Highlights
▸ Dopamine is essential for executive functioning. ▸ 7-repeat allele of the DRD4-48 base pair repeat gene leads to sub-sensitive postsynaptic D4 receptor. ▸ We investigated effects of genotype on executive functions with fMRI in children. ▸ 7-repeat allele influences brain activation patterns and connectivity patterns.
Abbreviations: ACC, anterior cingulate cortex; BL, baseline; BOLD, Blood Oxygen Level Dependency; bp, base pair; CER, cerebellum; FWE, family wise error; IC, Incompatibility Task; IFG, inferior frontal gyrus; k, cluster size; PPI, psychophysiological interactions; SPG, superior parietal gyrus; TD, typically developing; TT, Time Discrimination Task; VNTR, variable number of tandem repeats
Keywords: fMRI, Genetics, Attention, Dopaminergic system, Endophenotypes
Abstract
Genetic variants within the dopamine D4 receptor gene (DRD4) are among the strongest and most consistently replicated molecular genetic findings in attentional functioning as well as attention deficit hyperactivity disorder (ADHD). Functionally, the 7-repeat allele of the DRD4-48 base pair repeat gene leads to a sub-sensitive postsynaptic D4 receptor, which is expressed at a particularly high density in the frontal lobes. We used fMRI to investigate the influence of the 7-repeat allele on BOLD (Blood Oxygen Level Dependency) responses in 26 healthy children and adolescents while they performed a combined stimulus-response Incompatibility Task (IC) and a Time Discrimination Task (TT).
7-repeat non-carriers exhibited increased neural activation of the left middle and inferior frontal gyrus (IFG) in the IC and greater cerebellar activation in the TT. Furthermore, the 7-repeat non-carriers exhibited a stronger coupling in haemodynamic responses between left IFG and the anterior cingulate cortex (ACC) during the IC and between cerebellar activation and brain regions that have high DRD4 density, including the IFG and the ACC during the TT. Our results indicate that the 7-repeat allele influences both regional brain activation patterns as well as connectivity patterns between neural networks of incompatibility and temporal processing.
1. Introduction
Dopamine plays an important role in normal attention and disorders of attention such as in attention deficit/hyperactivity disorder (ADHD) (Thapar et al., 2005, Del Campo et al., 2011). The dopamine receptor genes in particular are of great interest given that they may contribute to diverse aspects of normal and abnormal human behaviour (Thapar et al., 2005). According to human post-mortem and studies with monkeys, the D4 receptor, which is a D2-like receptor (Strange, 1993), is expressed in several brain regions related to planning and reward (Simpson et al., 2010, Meador-Woodruff, 1994, Matsumoto et al., 1996, Mrzljak et al., 1996, Ariano et al., 1997, Sanyal and Van Tol, 1997). It plays an important role in the prefrontal cortex and in the anterior cingulate cortex (ACC) (see Oak et al., 2000, for a review). These brain regions are critical for regulating executive functions (Seeman et al., 1993). A frequently studied polymorphism of the DRD4 gene, which is located on chromosome 11p15.5, is a 48-base pair variable number of tandem repeat (VNTR) in exon III. This region of the DRD4 gene encodes the third cytoplasmic loop of the D4 receptor, which is responsible for the coupling of a G-protein and activates intracellular responses to dopamine release by changing intracellular cAMP levels (Oak et al., 2000). The 48-bp fragment can be repeated from 2 to 11 times (Van Tol et al., 1992). In functional terms, the DRD4 7-repeat allele seems to alter the function of the encoded receptor by making it less sensitive to dopamine compared to other numbers of repeats (Schoots and Van Tol, 2003, Asghari et al., 1995).
Most of the evidence concerning the relevance of differences in the expression of the DRD4 receptor and attentional functioning is based on research within the field of ADHD. The association between ADHD and the 48 bp repeat polymorphism of exon III of the DRD4 gene is the strongest and most consistently replicated molecular genetic finding in ADHD (Banaschewski et al., 2010). A meta-analysis of more than 30 studies found that the DRD4 7-repeat (DRD4-7r) allele increases the risk for ADHD, although this increase is only moderate with a pooled odds ratio of 1.34 (Faraone and Doyle, 2001, Li et al., 2006).
Studies investigating cognitive differences associated with the different DRD4-48 bp repeat genes in children and adults with ADHD have produced heterogeneous results. Some neuropsychological studies showed that participants carrying the 7-repeat allele indeed performed poorer on tasks of executive functions (Kieling et al., 2006, Langley et al., 2004) than those with other gene variants. In contrast, other studies have reported that children with ADHD who carry the 7-repeat allele have better performance on those tasks (Johnson et al., 2008, Swanson et al., 2000). However, some studies have failed to find any differences in attentional performance between carriers of the 7-repeat allele and those without it (Barkley et al., 2006, Konrad et al., 2010). There is an obvious lack of studies which deal with the effect of DRD4 gene variants on attentional and executive functions in healthy participants. In addition, to date, only a limited number of studies used neuroimaging to explore the relationship between the DRD4 7-repeat allele and differences in brain anatomy or function, although several studies have suggested that neuroimaging methods might be particularly powerful for unravelling gene-brain behavioural relationships (Weinberger et al., 2001). While no other study has yet investigated the impact of DRD4-risk alleles on neural networks associated with executive functions, there is first evidence that DRD4 impacts on brain circuits associated with neural responses in brain areas involved in reward processing such as insula and cingulate cortex (Camara et al., 2010, Forbes et al., 2009). Since, genetic variants may have a more direct effect on brain functions than on behavioural phenotypes (e.g., Goldberg and Weinberger, 2004), the aim of the current fMRI study was to explore how genetic variation in the dopamine-regulating gene DRD4 affects the pattern of neural activation associated with executive functions in typically developing children and adolescents. We decided to investigate children and adolescents since neural networks during development differ from those of adults (see Konrad et al., 2005, Durston and Casey, 2006) and genetically mediated disorders of attention (such as ADHD) typically have their onset during childhood. We analysed behavioural and BOLD responses in typically developing children using two tasks examining executive functions, a combined stimulus-response Incompatibility Task (IC) and a Time Discrimination Task (TT), with the same set of stimuli for both. The rationale for choosing these two tasks were to analyse neural mechanism underlying two different aspects of executive functions (Rubia and Smith, 2004) that are known to be modulated by dopamine (Konrad et al., 2004, Rubia et al., 2009) and which are known to be impaired in many subjects with attentional disorders (Rubia and Smith, 2004, Nigg, 2000, Vloet et al., 2009). We predicted that groups with and without the 7-repeat allele would display differences in neural activation patterns, particularly in brain regions with high dopaminergic receptor density such as the prefrontal cortex. Given the functional consequences of the 7-repeat allele (Asghari et al., 1995) and the association between the risk allele and ADHD (Li et al., 2006), one might hypothesise that 7-repeat-carriers show reduced BOLD responses in brain areas critical for EF task performance, although the unclear and contradicting results of previous neuropsychological studies and the lack of comparable neuroimaging studies hinder a precise prediction of the direction of this effect.
Consecutively, psychophysiological interactions were analysed to further investigate how the DRD4-48 bp repeat gene modulates functional connectivity within neural networks related to executive functions.
2. Materials and methods
2.1. Subjects
Twenty-six, typically developing 8–16-year-old Caucasian children and adolescents (17 boys, 9 girls, Mage = 11.4 ± 2 years) were recruited by board announcements in local primary and secondary schools. All subjects were carefully screened for childhood psychiatric disorders using a standardised semi-structured interview for the diagnosis of mental disorders in children (Unnewehr, 1995; Kaufman et al., 1997; Delmo et al., 2001) and were free of any past or present mental disorders (i.e., ADHD, pervasive developmental disorders, etc.). Each subject's IQ was also estimated based on a short version of the Wechsler Intelligence Scale for Children III (Tewes et al., 1999). An IQ below 85 resulted in exclusion from the study. All participants were screened for any contraindications against fMRI and were trained prior to scanning in a mock fMRI-scanner to familiarise them with the scanner environment. Please note that some data were included in Neufang et al. (2008). The inclusion in this study was based on the availability of blood samples from the participants as well as on the consent given by participants and parents to participate in genetic studies.
The study was carried out in accordance with the latest version of the Declaration of Helsinki. The protocol was reviewed and approved by the local ethics committee. Written informed consent was obtained after providing a complete description of the study to the subjects and their parents. Subjects were compensated for their expenses.
2.2. Genotyping
All participants were genotyped for the DRD4 48-bp repeat VNTR polymorphism.
Table 1 summarises the characteristics of the samples.
Table 1.
Characteristics of the DRD4 groups.
| DRD4-genotype | 7-repeat-absent |
7-repeat-present |
||
|---|---|---|---|---|
| N | Overall | 16 | Overall | 10 |
| Male | 10 | Male | 7 | |
| Female | 6 | Female | 3 | |
| Min–Max | M (SD) | Min–Max | M (SD) | |
| Age | 8.0–15.0 | 11.7 (2.4) | 9.1–4.1 | 11.0 (2.0) |
| IQ | 85–126 | 103.1 (12.0) | 83–118 | 102.0 (13.3) |
| IC reaction time (ms) | 628–1105 | 930 (153) | 601–1136 | 921 (148) |
| IC error rate (%) | 00–48.07 | 21.21 (19.95) | 3.57–41.14 | 23.75 (25.85) |
| TT reaction time (ms) | 758–1157 | 937 (90) | 718–1155 | 924 (151) |
| TT error rate (%) | 1.25–47.5 | 21.17 (22.59) | 1.25–47.5 | 19.38 (17.33) |
The groups did not differ significantly with regard to age (p = 0.420), IQ (p = 0.847) or sex distribution (p = 0.206). Genotyping was performed by the Department of Child and Adolescent Psychiatry, University of Duisburg-Essen, Germany. Polymerase chain reaction fragment length analysis was performed as described previously (Hinney et al., 1999). Briefly, the 48-bp repeat in the third exon of the DRD4 gene was amplified using primers D4-42 and D4-3 according to the guidelines outlined by Lichter et al. (1993). PCR products were run in ethidium bromide-stained 2.5% agarose gels. Positive controls for the variant alleles were run in each gel. All genotypes were scored independently by an experienced lab technician and subsequently by one scientist. Discrepancies were resolved unambiguously, either by reaching a consensus or by re-genotyping.
2.3. Task, experimental design
All participants were asked to perform a combined stimulus-response IC (adapted from Davidson et al., 2006) and a TT (adapted from Smith et al., 2003). In the combined paradigm, each stimulus consisted of two symbols, which were presented consecutively on the left or right side of the screen. The symbols could either be 1 square and 1 heart, or 2 squares or 2 hearts. All symbols were balanced for responses (i.e., the same number of right and left answers were given), symbols, and presentation order. The symbols were presented with a simultaneous onset, but for different durations, with the shorter-duration symbol being presented for 250 ms (reference item) and the longer-duration symbol being presented from 500 ms to 600 ms. Hence, one symbol was presented 250 to 350 ms longer than the other symbol (average difference = 300 ms). Responses were given by pressing one of two parallel buttons and communicated using a fibre optic response device. The buttons were situated beside the participant in the scanner and responses were given by using the index or the middle finger of the right hand respectively. In the TT, subjects were instructed to indicate on which side of the screen the symbol was presented longer (e.g. the left hand-side) by pressing the button on the same side (e.g. the left one of the two parallel buttons). In the IC task, subjects were asked to press the button on the same side of the screen on which the second symbol had appeared, if the symbols were identical. If two different symbols were presented, the subject had to press the button opposite to the position of the second symbol (i.e., a spatially incompatible response). The ratio of compatible/incompatible trials in the IC task, as well as the difference in symbol presentation length in the TT, was varied to create two levels of task difficulty. In the TT, the easier condition had a mean presentation difference of 350 ms (low task difficulty, TT_low), while the more difficult condition had a mean presentation difference of 250 ms (high task difficulty, TT_high) (Smith et al., 2003). In the IC task, the easier condition consisted of a slightly higher ratio of incompatible trials (60:40%, IC_low), while the more difficult condition had a much higher ratio of incompatible trials (80:20%, IC_high) (Casey et al., 2002). Block design was employed with blocks of either TT or IC trials. In addition, task difficulty was manipulated in a blockwise manner. However, since functional data analyses did not reveal a significant effect of task difficulty on neural activation patterns for both task conditions, blocks of high and low task difficulty were combined in further data analyses.
The experiment consisted of 4 runs; each run had 4 blocks of the same task. To minimise working memory load, the specific task symbol was presented for 1500 ms at the beginning of each block (every 10th stimulus). There were 15-sec breaks between each run serving as a low-level baseline condition. Stimuli were presented using a visual stimulation device (goggles, Silent Vision™, Avotec, FL, USA).
2.4. Data acquisition
A SONATA MRI system (Siemens, Erlangen, Germany) operating at 1.5 T was used to obtain T2*-weighted echoplanar (EPI) images with BOLD (Blood Oxygen Level Dependency) contrast (matrix size: 64 × 64; voxel size: 3.12 mm × 3.12 mm). In total, 266 volumes of 30 4-mm-thick axial slices were acquired sequentially with a 0.8-mm gap (repetition time = 3.2 s, echo time = 66 ms). The first five volumes were discarded to allow for T1 equilibration effects. Images were spatially realigned to the first volume to correct for inter-scan movement and were normalised to a standard EPI template (resampled to 3 mm × 3 mm × 3 mm voxels). The data were then smoothed with a Gaussian kernel of 8-mm, full-width at half-maximum to accommodate inter-subject anatomical variability. A high-pass filter (using a cut-off of 512 s) and a correction for temporal autocorrelation in the data (AR 1 + white noise) were applied to accommodate serial correlations. After the acquisition of the functional scan (∼14.2 min), high-resolution T1-weighed anatomical brain scans were collected using a rapid acquisition gradient-echo (MP-RAGE) pulse sequence (TE = 3.93 ms, TR = 2200 ms, α = 15°, FOV = 256 mm, matrix size = 256 × 256, voxel size = 1 mm × 1 mm × 1 mm, 160 slices, slice thickness = 1 mm).
2.5. Statistical analyses of the imaging data
Data were analysed using Statistical Parametric Mapping software (SPM2, Wellcome Department of Imaging Neuroscience, London (http://www.fil.ion.ucl.ac.uk/spm2.html; Friston et al., 1995) using random-effects models. There was no significant effect of task difficulty. Therefore, we performed a first-level analysis incorporating the five conditions (i.e., TT_high, TT_low, IC_high, IC_low, and the baseline condition or BL) into one design matrix. For each session, the five conditions were modulated as a boxcar function convolved with the synthetic haemodynamic response function (HRF). The six head movement parameters were included as confounds. Estimated motion parameters were examined on a subject-by-subject basis to ensure that the amount of absolute motion did not exceed 3 mm.
All subjects exhibited less than 1.5 mm of absolute motion over the course of the experiment and genotype groups did not differ with respect to the total amount of head movement during the scan.
The first-level analysis was primarily designed to identify neural networks that were activated by the two tasks. Thus, for each participant, condition-specific effects were estimated according to a general linear model, with parameter estimates passed into a second-level one-sample t-test with non-sphericity correction. The condition-specific effects were as follows: TT (TT_high + TT_low) > BL and IC (IC_high + IC_low) > BL. These random-effects analyses assessed the data based on inter-subject variance and thereby allowed for inferences about the population from which the subjects were drawn. Two-sample t-tests were performed on contrast images to investigate differences in neural activation between the 7-repeat present and the 7-repeat absent groups. We used a region-of-interest (ROI)-based approach with a height threshold of p < 0.05, FWE (family wise error)-corrected, restricting our analyses to those ROIs known to be activated by our executive tasks. These areas were the prefrontal cortex (Goghari and MacDonald, 2009, Neufang et al., 2008) the parietal cortex (Neufang et al., 2008) as well as the cerebellum (Neufang et al., 2008, Rubia and Smith, 2004). Anatomical ROIs were constructed using the WFU Pickatlas software (Tzourio-Mazoyer et al., 2002, Maldjian et al., 2003). For all ROI analyses, small volume corrections were applied across each respective region.
In addition, in order to determine gene effects on brain areas commonly involved in both, the IC and TT tasks, we performed a random effects conjunction analysis, based on inclusive masking, as suggested by Nichols et al. (2005). This conservative analysis corresponds to a logical AND operation, showing those voxels, which are significant in both the TT Task (TT_high + TT_low) > BL and IC Task (IC_high + IC_low) > BL. For this analysis, the threshold for each contrast entered into a conjunction analysis was set at p < 0.001 uncorrected. Between-group differences between genotypes were then reported at a threshold of p < 0.05, FWE (family wise error)-corrected across the whole brain.
Neural activation was localised using the anatomy toolbox developed by Eickhoff et al. (2005). In addition, SPM(T)images were overlaid on the averaged group T1-image, which was calculated from the subjects’ normalised T1-images.
To further explore the influences of genotype on the functional associations between different brain areas, we conducted psychophysiological interaction (PPI) analyses. Based on the results of the two-sample t-tests, the maxima of the genotype-dependent differential activation patterns obtained in the BOLD signal time course were used as seeds for a PPI analyses (see also Monk et al., 2008). A PPI analysis attempts to explain neural responses in one brain area in terms of its interactions with other brain regions and cognitive/sensory processes. Thus, a psychophysiological interaction can be interpreted as a condition-specific change in the coupling of neural activation between brain regions. The PPI analysis consists of a design matrix with three regressors: (i) the “psychological variable”, representing the cognitive/sensory process of interest, (ii) the “physiological variable”, representing the neural response in a given brain region, and (iii) the interaction between (i) and (ii).
The psychological variable used in the current study was a vector coding for the specific task convolved with the HRF. The individual time series for the left inferior frontal gyrus (IFG) and the cerebellum, respectively, were obtained by extracting the first principal component from all raw voxel time series in a sphere (8-mm radius) centred on the coordinates of the subject-specific activations. These time series were mean-corrected and high-pass filtered to remove low-frequency signal drifts. This process produced the physiological factor that was then multiplied by the psychological factor to produce the interaction term. PPI analyses were then carried out for each subject by creating a design matrix with the interaction term, the psychological factor, and the physiological factor as regressors. Subject-specific contrast images using the contrast [1 0 0], where the first column represented the interaction term, were then entered into a random-effects group analysis, comparing the groups according to genotype (7-repeat absent versus 7-repeat present). Task-specific ROI-analyses were performed (prefrontal cortex, parietal cortex, cerebellum) with a significance threshold set at p < 0.001, uncorrected for multiple comparisons, extent threshold k > 5. We decided for this more liberal threshold since PPI analyses are inherently less powered than their univariate counterparts (as only the unshared variance between the three regressors is attributed to the interaction term). Voxels that surpassed the threshold in these PPI analyses can be interpreted as showing a genotype-based difference in connectivity with the seed region as a function of EF task (Uncapher et al., 2011).
3. Results
3.1. Genotyping
Carriers of the 7-repeat allele were equally distributed across boys and girls (7-repeat absent: 10 males, 6 females; 7-repeat present: 7 males, 3 females). The groups did not differ significantly with respect to age or IQ (see Table 1). The n values were as follows: 7-repeat absent, N = 16 and 7-repeat present, N = 10; the most common DRD4 allele was the 4-repeat allele (84.6% with 50% homozygous), followed by the 7-repeat allele (38.5%) and the 2-repeat allele (19.2%). Other DRD4 gene variants were rare. Genotypes were in Hardy-Weinberg equilibrium.
3.2. Behavioural results
Mixed-model ANOVAs with the factors TASK and GROUP were calculated to analyse the percent correct responses and the reaction times (RTs).
Number of correct responses: ANOVA for the percent correct responses revealed no significant main effect of group (F1,24 = 0.003, p = 0.96) or task (F1,24 = 0.245, p = 0.625) as well as no significant interaction effect (F1,24 = 0.237, p = 0.631).
Reaction times: ANOVA models with RTs as dependent variables revealed no significant main effect of group (F1,24 = 0.061, p = 0.808) or task (F1,24 = 0.27, p = 0.872) as well as no significant interaction effect (F1,24 = 0.004, p = 0.949).
Since the groups did not differ in terms of behavioural performance, we could examine the effects of genotypes on brain activity independently of significant behavioural group differences.
3.3. Functional results
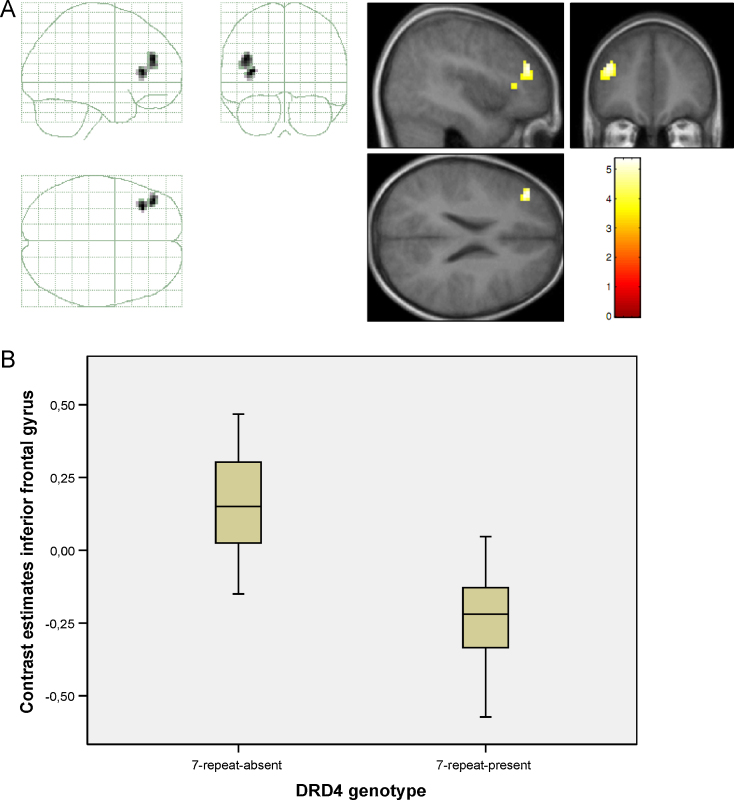
Two-sample t-tests were performed on contrast images to investigate group differences in neural activation between the DRD4 groups (7-repeat allele non-carrier versus 7-repeat allele carrier). The results stem from the ROI-analyses described under Section 2.5. Consistent with our hypothesis, the groups had significantly different BOLD responses during the IC task in frontal cortical areas involved in executive functions, specifically, in the left IFG and left middle frontal gyrus using a ROI-based analysis (x = −36, y = 30, z = 12; T = 5.28, p-FWE-corrected < 0.05, k = 40). The 7-repeat present group had significantly decreased haemodynamic responses compared to the 7-repeat absent group (see Fig. 1A and B).
Fig. 1.
(A) Genotype based analysis of the BOLD response during the Incompatibility Task. Contrast: 7-repeat absent > 7-repeat present. (B) Brain activation differences for the Incompatibility Task in the inferior frontal gyrus (x = −36, y = 30, z = 12).
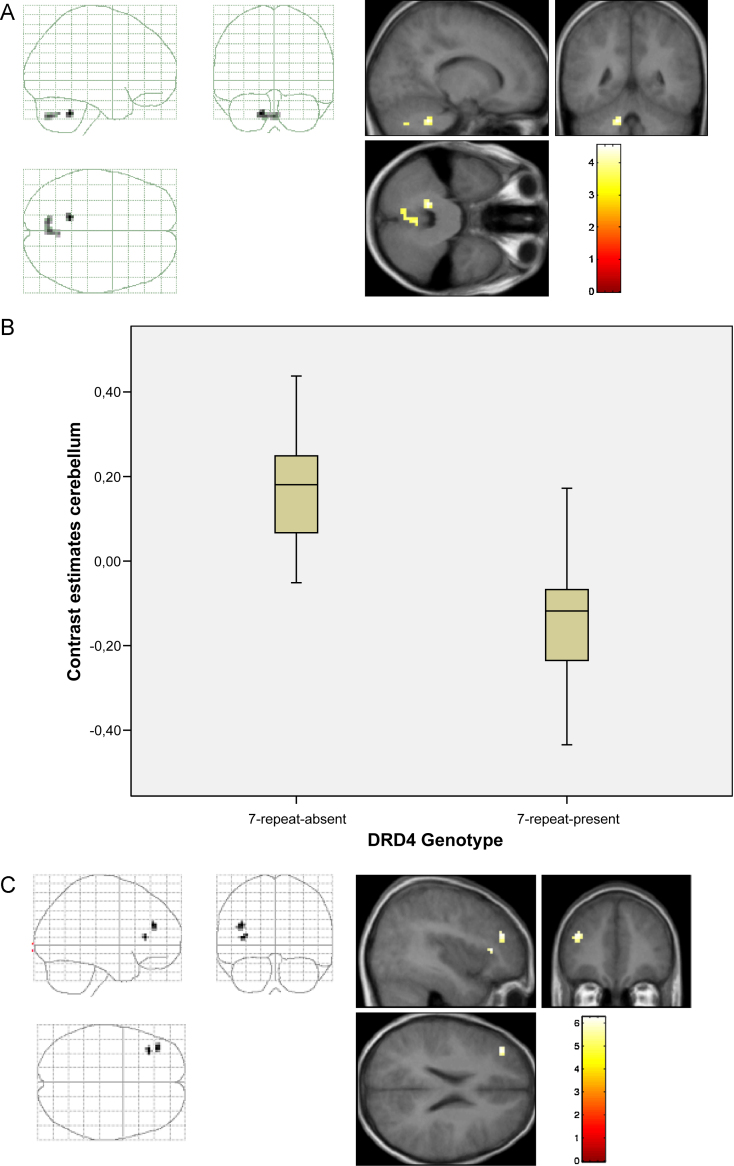
When individuals performed the TT, differences in neural activation between the different gene variants were located in the cerebellum in the ROI-based analysis (x = −15, y = −48, z = −36; T = 4.54, p-FWE-corrected < 0.05, k = 16). In accordance with the pattern of neural activation observed during the IC task, the 7-repeat present group had significantly decreased haemodynamic responses compared to the 7-repeat absent group (see Fig. 2A and B).
Fig. 2.
(A) Genotype based analysis of the BOLD response during the Time Discrimination Task. Contrast: 7-repeat absent > 7-repeat present. (B) Brain activation differences for the Time Discrimination Task in the cerebellum (x = −15, y = −48, z = −36). (C) Brain activation differences for the conjunction of both tasks (Incompatibility and Time Discrimination Task) in the middle frontal gyrus (x = −39, y = 39, z = 24, x = −36, y = 30, z = 9). Contrast: 7-repeat absent > 7-repeat present.
A conjunction analysis (whole brain analysis) across both tasks confirmed DRD4-genotype effects on haemodynamic responses in two clusters in the left middle frontal gyrus.
Please see Table 2 for a summary of all resulting contrasts.
Table 2.
Summary of results. Contrast: 7-repeat absent > 7-repeat present.
| Task | Analysis | k | p-FWE-corrected | T | x y z | Anatomical region |
|---|---|---|---|---|---|---|
| IC | ROI | 40 | 0.001 | 5.28 | −36 30 12 | Left inferior frontal gyrus |
| TT | ROI | 16 | 0.001 | 4.54 | 15 −48 −36 | Cerebellum |
| Conjunction IC∩TT | Wholebrain | 18 | 0.002 | 6.25 | −39 39 24 | Left middle frontal gyrus |
| 11 | 0.003 | 6.10 | −36 30 9 | Left middle frontal gyrus | ||
3.4. PPI analysis
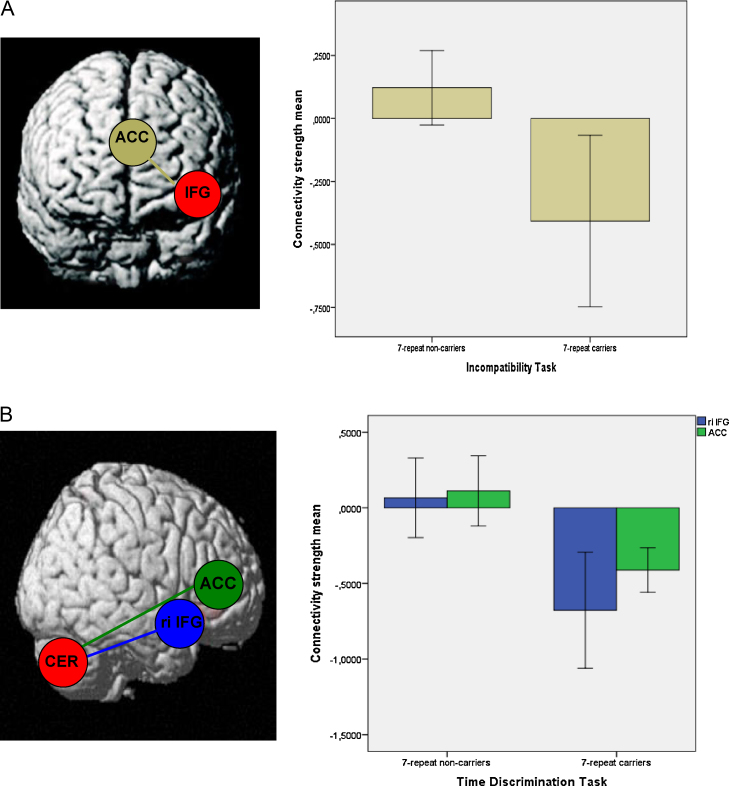
Based on the genotype-dependent between-group differences, the BOLD signal time courses at the local maxima in specific regions within the IFG (x = −36, y = 30, z = 12) for the IC task and within the cerebellum (x = −15, y = −48, z = −36) for the TT were entered into PPI analyses at the second level. ROI analyses were performed (see Section 2.5) and are presented in the following. During the IC task, the connectivity analysis revealed that there were differences between the DRD4 genotypes in the association between haemodynamic responses in the left IFG and the ACC (ROI-analysis: x = 6, y = 27, z = 24; T = 3.60, p-uncorrected < 0.001, k = 50), as shown in Fig. 3A. Carriers of the DRD4-7 repeat allele showed reduced coupling between the interhemispheric inferior frontal brain regions compared to non-carriers of the risk allele.
Fig. 3.
(A) Genotype based group comparison of the psychophysiological interactions associated with the left inferior frontal gyrus and connectivity strength between the left IFG and the ACC during the Incompatibility Task. Contrast: 7-repeat absent > 7-repeat present. (B) Genotype based group comparison psychophysiological interactions associated with the left inferior frontal gyrus and connectivity strength between the cerebellum and associated brain regions during the Time Discrimination Task. Contrast: 7-repeat absent > 7-repeat present.
During the TT, the correlation between haemodynamic responses in the cerebellum, the right IFG (ROI-analysis: x = 45, y = 24, z = −15; T = 3.61, p-uncorrected < 0.001, k = 5) and the ACC (ROI-analysis: x = −3, y = 36, z = −6; T = 3.54, p-uncorrected < 0.001, k = 25) was different with respect to DRD4 genotype, as evident in Fig. 3B. Again, there was a reduction of the correlation of haemodynamic responses in the 7-repeat present group relative to the 7-repeat absent group.
4. Discussion
The results of the present study confirmed that the genotype of the DRD4-48 bp repeat affects neural activation patterns in children and adolescents during both the response IC and the TT, two tasks that tap the executive domain. In line with our hypothesis, we found that carriers of the DRD-4 risk allele showed reduced prefrontal and cerebellar brain activation during EF tasks, similarly to findings previously described for subjects with ADHD (see Vloet et al., 2009, Dickstein et al., 2006 for a meta-analysis). During the IC, we found DRD4-48 bp repeat gene-dependent differences in neural activation patterns in the left IFG. Coupling differences were observed between left IFG and ACC. The location of genotype dependent differences in neural activation patterns during the IC task is in line with other studies showing that the IFG and ACC are important for individual differences in executive functioning (Osaka et al., 2004, Morimoto et al., 2008). The conjunction analysis also confirmed main gene effects on left prefrontal cortex activity located in the middle frontal gyrus. During TT, there were DRD4-48 bp repeat gene-dependent differences in neural activation patterns in the cerebellum, as well as differences in neural connectivity between the ACC, IFG and SPG. For both the IC and TT, haemodynamic responses and connectivity were higher in the group with 7-repeat allele non-carriers compared to the group of 7-repeat allele carriers. The analysis of psychophysiological interactions revealed that there were genotype-based differences in the functional coupling of haemodynamic responses between brain regions that are known to interact during executive functioning (Neufang et al., 2008). These differences in functional associations indicate that a specific genotype not only modulates activity in circumscribed brain regions but also has a significant effect on connectivity between brain regions.
At first glance, the genotype-dependent differences in neural activation patterns during the TT may seem unexpected given that the cerebellum does not have a high density of D4 receptors (Moreland et al., 2004). In general, cerebellar activation is observed during time processing tasks, and cognitive models of time perception have implicated the cerebellum in the regulation of timing mechanisms in particular if very small time difference (<1 s) have to be detected (Ivry and Fiez, 2000). However, Monuteaux et al. (2008) found differences in cerebellar volume between 7-repeat allele carriers and those without this gene variant in 24 participants with ADHD, suggesting that there are DRD4-48 bp repeat gene-dependent effects on cerebellar structures. To further explore this issue, psychophysiological interactions were calculated. The results revealed that the 7-repeat non-carriers exhibited greater coupling between cerebellar activity and haemodynamic responses in the ACC and IFG, than did the 7-repeat carriers. This difference in neural connectivity between the cerebellum and brain regions with dense DRD4 receptor expression may help to explain the influence of the DRD4-48 bp repeat gene on cerebellar activity during the TT.
Unfortunately, to date no other functional neuroimaging study has investigated the association between the same DRD4 polymorphism and neural networks of executive functions. Therefore, replication of this study is urgently needed. However, the results are in line with previous fMRI studies that demonstrated the importance of genetic differences of D2-type receptors, to which the D4 receptor belongs on neural networks associated with planning, feedback-based learning and reward (Camara et al., 2010, Fan et al., 2003, Klein et al., 2007). For example, Fan et al. (2003) studied the insertion/deletion of a guanosine residue at the upstream position −1217 of the DRD4 gene and found greater conflict-related brain activity in the ACC in participants carrying the insertion variant of the polymorphism. Moreover, Klein et al. (2007) demonstrated that presence of the A1-allele, leading to a reduced receptor density, is associated with a reduced BOLD response to negative feedback in the medial prefrontal cortex. In line with this, carriers of the DRD4 allele in our study showed a reduced response to the cognitive stimuli of both our tasks, which might be related to the 7-repeat allele's modulation of subsensitivity of the DRD4 receptor. Although the transcriptional effects of this polymorphism are not completely known (Ogawa, 1995, Kereszturi et al., 2006) one might speculate that a subsensitive dopamine receptor might result in a reduced neural response to cognitive stimuli as well as altered connectivity between different brain regions. This line of argumentation could also support those neuropsychological findings that demonstrated the presence of the 7-repeat allele as a risk factor for impairments in executive functions and may go hand in hand with the associations between this polymorphism and ADHD (Faraone and Doyle, 2001, Li et al., 2006).
However, in our study we could not replicate the behavioural results found in neuropsychological studies that investigated differences between carriers of the DRD4 7-repeat allele and carriers of other gene variants. This lack of behavioural differences could probably stem from the lack of power or could be due to the fact that our sample was composed of healthy subjects whereas other neuropsychological studies were conducted mainly with patients with ADHD. However, given the fact that behavioural results have been contradictory so far, the lack of behavioural differences in the present study gave us the opportunity to focus on functional differences in haemodynamic responses without accounting for behavioural variation. As has been shown earlier (Fink et al., 1996, Fink et al., 2002) functional neuroimaging can unravel differences in neural processes that remain undetected by neuropsychological measures. Additionally, it has to be noted that in previous studies children or adolescents with ADHD were compared to each other or healthy controls regarding their genotype whereas we compared a sample of healthy participants. Thus, we avoided to confound diagnosis with genotype effects.
Our results confirmed the hypothesis on the influence of the DRD4-48 bp repeat gene on neural activation patterns and task-dependent connectivity patterns between brain areas associated with executive functions. Modulation of cerebellar activity by the DRD4-48 bp repeat gene could be explained by genotype-dependent differences in functional coupling between the cerebellum and the ACC, IFG. Thus, our connectivity analyses provide important information by showing that there is an association between genotype and the functional connectivity between cerebellar activation and activation in brain areas that have a high density of DRD4 receptors. However, given the more liberal threshold chosen for the connectivity analyses (not corrected for multiple comparisons), our findings have to be considered with caution and require replication in independent samples.
Overall, our findings confirm an influence of DRD4-genotype on prefrontal functioning during typical development which has not been shown consistently in neuropsychological measures. Thus, the inclusion of neuroimaging will remain important and indispensable to further explore the relationship between genotype and executive functions.
Conflict of interest
The authors declare that they have no conflicts of interest.
Acknowledgement
This study was supported by a grant to K.K. and B.H.-D. by the Deutsche Forschungsgemeinschaft (DFG-KFO112-II, TP 5).
References
- Ariano M.A., Wang J., Noblett K.L., Larson E.R., Sibley D.R. Cellular distribution of the rat D4 dopamine receptor protein in the CNS using anti-receptor antisera. Brain Research. 1997;752:26–34. doi: 10.1016/s0006-8993(96)01422-9. [DOI] [PubMed] [Google Scholar]
- Asghari V., Sanyal S., Buchwaldt S., Paterson A., Jovanovic V., Van Tol H.H. Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. Journal of Neurochemistry. 1995;65:1157–1165. doi: 10.1046/j.1471-4159.1995.65031157.x. [DOI] [PubMed] [Google Scholar]
- Banaschewski T., Becker K., Scherag S., Franke B., Coghill D. Molecular genetics of attention-deficit/hyperactivity disorder: an overview. European Child and Adolescent Psychiatry. 2010;19:237–257. doi: 10.1007/s00787-010-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley R.A., Smith K.M., Fischer M., Navia B. An examination of the behavioral and neuropsychological correlates of three ADHD candidate gene polymorphisms (DRD4 7+, DBH TaqI A2, and DAT1 40 bp VNTR) in hyperactive and normal children followed to adulthood. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2006;141 doi: 10.1002/ajmg.b.30326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara E., Krämer U.M., Cunillera T., Marco-Pallares J., Cucurell D., Nager W., Mestres-Misse A., Bauer P., Schüle R., Schöls L., Tempelmann C., Rodriguez-Fornells A., Münte T.F. The effects of COMT (Val108/158Met) and DRD4 (SNP −521) dopamine genotype on brain activations related to valence and magnitude of rewards. Cerebral Cortex. 2010;20:1985–1996. doi: 10.1093/cercor/bhp263. [DOI] [PubMed] [Google Scholar]
- Casey B.J., Thomas K.M., Davidson M.C., Kunz K., Franzen P.L. Dissociating striatal and hippocampal function developmentally with a stimulus-response CT. Journal of Neuroscience. 2002;22:8647–8652. doi: 10.1523/JNEUROSCI.22-19-08647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson M.C., Amso D., Anderson L.C., Diamond A. Development of cognitive control and executive functions from 4 to 13 years: evidence from manipulations of memory, inhibition, and task switching. Neuropsychologia. 2006;44:2037–2078. doi: 10.1016/j.neuropsychologia.2006.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo N., Chamberlain S.R., Sahakian B.J., Robbins T.W. The roles of dopamine and noradrenaline in the pathophysiology and treatment of attention-deficit/hyperactivity disorder. Biological Psychiatry. 2011:15145–15157. doi: 10.1016/j.biopsych.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Dickstein S.G., Bannon K., Castellanos F.X., Milham M.P. The neural correlates of attention deficit hyperactivity disorder: an ALE meta-analysis. Journal of Child Psychology and Psychiatry and Allied Disciplines. 2006;47:1051–1062. doi: 10.1111/j.1469-7610.2006.01671.x. [DOI] [PubMed] [Google Scholar]
- Durston S., Casey B.J. What have we learned about cognitive development from neuroimaging? Neuropsychologia. 2006;44:2149–2157. doi: 10.1016/j.neuropsychologia.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Eickhoff S.B., Stephan K.E., Mohlberg H., Grefkes C., Fink G.R., Amunts K., Zilles K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Fan J., Fossella J., Sommer T., Wu Y., Posner M.I. Mapping the genetic variation of executive attention onto brain activity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7406–7411. doi: 10.1073/pnas.0732088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone S.V., Doyle A.E. The nature and heritability of attention-deficit/hyperactivity disorder. Child and Adolescent Psychiatric Clinics of North America. 2001;10:299–316. [PubMed] [Google Scholar]
- Fink G.R., Halligan P.W., Marshall J.C., Frith C.D., Frackowiak R.S., Dolan R.J. Where in the brain does visual attention select the forest and the trees? Nature. 1996;382:626–628. doi: 10.1038/382626a0. [DOI] [PubMed] [Google Scholar]
- Fink G.R., Marshall J.C., Weiss P.H., Toni I., Zilles K. Task instructions influence the cognitive strategies involved in line bisection judgements: evidence from modulated neural mechanisms revealed by fMRI. Neuropsychologia. 2002;40:119–130. doi: 10.1016/s0028-3932(01)00087-2. [DOI] [PubMed] [Google Scholar]
- Forbes E.E., Brown S.M., Kimak M., Ferrell R.E., Manuck S.B., Hariri A.R. Genetic variation in components of dopamine neurotransmission impacts ventral striatal reactivity associated with impulsivity. Molecular Psychiatry. 2009;14:60–70. doi: 10.1038/sj.mp.4002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J., Holmes A.P., Poline J.B., Grasby P.J., Williams S.C., Frackowiak R.S., Turner R. Analysis of fMRI time-series revisited. NeuroImage. 1995;2:45–53. doi: 10.1006/nimg.1995.1007. [DOI] [PubMed] [Google Scholar]
- Goghari V.M., MacDonald A.W., III The neural basis of cognitive control: response selection and inhibition. Brain and Cognition. 2009;71:72–83. doi: 10.1016/j.bandc.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg T.E., Weinberger D.R. Genes and the parsing of cognitive processes. Trends in Cognitive Sciences. 2004;8:325–335. doi: 10.1016/j.tics.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Hinney A., Schneider J., Ziegler A., Lehmkuhl G., Poustka F., Schmidt M.H., Mayer H., Siegfried W., Remschmidt H., Hebebrand J. No evidence for involvement of polymorphisms of the dopamine D4 receptor gene in anorexia nervosa, underweight and obesity. American Journal of Medical Genetics. 1999;88:594–597. [PubMed] [Google Scholar]
- Ivry R.B., Fiez J.A. Cerebellar contributions to cognition and imagery. In: Gazzaniga M., editor. The Cognitive Neurosciences. second edition. MIT Press; Cambridge, MA: 2000. pp. 999–1011. [Google Scholar]
- Johnson K.A., Kelly S.P., Robertson I.H., Barry E., Mulligan A., Daly M., Lambert D., McDonnell C., Connor T.J., Hawi Z., Gill M., Bellgrove M.A. Absence of the 7-repeat variant of the DRD4 VNTR is associated with drifting sustained attention in children with ADHD but not in controls. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;88:594–597. doi: 10.1002/ajmg.b.30718. [DOI] [PubMed] [Google Scholar]
- Kaufman J., Birmaher B., Brent D., Rao U., Flynn C., Moreci P., Williamson D., Ryan N. Schedule for affective disorders and schizophrenia for school-age children-present and lifetime version (K-SADS-PL): initial reliability and validity data. Journal of the American Academy of Child and Adolescent Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
- Kereszturi E., Kiraly O., Barta C., Molnar N., Sasvari-Syekely M., Csapo Z. No direct effect of the −521 C\T polymorphism in the human dopamine D4 receptor gene promoter on transcriptional activity. BMC Molecular Biology. 2006;7:18. doi: 10.1186/1471-2199-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieling C., Roman T., Doyle A.E., Hutz M.H., Rohde L.A. Association between DRD4 gene and performance of children with ADHD in a test of sustained attention. Biological Psychiatry. 2006;60:1163–1165. doi: 10.1016/j.biopsych.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Klein T.A., Neumann J., Reuter M., Hennig J., von Cramon D.Y., Ullsperger M. Genetically determined differences in learning from errors. Science. 2007;318:1642–1645. doi: 10.1126/science.1145044. [DOI] [PubMed] [Google Scholar]
- Konrad K., Gunther T., Hanisch C., Herpertz-Dahlmann B. Differential effects of methylphenidate on attentional functions in children with attention-deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2004;43:191–198. doi: 10.1097/00004583-200402000-00015. [DOI] [PubMed] [Google Scholar]
- Konrad K., Neufang S., Thiel C.M., Specht K., Hanisch C., Fan J., Herpertz-Dahlmann B., Fink G.R. Development of attentional networks: an fMRI study with children and adults. Neuroimage. 2005;28:429–439. doi: 10.1016/j.neuroimage.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Konrad K., Dempfle A., Friedel S., Heiser P., Holtkamp K., Walitza S., Sauer S., Warnke A., Remschmidt H., Gilsbach S., Schäfer H., Hinney A., Hebebrand J., Herpertz-Dahlmann B. Familiarity and molecular genetics of attention networks in ADHD. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153:148–158. doi: 10.1002/ajmg.b.30967. [DOI] [PubMed] [Google Scholar]
- Langley K., Marshall L., van den B.M., Thomas H., Owen M., O’Donovan M., Thapar A. Association of the dopamine D4 receptor gene 7-repeat allele with neuropsychological test performance of children with ADHD. American Journal of Psychiatry. 2004;161:133–138. doi: 10.1176/appi.ajp.161.1.133. [DOI] [PubMed] [Google Scholar]
- Li D., Sham P.C., Owen M.J., He L. Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD) Human Molecular Genetics. 2006;15:2276–2284. doi: 10.1093/hmg/ddl152. [DOI] [PubMed] [Google Scholar]
- Lichter J.B., Barr C.L., Kennedy J.L., Van Tol H.H., Kidd K.K., Livak K.J. A hypervariable segment in the human dopamine receptor D4 (DRD4) gene. Human Molecular Genetics. 1993;2:767–773. doi: 10.1093/hmg/2.6.767. [DOI] [PubMed] [Google Scholar]
- Maldjian J.A., Laurienti P.J., Kraft R.A., Burdette J.H. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. NeuroImage. 2003;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Hidaka K., Akiho H., Tada S., Okada M., Yamaguchi T. Low stringency hybridization study of the dopamine D4 receptor revealed D4-like mRNA distribution of the orphan seven-transmembrane receptor, APJ, in human brain. Journal of Neurochemistry. 1996;66:915–919. doi: 10.1016/s0304-3940(96)13198-0. [DOI] [PubMed] [Google Scholar]
- Meador-Woodruff J.H. Update on dopamine receptors. Annals of Clinical Psychiatry. 1994;6:79–90. doi: 10.3109/10401239409148986. [DOI] [PubMed] [Google Scholar]
- Monk C.S., Telzer B.A., Mogg K., Bradley B.P., Mai X., Louro H.M.C., Chen G., McClure-Tone E.B., Ernst M., Pine D.S. Amygdala and ventrolateral prefrontal cortex activation to masked angry faces in children and adolescents with generalized anxiety disorder. Archives of General Psychiatry. 2008;65:568–576. doi: 10.1001/archpsyc.65.5.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuteaux M.C., Seidman L.J., Faraone S.V., Makris N., Spencer T., Valera E., Brown A., Bush G., Doyle A.E., Hughes S., Helliesen M., Mick E., Biederman J. A preliminary study of dopamine d4 receptor genotype and structural brain alterations in adults with ADHD. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147B:1436–1441. doi: 10.1002/ajmg.b.30870. [DOI] [PubMed] [Google Scholar]
- Moreland R.B., Terranova M.A., Chang R., Uchic M.E., Matulenko M.A., Surber B.W., Stewart A.O., Brioni J.D. [3H] A-369508 ([2-[4-(2-cyanophenyl)-1-piperazinyl]-N-(3-methylphenyl)acetamide): an agonist radioligand selctive for the dopamine D4 receptor. European Journal of Pharmacology. 2004;297:147–154. doi: 10.1016/j.ejphar.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Morimoto H.M., Hirose S., Chikazoe J., Jimura K., Asari T., Yamashita K.I., Miyashita Y., Konishi S. On verbal/nonverbal modality dependence of left and right inferior prefrontal activation during performance of flanker interference task. Journal of Cognitive Neuroscience. 2008;20:2006–2014. doi: 10.1162/jocn.2008.20138. [DOI] [PubMed] [Google Scholar]
- Mrzljak L., Bergson C., Pappy M., Huff R., Levenson R., Goldman-Rakic P.S. Localization of dopamine D4 receptors in GABAergic neurons of the primate brain. Nature. 1996;381:245–248. doi: 10.1038/381245a0. [DOI] [PubMed] [Google Scholar]
- Neufang S., Fink G.R., Herpertz-Dahlmann B., Willmes K., Konrad K. Developmental changes in neural activation and psychophysiological interaction patterns of brain regions associated with interference control and time perception. NeuroImage. 2008;43:399–409. doi: 10.1016/j.neuroimage.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Nichols T., Brett M., Andersson J., Wager T., Poline J.B. Valid conjunction inference with the minimum statistic. NeuroImage. 2005;25:653–660. doi: 10.1016/j.neuroimage.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Nigg J.T. On inhibition/disinhibition in developmental psychopathology: views from cognitive and personality psychology and a working inhibition taxonomy. Psychological Bulletin. 2000;126:220–246. doi: 10.1037/0033-2909.126.2.220. [DOI] [PubMed] [Google Scholar]
- Oak J.N., Oldenhof J., Van Tol H.H. The dopamine D4 receptor: one decade of research. European Journal of Pharmacology. 2000;405:303–327. doi: 10.1016/s0014-2999(00)00562-8. [DOI] [PubMed] [Google Scholar]
- Ogawa N. Molecular and chemical neuropharmacology of dopamine receptor subtypes. Acta Medica Okayama. 1995;49:1–11. doi: 10.18926/AMO/30418. [DOI] [PubMed] [Google Scholar]
- Osaka N., Osaka M., Kondo H., Morishita M., Fukuyama H., Shibasaki H. The neural basis of executive function in working memory: an fMRI study based on individual differences. NeuroImage. 2004;21:623–631. doi: 10.1016/j.neuroimage.2003.09.069. [DOI] [PubMed] [Google Scholar]
- Rubia K., Smith A. The neural correlates of cognitive time management: a review. Acta Neurobiologiae Experimentalis. 2004;64:329–340. doi: 10.55782/ane-2004-1517. [DOI] [PubMed] [Google Scholar]
- Rubia K., Halari R., Christakou A., Taylor E. Impulsiveness as a timing disturbance: neurocognitive abnormalities in attention-deficit hyperactivity disorder during temporal processes and normalization with methylphenidate. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences. 2009;364:1919–1931. doi: 10.1098/rstb.2009.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S., Van Tol H.H. Review the role of dopamine D4 receptors in schizophrenia and antipsychotic action. Journal of Psychiatric Research. 1997;31:219–232. doi: 10.1016/s0022-3956(96)00039-8. [DOI] [PubMed] [Google Scholar]
- Schoots O., Van Tol H.H. The human dopamine D4 receptor repeat sequences modulate expression. Pharmacogenomics. 2003:343–348. doi: 10.1038/sj.tpj.6500208. [DOI] [PubMed] [Google Scholar]
- Seeman P., Guan H.C., Van Tol H.H. Dopamine D4 receptors elevated in schizophrenia. Nature. 1993;365:441–445. doi: 10.1038/365441a0. [DOI] [PubMed] [Google Scholar]
- Simpson J., Vetuz G., Wilson M., Brookes K.J., Kent L. The DRD4 receptor Exon 3 VNTR and 5′ SNP variants and mRNA expression in human post-mortem brain tissue. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153:1228–1233. doi: 10.1002/ajmg.b.31084. [DOI] [PubMed] [Google Scholar]
- Smith A., Taylor E., Lidzba K., Rubia K. A right hemispheric frontocerebellar network for time discrimination of several hundreds of milliseconds. NeuroImage. 2003;20:344–350. doi: 10.1016/s1053-8119(03)00337-9. [DOI] [PubMed] [Google Scholar]
- Strange P.G. Dopamine receptors in the basal ganglia: relevance to Parkinson's disease. Movement Disorders. 1993;8:263–270. doi: 10.1002/mds.870080303. [DOI] [PubMed] [Google Scholar]
- Swanson J., Oosterlaan J., Murias M., Schuck S., Flodman P., Spence M.A., Wasdell M., Ding Y., Chi H.C., Smith M., Mann M., Carlson C., Kennedy J.L., Sergeant J.A., Leung P., Zhang Y.P., Sadeh A., Chen C., Whalen C.K., Babb K.A., Myzis R., Posner M.I. Attention deficit/hyperactivity disorder children with a 7-repeat allele of the dopamine receptor D4 gene have extreme behavior but normal performance on critical neuropsychological tests of attention. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4754–4759. doi: 10.1073/pnas.080070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewes U., Rossman P., Schallberger U. Huber; Bern: 1999. Hamburg-Wechsler-Intelligenztest für Kinder (HAWIK-IIIR) [Google Scholar]
- Thapar A., O’Donovan M., Owen M.J. The genetics of attention deficit hyperactivity disorder. Human Molecular Genetics. 2005;14:R275–R282. doi: 10.1093/hmg/ddi263. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Uncapher M.R., Hutchinson J.B., Wagner A.D. Dissociable effects of top-down and bottom-up attention during episodic encoding. Journal of Neuroscience. 2011;31:12613–12628. doi: 10.1523/JNEUROSCI.0152-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tol H.H., Wu C.M., Guan H.C., Ohara K., Bunzow J.R., Civelli O., Kennedy J., Seeman P., Niznik H.B., Jovanovic V. Multiple dopamine D4 receptor variants in the human population. Nature. 1992;358:149–152. doi: 10.1038/358149a0. [DOI] [PubMed] [Google Scholar]
- Vloet T.D., Gilsbach S., Neufang S., Fink G.R., Herpertz-Dahlmann B., Konrad K. Neural mechanisms of interference control and time discrimination in attention-deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2009;49:356–367. [PubMed] [Google Scholar]
- Weinberger D.R., Egan M.F., Bertolino A., Callicott J.H., Mattay V.S., Lipska B.K., Berman K.F., Goldberg T.E. Prefrontal neurons and the genetics of schizophrenia. Biological Psychiatry. 2001;50:825–844. doi: 10.1016/s0006-3223(01)01252-5. [DOI] [PubMed] [Google Scholar]