targets. Infect. Immun. 2013;81:226–237. doi: 10.1128/IAI.00785-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Määttänen P, et al. Divergent immune responses to Mycobacterium avium subsp. paratuberculosis infection correlate with kinome responses at the site of intestinal infection. Infect. Immun. 2013;81:2861–2872. doi: 10.1128/IAI.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Régnier Marion, Gourbeyre Pascal, Pinton Philippe, Napper Scott, Laffite Joëlle, Cossalter Anne-Marie, Bailly Jean-Denis, Lippi Yannick, Bertrand-Michel Justine, Bracarense Ana Paula F.R.L., Guillou Hervé, Loiseau Nicolas, Oswald Isabelle P. Identification of Signaling Pathways Targeted by the Food Contaminant FB1: Transcriptome and Kinome Analysis of Samples from Pig Liver and Intestine. Molecular Nutrition & Food Research. 2017;61(12):1700433. doi: 10.1002/mnfr.201700433. [DOI] [PubMed] [Google Scholar]
- 38.Arsenault RJ, Napper S, Kogut MH. Salmonella enterica Typhimurium infection causes metabolic changes in chicken muscle involving AMPK, fatty acid and insulin/mTOR signaling. Vet. Res. 2013;44:35. doi: 10.1186/1297-9716-44-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trost B, et al. Kinotypes: stable species- and individual-specific profiles of cellular kinase activity. BMC Genomics. 2013;14:854. doi: 10.1186/1471-2164-14-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flenniken Michelle L., Andino Raul. Non-Specific dsRNA-Mediated Antiviral Response in the Honey Bee. PLoS ONE. 2013;8(10):e77263. doi: 10.1371/journal.pone.0077263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans J. D., Aronstein K., Chen Y. P., Hetru C., Imler J.-L., Jiang H., Kanost M., Thompson G. J., Zou Z., Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Molecular Biology. 2006;15(5):645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brutscher, L. M., Daughenbaugh, K. F. & Flenniken, M. L. Antiviral defence mechanisms in honey bees. Curr. Opin. Insect Sci. 10, 71–82 (201). [DOI] [PMC free article] [PubMed]
- 43.Chen Y, et al. Investigation of the physiological, behavioral, and biochemical responses of cattle to restraint stress. J. Ani. Sci. 2016;94:3240–3254. doi: 10.2527/jas.2016-0549. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Arsenault R, Napper S, Griebel P. Models and methods to investgate acute stress response in cattle. Animals. 2015;5:1268–1295. doi: 10.3390/ani5040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trost B, Kindrachuk J, Määttänen P, Napper S, Kusalik A. PIIKA 2: an expanded, web-based platform for analysis of Kinome microarray data. PLoS One. 2013;8:e80837. doi: 10.1371/journal.pone.0080837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynn DJ, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]