Summary
The mammalian brain contains few niches for neural stem cells (NSCs) capable of generating new neurons, whereas other regions are primarily gliogenic. Here we leverage the spatial separation of the sub-ependymal zone NSC niche and the olfactory bulb, the region to which newly generated neurons from the sub-ependymal zone migrate and integrate, and present a comprehensive proteomic characterization of these regions in comparison to the cerebral cortex, which is not conducive to neurogenesis and integration of new neurons. We find differing compositions of regulatory extracellular matrix (ECM) components in the neurogenic niche. We further show that quiescent NSCs are the main source of their local ECM, including the multi-functional enzyme transglutaminase 2, which we show is crucial for neurogenesis. Atomic force microscopy corroborated indications from the proteomic analyses that neurogenic niches are significantly stiffer than non-neurogenic parenchyma. Together these findings provide a powerful resource for unraveling unique compositions of neurogenic niches.
Keywords: extracellular matrix, tissue stiffness, transglutaminase2, C1ql3, S100a6, neuroblast, transit-amplifying progenitor, cerebral cortex, olfactory bulb, subventricular zone
Graphical Abstract
Highlights
-
•
Proteomics define the NSC niche-specific extracellular matrix
-
•
Detergent-solubility profiling reveals extracellular matrix architecture
-
•
Transglutaminase 2 regulates neurogenesis
-
•
Stiffness is increased in the neurogenic niches of the brain
The physical properties of stem cell niches are thought to mediate important regulatory functions. Here we provide a proteomic resource of the neural stem cell niche in comparison to gliogenic brain parenchyma, highlighting stiffness and the enzyme transglutaminase 2 as key regulators of neurogenesis.
Introduction
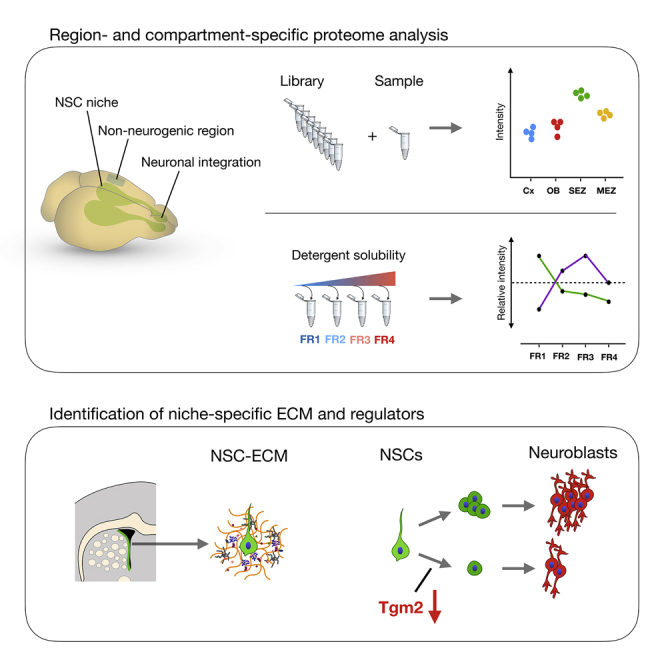
Adult stem cell niches provide complex regulatory architectures that maintain the stem cell population and prevent terminal differentiation (Crane et al., 2017, Donnelly et al., 2018, Gonzales and Fuchs, 2017, Meran et al., 2017, Vining and Mooney, 2017). In the adult mammalian brain, the neural stem cell (NSC) niches are unique in supporting neurogenesis (Conover and Todd, 2017, Ruddy and Morshead, 2018), as only gliogenesis takes place in the remainder of the brain (see e.g., Götz et al., 2016, Lim and Alvarez-Buylla, 2016). The brain parenchyma outside these niches (e.g., neocortex; Figure 1A) even limits the neurogenic potential of transplanted NSCs or neuroblasts (Barker et al., 2018, Winkler et al., 1998, Englund et al., 2002, Fricker et al., 1999, Seidenfaden et al., 2006). The largest NSC niche, the sub-ependymal zone (SEZ), generates neuroblasts that migrate to the distant olfactory bulb (OB), where they differentiate and integrate (Figures 1B and 1C) (Lim and Alvarez-Buylla, 2016). This spatial separation allows the determination of the composition of the NSC niche (SEZ) that maintains the stem cells and generation of neuroblasts in comparison to the OB niche as an environment fostering neuronal differentiation and integration of new neurons.
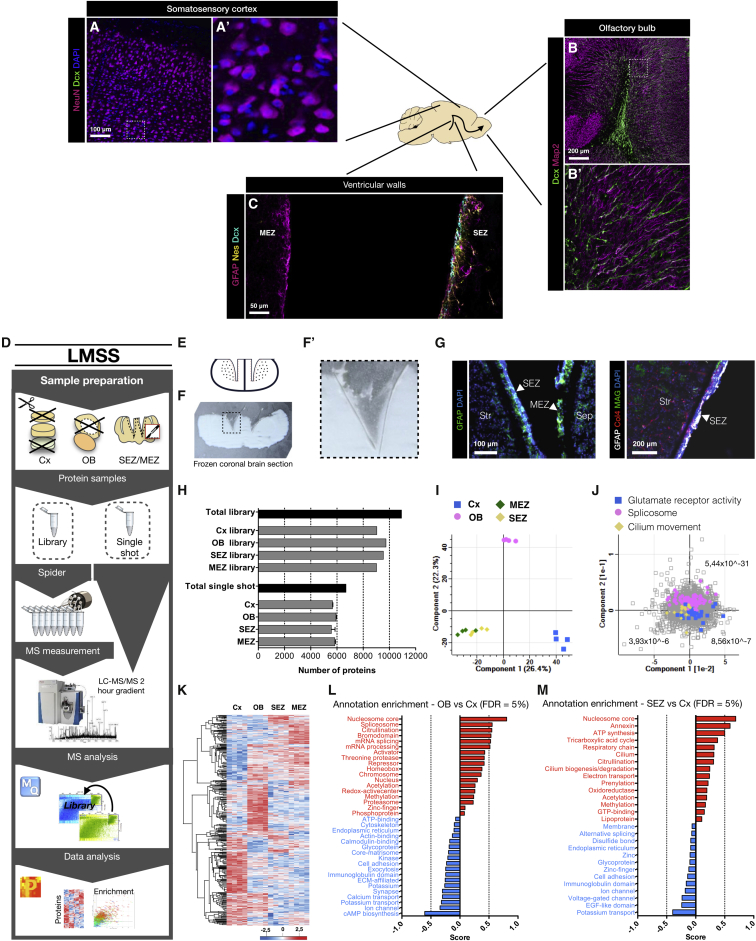
Figure 1.
High-Resolution Proteome of the Somatosensory Cortex and Neurogenic Niches
(A–C) The schematic drawing indicates a sagittal section of the adult murine brain with example photomicrographs of the regions used in this analysis—the non-neurogenic somatosensory cortex (A), the olfactory bulb (OB), where new neurons (labeled for doublecortin [Dcx]) integrate (B), and the lateral sub-ependymal zone (SEZ) where most NSCs reside, whereas only a few are located in the medial sub-ependymal zone (MEZ) (C). Sections were immunostained as indicated in the panels and are confocal z stacks.
(D) Experimental workflow using loss-less nano-fractionation for library-matched single shot measurements.
(E) Schematic of the high-precision cryo-dissection of the SEZ and the MEZ.
(F) Picture of a 50-μm frozen coronal section (white, ventral down) with cortex, corpus callosum, and choroid plexus removed. (F′) shows magnification of the dissected region visible as a thin gray line.
(G) Photomicrographs of cryo-dissected SEZ and MEZ (separated from striatum [Str] and septum [Sep]) stained for GFAP and DAPI (left panel) and cryo-dissected SEZ stained for GFAP, collagen 4 (Col4), myelin-associated glycoprotein (MAG), and DAPI (right panel).
(H) Number of proteins quantified in the library sample measurements and the library-matched single shot (LMSS) sample measurements for each region. Data are shown as mean ± standard deviation (n = 1 library sample per region, n = 4 single shot samples per region). See also Figures S1A–S1D.
(I) Principal component analysis (PCA) for each brain region. Components 1 and 2 separate the main regions. The SEZ and the MEZ are similar in these components.
(J) Colors indicate three categories that are enriched, respectively, in the Cx, the OB, and both the SEZ and the MEZ (FDR is presented for each category).
(K) Heatmap of 4,786 proteins found to be of different abundance comparing the four brain regions (n = 4 per region). Intensities are based on label-free quantification (LFQ) intensities after unsupervised hierarchical clustering (ANOVA with Benjamin-Hochberg post hoc test, FDR = 0.05).
(L) The datasets were annotated with Uniprot keywords and the matrisome annotation (see STAR Methods). Enriched features of the OB in comparison to the Cx were then scored (0 to 1) and are displayed in a bar graph (1D-annotation enrichment, FDR = 0.05). Conversely, features with a negative score (0 to −1) are enriched in the Cx compared to the OB.
(M) Enriched features of the SEZ in comparison to the Cx were analyzed in the same manner (1D-annotation enrichment, FDR = 0.05).
Scale bars as indicated in the panels.
The SEZ, located at the lateral wall of the lateral ventricles that are lined by ependymal cells, harbors nestin+ quiescent NSCs that can be activated and give rise to transit-amplifying progenitors (TAPs) that then generate doublecortin (Dcx)-positive neuroblasts (Figure 1C). The medial side of the lateral ventricle, the medial sub-ependymal zone (MEZ), also shares ependymal cell lining and access to the cerebrospinal fluid (CSF). As it contains many fewer NSCs and Dcx+ neuroblasts (Figure 1C) than the SEZ, this region provides a useful comparative tissue to identify specific components of the NSC and neurogenesis-associated environment. Individual proteins and ECM structures (Kerever et al., 2007) have been described as specific for the NSC niches in the adult murine brain, such as the ECM proteins Tenascin-C (Tnc) and Thrombospondin 4 (Thbs4) that both may regulate aspects of migration (Faissner et al., 2017, Garcion et al., 2001, Girard et al., 2014, Kazanis et al., 2007) or the matrix-associated protein Plexin-b2 that regulates proliferation and migration (Saha et al., 2012). However, no comprehensive proteomic analysis of this important niche has yet been performed. Likewise, in the OB, two matrix proteins, Reelin (Reln) and Tenascin-R (Tnr), have been reported to regulate the change from tangential to radial neuroblast migration (Figure 1B) (David et al., 2013, Hack et al., 2002), and, besides neuronal activity, little is known about factors allowing the integration of new neurons into pre-existing networks in the adult brain (Hardy and Saghatelyan, 2017, Lledo and Valley, 2016).
In contrast to the above-described niches, the mature brain parenchyma allows neither neurogenesis nor integration of new neurons in the absence of injury (Frisén, 2016). It contains a specialized extracellular matrix called “perineuronal nets” (PNNs) that is built up around neurons late in development and serves to restrict neuronal plasticity (Deepa et al., 2006). As the NSC niche contains no neurons, and hence no PNNs, its ECM composition is expected to differ from the brain parenchyma, but the actual composition is largely unknown, as are potential differences in ECM composition between the OB and brain parenchyma.
The ECM, the “matrisome,” consists of core ECM proteins often forming ECM structures, such as the basement membrane (BM) and associated ECM proteins, many with primarily protein-regulatory functions. The latter is essential for the enrichment and function of many growth factors and other signaling factors and, hence, is of key interest to mediate regulatory functions of a local niche. The proteins associated with the structural ECM can be enriched through detergent de-cellularization (Naba et al., 2012), but this has not yet been done in the NSC niches.
In contrast to the paucity of proteome data, the transcriptomes of SEZ NSCs, their progeny, and the surrounding niche cells are well studied (Beckervordersandforth et al., 2010, Codega et al., 2014, Kalamakis et al., 2019, Llorens-Bobadilla et al., 2015). However, gene expression analysis has proven inadequate to fully describe the proteome, including the tissue matrix environment (Angelidis et al., 2019, Schiller et al., 2015). For example, neurogenic and neuronal mRNAs are upregulated in NSCs and progenitors, but translation is inhibited to avoid premature differentiation and allow amplification of the lineage, a phenomenon referred to as “lineage priming” (Baser et al., 2019, Beckervordersandforth et al., 2010, Götz et al., 2016, Lepko et al., 2019). Moreover, proteins may be derived from the CSF that is contributing to the maintenance of the SEZ stem cell niche (Lepko et al., 2019, Silva-Vargas et al., 2016). Determining the composition of the proteome, including the matrisome of the neurogenic niches, is thus important, particularly given the influence of the ECM composition on mechanical tissue properties, which regulate fate decisions of adult stem cells, such as muscle or mesenchymal SCs (Engler et al., 2006, Gilbert et al., 2010, Vining and Mooney, 2017). Neural crest SCs have been found to differentiate into smooth muscle cells on stiffer substrates and glial cells on softer substrates (Zhu et al., 2019), consistent with CNS tissue belonging to the softest tissues in our body (Franze et al., 2013). This softness of CNS tissue is most likely due to low expression of certain structural matrix constituents, such as collagen I, and the soft nuclear matrix of neurons, given their low content of lamin A (Swift et al., 2013). Tissue stiffness has been shown to potently influence neurite outgrowth (Koser et al., 2016, Stukel and Willits, 2018), and NSCs possess mechanosensitive ion channels whose activity affects their self-renewal and differentiation (Pathak et al., 2014, Petrik et al., 2018). However, the mechanical properties of the adult stem cell niche are currently unknown.
Here we set out to provide a first in-depth characterization of what makes the composition and architecture of the adult brain’s neurogenic niche uniquely different from the non-neurogenic brain parenchyma.
Results
High-Resolution Proteome Defines Niche-Specific Features
For ultra-deep proteomic assessment of the neurogenic niches, we used library-matched single shot (LMSS) proteomics (see STAR Methods). Cerebral cortex (Cx) gray matter (GM, omitting the white matter [WM] and meninges) was compared with the OB (also omitting the meninges), the SEZ, and the MEZ (Figure 1D). To obtain SEZ and MEZ samples, we performed cryo-micro-dissections on 100-μm-thick tissue sections after removing cortex with corpus callosum and choroid plexus (10 sections per sample) (Figures 1E, 1F, and 1F′). The combination of methods allowed very high precision and proteome-measurement depth of the small SEZ and MEZ regions, avoiding the myelin-associated-glycoprotein-rich (MAG+) brain parenchyma (Figure 1G).
We identified a total of 10,923 proteins in the library samples (four pooled tissue samples from each region) and 6,690 in LMSS samples (n = 4 per region) (Figure 1H; Table S1). Among the library-exclusive proteins, transcription factors (p = 1.68 × 10−6), cytokines, and neurogenesis-associated mitogens (e.g., interleukin-18, insulin growth factor, Vegf-α, etc.) were significantly enriched. As these are low-abundance proteins, in vivo proteome measurements of such factors have previously been unattainable. Our library measurements demonstrate that the mitogens and transcription factors known to be required for neurogenesis (e.g., Pax6) (Ninkovic et al., 2013) can be revealed and quantified in vivo with a proteome depth of 10,000 proteins (Figures S1A–S1D; Table S1).
The principal component analysis (PCA) of the four regions revealed that the SEZ and the MEZ have a more similar proteome than the other two regions (Figure 1I). An enriched common category was cilium movement (p = 3.93 × 10−6) (Figure 1J), highlighting that proteins from a single cell layer, the ependymal cells lining the ventricle, can be detected: e.g., Tektin (Tek1), a protein exclusive to ependymal cells and NSCs at the SEZ (https://shiny.mdc-berlin.de/SVZapp/).
In total, 4,786 proteins had a differential abundance among the four regions (ANOVA, FDR = 0.05) (Figure 1K). To identify features enriched in the neurogenic niche, we analyzed differences in protein abundance for either the OB or the SEZ in comparison to the Cx. Proteins were annotated with Uniprot keywords and the improved ECM annotation (http://matrisome.org; see STAR Methods). Enriched features of the OB included several nuclear and gene-regulatory processes (1D-annotation enrichment, FDR = 0.05) (Figures 1L and S1F; Table S2). This suggested that the OB has a larger proportion of gene-regulatory proteins, possibly because of the large population of maturing neuroblasts. Processes less pronounced in the OB compared to the Cx included synapse-associated features and core-matrisome proteins.
Proteins enriched in the SEZ, like in the OB, were associated with gene regulation and also oxidative phosphorylation (Figures 1M and S1E; Table S2), which is consistent with the fact that NSCs are largely glycolytic and the metabolism has to change as they differentiate into neuroblasts (Beckervordersandforth, 2017, Knobloch and Jessberger, 2017). Annexin-family proteins were found enriched in the SEZ compared to the Cx (Figure 1M), a notable observation given their importance in regulating the proliferation and migration of cancer cells (Lauritzen et al., 2015). Core matrisome proteins demonstrated the highest abundance in Cx (p ≤ 0.0001, Kruskal-Wallis test with Dunn’s multiple comparison test) (Figure 2A), and several proteins of the PNNs had higher abundance in the Cx and the MEZ compared to the SEZ and the OB (Figures 1L and 1M). None of the proteoglycans associated with migration during developmental neurogenesis, such as neurocan, aggrecan, or versican (Long and Huttner, 2019, Maeda, 2015), were an enriched component of the matrisome at the SEZ; instead, aggrecan was enriched in the OB in line with an association to neuroblasts (Figures 2B, 2C, and S2A–S2C). These data validate the quality of the dataset and provide a rich resource that can be accessed as a web-based database (https://neuronicheproteome.org).
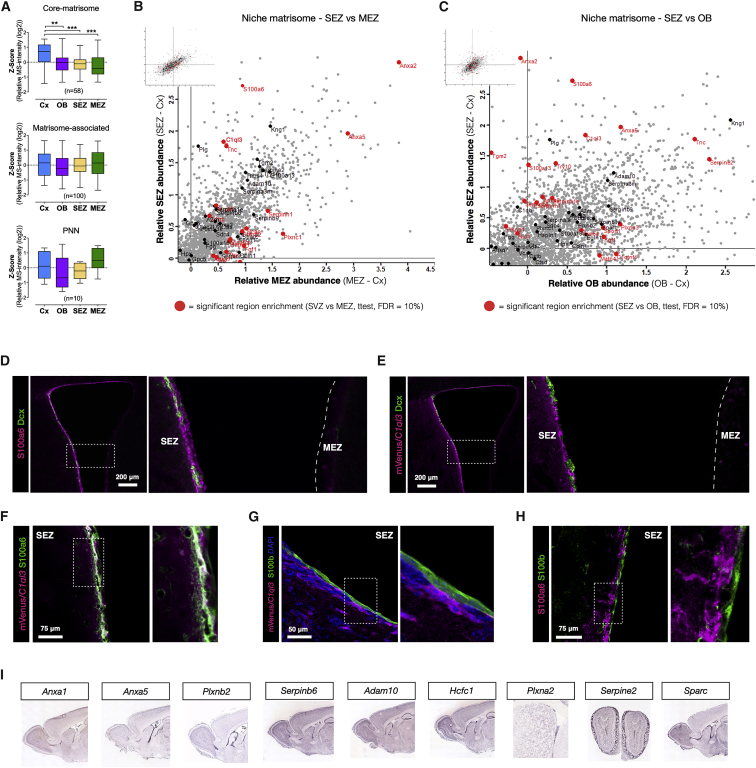
Figure 2.
Niche-Specific ECM and NSC Markers
(A) Distribution plots of each brain region in the different categories of the matrisome as indicated. Average LFQ intensities for each protein were Z scored and displayed in whisker plots (ANOVA, Kruskal-Wallis test with Dunn’s multiple comparison test, ∗p = 0.05, ∗∗p = 0.01, and ∗∗∗p = 0.001).
(B) Scatterplot with the matrisome (black) and matrisome significantly different (FDR ≤ 0.1) comparing the SEZ and the MEZ (red) highlighted.
(C) Scatterplot with the relative SEZ and OB values and significant differences (FDR ≤ 0.1) between intensities for the SEZ and the OB. Both plots highlight S100a6 and C1ql3 as enriched in the SEZ. See also Figures S2B and S2C.
(D–H) Photomicrographs of the ventricle and the SEZ and the MEZ from coronal brain sections of C57BL/6J mouse or mVenus/C1ql3 transcriptional reporter mouse immunostained as indicated. Note that S100a6 and C1ql3 are not found in Dcx+ neuroblasts or parenchymal astrocytes and, typically, neither in ependymal cells. Scale bars as indicated, and (D) and (E) are z stacks of confocal images, while (F)–(H) are single optical sections. See also Figures S3 and S4A–S4I.
(I) In situ hybridization shows mRNA expression in the SEZ. Image credit: Allen Institute for Brain Science.
Identification of Niche-Specific Marker Proteins
The abundance of neurogenesis-associated proteins was lower in the MEZ compared to the SEZ but still detectable, further supporting the depth of our analysis. One such protein is Tnc, the neurogenic niche-associated ECM protein discussed above (Roll and Faissner, 2014) that was enriched in the SEZ versus the MEZ, as expected (Figures 2A and 2B). We then compared the relative abundance of proteins in the SEZ, the MEZ, and the OB to the Cx as a control. These region-specific comparisons determined, S100a6 and C1ql3 among others, to be distinctly enriched among matrisome-associated proteins of the SEZ (Figures 2B, 2C, S2B, and S2C). We therefore aimed to determine which cells produce these SEZ-enriched proteins. Immunostaining for S100a6 was high in the SEZ and comparably absent in the MEZ (Figure 2D) and low to absent in astrocytes in the Cx parenchyma (Figures S3G and S3H), in agreement with our analysis. Excitingly, however, S100a6 staining labeled NSCs (slow dividing nestin+ cells in the SEZ; Figures S3B, S3C, and S3F), but not neuroblasts (Dcx+; Figures 2D, 2G, and S3A–S3C). Likewise, S100a6 staining also extends into the rostral migratory stream (RMS), co-localizing with GFAP+ cells, but not Dcx+ neuroblasts (Figures S3I and S3K). It is worth noting that NSCs, expressing GFAP, have been identified also at these more rostral positions (Alonso et al., 2008). Moreover, we also found S100a6+ GFAP+ cells in the WM (Figures S3E and S3J), consistent with the presence of some NSCs there (Lim and Alvarez-Buylla, 2016). Thus, S100a6 not only labels NSCs in the dentate gyrus (DG) (Yamada and Jinno, 2014) but more broadly allows distinguishing NSCs from astrocytes, which is important as especially reactive astrocytes and NSCs share most of the so-called “astrocyte markers” (Beckervordersandforth et al., 2010, Götz et al., 2015).
Interestingly, single-cell RNA sequencing (scRNA-seq) data from the SEZ (Kalamakis et al., 2019) also showed enrichment of S100a6 specifically in a primed-quiescent subtype of NSCs, qNSC2 (Figure S6A), identifying these cells as the main source for the specific enrichment of S100a6 in the SEZ. Most notably, this proved to be the case for most mRNAs encoding for matrix proteins that are highest in quiescent NSCs, whereas activated NSCs, TAPs, and neuroblasts express very few ECM components (Figure S6A). Thus, NSCs contribute to the composition of their own niche (see also Faissner et al., 2017).
Some proteins enriched in the SEZ were also enriched at mRNA levels as seen by in situ hybridizations in the Allen Brain Atlas (Figures 2I and S4J). The systematic comparison of RNA and protein enrichment in the SEZ compared to the Cx and the OB, however, showed profound differences (Figures S6B and S6C; Table S5). For example, some RNAs were expressed at significantly higher levels in the SEZ compared to the Cx, but the respective proteins were lower abundance or not much different between the SEZ compared to the Cx (Figure S6B; Table S6). This is to be expected from the “lineage priming” as introduced above (Beckervordersandforth et al., 2010, Götz et al., 2016, Lepko et al., 2019) and would lead to “false positives” if relying on RNA data only. Even more misleading could be the class of proteins that we found enriched in the SEZ compared to the Cx, which had lower mRNA levels in SEZ compared to the Cx (Figure S6B). For example, the C1ql3 protein is enriched at protein level in the SEZ, but mRNA is lower in the SEZ compared to the Cx. Moreover, C1ql3 was not detectable among the stem cell stages in the published scRNA-seq data (Figure S6C). This prompted us to examine the source of this protein in the SEZ.
To do so, we took advantage of the C1ql3-mVenus reporter mouse line (Martinelli et al., 2016). With this allele, any cell that expresses C1ql3 is marked by expression of cytoplasmic mVenus (not a fusion protein). In brain, C1ql3 has thus far been identified as a synaptic protein and, accordingly, mVenus+ cells in this reporter line in the brain parenchyma are mostly neurons (Martinelli et al., 2016). In the SEZ, however, C1ql3-mVenus+ cells were Dcx negative, i.e., not neuroblasts but nestin+ and S100a6+ NSCs (Figures 2E, 2F, S4A–S4C, and S4F). Conversely, we could not detect mVenus+ astrocytes in the brain parenchyma and the OB (Figures S4D, S4E, and S4G–S4I). Ependymal cells located at the ventricle and S100b+ did not contain S100a6 levels above background nor co-localized with C1ql3-mVenus (Figures 2G and 2H). Hence our analysis not only identified two SEZ-enriched NSC proteins whose role may extend beyond being niche-specific marker proteins but also showed a multitude of SEZ enriched proteins that were not detected by RNA analysis (Figures S6B and S6C).
Biochemical Profiling of the Neurogenic Niche-Specific Matrisome
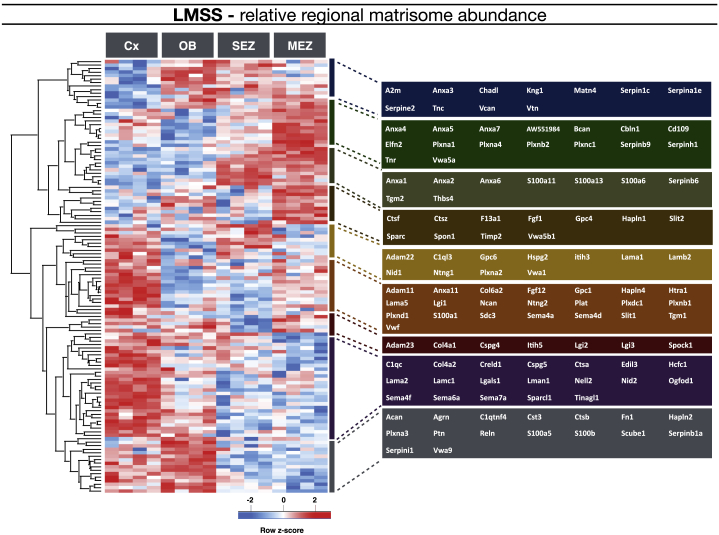
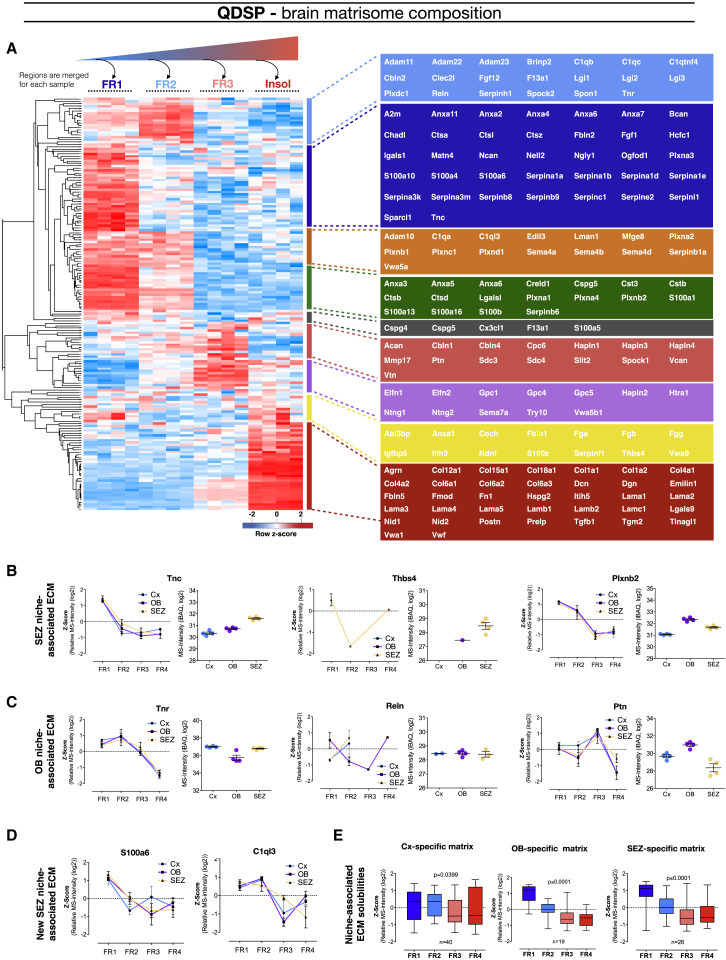
Here we present an overview of the regional distribution of all matrisome proteins with significant abundance differences across conditions (Figure 3; unsupervised hierarchical clustering, Pearson correlation). The OB and the SEZ are particularly rich in serpins, vitronectin, and Tnc, whereas the SEZ and the MEZ share high levels of annexin and several S100 proteins (Figure 3). Interestingly, annexin and S100 proteins interact and regulate a variety of processes, including membrane fusion and repair (Jaiswal and Nylandsted, 2015). These may be of particular relevance in migrating cells present in both of these regions. Additional ECM-associated proteins in both the SEZ and the MEZ included the well-known Thrombospondin 4 (Thbs4) and the ECM cross-linking protein Transglutaminase 2 (Tgm2) that has so far not been described at protein level in the neurogenic niches (see below).
Figure 3.
Regional Matrisome Distribution and Neurogenic Niche-Specific Matrisome
We compared 158 matrisome proteins and 78 of these had a significantly different distribution in the respective regions, of which the somatosensory cortex was found to be most abundant with extracellular matrix proteins. The heatmap displays unsupervised hierarchical clustering of the matrisome proteins with significantly different abundance when comparing the four brain regions (ANOVA with Benjamin-Hochberg post hoc test, FDR = 0.05). Members of different clusters (indicated by bars on the right of the heatmap) are listed on the further right of the heatmap in colored areas.
Matrisome proteins provide structural support and tether cell-surface proteins or soluble growth factors and thereby regulate their signaling functions. A way of experimentally enriching ECM proteins is to de-cellularize tissue using detergents and analyze the detergent-insoluble ECM components (Naba et al., 2012). However, such a method inherently loses ECM-affiliated proteins that associate with weaker affinity with the ECM. In contrast, methods that use protein-abundance correlation across fractions of protein derived by stepwise centrifugation or differential detergent extraction can be used to assess biochemical properties of proteins, including their association strength with ECM, in a proteome-wide fashion. We therefore used the quantitative detergent solubility profiling (QDSP) method (Schiller et al., 2015) and sequentially separated Cx, OB, and SEZ tissue samples into four protein fractions.
Tissue proteins were separated by extraction with increasing stringency using consecutive extraction with four distinct detergent mixtures (Figure 4A), leading to the identification of 8,308 proteins (Figure 4B; Table S3). For comparison of protein solubility profiles, their intensities were Z scored within each region and relative protein abundance was compared in the four fractions by averaging the profile between regions to permit “brain” solubility profile comparisons for specific categories. As expected, we found “brain” core matrisome proteins, such as collagens and laminins, to be enriched in the fourth fraction and thus more insoluble, whereas matrisome-associated proteins, such as S100 proteins and serpins, were most abundant in the first fraction and thus more soluble (p ≤ 0.0001, Kruskal-Wallis test) (Figures 4C, S5A, and S5B). Notably, the brain-specific ECM proteins associated with the PNNs (Figures 4C and S2G) form a soft structural matrix in the Cx, since they typically enrich in fraction three with much lower abundance in the insoluble fraction four (p = 0.0002, Kruskal-Wallis test). The PNN proteins neurocan (Ncan) and hyaluronan and proteoglycan link protein 1 (Hapln1) were distinctly more soluble in the OB compared to other regions (Figure S2G), an observation possibly related to the integration of new neurons into the network and a high degree of synaptogenesis.
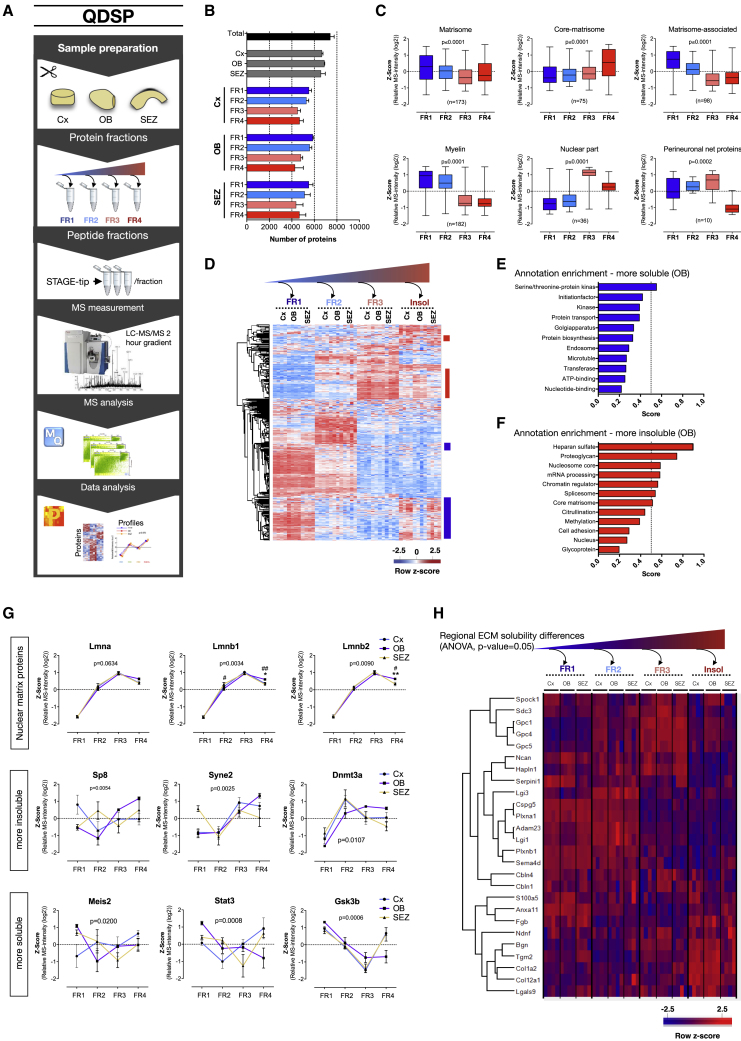
Figure 4.
Compartment Analysis with In-Depth Quantitative Proteomes of the Somatosensory Cortex and the Neurogenic Niches
(A) With stepwise de-cellularization we determined insoluble and various diffusible grades of ECM and other cellular compartment-associated proteins.
(B) Total number of quantified proteins for all regions (top, black and gray) and proteins quantified in each detergent fraction from each of the three brain regions (bottom, color). Each sample fraction is shown as mean ± standard deviation (n = 4 in each brain region).
(C) Solubility profile overview and distribution plot for the proteins in the displayed categories. Abundances were Z scored and then averaged for each protein in these categories shown in whisker plots with number of proteins in each category displayed in the graphs. Insoluble proteins distribute more toward fraction four and soluble proteins distribute toward fraction one (significance analyzed with Kruskal-Wallis test with Dunn’s multiple comparison test). See also Figures S2G, S5A, and S5B.
(D) Heatmap of 1,216 proteins with significantly different solubility among our three regions (FDR ≥ 0.05).
(E and F) Significantly enriched features among the more soluble (E) and insoluble (F) proteins in the OB when compared to the Cx using the relative difference of the LFQ intensities in the fourth fraction (1D-annotation enrichment, FDR = 0.05). The dataset was annotated with Uniprot keywords, matrisome, and a custom perineural nets annotation (see STAR Methods).
(G) From the relatively more soluble and insoluble proteins in the OB, we display the quantitative profile of lamins of the nuclear matrix and neurogenesis-associated proteins (two-way ANOVA). Data are presented as mean ± SEM.
(H) Matrisome proteins with significantly different solubility profiles comparing the three brain regions (Z scored LFQ intensity values, two-way ANOVA, p ≥ 0.05). Rows have undergone unsupervised hierarchical clustering.
Comparing all solubility profiles between the three regions, we found 1,208 proteins to have significantly different profiles (two-way ANOVA, p ≤ 0.05) (Figure 4D). Unsupervised hierarchical cluster analysis revealed the OB to have many profiles that differ in their solubility from Cx and the SEZ, in particular in the fourth fraction. Since such solubility shifts may contain information concerning protein functions, we assessed the cellular features associated with proteins either more or less soluble (only fourth fraction) in the OB compared to the Cx (1D-annotation analysis, FDR = 0.05). Enriched features among the more insoluble proteins in the OB were associated with gene regulation and cell adhesion, as well as core-matrisome proteins and proteoglycans (Figure 4F; Table S4). Lamin b1 and Lamin b2 were significantly more insoluble in the OB, suggesting that some or many cells have a nuclear matrix that is more insoluble (Figure 4G; Table S4). It should be noted that many, but not all, gene regulatory-associated proteins had a more insoluble profile in the OB, highlighting possible differences in nuclear architecture that remain to be explored. Along these lines, we observed that the presence of transcriptional and epigenetic regulators in different solubility fractions with tissue specificity, e.g., Sp8 and Dnmt3a, were less soluble, whereas Stat3, Meis2, and Gsk3b were more soluble in SEZ tissue (Figure 4G). Meis2 is an important regulator of neurogenesis in the SEZ (Kolb et al., 2018), prompting the suggestion that different solubility may reveal tissue-specific differences in transcriptional function. Another category in the more insoluble group of the OB included matrisome proteins, e.g., all three detected glypicans (Gpc1, Gpc4, and Gpc5) (Figure 4H). Thus, its less soluble ECM and nuclear lamina predict potentially higher tissue stiffness of the OB, a prediction that we tested and confirmed below.
Next, we focused on the solubility profiles of ECM components, providing a heatmap of the “brain” matrisome solubility profiles (Figure 5A; unsupervised hierarchical clustering, Pearson correlation). This highlights that familiar neurogenic niche ECM proteins are surprisingly soluble (Figures 5B and 5C). Interestingly, the solubility profile of C1ql3 in the SEZ is distinctly different from the one in the OB and the Cx (Figure 5D), suggesting its localization in a different more soluble compartment compared to its normal synaptic membrane association. Indeed, matrisome proteins were generally more enriched in the soluble compartments of neurogenic niche-specific ECM-associated proteins (p ≤ 0.0001, Kruskal-Wallis test) in the SEZ (n = 26) or the OB (n = 19) compared to the Cx in the LMSS data (FDR = 0.1) (Figure 5E). The solubility of proteins enriched in the Cx (n = 40) compared to either the SEZ or the OB (FDR = 0.1) in contrast had either a more membrane-associated or non-diffusible solubility profile. The structural ECM is thus not enriched in the neurogenic niche, including the typical “soft” structural brain ECM, although individual proteins with these properties can be found enriched in both the SEZ and the OB. Reln is uniquely insoluble in the OB (Figure 5C), whereas Tgm2 is one of the few proteins that was enriched in the SEZ (and the MEZ) that has a largely insoluble profile (Figure 5A).
Figure 5.
Brain- and Niche-Matrisome Composition
(A) The matrisome protein solubility profiles are displayed using unsupervised hierarchical clustering of the detergent solubility profiles derived from averaged Z scores from each brain region (the Cx, the OB, and the SEZ).
(B–D) Detergent solubility profiles for the SEZ-associated ECM proteins (B) Tenascin-C (Tnc), Thrombospondin-4 (Thbs4), and Plexin-b2 (Plxnb2); the OB-associated proteins (C) Tenascin-R (Tnr), Reelin (Reln), and Pleiotrophin (Ptn); and the two neurogenic niche-specific proteins (D) S100a6 and C1ql3 (p = 0.0948). Data are presented as mean ± SEM.
(E) Solubility profiles for Cx-, SEZ-, and OB-enriched matrisome proteins shown in whisker plots (ANOVA, p values in graphs).
Transglutaminase 2 Regulates Adult Neurogenesis
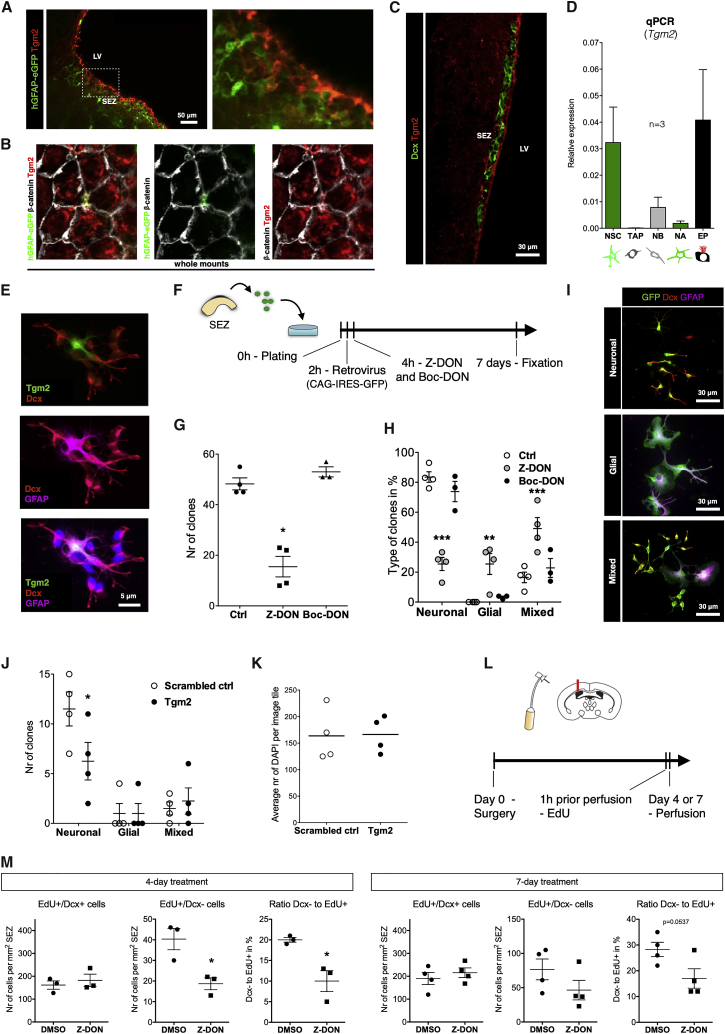
Given the specific enrichment and solubility profile of Tgm2 in both the SEZ and the MEZ (Figure S5D), we determined its cellular origin. Tgm2 immunoreactivity was found in ependymal cells (Figures 6A and 6B) and NSCs (hGFAP-GFP+; see Beckervordersandforth et al., 2010, Codega et al., 2014) that extend an apical process to the ventricle (Figure 6B), but not in neuroblasts (Dcx+) (Figure 6C). Consistent with ependymal cells and NSCs containing the highest protein levels, these cells isolated by fluorescence-activated cell sorting (Beckervordersandforth et al., 2010, Fischer et al., 2011) also had high Tgm2 mRNA levels, whereas TAPs and neuroblasts contained very low mRNA levels (Figure 6D). Notably, Tgm2 is the only family member detectable in the SEZ (Tgm1, Tgm3, Tgm5, and Tgm6 were not detectable). The expression of Tgm2 in ependymal cells explains its similar levels in the SEZ and the MEZ (Figure S5D), but its additional presence in NSCs implies possible autocrine or cell-intrinsic effects that we explored next.
Figure 6.
Transglutaminase 2 Promotes Neurogenesis
(A) NSCs were identified as hGFAP-GFP+ cells in the SEZ in sagittal sections counterstained with Tgm2 and inserted to the right indicated by the dashed line in the lower magnification picture on the left. Both NSCs and ependymal cells were labeled with Tgm2. LV, lateral ventricle.
(B) Whole-mount section of the SEZ showing an hGFAP-GFP+ Tgm2+ apical endfoot between ependymal cells delineated by β-catenin+ junctions.
(C) Single-plane confocal picture of the coronal section of the SEZ immunostained for Dcx and Tgm2 showing no double-positive cells.
(D) Tgm2 expression analysis by qRT-PCR in cells isolated from the SEZ by fluorescence-activated cell sorting (FACS). NSCs were identified by hGFAP-eGFP+ and the apical membrane marker CD133+, ependymal cells (EP) as hGFAP-GFP-/CD133+ and hGFAP-GFP+, and CD133-, PSA-NCAM-, EGFR- cells as niche astrocytes (NA). Note that NSCs and ependymal cells express high levels of Tgm2 mRNA. The direct progeny of NSCs, the transit-amplifying progenitors (TAPs), isolated as EGFR+, CD133-, PSA-NCAM-, and neuroblasts, isolated as PSA-NCAM+ also hardly expressed Tgm2. Data are presented as mean ± standard deviation.
(E) Primary culture from the SEZ stained as indicated showing that Tgm2+ cells were also GFAP+.
(F) Experimental setup for the primary SEZ culture and clonal analysis following Tgm2 inhibition with Z-DON (irreversible Tgm2 inhibitor) or Boc-DON (cell membrane impermeable and irreversible Tgm2 inhibitor).
(G) 10-μM Z-DON treatment at 4 h after plating significantly reduced the number of retrovirally labeled cell clusters (clones, i.e., a cluster of cells sharing the cell of origin), whereas 100-μM Boc-DON did not alter the number of clones. Data are presented as mean ± SEM. ∗p ≤ 0.05, two-tailed Mann-Whitney test.
(H) With Z-DON, but not Boc-DON, the proportion of GFP+ clones containing newly generated neuroblasts (Dcx+) was reduced, whereas the proportion of mixed and glial clones arising from NSCs was conversely increased. Data are presented as mean ± SEM, two-way ANOVA with Bonferroni’s multiple comparison test, ∗∗p ≤ 0.01 and ∗∗∗p ≤ 0.001.
(I) Examples of retrovirally labeled (CAG-IRES-GFP) clones composed of neuronal, glial, and mixed cell types stained as indicated. Scale bars as indicated.
(J) Primary SEZ cultures were treated with 10-nM siRNAs against Tgm2 and showed a reduced number of neuronal clones compared to the control (scrambled siRNA) (n = 4, Data are presented as mean ± SEM, two-way ANOVA with Bonferroni’s multiple comparison test, ∗p ≤ 0.05).
(K) Countings of DAPI stainings from representative tiles (n = 4, with nine tiles counted in each n).
(L) Experimental setup for osmotic pump experiment with two time-points, 4 and 7 days, with continuous intra-ventricular infusion of 100-μM Z-DON in artificial CSF.
(M) On the contralateral side of the infusion, we quantified EdU+ cells that were either Dcx+ or Dcx− at the SEZ. After 4 days Z-DON treatment, we found a significant reduction in TAPs (EdU+/Dcx−), whereas proliferating neuroblasts (EdU+/Dcx+) remained similar to control (Data are presented as mean ± SEM. ∗p ≤ 0.05, two-tailed t test). This trend continued after 7 days treatment (Data are presented as mean ± SEM. p = 0.0537, two-tailed t test). Confocal image stacks from 6 sections were quantified per brain.
To do so, we used primary SEZ cultures as described before (Costa et al., 2011); cells were cultured without growth factors and in the absence of matrix proteins (Figure 6F). Often a single GFAP+ NSC is surrounded by the Dcx+ neuroblast progeny (Figures 6E and 6I) with the former Tgm2+, whereas neuroblasts were negative, consistent with the data shown above (Figures 6C and 6D). To probe Tgm2 function in this culture system, we used 10 μM of the Tgm2 inhibitor Z-DON. Cells were transduced 2 h after plating with retroviral vectors containing CAG-IRES-GFP at low titer to label few cells and allow detection of the progeny of a single cell as a distinct cluster of cells, i.e., a clone (Ortega et al., 2011). Notably, a single dose of Z-DON (at 4 h after plating) drastically reduced the number of clones (Figure 6G; comprising all clusters of GFP+ cells irrespective of their identity), suggesting a possible role of Tgm2 in promoting proliferation or survival. When we analyzed the composition of the clones, we noted a specific effect of the Tgm2 inhibitor on the clones comprising only neuroblasts (referred to as “neuronal clones” in Figures 6H and 6I) that are generated by proliferating TAPs or neuroblasts in these cultures (Costa et al., 2011). Conversely, NSC clones (containing one or few GFAP+ cells and Dcx+ neuroblasts, referred to as “mixed” in Figures 6H and 6I) or clones containing only GFAP+ cells (referred to as “glial” in Figures 6H and 6I) were favored by Tgm2 inhibition (Figure 6H).
Tgm2 has multiple modes of action, including intracellular and extracellular functions (Eckert et al., 2014, Lee and Park, 2017), and the above inhibitor blocks all of them. The inhibitor Boc-DON cannot enter the cells and hence specifically blocks extracellular Tgm2 but had no effects in these cultures (Figures 6G and 6H), suggesting that on this artificial glass substrate with high abundant media volume, extracellular functions of Tgm2 play no roles. Importantly, we confirmed the specific role of Tgm2 using Tgm2 small interfering RNA (siRNA) (Figure 6J). Notably, the reduced number of neuronal progeny after knockdown of Tgm2 was not due to cell death, as the control and knockdown condition had equal numbers of cells (Figure 6K).
Since Z-DON was as effective as siRNA in vitro, we sought to determine whether it would have a similar effect in vivo. Osmotic minipumps loaded with 100 μM Z-DON in artificial CSF were implanted intra-ventricular in mice and inhibitor was administered for either 4 or 7 days. A pulse of EdU (1 h prior perfusion) allowed quantification of proliferating neuroblasts (Dcx+ and EdU+ cells) or TAPs (Dcx− and EdU+ cells) at the SEZ. Interestingly, blocking Tgm2 for 4 days affected mostly the number of TAPs, and this trend was largely maintained after 7 days of administrating Z-DON (Figure 6M). This phenotype obtained in vivo is consistent with reduced clone numbers (Figure 6G) and effects on pure neuronal clones (Figures 6H and 6J) in vitro, as TAPs almost exclusively generate neuroblasts. Thus, the proteome analysis allowed the identification of Tgm2 in promoting neurogenesis from NSCs.
Higher Tissue Stiffness in Neurogenic Niches
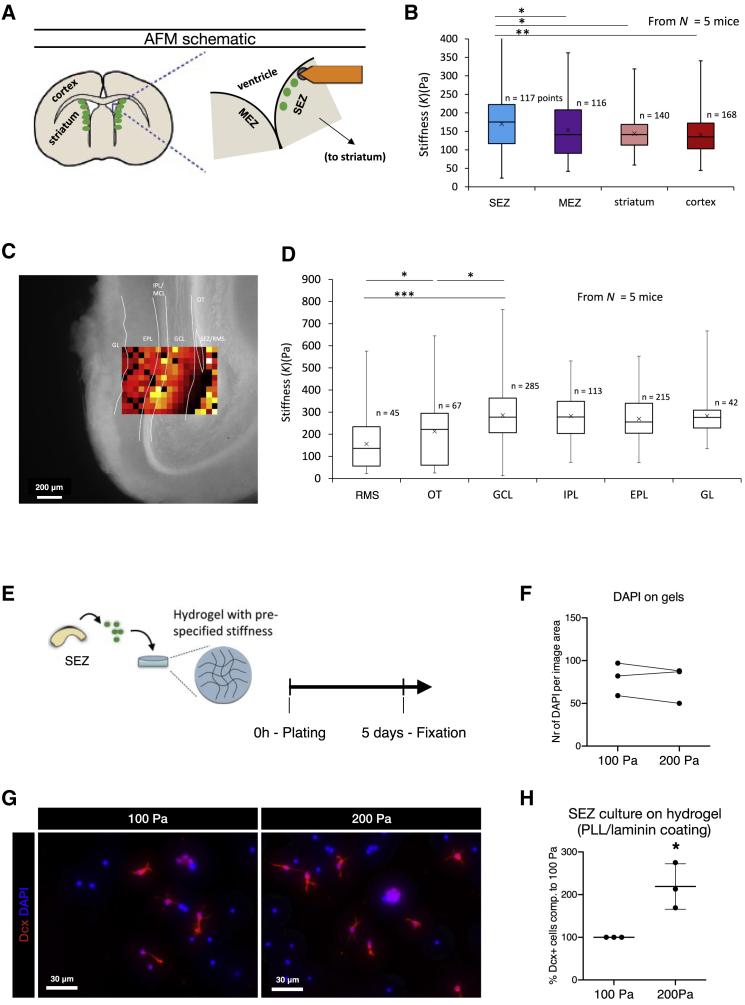
The above-described tissue-specific distribution of potentially mechanically important proteins and the susceptibility of adult NSCs to mechanical signals (Pathak et al., 2014, Petrik et al., 2018) prompted us to examine the stiffness of these regions in coronal slices using atomic force microscopy (AFM) (Figure 7A). The brain parenchymal regions, the cortex (GM) and the striatum, had a similar stiffness (Figure 7B). Both the SEZ and the MEZ regions were significantly stiffer (Figure 7B), and the SEZ, the main site of neurogenesis, was significantly stiffer than the MEZ, where few neuroblasts arise (Bordiuk et al., 2014) (p ≤ 0.05, Mann-Whitney, two tailed). These data suggested a close correlation between higher tissue stiffness and more NSCs dedicated to adult neurogenesis. To examine the extent to which the stiffness differences of about 100 Pa are relevant for neurogenesis, we cultured the above-described primary SEZ cells on gels with the stiffness of 100 and 200 Pa. Cells were fixed and stained after 5 days, and no difference in total cell number was observed (Figures 7E and 7F). However, more than double the number of neuroblasts was detected on the stiffer 200-Pa substrate (Figures 7G and 7H), even though the outcome of this difference appears to depend on cell density (data not shown). In summary, these data support the concept that neurogenesis is responsive to such differences in stiffness.
Figure 7.
Higher Stiffness of the Neurogenic Niches
(A) Schematic drawing of the stiffness measurements on coronal slices (300 μm) with AFM.
(B) Stiffness was assessed in the SEZ, the MEZ, the striatum, and the Cx. Both ventricular regions are significantly stiffer than the Cx and the striatum that both have similar tissue stiffness. The SEZ was significantly stiffer than the MEZ. Data shown as whisker plots, ∗p = 0.05 and ∗∗p = 0.01.
(C) Representative tissue heatmap of OB measurements with scale bar as indicated.
(D) In the OB, the end of the RMS was less stiff in comparison to the adjacent olfactory tract. The granule cell layer (GCL) was even stiffer still, as well as the internal and external plexiform layer (IPL/EPL) and the glomerular layer (GL). Data shown as whisker plots, Mann-Whitney test (two tailed), ∗p = 0.05 and ∗∗∗p = 0.001.
(E) Experimental setup for the primary SEZ culture plated on hydrogels with 100- or 200-Pa stiffness.
(F) Number of DAPI cells was similar at the end of the 5-day experimental period.
(G) Representative images of the Dcx+ cells at 5 days after plating.
(H) Hydrogels with 200-Pa stiffness significantly increased the percentage of Dcx+ cells in comparison to the same primary SEZ culture on hydrogels with 100 Pa stiffness. Data are presented as mean ± standard deviation. ∗p = 0.05, paired t test.
Assessing the OB, we also found an increase in stiffness from the RMS toward the granule cell layer (GCL), which was stiffer throughout the OB parenchyma (Figures 7C and 7D). Thus, both neurogenic niches were significantly stiffer than the brain parenchyma, suggesting that components of the proteome contribute to regulate these niche-specific mechanical properties, which affects neurogenesis.
Discussion
Here we provide a comprehensive characterization of the neurogenic niche proteome compared to normal brain parenchyma and make the data available on an easy-to-use webpage (https://neuronicheproteome.org). Our two proteome datasets (LMSS and QDSP) allowed expanding the set of neurogenic niche-specific proteins and defining their region-specific compartment association. These data are an important complement to the RNA expression data, as RNA and protein enrichment are only partially congruent. This has been described in detail when comparing scRNA-seq and deep proteome data in the aging lung (Angelidis et al., 2019) and was also apparent in our comparison of RNA and proteome data (Figure S6). The possible mechanisms for this are multiple. RNA stability and RNA-binding proteins may differ between brain regions, and the latter (including microRNAs [miRNAs]) may differentially regulate translation, leading to different abundances of newly produced proteins. Conversely, protein degradation may be regulated differently between regions, such that RNA could be enriched, but protein may be fast degraded and hence reduced. Likewise, especially for the matrisome proteins, the secretory pathways leading to their final localization in the ECM could be differentially regulated, causing a discrepancy between the mRNA coding for the protein and its amount in the ECM. Lastly, the location of the SEZ at the ventricle puts NSCs in direct contact with the CSF, and several factors released by the choroid plexus into the CSF have already been identified to regulate adult neurogenesis (Lepko et al., 2019, Silva-Vargas et al., 2016), including miRNAs (Lepko et al., 2019) or proteins (Silva-Vargas et al., 2016). In the latter case, we would detect the protein in our proteome analysis, but no RNA would be found, which is the case for many. Interestingly, especially proteins of the matrisome enriched in the SEZ were not enriched at RNA levels (Figure S7A), consistent with data obtained in lung (Angelidis et al., 2019). Prominent categories of proteins enriched in the OB compared to the cortex, not enriched at RNA level, were related to splicing (Figure S7B). Therefore, this proteome analysis provides an important resource, as proteins play key roles in the function of this unique niche.
Specific Matrisome of the NSC Niche
Various soluble factors are the most explored cell-extrinsic signals that regulate the adult NSC. Indeed, many matrix-associated proteins are more soluble in the neurogenic niches, and we even found several core-matrix proteins to be more soluble in the neurogenic niches compared to other brain regions and tissues (Figure S5C). This includes Hapln1, Tnc, and Thbs4. In addition to binding other core-matrisome proteins, such as collagens and fibronectin, Tnc interacts with a diverse set of ligands, such as growth factors (e.g., Wnt3a and transforming growth factor β [Tgf-β]) and receptors (e.g., Toll-like receptor 4 [TLR-4] and Rptpβ) (De Laporte et al., 2013; Midwood et al., 2016). Thbs4 may act as, e.g., a voltage-gated ion-channel blocker or have intracellular functions (Brody et al., 2015, Girard et al., 2014, Lana et al., 2016, Narouz-Ott et al., 2000). The other niche-specific ECM proteins with a soluble profile include serpins, S100 proteins, and annexins that form a core interaction hub in the SEZ-enriched matrisome (Figure S7C). Soluble ECM proteins such as these may stem from the various cell types in the brain or blood (Geyer et al., 2016). However, we found no evidence for a general increase in blood proteins from allegedly leaky vessels (Tavazoie et al., 2008) in the SEZ tissue (Figures S2D and S2E) but rather expression of these genes (e.g., serpinb6, annexin 1, and annexin 5) by cells in the SEZ (Figure 2I), mostly qNSCs (Figure S6A). Another interesting protein in this category enriched in both the SEZ and the OB is Kininogen 1 (Kng1). Kng1 is a precursor for bradykinin (Figures 2B and 2C), which has been found to promote neurogenesis versus gliogenesis in vitro (Pillat et al., 2016). Interaction analysis of the niche-specific matrix protein in the SEZ highlights annexin-S100 protein interactions and calcium-binding and catalytic activity regulation, whereas in the OB, several serpins have known interactions with other negative regulators of endopeptidase activity (Figures S7C and S7D).
Notably, the SEZ-enriched matrisome unraveled here differs profoundly from the enrichments found recently by RNA-seq of NSC niches in the developing Cx of murine or human samples (Fietz et al., 2012). Although RNA and proteome discrepancies may contribute, it is important to note that some of the crucial core matrisome proteins regulating embryonic SVZ expansion and cortex folding are not enriched in the adult NSC niche (Long et al., 2018, Long and Huttner, 2019). Indeed, many of the SEZ-enriched proteins are upregulated at early postnatal stages when the adult NSC niche forms (F.V., P.S., and M. Götz, unpublished data). Thus, as in many other organs, the niche maintaining adult stem cells differs profoundly from the niche regulating expansion in development.
S100a6 and C1ql3 were both soluble ECM-associated proteins that we found enriched in NSCs of the SEZ. Interestingly, S100a6 also marks NSCs in the DG (Yamada and Jinno, 2014) and is hence common to NSCs across regions. S100a6 has extracellular and intracellular functions (Donato et al., 2017); some of the latter are calcium dependent and involved in promoting proliferation in various cancer cells (Lerchenmüller et al., 2016, Li et al., 2015). This may explain its higher levels in late-stage quiescent NSCs compared to postmitotic parenchymal astrocytes. Moreover, S100a6 may be involved in the signal transduction cascade of flow- or stretch-sensitive channels, such as the epithelial sodium channel that is also absent from non-proliferating astrocytes but present in NSCs and increases the frequency of Ca signals when promoting proliferation (Petrik et al., 2018).
In brain, C1ql3 has thus far only been identified as a synapse-associated protein (Chew et al., 2017, Martinelli et al., 2016, Matsuda et al., 2016). Here we found it enriched with a specific solubility profile in the SEZ-niche and identified NSCs as the major source. Thus, C1ql3 also allows the discrimination of NSCs from parenchymal astrocytes, an urgent need given the labeling of NSCs by most astrocyte markers (see also Beckervordersandforth et al., 2010). Notably, C1ql3 is enriched in the SEZ only at the protein level and was hence not identified as a NSC marker in RNA analysis (Figure S6B). C1ql3’s differential solubility at the SEZ suggests its localization in a different compartment that is unrelated to its reported synapse function in other brain regions. Outside the brain, extracellular C1ql3 has been reported to control cellular glucose homeostasis (Wei et al., 2011), which makes C1ql3 interesting as a potential regulator of NSC metabolism and perhaps part of the metabolism-enriched machinery we identified in the SEZ (Figure 1M). In order to further demonstrate the functional relevance of the proteome differences detected in this resource, we showed that the niche-candidate Tgm2 regulates proliferation and neurogenesis in vitro and in vivo. Tgm2 is a multifunctional enzyme and may regulate neurogenesis by intracellular and extracellular mechanisms in vivo. Worth noting is its predominant insoluble profile in the SEZ. Most importantly, it serves to substantiate the relevance of proteins found to be enriched in the SEZ.
Specific Matrisome of the OB, the Niche for Neuronal Integration
A key aspect of adult neurogenesis is the integration of the new neurons into pre-existing circuitry. However, the niche conditions allowing this integration in the DG and the OB are not characterized. PNNs in the brain parenchyma have spurred much interest because of their role in plasticity (Sorg et al., 2016). Overall, many of the PNN-associated proteins are present at a lower degree in the SEZ and the OB compared to the Cx and the MEZ. In the OB, we found two PNN proteins (Hapln1 and Ncan) to be substantially more soluble than in the Cx and the SEZ, which suggests that these proteins may contribute less to PNNs in the OB compared to the Cx and hence constitute a composition of PNN proteins that may be involved in allowing constant synaptic plasticity in the OB. Indeed, Hapln1 is crucial for the formation of new PNNs and is key in mediating plasticity (Carulli et al., 2010). Ncan can act as repellent signal for PSA-NCAM and Epha3 signaling and may be integral to neuroblast migration and circuitry integration (Sullivan et al., 2018). We could further corroborate the absence of a typical PNN composition in the OB by staining with Wisteria floribunda agglutinin (WFA), a lectin-binding carbohydrate of the PNN, e.g., in the Cx, although it does not mark any nets around neurons of the OB (Figure S4K). The limited matrix association of PNN-associated proteins in the OB and the absence of typical PNNs makes them key candidates for the OB’s distinct capacity to permit the integration of new neurons into the pre-existing circuitry.
Unique Stiffness of the Neurogenic Niches
An ECM that increases tissue stiffness typically contains insoluble proteins, such as the BM proteins, including collagens and laminins (Swift et al., 2013). Our solubility profiles show that the neuron-associated ECM clearly has a different composition and architecture compared to the insoluble BM proteins, and our data suggest it has very little contribution to tissue stiffness. Conversely, we found more insoluble ECM components that may relate to the increased stiffness of the SEZ, such as Laminin-b2 (Lamb2), Nidogen-1 (Nid1), and Perlecan (Hspg2) in the SEZ compared to the MEZ (Figure S2B), which suggests that the SEZ contains more or larger BM structures. This is in line with previous observations suggesting the specific BM structures in the SEZ, referred to as fractones, as sites for growth-factor accumulation (Kerever et al., 2007). Moreover, the higher expression of the ECM cross-linker Tgm2 originating from ependymal cells and NSCs may further contribute to the increased mechanical stiffness of this niche, besides its possible intracellular functions shown here in vitro. Transglutaminases have previously been shown to stiffen tissue (Majkut et al., 2013), and increased substrate stiffness promotes neurogenesis in vitro shown here and previously (Pathak et al., 2014).
We found no general enrichment of the insoluble ECM in the OB parenchyma, except for Reln, which suggests unique interconnections here that may be linked to its role in promoting neuroblast chain-migration detachment (Hack et al., 2002). Nevertheless, the OB was the stiffest among the investigated brain regions. The OB is altogether a cell dense region and cell density is known to correlate with tissue stiffness (Koser et al., 2015, Thompson et al., 2019). However, we also found all lamins of the nuclear matrix to be enriched in the OB, and lamin A correlates with tissue stiffness (Swift et al., 2013) (Figure S2F). The lamins were also more insoluble in the OB, which suggests they may have a different composition or associate to cytoskeletal proteins through proteins such as Nesprin-2 (Syne2), which had similar distinct solubility characteristics in the OB. Nesprin-2 regulates nuclear movement during neurogenesis (Zhang et al., 2009) and may hence be involved in neuroblast migration in the OB. Cell migration is also regulated by tissue stiffness, and durotaxis (migration toward stiff substrate) has been described for several stem cells (Choi et al., 2012, Vincent et al., 2013). Moreover, the nuclear envelope can act as a mechanosensor (Donnaloja et al., 2019), highlighting the importance of nuclear envelope and cytoskeletal interactions. Importantly, stiffness has been found to regulate NSC differentiation in vitro (Saha et al., 2008), and we showed here the relevance of the 100-Pa difference measured in vivo exerting effects on neurogenesis in vitro. Thus, the unique properties of the SEZ niche are functionally relevant. Therefore, the niche-specific proteome described here provides a rich resource for a deeper understanding of the unique properties of this NSC niche in comparison to other adult stem cell niches.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Beta-catenin (1/2000) | BD bioscience | Cat# 610153; RRID:AB_397554 |
| Collagen 4 (1/100) | Millipore | AB756P; RRID:AB_2276457 |
| DCX (1/500) | Millipore | Cat# AB2253; RRID:AB_1586992 |
| GFAP (rb) (1/500) | DAKO | Cat# Z0334; RRID:AB_10013382 |
| GFAP (ms) (1/500) | Sigma | Cat# G3893; RRID:AB_477010 |
| GFAP (goat) (1/500) | abcam | Cat# ab53554; RRID:AB_880202 |
| GFP (1/1000) | Millipore | Cat# MAB3580; RRID:AB_94936 |
| GFP (1/2000-4000) | Aves lab | Cat# GFP-1020; RRID:AB_10000240 |
| MAG (1/400) | Millipore | Cat# MAB1567; RRID:AB_2137847 |
| MAP2 (1/1000) | Sigma | Cat# M4403; RRID:AB_477193 |
| Nestin (1/100) | Millipore | Cat# MAB353; RRID:AB_94911 |
| NeuN (1/100) | Millipore | Cat# MAB377; RRID:AB_2298772 |
| S100a6 (1/500) | Abcam | Cat# ab181975 |
| S100B (ms) 1/500) | Sigma | Cat# S2532; RRID:AB_477499 |
| Tgm2 (1/100) | Labvision | Cat# MS-224-B0; RRID:AB_62201 |
| WFA (biotin conjugated) (1/500) | Sigma | Cat# L1516-2MG; RRID:AB_2620171 |
| Secondary antibodies | ||
| Alexa Fluor® secondary antibodies (488, 555, 647) (1/1000) | ThermoFisher Scientific | Cat# A-11001; RRID:AB_2534069 |
| 555 Alexa Fluor® conjugated streptavidin | ThermoFisher Scientific | Cat# S32355; RRID:AB_2571525 |
| FACS antibodies | ||
| CD133-PE (1/250) | eBioscience | Cat# 12-1331-82; RRID:AB_465849 |
| EGF–Alexa Fluor 647 (1/300) | Molecular Probes | Cat# E-35351 |
| Anti-PSA-NCAM-PE (1/250) | Miltenyi | Cat# 130-093-274; RRID:AB_1036069 |
| Rat IgG1 K isotype control PE (1/250) | eBioscience | Cat# 12-4301-81; RRID:AB_470046 |
| Mouse IgM-PE antibody (isotype control (1/250)) | Miltenyi | Cat# 130-093-177; RRID:AB_871723 |
| Chemicals, Peptides, Recombinant Proteins and Kits | ||
| IGPAL-CA-630 | Sigma | Cat# I8896 |
| Phosphatase inhibitors | Roche | Cat# 04906837001 |
| Benzonase | Merck | Cat# 70746-3 |
| Protease inhibitors (+EDTA) | LifeTech | Cat# 78430 |
| Sodium deoxycholate | Sigma | Cat# D6750 |
| TCEP | Sigma | Cat# 646547 |
| 2-Chloroacetamide | Sigma | Cat# C0267 |
| Ammonium acetate | Sigma | Cat# V800034 |
| Formic acid | Sigma | Cat# 543804 |
| Gaudinium | Sigma | Cat# G4505 |
| Thio-urea | Sigma | Cat# T8656 |
| SDS | Sigma | Cat# L4509 |
| HEPES | Sigma | Cat# H3375 |
| Trypsin | Sigma | Cat# T9201 |
| Hyaluronidase | Sigma | Cat# H3884 |
| DMEM/F12 | Life Technologies | Cat# 21331020 |
| B27 Supplement | Life Technologies | Cat# 17504044 |
| Poly-D-lysine hydrobromide | Sigma | Cat# P0899 |
| GlutaMax | Life Technologies | Cat# 35050038 |
| Trypsin-EDTA (0.05%) | ThermoFisher Scientific | Cat# 25300054 |
| Poly-L-ornithin | Sigma | Cat# P4957 |
| Laminin | Roche | Cat# 11243217001 |
| TSA Tetramethylrhodamine kit | PerkinElmer | Cat# NEL702001KT |
| Acetonitrile | Sigma | Cat# 271004 |
| Trifluoroacetic acid | Sigma | Cat# 302031 |
| Micro BCA protein assay kit | ThermoFisher Scientific | Cat# 23235 |
| Goat Serum | ThermoFisher Scientific | Cat# 16210072 |
| RLT lysis buffer | QIAGEN | Cat# 79216 |
| Triton X-100 | Sigma | Cat# T8787 |
| RLT lysis buffer | QIAGEN | Cat# 79216 |
| RNeasy Micro Kit | QIAGEN | Cat# 74004 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| SuperScript III | Invitrogen | Cat# 18080093 |
| iQ SYBR Green Supermix | BIO-RAD | Cat# 1708880 |
| IsoFlo | Abbott Laboratories | Cat# NDC 0044-5260-03 |
| DMSO | Sigma | Cat# D2438-10ML |
| ZDON | Zedira | Cat# Z006 |
| BocDON | Zedira | Cat# B003 |
| EdU | ThermoFisher Scientific | Cat# E10187 |
| Click-iT EdU Alexa fluor 647 Imaging kit | Invitrogen | Cat# C10340 |
| HiPerFect® transfection reagent | QIAGEN | Cat# 301704 |
| Allstars Negative control siRNA | QIAGEN | Cat# 1027280 |
| FlexiTube GeneSolution for Tgm2 | QIAGEN | Cat# GS21817 |
| Glutaraldehyde | Sigma | Cat# G5882 |
| (3-Aminopropyl) trimethoxysilane (APTMS) | Sigma | Cat# 281775 |
| Acrylamide | Sigma | Cat# A4058 |
| N-Hydroxyethyl-acrylamide | Sigma | Cat# 697931 |
| Bis-acrylamide | Fisher Scientific | Cat# BP1404-250 |
| Experimental Models: Organisms/Strains | ||
| C57Bl6/J mice | In-house breeding | N/A |
| C1ql3flox; C1ql3tm1.1Sud | Martinelli et al., 2016 | RRID: MGI_5779515 |
| hGFAP-GFP mice (Tg(GFAP-EGFP)1Hket) | Nolte et al., 2001 | MGI:6188855 |
| Deposited Data | ||
| Proteome dataset | ProteomeXchange Consortium at http://proteomecentral.proteomexchange.org | PXD016632 |
| Supplementary tables (of in-article analyzed data) | The journal | N/A |
| Proteome dataset | https://neuronicheproteome.org | N/A |
| Microarray dataset | Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/ | accession number GPL15692 |
| Software and Algorithms | ||
| MATLAB, including custom-written scripts to analyze AFM data | Mathworks: https://www.mathworks.com/products/matlab/Custom MATLAB scripts: https://github.com/FranzeLab | Version R2018b |
| Maxquant | http://www.coxdocs.org/doku.php?id=maxquant:start | Version 1.6.0.16 |
| Perseus | http://www.coxdocs.org/doku.php?id=perseus:start | Version 1.6.0.7 |
| Prism | Graphpad | Version 5 |
| Microarray analysis | GeneSpring GX v11.5.1 software package | Agilent Technologies |
| String | https://string-db.org/ | N/A |
| ZEN imaging software | Carl Zeiss | N/A |
| Other | ||
| CellHesion 200 AFM head | JPK Instruments | N/A |
| PetriDishHeater for maintaining constant temperature during AFM-based stiffness measurements | JPK Instruments | N/A |
| Tipless silicon cantilevers for AFM-based stiffness measurements | NanoWorld | Arrow TL-1 |
| Spherical probes for AFM cantilevers | microParticles GMBH | 37.28 μm polystyrene beads, PS-R-37.0 |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Prof. Magdalena Götz (magdalena.goetz@helmholtz-muenchen.de). This study did not generate new unique reagents.
Experimental Model and Subject Details
WT Mice (Proteomics, Cell Culture, Osmotic Pumps, IHC, and AFM) and hGFAP-eGFP mice (FACS, IHC)
All experimental procedures in this study done at LMU Muenchen were performed in accordance with German and European Union guidelines and were approved by the government of upper Bavaria. In addition, all procedures performed in Cambridge were carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 and with university guidelines. For the proteomics experiments, only male C57BL/6J mice between 8-10 weeks were used, for osmotic pump experiments, only male C57Bl6/J mice between 10-12 weeks were used, and for AFM experiments, only male C57Bl6/J mice aged 8 weeks were used (to reduce any potential variability). For other experiments both male and female C57BL/6J mice were used between 8-12 weeks. For FACS and IHC experiments, we used both male and female 8-12 week old hGFAP-eGFP mice (Tg(GFAP-EGFP)1Hket, Nolte et al., 2001). Mice were fed ad libitum and housed with a 12/12 h light and dark cycle and kept under specific-pathogen-free (SPF) conditions.
C1ql3 Reporter Mice
The analysis of C1ql3 gene expression was performed using the IRES-mVenus knockin reporter allele (C1ql3flox; C1ql3tm1.1Sud RRID: MGI_5779515) described in (Martinelli et al., 2016). The background strain of the mice was C57BL/6. All procedures involving the use of mice at the University of Connecticut Health Center were approved by the Institutional Animal Care and Use Committee, and in accordance with guidelines set forth by the National Research Council of the National Academies Guide for the Care and Use of Laboratory Animals.
Primary SEZ Culture
The sub-ependymal zones (SEZ) of 8 - 12 weeks old C57BL/6J wild-type mice were dissected in Hank’s Balanced Salt Solution (HBSS) with 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) and after removing the dissection medium incubated in 5 mL dissociation solution (HBSS containing 15 mM HEPES, 5.4 Glucose, 3.4 mg trypsin powder and 3.5 mg hyaluronidase powder) at 37°C for 15 min. The SEZ pieces were triturated 10 times using a 5 mL glass pipette and incubated for another 15 min at 37°C. 5 mL ice-cold solution 3 (solution 3: EBSS (Earle’s Balanced Salt Solution) containing 20 mM HEPES and 0,04 g/mL BSA (Bovine Serum Albumin); pH 7.5) were added and solutions were mixed by pipetting 10 times with a 10 mL pipette. The cell suspension was filtrated through a 70-μm cell strainer and centrifuged at 250 g for 5 min at 4°C. The supernatant was discarded and cells were resuspended in 10 mL ice-cold solution 2 (solution 2: HBSS containing 0.9 M Sucrose; pH 7.5) and centrifuged at 650 g for 10 min at 4°C. After discarding the supernatant, cells were resuspended in 2 mL ice-cold solution 3. 2 mL of the cell suspension were added to a 15 mL falcon tube with 12 mL ice-cold solution 3 and centrifuged at 350 g for 7 min at 4°C. The supernatant was carefully removed and cells were resuspended in B27-supplemented culture medium (culture medium: DMEM/F12 (1:1) with 1x GlutaMAX containing 1x B27 serum-free supplement, 100 units/mL penicillin, 100 mg/mL streptomycin, 8 mM HEPES) and seeded (2 SEZs per well) onto poly-D-lysine - coated (PDL) coverslips in 24-well tissue culture plates (Ortega et al., 2011). Cells were kept in the incubator for 7d at 37°C and 5% CO2, with the exception of cultures on hydrogels (see Hydroxy-Acrylamid Gel Preparation for Cell Culture) that were kept for 5 d.
Method Details
Sample Preparation for Proteome Analysis
Library-Matched Single Shot (LMSS) Method. Mice were sacrificed by cervical dislocation and brains were subsequently extracted and put into cold phosphate buffered saline (PBS). The ventricular walls were laid bare by removing the dorsal ventricular wall and all tissue above it, as well as the choroid plexus. Brains were then snap-frozen on dry ice and cut into 100 μm sections on a cryostat (Leica CM1000S). The medial (MEZ) and lateral ventricular (SEZ) walls were then manually dissected under a light microscope (Leica MZ6). 8-10 sections from each animal were collected per sample (n = 8) and kept on dry ice until tissue lysis. Somatosensory cortex (Cx) samples were dissected by removing corpus callosum and top layer of cortex (including meninges). Olfactory bulb (OB) was dissected by cutting out the core of the OB approximately along the external plexiform layer. Both Cx and OB samples were subsequently snap-frozen on dry ice (n = 8). Tissues were lysed in buffer containing 0.1 M Tris-HCl (pH 8.5), 1% (w/v) Sodium Deoxycholate, 10 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and 40 mM 2-Chloroacetamide at 99°C for 10 min. The lysates were sonicated for 2 × 15 min (or until homogeneous) (Bioruptor, model UCD-200, Diagenode) and protein concentration was determined using the BCA method (Micro BCA protein assay kit) according to manufacturer instructions. 25 μg of protein was digested with LysC and trypsin overnight at 37°C. 250 μl Isopropanol with trifluoroacetic acid (TFA) (each sample contains 1% TFA) was added to each sample and then mixed strongly. Samples were desalted using the StageTip method with SDB-RPS (styrene-divinyl-benzene reverse phase sulfonate; 3M, #2241) filters (Kulak et al., 2014). Therefore, filters were activated with acetonitrile (ACN) and equilibrated with 30% methanol (MeOH) and 1% TFA. After washing with 0.2% TFA, the samples were added to the stage tips and then filters were washed again. Elution was done using 1% Ammonia and 80% ACN. Four of the peptide samples from each region were combined and used for the library proteome of each region. These pooled peptide samples were divided into 8 fractions with nano-fractionation using a high pH reversed-phase fractionator that switches the elution flow every 90 s using a rotating valve (Kulak et al., 2017). The four remaining single-shot samples from each region were analyzed without fractionation.
Quantitative Detergent Solubility Profile (QDSP) Method. Mice were sacrificed by cervical dislocation and brains were subsequently extracted and put into cold PBS. The OB was removed by dissection at its base. Somatosensory cortex (Cx) was dissected using a 2.5 mm biopsy punch and the white matter was removed. Both subependymal zones (SEZ and MEZ) were dissected (Ortega et al., 2011). All samples were homogenized using a (100 μl) dounce homogenizer (Wheaton #357844) in 100 μl PBS (with protease inhibitor cocktail and Ethylenediaminetetraacetic acid (EDTA)) and directly frozen in liquid nitrogen and stored at −80°C until tissue protein fractionation. Tissue lysates from 3 animals were pooled, resulting in 5 samples per region and then processed simultaneously (a total of 15 mice). Following centrifugation, we collected the supernatant (protein fraction 0) and then sequentially extracted proteins using the MS analysis adapted de-cellularization protocol of Schiller et al. (Schiller et al., 2015). Therefore, we resuspended the pellet in three buffers, each followed by a centrifugation for 20 min at 16,000 g. The samples were incubated in buffer 1 (150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5% glycerol, 1% IGEPAL, 1 mM MgCl2, protease inhibitors (+EDTA), 1% benzonase, 1 × phosphatase inhibitors) and buffer 2 (150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5% glycerol, 1.0% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 1 × protease inhibitors (+EDTA), and 1% benzonase) for 20 min on ice, and in buffer 3 (500 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5% glycerol, 1.0% IGEPAL, 2% sodium deoxycholate, 1% SDS, 1 × protease inhibitors (+EDTA), and 1% benzonase) for 20 min at RT. Each of the supernatant from the buffer treatment resulted in fraction 1, 2, and 3, with the residual insoluble material resulting in fraction 4. Fraction 0 and 1 were combined to generate our first fraction. All four fractions were precipitated in 80% acetone and sonicated for 5x30 s (Bioruptor, model UCD-200, Diagenode). Afterward, samples were incubated at −20°C for a minimum of 1 h and were then centrifuged. The precipitation was repeated once in order to remove any residual detergent. Alkylation/reduction buffer (100 mM Tris-HCl (pH 8.5), 6M GDmCl, 10 mM TCEP, and 50 mM 2-chloroacetamide) was added to the samples and then boiled at 99°C for 15 min, followed by sonication for 10 × 30 s. Protein concentration was determined using the BCA method (Micro BCA protein assay kit) according to manufacturer instructions. Enzymatic digestion was done in two steps. First, samples were incubated at 37°C for 2 h with LysC (1/50) and then with LysC (1/50) and Trypsin (1/25) overnight. Both digestions were aided by 10 × 30 s sonification. Samples were then acidified by adding 1% TFA followed by desalting using the StageTip method with SDB-RPS filters (Kulak et al., 2014). Therefore, filters were activated with acetonitrile (ACN) and equilibrated with 30% methanol (MeOH) and 1% TFA. After washing with 0.2% TFA, the samples were added to the stage tips and then filters were washed again. Each protein lysate was eluted into three peptide fractions using three buffers (buffer 1: 150 mM NH4HCO2, 40% acetonitrile, 0.5% Formic acid (FA); buffer 2: 150 mM NH4HCO2, 60% acetonitrile, 0.5% FA and buffer 3: 5% ammonia (from 25% stock solution) and 80% acetonitrile) resulting in a total of 12 fractions per sample.
Mass Spectrometry
For both the LMSS (including each library sample) and QDSP samples, we loaded approximately 2 μg of peptides in buffer A (0.1% (v/v) formic acid). We separated peptides by a 2 h gradient in a 50 cm long C18 column (75 μm inner diameter filled in house with ReproSil-Pur C18-AQ 1.9-lm resin (Dr. Maish GmbH)). Samples were eluted in 5%–60% buffer B (0.1% (v/v) formic acid, 80% (v/v) acetonitrile) at a flow rate of 250 nL/ min using a nanoflow UHPLC (Easy nLC, Thermo Fisher Scientific) online coupled to the mass spectrometer (Q Exactive HF Orbitrap, Thermo Fisher Scientific). Each gradient was followed by a wash with buffer B and recalibration with buffer A. Survey scans had a resolution of 70,000 at m/z 400 with a maximum injection time of 20 ms. Target value for the full scan MS spectra was 3 × 106 and isolation window of 1.6 m/z with 10 most abundant precursor ions chosen for fragmentation. MS/MS scans had a resolution of 17,500 at m/z 400 with a maximum injection time of 120 ms. Ion target value for the MS/MS scan was 1 × 105.
Immunohistochemistry
Brain Sections. For obtaining brain sections, mice (hGFAP-eGFP, mVenus/C1ql3, or C57BL/6J) were anaesthetized by intraperitoneal injection of ketamine (100mg per kg of body weight) and xylazine (10mg per body weight) and then transcardially perfused first with PBS followed by 4% Paraformaldehyde (PFA). Brains were dissected and cut at 80-100 μm thickness at the vibratome (Leica VT1000S) or 30 μm at the cryostat (Leica CM3050S). Sections were stained with primary antibodies in PBS containing 0.1% Triton X-100 and 10% normal goat serum (NGS) overnight at 4°C, washed and incubated with secondary antibodies in PBS containing 10% NGS for 2 h at RT. mVenus/C1ql3 expression was detected using chicken anti-GFP (Aves, 1:4000).
Perfused brains from the osmotic pump experiments were sectioned coronally (20 μm) along the ventricles. Sections were stained for Dcx and EdU according to manufacturer instructions (Click-iT™). Photomicrographs of the SEZ contralateral to the pump implantation site for quantifications were acquired using confocal microscope LSM 710 (Zeiss).
Whole Mounts. Whole mounts of the SEZ from three months old hGFAP-eGFP transgenic mice were dissected (Mirzadeh et al., 2010) and fixed for 15 min with 2% PFA. After washing with PBS, the tissue was stained for 48 h with primary antibodies in PBS containing 0.1% Triton X-100 and 10% normal goat serum (NGS). Whole mounts were washed three times with PBS at room temperature and incubated with the secondary antibodies in PBS containing 10% NGS overnight at 4°C. After three washings in PBS, DAPI was added for 5 min and washed again. Primary antibodies used were: mouse IgG1 anti-Tgm2 (Labvision, 1:100), rabbit anti-β-catenin (Sigma, 1:2000) and chicken anti-GFP (Aves, 1:2000). Tyramide Signal Amplification was used to enhance the Tgm2 fluorescence signal according to the manufacturer instructions (TSA Tetramethylrhodamine kit, PerkinElmer). Photomicrographs were acquired using confocal microscope LSM 710 (Zeiss).
Primary SEZ Cultures. Primary SEZ cultures were fixed with 400 μl of 4% (wt/vol) PFA for 15 min at room temperature (RT) and afterward washed twice with PBS. The staining solution contained primary antibodies in PBS with 0.5% Triton X-100 and 10% NGS. Primary antibodies used were: mouse IgG1 anti-Tgm2 (Labvision, 1:100), chicken anti-GFP (Aves, 1:2000), rabbit anti-GFAP (Dako, 1:500), and guinea pig anti-Dcx (Millipore, 1:500). Cells were incubated with the primary antibodies at 4°C overnight, washed with PBS twice and incubated with secondary antibodies in PBS with 10% NGS for 2 h at RT. Photomicrographs were acquired using fluorescent microscope AXIO Imager.Z1 (Zeiss).
All primary and secondary antibodies used can be found in the KEY RESOURCES TABLE (including the used concentration).
Fluorescence-activated Cell Sorting
SEZ from heterozygous hGFAP-eGFP (Nolte et al., 2001) and C57BL/6J (WT) mice were dissected in dissection medium (HBSS containing 10mM HEPES) on ice and transferred into a 15 mL falcon tube containing 5 mL of solution 1 (solution 1 (HBSS-glucose), see above, 0,81% glucose, 15 mM HEPES in HBSS; pH 7.5). The tissue was mechanically dissociated by pipetting up and down ten times with a fire-polished glass Pasteur pipette. 100 μl of 0.05% trypsin was added to the sample, followed by an incubation step at 37°C for 15 min. Trituration was repeated after 15 min with a fire-polished Pasteur pipette and cells were incubated for additional 15 min. Final trituration was done ten times at the end of the incubation. 5 mL of ice-cold solution 3 (solution 3 (BSA-EBSS-HEPES) 20 mM HEPES, 0.04 g/mL BSA in EBSS; pH 7.5) was added and solutions were mixed by pipetting several times up and down. The cell suspension was filtrated using a 70-μm cell strainer and centrifuged at 180 g for 5 min at 4°C. The supernatant was removed and cells were resuspended in 10 mL of ice-cold solution 2 (solution 2 (saccharose-HBSS) 0.9 M saccharose) in HBSS; pH 7.5) and centrifuged at 510 g for 20 min at 4°C. The pellet was resuspended in 2 mL of ice-cold solution 3 and pured on top of 12 mL of ice-cold solution 3 and centrifuged at 290 g for 12 min at 4°C. Staining solution (0.02% sodium azide, 10%, FBS in PBS) was added to the pellet and cells were incubated with primary antibodies for 30 min at 4°C. After washing with PBS, cells were resuspended in PBS and sorted using the FACS Aria III (BD). Gates were set by the use of isotype controls (Fischer et al., 2011).
To collect neural stem cells (NSC), transient amplifying progenitors (TAP), neuroblasts (NB), ependymal cells (EC) and niche astrocytes (NA) we stained the cells as follows: tube 1, cells from hGFAP-eGFP mice with CD133-PE (1:250); tube 2, cells from hGFAP-eGFP mice with CD133-PE (1:250), EGF–Alexa Fluor 647 (1:300) and Anti-PSA-NCAM-PE (1:250) and tube 3, cells from hGFAP-eGFP mice with Anti-PSA-NCAM-PE (1:250). Controls to set the gates were prepared as follows: tube 4, cells from WT mice lacking any antibody; tube 5, cells from WT with rat IgG1 K isotype control PE (1:250); tube 6, cells from WT mice with mouse IgM-PE isotype control (1:250).
qPCR
FACS sorted cells were directly collected into RLT lysis buffer (QIAGEN) during the sorting procedure. Total RNA was isolated with the RNeasy MICRO kit (QIAGEN) according to the manufacturer’s instructions. Quality and concentration of total RNA was examined with the Agilent Bioanalyzer. Subsequently, cDNA was synthesized with SuperScript III (Invitrogen) as per manufacturer’s instructions. qPCR was performed on an Opticon (BIO-RAD) with iQ SYBR Green Supermix (BIO-RAD) and expression levels were normalized to GAPDH.
Clonal Analysis in Primary SEZ Cultures
The primary SEZ cultures were prepared for cluster analysis according to the protocol above (Ortega et al., 2011). The SEZ from two mice were pooled in order to provide an n = 1 and the cells were plated in two wells (24-well plate) onto poly-D-Lysin coated coverslips. An n = 4 was prepared and analyzed for these experiments. Low titer of CAG-IRES-GFP retrovirus was added at 2 h after plating. Tgm2 inhibitor Z-DON (Zedira) or Boc-DON (Zedira) was added 4 h after plating at a concentration of 10 μM and 100 μM, respectively. Control samples had the equivalent volume of DMSO as the Boc-DON samples added to them (2 μl/well). Cells were incubated at 37°C with 5% CO2 for 7 days until fixation with 4% PFA for 15 min at RT. Immunocytochemistry was performed as described above. Cluster counting and analysis was performed after photomicrograph acquisition of the whole coverslips (Axio Imager M2m, Zeiss). SiRNA experiments were performed in the same manner as the in vitro inhibitor experiments, except that the siRNA (QIAGEN) was added to the cultures immediately after plating together with an equal part Hitransfect (QIAGEN) (mixed 30 min prior to use). Each well was transfected with a total concentration of 10 nM siRNA containing four Tgm2 siRNAs (Flexitube, QIAGEN) or the same concentration of scrambled siRNA control (Allstar negative control, QIAGEN).
Hydroxy-Acrylamid Gel Preparation for Cell Culture
In order to prepare gels with different stiffness we used the method developed by Bollmann et al., 2015. Glass bottom Petri dishes were washed first with 70% Ethanol and then 0.1% sodium hydroxide (NaOH). The NaOH treated surface was treated with (3-Aminopropyl) trimethoxysilane (APTMS) for 3 min, washed and subsequently 0.5% glutaraldehyde was applied for 30 min. The gel solution was prepared by adding 500 μl 40% acrylamide, 65 μl 100% hydroxyl-acrylamide and 250 μl 2% bis-acrylamide. 89.4% and 88.8% PBS were added to the premix to obtain the desired shear moduli of 100 Pa and 200 Pa, respectively. The desired gel-stiffness was confirmed with Atomic force microscopy (AFM). Finally, gels were coated with Poly-L-ornithin (Sigma, P4957) for 2 h followed by Laminin coating (Roche, #11243217001) for another 2 h.
Osmotic Pump Preparation and Surgery
Osmotic pumps (model 2001, ALZET®) were prepared the day before surgery according to the vendors instructions. Pumps were filled with 100 μM Z-DON in artificial cerebrospinal fluid (aCSF) or DMSO in an equivalent concentration (0.4%) as in the treatment group. Pumps were kept in sterile PBS at 37°C until surgery. The experimental procedure was approved by the government of upper Bavaria. Mice were anaesthetized by intraperitoneal injection of fentanyl (0.05 mg/kg), midazolam (5 mg/kg), and medetomidine (0.5 mg/kg) and after the surgery the anesthesia was antagonized by injection of buprenorphine (0.1 mg/kg), atipamezole (2.5 mg/kg), and flumazenil (0.5 mg/kg). Intra ventricular osmotic pump implantation (Brain kit 2, ALZET®) was performed at the coordinates 1.2 mm laterally to and 0.5 mm posterior to the bregma (right side) (Lepko et al., 2019). The experimental endpoints were 4 (n = 3) and 7 days (n = 4) after surgery. 5 mg/mL EdU in saline was administered i.p. at a volume of 10 ul per gram of the mouse’s weight 1 h h prior perfusion. The mice were perfused with 4% PFA and the brains were post-fixated overnight.
Stiffness Measurements
Sample Preparation. Male C57BL6/J mice (N = 5 animals), aged 8 weeks, were anaesthetized using 5% isofluorane (IsoFlo, Abbott Laboratories) and euthanized by intraperitoneal injection of a lethal dose of pentobarbital (Pentoject, Animalcare UK), followed by cardiac perfusion with cold slicing aCSF. Brains were immediately dissected out into cold slicing aCSF, keeping the olfactory bulbs intact, and sections prepared for stiffness measurements using the ex vivo acute CNS slice protocol described previously in Koser et al. (2015) and Moeendarbary et al. (2017). Hence, each brain was embedded in 4% low melting point agarose (Sigma; in 1 × PBS), the agarose block containing the tissue was glued onto a vibratome platform (Leica), and 300 mm thick coronal sections were cut in cold slicing aCSF. Sections containing the anatomical regions of interest were transferred to a collection chamber containing measuring aCSF at room temperature (approx. 20°C) and allowed to equilibrate for ∼5-10 min. Sections were then attached to 35 mm Petri dishes (TPP) using small dabs of superglue at the outer corners of the surrounding agarose, immediately covered with fresh measuring aCSF, and transferred to the AFM set-up for measurement. The time elapsed between euthanasia of the animals and the beginning of slicing was ∼30 min.
The compositions of the buffers used in this protocol were as follows: for slicing aCSF, 191 mM sucrose, 0.75 mM K-gluconate, 1.25mM KH2PO4, 26 mM NaHCO3, 4mM MgSO4, 1mM CaCl2, 20 mM glucose, 2 mM kynurenic acid, 1 mM (+)-sodium L-ascorbate, 5 mM ethyl pyruvate, 3 mM myo-inositol, and 2 mM NaOH; and for measuring aCSF, 121 mM NaCl, 3mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 1.1 mM MgCl2, 2.2 mM CaCl2, 15mM glucose, 1mM (+)-sodium L-ascorbate, 5mM ethyl pyruvate, and 3mM myo-inositol. Both solutions were freshly prepared before each experiment and bubbled with 95% O2 and 5% CO2, beginning at least 30 min before first use and continuing throughout the procedure (Koser et al., 2015, Moeendarbary et al., 2017).
Atomic Force Microscopy (AFM) Setup. A JPK CellHesion 200 atomic force microscope (JPK Instruments) was mounted on an inverted optical microscope (AxioObserver.A1, Zeiss) with a motorised x-y stage (JPK Instruments). For stiffness measurements, the spring constant k of tipless silicon cantilevers (Arrow-TL1, NanoWorld) were determined using the thermal noise method (Hutter and Bechhoefer, 1993) and cantilevers with k of 0.01-0.04 N/m (for olfactory bulb (OB) measurements) or 0.05-0.07 N/m (for SEZ/MEZ measurements) were selected. Spherical monodisperse polystyrene beads (diameter = 37.28 ± 0.34 μm (for olfactory bulb) or 19.3 ± 0.34 μm (for SEZ/MEZ)) (microParticles GmbH) were used as probes and attached to the ends of the cantilevers using heat-curing glue (M-Bond 610, MicroMeasurements). The use of spherical probes ensured a consistent contact area with the sample surface and prevented damage to the slices. A PetriDishHeater (JPK Instruments) was set up on the motorised stage and used to maintain samples at a set temperature of 34°C for the duration of AFM measurements.
AFM Measurements. Prepared sections were placed in the sample holder and allowed to equilibrate for ∼5 min while brightfield images of the slices were collected with an Andor Zyla 4.2 CMOS camera (connected to a modified upright AxioZoom V.16 system (Zeiss)). Force-distance curves (set force: 10 nN, approach speed: 5 μm/s, sampling rate: 1,000 Hz) were either taken manually (for SEZ/MEZ measurements), or automatically every 30-40 μm apart in a raster scan (for OB), using a custom-written script (Koser et al., 2015, Koser et al., 2016) generating a ‘stiffness map’ of the area. For stiffness maps, images of the upper right and lower left corners of the measured area were also collected to identify the region of the slice mapped by the AFM.
Microarray Analysis
After dissecting the OB, SEZ and Cx the RNA was extracted using the RNeasy Mini Kit from QIAGEN following the manufacturer’s instructions. Sample preparation and microarray analysis was carried out by Arraystar applying the Agilent Array platform. The sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols with minor modifications (Agilent Low Input Quick Amp Labeling Kit). Briefly, the sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method. The labeled cRNAs were hybridized onto the Mouse LncRNA Array v2.0 (8 × 60K, Arraystar). After having washed the slides, the arrays were scanned by the Agilent Scanner G2505B.
Quantification and Statistical Analysis
Mass Spectrometric Data Analysis
We processed the mass spectra using MaxQuant (http://coxdocs.org/doku.php) (Cox and Mann, 2008). Using the Andromeda search engine, the spectra were searched against the mouse Uniprot sequence database (https://www.uniprot.org). Cysteine carbamidomethylation was set as fixed modification while variable modifications included hydroxylation of proline and methionine oxidation. For both protein and peptide level we set FDR to 0.01 and only peptides with an amino-acid length of seven or more were considered. The peptide identifications among the library samples from the four brain regions (Cx, OB, SEZ, MEZ) in the LMSS dataset were used to additionally identify peptides in the single shot samples based on similar mass and retention time. The matching between runs feature (a feature of the Max quant software) thus allows identification of proteins in the single shot samples (that were identified in library samples) when precursor peptides were found in MS1, but not selected for fragmentation and identification in MS2. Single shots and library samples were also matched within themselves, which was also the case for samples in the QDSP dataset. The label-free protein quantification was restricted to proteins identified with at least two unique peptides. Label-free quantification (LFQ) algorithm was used for protein quantifications (Cox et al., 2014). LFQ intensities are normalized median mass spectra intensity values that allow this quantification to be performed with any peptide and protein fractionation while maintaining high accuracy (Cox et al., 2014). For a protein to be considered valid, two peptide ratios are needed. Among the QDSP samples, there was one sample with two of the brain regions that had protein fraction with very low protein content and several fractions with lower protein identification. Thus, we omitted the whole sample from further bioinformatics analysis. This resulted in 4 samples per region (each with their respective four solubility fractions).
Bioinformatic Analysis and Statistics
Bioinformatic analysis was performed primarily with the Perseus software (coxdocs.org/doku.php) (Tyanova et al., 2016). For all datasets, we have used log2 LFQ intensities for analysis and comparisons. Plots of selected categories or individual proteins were done in Graphpad Prism (version 5).
Bioinformatic Analysis of LMSS Dataset
The presented library proteomes depth signifies number of identified proteins (n = 1/region). The presented library-matched single shot sample measurements are filtered for at least 3 values per region (n = 4/region). This filtering was also used for data imputation. Imputation of missing values was done by random selection according to a normal distribution with negative shift of 1.8 standard deviations from the mean and with a width of 0.3 standard deviations. These log2 LFQ intensities values for all proteins were then used for PCA, statistical analysis, gene ontology analysis and heatmap presentations. Proteome comparisons of regions were done with one-way ANOVA and p values were used for filtering significant regional abundance differences after FDR correction. Heatmaps display proteins with an FDR of 0.05 (Figure 1K). 1D and 2D enrichment analysis of annotated terms was done in Perseus (Figures 1L, 1M, 4E, 4F, 1SE, and 1SF) with an FDR of 0.05 and full annotation enrichment lists can be found in Table S2, and 4. Gene ontology analysis for library exclusive proteins was done in Panther (pantherdb.org) (Thomas et al., 2003) (Figures S1B and S1C). Matrisome distribution plots (Wisker plots) comprise of z-scored mean LFQ intensities for proteins of the respective category (number of proteins for each category is presented in the respective graphs). This data was analyzed using Kruskal-Wallis test with Dunn’s multiple comparison test and p ≤ 0.05 was considered significant. For t test comparisons between brain regions an FDR ≤ 0.1 was considered significant.
Bioinformatic Analysis of QDSP Dataset
The presented total number of proteins from all regions signifies number of identified proteins (n = 4/region). The total number of identifications from each region is instead filtered for at least 3 values in a region. We filtered the data for at least 8 values in a region prior intensity imputation. Imputation of missing values for the protein fractions of each region was done by random selection according to a normal distribution with negative shift of 1.7 standard deviations from the mean and with a width of 0.4 standard deviations. These log2 LFQ intensities values for all fractions were then used for PCA, annotation enrichment analysis, category solubility profiles, and heatmap presentations. 1D enrichment analysis of annotated terms (Figures 4E and 4F) used an FDR of 0.05. Category solubility profile plots (Whisker plots) comprise of z-scored mean LFQ intensities for protein fractions of the respective category (number of proteins for each category is presented in the respective graphs). We present the mean value of the three regions in all “brain” solubility profiles for different categories (z-scoring was always done within regions). This data was analyzed using Kruskal-Wallis test with Dunn’s multiple comparison test and p ≤ 0.05 was considered significant. Categories in the PCA were considered significant with a FDR ≤ 0.05. Differences between brain region solubility profiles were determined using the z-scored LFQ intensities and two-way ANOVA with FDR correction (p value ≤ 0.05 was considered significant). The matrisome proteins with differential solubility (Figure 4H) (p value ≤ 0.05) were filtered from the analysis of all proteins (in heatmap, Figure 4D). In Figure S5C, we compared the brain matrisome data to previously published datasets (using the QDSP method) in aorta and lung tissue (Schiller et al., 2015, Wierer et al., 2018). The averaged solubility profiles here are comprised of an average from all experimental groups of each study.
Analysis and Presentation of AFM Data
Analysis of Raw AFM Data. First, the force-distance curves collected from AFM measurements were analyzed using a custom-written MATLAB script (described previously in Koser et al. [2015] and Koser et al. [2016]) to obtain the reduced apparent elastic modulus K, a measure of tissue stiffness. Raw AFM data were fitted to the Hertz model,
with the applied force F, the reduced apparent elastic modulus K = E/(1-ν2), with E being the Young’s modulus and ν the Poisson’s ratio, R the radius of the probe, and δ the indentation depth (Hertz, 1881). Force-distance curves were analyzed at the maximum applied force of 10 nN. Points where the AFM data was not analyzable were excluded from further analysis. Criteria for excluding individual force-distance curves were the inability to apply linear fits through the baseline of the curve, e.g., due to noise, and the inability to apply good-quality Hertz-fits to the indentation region. K values were color-coded and converted to 8-bit scale colormaps, using the MATLAB ‘hot’ colormap pre-set. The resulting two-dimentional ‘stiffness maps’ were overlaid onto images of the samples using custom-written MATLAB scripts (Koser et al., 2015, Koser et al., 2016). Local brain region stiffness was statistically analyzed using Mann-Whitney test (two-tailed).
Bioinformatic Analysis Microarray Data
Agilent Feature Extraction software (version 10.7.3.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were carried out by Arraystar using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After quantile normalization of the raw data, mRNAs that at least 6 out of 12 samples have flags in Present or Marginal (“All Targets Value”) were chosen for further data analysis. Differentially expressed mRNAs were identified through Volcano Plot filtering.
Data and Code Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and the accession number for the proteomes reported in this paper is ProteomeXchange: PXD016632 (http://proteomecentral.proteomexchange.org). We also provide excel tables with the analyzed proteomics data for easy access. Furthermore, the two proteomes are available with pre-made graphs for each protein on the webpage https://neuronicheproteome.org. The microarray dataset is accessible at GEO: GPL15692. Custom-written scripts used for motorised stage control, processing of AFM raw data, and the generation and alignment of colormaps can be found at https://github.com/FranzeLab.
Acknowledgments
We wish to deeply thank Matthias Mann for allowing us to perform the proteome part of this study in his lab. We thank Fabian Coscia and Fabian Hosp for their help with the proteomics and Christoph Schaab for QDSP graphs. We are very grateful to Andrea Steiner-Mezzadri, Tatiana Simon-Ebert, Korbinian Mayr, and Igor Paron for their technical help. We also thank David Petrik for providing the mouse brain sections from EdU-treated mice and Stefanie Hauck for feedback on the manuscript.
We also gratefully acknowledge funding from the German Research Council (SFB870, SPP1751, and SPP1738 to M.G. and J.N.), the EU (Eranet S-700982-5008-001 to M.G.) and the ERC (Consolidator Grant 772426 to K.F. and Advanced Grant 340793 to M.G.), and the Swedish Society for Medical Research (SSMF) postdoctoral grant (to J.K.).
Author Contributions
M.G. conceived the project. J.K., H.B.S., M.G., and K.F. conceptualized and planned the project. J.K., J.F.-S., H.B.S., K.F., A.J.T., and M.G. designed experiments. J.K. performed all experiments and analyzed all results other than the following: J.F.-S. performed Tgm2 experiments including FACS sorting, whole-mount stainings, qPCR, and sample collection for the microarrays; J.N. performed osmotic minipump implantation and microarray analysis; A.J.T. performed AFM and its analysis and conceptualized these experiments with K.F.; C.F. performed SEZ cryo-dissections and SEZ stainings; M.J.S. and D.C.M. provided C1ql3-mVenus reporter mice. J.K. and M.G. wrote the paper with feedback from the co-authors.
Declaration of Interests
The authors declare no competing interests.
Published: February 6, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2020.01.002.
Supplemental Information
The dataset has been filtered and missing values have been imputed (see STAR Methods). Library-exclusive proteins are provided in a separate sheet. The data are related to Figures 1A–1H and S1A–S1D.
1- and 2-dimentional enrichment analysis for the Uniprot “keyword”-annotation performed in Perseus (see STAR Methods). All Niche enrichment analysis has been done with values of each region’s protein abundance difference to that of the Cx samples. The data are related to Figures 1L, 1M, S1E, and S1F.
The dataset has been filtered and missing values have been imputed (see STAR Methods). The data are related to Figures 4, S5A, and S5B.
1-dimentional enrichment analysis for the Uniprot “keyword”-annotation performed in Perseus (see STAR Methods). The data are only analyzed for the proteins in the OB highlighted in Figure 4D and are directly related to Figures 4E and 4F.
The dataset is based on RNA anlysis of Cx, OB, and SVZ (n = 3 per region). The data are related to Figures 6B and 6C.
The data display protein that diverge in their expression comparing the proteome and microarray data (significantly higher or lower) and 2-dimentional enrichment analysis for the Uniprot “keyword”-annotation comparing the two data sets. The data are related to Figure 2, S7A, and S7B.
References
- Alonso M., Ortega-Pérez I., Grubb M.S., Bourgeois J.P., Charneau P., Lledo P.M. Turning astrocytes from the rostral migratory stream into neurons: a role for the olfactory sensory organ. J. Neurosci. 2008;28:11089–11102. doi: 10.1523/JNEUROSCI.3713-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidis I., Simon L.M., Fernandez I.E., Strunz M., Mayr C.H., Greiffo F.R., Tsitsiridis G., Ansari M., Graf E., Strom T.M. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019;10:963. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker R.A., Götz M., Parmar M. New approaches for brain repair-from rescue to reprogramming. Nature. 2018;557:329–334. doi: 10.1038/s41586-018-0087-1. [DOI] [PubMed] [Google Scholar]
- Baser A., Skabkin M., Kleber S., Dang Y., Gülcüler Balta G.S., Kalamakis G., Göpferich M., Ibañez D.C., Schefzik R., Lopez A.S. Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature. 2019;566:100–104. doi: 10.1038/s41586-019-0888-x. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth R., Tripathi P., Ninkovic J., Bayam E., Lepier A., Stempfhuber B., Kirchhoff F., Hirrlinger J., Haslinger A., Lie D.C. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth R. Mitochondrial Metabolism-Mediated Regulation of Adult Neurogenesis. Brain plasticity. 2017;3:73–87. doi: 10.3233/BPL-170044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollmann L., Koser D.E., Shahapure R., Gautier H.O., Holzapfel G.A., Scarcelli G., Gather M.C., Ulbricht E., Franze K. Microglia mechanics: immune activation alters traction forces and durotaxis. Front. Cell. Neurosci. 2015;9:363. doi: 10.3389/fncel.2015.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordiuk O.L., Smith K., Morin P.J., Semënov M.V. Cell proliferation and neurogenesis in adult mouse brain. PLoS ONE. 2014;9:e111453. doi: 10.1371/journal.pone.0111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody M.J., Schips T.G., Vanhoutte D., Kanisicak O., Karch J., Maliken B.D., Blair N.S., Sargent M.A., Prasad V., Molkentin J.D. Dissection of Thrombospondin-4 Domains Involved in Intracellular Adaptive Endoplasmic Reticulum Stress-Responsive Signaling. Mol. Cell. Biol. 2015;36:2–12. doi: 10.1128/MCB.00607-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulli D., Pizzorusso T., Kwok J.C., Putignano E., Poli A., Forostyak S., Andrews M.R., Deepa S.S., Glant T.T., Fawcett J.W. Animals lacking link protein have attenuated perineuronal nets and persistent plasticity. Brain. 2010;133:2331–2347. doi: 10.1093/brain/awq145. [DOI] [PubMed] [Google Scholar]
- Chew K.S., Fernandez D.C., Hattar S., Südhof T.C., Martinelli D.C. Anatomical and Behavioral Investigation of C1ql3 in the Mouse Suprachiasmatic Nucleus. J. Biol. Rhythms. 2017;32:222–236. doi: 10.1177/0748730417704766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Vincent L.G., Lee A.R., Kretchmer K.C., Chirasatitsin S., Dobke M.K., Engler A.J. The alignment and fusion assembly of adipose-derived stem cells on mechanically patterned matrices. Biomaterials. 2012;33:6943–6951. doi: 10.1016/j.biomaterials.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P., Silva-Vargas V., Paul A., Maldonado-Soto A.R., Deleo A.M., Pastrana E., Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover J.C., Todd K.L. Development and aging of a brain neural stem cell niche. Exp. Gerontol. 2017;94:9–13. doi: 10.1016/j.exger.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.R., Ortega F., Brill M.S., Beckervordersandforth R., Petrone C., Schroeder T., Götz M., Berninger B. Continuous live imaging of adult neural stem cell division and lineage progression in vitro. Development. 2011;138:1057–1068. doi: 10.1242/dev.061663. [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Crane G.M., Jeffery E., Morrison S.J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 2017;17:573–590. doi: 10.1038/nri.2017.53. [DOI] [PubMed] [Google Scholar]
- David L.S., Schachner M., Saghatelyan A. The extracellular matrix glycoprotein tenascin-R affects adult but not developmental neurogenesis in the olfactory bulb. J. Neurosci. 2013;33:10324–10339. doi: 10.1523/JNEUROSCI.5728-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laporte L., Rice J.J., Tortelli F., Hubbell J.A. Tenascin C Promiscuously Binds Growth Factors via Its Fifth Fibronectin Type III-Like Domain. PloS One. 2013;8:e62076. doi: 10.1371/journal.pone.0062076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepa S.S., Carulli D., Galtrey C., Rhodes K., Fukuda J., Mikami T., Sugahara K., Fawcett J.W. Composition of perineuronal net extracellular matrix in rat brain: a different disaccharide composition for the net-associated proteoglycans. J. Biol. Chem. 2006;281:17789–17800. doi: 10.1074/jbc.M600544200. [DOI] [PubMed] [Google Scholar]
- Donato R., Sorci G., Giambanco I. S100A6 protein: functional roles. Cell. Mol. Life Sci. 2017;74:2749–2760. doi: 10.1007/s00018-017-2526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnaloja F., Jacchetti E., Soncini M., Raimondi M.T. Mechanosensing at the Nuclear Envelope by Nuclear Pore Complex Stretch Activation and Its Effect in Physiology and Pathology. Front. Physiol. 2019;10:896. doi: 10.3389/fphys.2019.00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly H., Salmeron-Sanchez M., Dalby M.J. Designing stem cell niches for differentiation and self-renewal. J. R. Soc. Interface. 2018;15:20180388. doi: 10.1098/rsif.2018.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R.L., Kaartinen M.T., Nurminskaya M., Belkin A.M., Colak G., Johnson G.V., Mehta K. Transglutaminase regulation of cell function. Physiol. Rev. 2014;94:383–417. doi: 10.1152/physrev.00019.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Englund U., Fricker-Gates R.A., Lundberg C., Björklund A., Wictorin K. Transplantation of human neural progenitor cells into the neonatal rat brain: extensive migration and differentiation with long-distance axonal projections. Exp. Neurol. 2002;173:1–21. doi: 10.1006/exnr.2001.7750. [DOI] [PubMed] [Google Scholar]
- Faissner A., Roll L., Theocharidis U. Tenascin-C in the matrisome of neural stem and progenitor cells. Mol. Cell. Neurosci. 2017;81:22–31. doi: 10.1016/j.mcn.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Fietz S.A., Lachmann R., Brandl H., Kircher M., Samusik N., Schröder R., Lakshmanaperumal N., Henry I., Vogt J., Riehn A. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. USA. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J., Beckervordersandforth R., Tripathi P., Steiner-Mezzadri A., Ninkovic J., Götz M. Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nat. Protoc. 2011;6:1981–1989. doi: 10.1038/nprot.2011.412. [DOI] [PubMed] [Google Scholar]
- Franze K., Janmey P.A., Guck J. Mechanics in neuronal development and repair. Annu. Rev. Biomed. Eng. 2013;15:227–251. doi: 10.1146/annurev-bioeng-071811-150045. [DOI] [PubMed] [Google Scholar]
- Fricker R.A., Carpenter M.K., Winkler C., Greco C., Gates M.A., Björklund A. Site-specific migration and neuronal differentiation of human neural progenitor cells after transplantation in the adult rat brain. J. Neurosci. 1999;19:5990–6005. doi: 10.1523/JNEUROSCI.19-14-05990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisén J. Neurogenesis and Gliogenesis in Nervous System Plasticity and Repair. Annu. Rev. Cell Dev. Biol. 2016;32:127–141. doi: 10.1146/annurev-cellbio-111315-124953. [DOI] [PubMed] [Google Scholar]
- Garcion E., Faissner A., ffrench-Constant C. Knockout mice reveal a contribution of the extracellular matrix molecule tenascin-C to neural precursor proliferation and migration. Development. 2001;128:2485–2496. doi: 10.1242/dev.128.13.2485. [DOI] [PubMed] [Google Scholar]
- Geyer P.E., Kulak N.A., Pichler G., Holdt L.M., Teupser D., Mann M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016;2:185–195. doi: 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Gilbert P.M., Havenstrite K.L., Magnusson K.E., Sacco A., Leonardi N.A., Kraft P., Nguyen N.K., Thrun S., Lutolf M.P., Blau H.M. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard F., Eichenberger S., Celio M.R. Thrombospondin 4 deficiency in mouse impairs neuronal migration in the early postnatal and adult brain. Mol. Cell. Neurosci. 2014;61:176–186. doi: 10.1016/j.mcn.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Gonzales K.A.U., Fuchs E. Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev. Cell. 2017;43:387–401. doi: 10.1016/j.devcel.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M., Sirko S., Beckers J., Irmler M. Reactive astrocytes as neural stem or progenitor cells: In vivo lineage, In vitro potential, and Genome-wide expression analysis. Glia. 2015;63:1452–1468. doi: 10.1002/glia.22850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M., Nakafuku M., Petrik D. Neurogenesis in the Developing and Adult Brain-Similarities and Key Differences. Cold Spring Harb. Perspect. Biol. 2016;8:a018853. doi: 10.1101/cshperspect.a018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack I., Bancila M., Loulier K., Carroll P., Cremer H. Reelin is a detachment signal in tangential chain-migration during postnatal neurogenesis. Nat. Neurosci. 2002;5:939–945. doi: 10.1038/nn923. [DOI] [PubMed] [Google Scholar]
- Hardy D., Saghatelyan A. Different forms of structural plasticity in the adult olfactory bulb. Neurogenesis (Austin) 2017;4:e1301850. doi: 10.1080/23262133.2017.1301850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz H. Über die Berührung fester elastischer Körper. J. Reine Angew. Math. 1881;92:156–171. [Google Scholar]
- Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- Jaiswal J.K., Nylandsted J. S100 and annexin proteins identify cell membrane damage as the Achilles heel of metastatic cancer cells. Cell Cycle. 2015;14:502–509. doi: 10.1080/15384101.2014.995495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamakis G., Brüne D., Ravichandran S., Bolz J., Fan W., Ziebell F., Stiehl T., Catalá-Martinez F., Kupke J., Zhao S. Quiescence modulates stem cell maintenance and regenerative capacity in the aging brain. Cell. 2019;176:1407–1419.e14. doi: 10.1016/j.cell.2019.01.040. [DOI] [PubMed] [Google Scholar]
- Kazanis I., Belhadi A., Faissner A., Ffrench-Constant C. The adult mouse subependymal zone regenerates efficiently in the absence of tenascin-C. J Neuroscience. 2007;27:13991–13996. doi: 10.1523/JNEUROSCI.3279-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerever A., Schnack J., Vellinga D., Ichikawa N., Moon C., Arikawa-Hirasawa E., Efird J.T., Mercier F. Novel extracellular matrix structures in the neural stem cell niche capture the neurogenic factor fibroblast growth factor 2 from the extracellular milieu. Stem Cells. 2007;25:2146–2157. doi: 10.1634/stemcells.2007-0082. [DOI] [PubMed] [Google Scholar]
- Knobloch M., Jessberger S. Metabolism and neurogenesis. Curr. Opin. Neurobiol. 2017;42:45–52. doi: 10.1016/j.conb.2016.11.006. [DOI] [PubMed] [Google Scholar]
- Kolb J., Anders-Maurer M., Müller T., Hau A.C., Grebbin B.M., Kallenborn-Gerhardt W., Behrends C., Schulte D. Arginine Methylation Regulates MEIS2 Nuclear Localization to Promote Neuronal Differentiation of Adult SVZ Progenitors. Stem Cell Reports. 2018;10:1184–1192. doi: 10.1016/j.stemcr.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser D.E., Moeendarbary E., Hanne J., Kuerten S., Franze K. CNS cell distribution and axon orientation determine local spinal cord mechanical properties. Biophys. J. 2015;108:2137–2147. doi: 10.1016/j.bpj.2015.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser D.E., Thompson A.J., Foster S.K., Dwivedy A., Pillai E.K., Sheridan G.K., Svoboda H., Viana M., Costa L.D., Guck J. Mechanosensing is critical for axon growth in the developing brain. Nat. Neurosci. 2016;19:1592–1598. doi: 10.1038/nn.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Kulak N.A., Geyer P.E., Mann M. Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol. Cell. Proteomics. 2017;16:694–705. doi: 10.1074/mcp.O116.065136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lana B., Page K.M., Kadurin I., Ho S., Nieto-Rostro M., Dolphin A.C. Thrombospondin-4 reduces binding affinity of [(3)H]-gabapentin to calcium-channel α2δ-1-subunit but does not interact with α2δ-1 on the cell-surface when co-expressed. Sci. Rep. 2016;6:24531. doi: 10.1038/srep24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen S.P., Boye T.L., Nylandsted J. Annexins are instrumental for efficient plasma membrane repair in cancer cells. Semin. Cell Dev. Biol. 2015;45:32–38. doi: 10.1016/j.semcdb.2015.10.028. [DOI] [PubMed] [Google Scholar]
- Lee C.S., Park H.H. Structural aspects of transglutaminase 2: functional, structural, and regulatory diversity. Apoptosis. 2017;22:1057–1068. doi: 10.1007/s10495-017-1396-9. [DOI] [PubMed] [Google Scholar]
- Lepko T., Pusch M., Müller T., Schulte D., Ehses J., Kiebler M., Hasler J., Huttner H.B., Vandenbroucke R.E., Vandendriessche C. Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain. EMBO J. 2019;38:e100481. doi: 10.15252/embj.2018100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerchenmüller C., Heißenberg J., Damilano F., Bezzeridis V.J., Krämer I., Bochaton-Piallat M.L., Hirschberg K., Busch M., Katus H.A., Peppel K. S100A6 Regulates Endothelial Cell Cycle Progression by Attenuating Antiproliferative Signal Transducers and Activators of Transcription 1 Signaling. Arterioscler. Thromb. Vasc. Biol. 2016;36:1854–1867. doi: 10.1161/ATVBAHA.115.306415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wagner E.R., Yan Z., Wang Z., Luther G., Jiang W., Ye J., Wei Q., Wang J., Zhao L. The Calcium-Binding Protein S100A6 Accelerates Human Osteosarcoma Growth by Promoting Cell Proliferation and Inhibiting Osteogenic Differentiation. Cell. Physiol. Biochem. 2015;37:2375–2392. doi: 10.1159/000438591. [DOI] [PubMed] [Google Scholar]
- Lim D.A., Alvarez-Buylla A. The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb. Perspect. Biol. 2016;8:a018820–a018834. doi: 10.1101/cshperspect.a018820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledo P.M., Valley M. Adult Olfactory Bulb Neurogenesis. Cold Spring Harb. Perspect. Biol. 2016;8:a018945. doi: 10.1101/cshperspect.a018945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Long K.R., Newland B., Florio M., Kalebic N., Langen B., Kolterer A., Wimberger P., Huttner W.B. Extracellular matrix components HAPLN1, Lumican and collagen I cause hyaluronic acid-dependent folding of the developing human neocortex. Neuron. 2018;99:702–719.e6. doi: 10.1016/j.neuron.2018.07.013. [DOI] [PubMed] [Google Scholar]
- Long K.R., Huttner W.B. How the extracellular matrix shapes neural development. Open Biol. 2019;9:180216. doi: 10.1098/rsob.180216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N. Proteoglycans and neuronal migration in the cerebral cortex during development and disease. Front. Neurosci. 2015;9:98. doi: 10.3389/fnins.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majkut S., Idema T., Swift J., Krieger C., Liu A., Discher D.E. Heart-specific stiffening in early embryos parallels matrix and myosin expression to optimize beating. Curr. Biol. 2013;23:2434–2439. doi: 10.1016/j.cub.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli D.C., Chew K.S., Rohlmann A., Lum M.Y., Ressl S., Hattar S., Brunger A.T., Missler M., Südhof T.C. Expression of C1ql3 in Discrete Neuronal Populations Controls Efferent Synapse Numbers and Diverse Behaviors. Neuron. 2016;91:1034–1051. doi: 10.1016/j.neuron.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K., Budisantoso T., Mitakidis N., Sugaya Y., Miura E., Kakegawa W., Yamasaki M., Konno K., Uchigashima M., Abe M. Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron. 2016;90:752–767. doi: 10.1016/j.neuron.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Meran L., Baulies A., Li V.S.W. Intestinal Stem Cell Niche: The Extracellular Matrix and Cellular Components. Stem Cells Int. 2017;2017:7970385. doi: 10.1155/2017/7970385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midwood K.S., Chiquet M., Tucker R.P., Orend G. Tenascin-C at a glance. J. Cell Sci. 2016;129:4321–4327. doi: 10.1242/jcs.190546. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z., Doetsch F., Sawamoto K., Wichterle H., Alvarez-Buylla A. The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 2010;6:1938. doi: 10.3791/1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeendarbary E., Weber I.P., Sheridan G.K., Koser D.E., Soleman S., Haenzi B., Bradbury E.J., Fawcett J., Franze K. The soft mechanical signature of glial scars in the central nervous system. Nat. Commun. 2017;8:14787. doi: 10.1038/ncomms14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., Hynes R.O. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014647. M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narouz-Ott L., Maurer P., Nitsche D.P., Smyth N., Paulsson M. Thrombospondin-4 binds specifically to both collagenous and non-collagenous extracellular matrix proteins via its C-terminal domains. J. Biol. Chem. 2000;275:37110–37117. doi: 10.1074/jbc.M007223200. [DOI] [PubMed] [Google Scholar]
- Ninkovic J., Steiner-Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., von Holst A., Beckers J., Lie C.D. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte C., Matyash M., Pivneva T., Schipke C.G., Ohlemeyer C., Hanisch U.K., Kirchhoff F., Kettenmann H. GFAP promoter-controlled EGFP-expressing transgenic mice: a tool to visualize astrocytes and astrogliosis in living brain tissue. Glia. 2001;33:72–86. [PubMed] [Google Scholar]
- Ortega F., Costa M.R., Simon-Ebert T., Schroeder T., Götz M., Berninger B. Using an adherent cell culture of the mouse subependymal zone to study the behavior of adult neural stem cells on a single-cell level. Nat. Protoc. 2011;6:1847–1859. doi: 10.1038/nprot.2011.404. [DOI] [PubMed] [Google Scholar]
- Pathak M.M., Nourse J.L., Tran T., Hwe J., Arulmoli J., Le D.T., Bernardis E., Flanagan L.A., Tombola F. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:16148–16153. doi: 10.1073/pnas.1409802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik D., Myoga M.H., Grade S., Gerkau N.J., Pusch M., Rose C.R., Grothe B., Götz M. Epithelial Sodium Channel Regulates Adult Neural Stem Cell Proliferation in a Flow-Dependent Manner. Cell Stem Cell. 2018;22:865–878.e8. doi: 10.1016/j.stem.2018.04.016. [DOI] [PubMed] [Google Scholar]
- Pillat M.M., Lameu C., Trujillo C.A., Glaser T., Cappellari A.R., Negraes P.D., Battastini A.M.O., Schwindt T.T., Muotri A.R., Ulrich H. Bradykinin promotes neuron-generating division of neural progenitor cells through ERK activation. J. Cell Sci. 2016;129:3437–3448. doi: 10.1242/jcs.192534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll L., Faissner A. Influence of the extracellular matrix on endogenous and transplanted stem cells after brain damage. Front. Cell. Neurosci. 2014;8:219. doi: 10.3389/fncel.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddy R.M., Morshead C.M. Home sweet home: the neural stem cell niche throughout development and after injury. Cell Tissue Res. 2018;371:125–141. doi: 10.1007/s00441-017-2658-0. [DOI] [PubMed] [Google Scholar]
- Saha K., Keung A.J., Irwin E.F., Li Y., Little L., Schaffer D.V., Healy K.E. Substrate modulus directs neural stem cell behavior. Biophys. J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B., Ypsilanti A.R., Boutin C., Cremer H., Chédotal A. Plexin-B2 regulates the proliferation and migration of neuroblasts in the postnatal and adult subventricular zone. J. Neurosci. 2012;32:16892–16905. doi: 10.1523/JNEUROSCI.0344-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H.B., Fernandez I.E., Burgstaller G., Schaab C., Scheltema R.A., Schwarzmayr T., Strom T.M., Eickelberg O., Mann M. Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol. Syst. Biol. 2015;11:819. doi: 10.15252/msb.20156123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidenfaden R., Desoeuvre A., Bosio A., Virard I., Cremer H. Glial conversion of SVZ-derived committed neuronal precursors after ectopic grafting into the adult brain. Mol. Cell. Neurosci. 2006;32:187–198. doi: 10.1016/j.mcn.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Silva-Vargas V., Maldonado-Soto A.R., Mizrak D., Codega P., Doetsch F. Age-Dependent Niche Signals from the Choroid Plexus Regulate Adult Neural Stem Cells. Cell Stem Cell. 2016;19:643–652. doi: 10.1016/j.stem.2016.06.013. [DOI] [PubMed] [Google Scholar]
- Sorg B.A., Berretta S., Blacktop J.M., Fawcett J.W., Kitagawa H., Kwok J.C.F., Miquel M. Casting a Wide Net: Role of Perineuronal Nets in Neural Plasticity. J. Neurosci. 2016;36:11459–11468. doi: 10.1523/JNEUROSCI.2351-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukel J.M., Willits R.K. The interplay of peptide affinity and scaffold stiffness on neuronal differentiation of neural stem cells. Biomed. Mater. 2018;13:024102. doi: 10.1088/1748-605X/aa9a4b. [DOI] [PubMed] [Google Scholar]
- Sullivan C.S., Gotthard I., Wyatt E.V., Bongu S., Mohan V., Weinberg R.J., Maness P.F. Perineuronal Net Protein Neurocan Inhibits NCAM/EphA3 Repellent Signaling in GABAergic Interneurons. Sci. Rep. 2018;8:6143. doi: 10.1038/s41598-018-24272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.W., Tewari M. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M., Van der Veken L., Silva-Vargas V., Louissaint M., Colonna L., Zaidi B., Garcia-Verdugo J.M., Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Pillai E.K., Dimov I.B., Foster S.K., Holt C.E., Franze K. Rapid changes in tissue mechanics regulate cell behaviour in the developing embryonic brain. eLife. 2019;8:e39356. doi: 10.7554/eLife.39356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Vincent L.G., Choi Y.S., Alonso-Latorre B., del Álamo J.C., Engler A.J. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol. J. 2013;8:472–484. doi: 10.1002/biot.201200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining K.H., Mooney D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Peterson J.M., Wong G.W. Metabolic regulation by C1q/TNF-related protein-13 (CTRP13): activation OF AMP-activated protein kinase and suppression of fatty acid-induced JNK signaling. J. Biol. Chem. 2011;286:15652–15665. doi: 10.1074/jbc.M110.201087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierer M., Prestel M., Schiller H.B., Yan G., Schaab C., Azghandi S., Werner J., Kessler T., Malik R., Murgia M. Compartment-resolved Proteomic Analysis of Mouse Aorta during Atherosclerotic Plaque Formation Reveals Osteoclast-specific Protein Expression. Mol. Cell. Proteomics. 2018;17:321–334. doi: 10.1074/mcp.RA117.000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C., Fricker R.A., Gates M.A., Olsson M., Hammang J.P., Carpenter M.K., Björklund A. Incorporation and glial differentiation of mouse EGF-responsive neural progenitor cells after transplantation into the embryonic rat brain. Mol. Cell. Neurosci. 1998;11:99–116. doi: 10.1006/mcne.1998.0674. [DOI] [PubMed] [Google Scholar]
- Yamada J., Jinno S. S100A6 (calcyclin) is a novel marker of neural stem cells and astrocyte precursors in the subgranular zone of the adult mouse hippocampus. Hippocampus. 2014;24:89–101. doi: 10.1002/hipo.22207. [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li X., Janairo R.R.R., Kwong G., Tsou A.D., Chu J.S., Wang A., Yu J., Wang D., Li S. Matrix stiffness modulates the differentiation of neural crest stem cells in vivo. J. Cell. Physiol. 2019;234:7569–7578. doi: 10.1002/jcp.27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The dataset has been filtered and missing values have been imputed (see STAR Methods). Library-exclusive proteins are provided in a separate sheet. The data are related to Figures 1A–1H and S1A–S1D.
1- and 2-dimentional enrichment analysis for the Uniprot “keyword”-annotation performed in Perseus (see STAR Methods). All Niche enrichment analysis has been done with values of each region’s protein abundance difference to that of the Cx samples. The data are related to Figures 1L, 1M, S1E, and S1F.
The dataset has been filtered and missing values have been imputed (see STAR Methods). The data are related to Figures 4, S5A, and S5B.
1-dimentional enrichment analysis for the Uniprot “keyword”-annotation performed in Perseus (see STAR Methods). The data are only analyzed for the proteins in the OB highlighted in Figure 4D and are directly related to Figures 4E and 4F.
The dataset is based on RNA anlysis of Cx, OB, and SVZ (n = 3 per region). The data are related to Figures 6B and 6C.
The data display protein that diverge in their expression comparing the proteome and microarray data (significantly higher or lower) and 2-dimentional enrichment analysis for the Uniprot “keyword”-annotation comparing the two data sets. The data are related to Figure 2, S7A, and S7B.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and the accession number for the proteomes reported in this paper is ProteomeXchange: PXD016632 (http://proteomecentral.proteomexchange.org). We also provide excel tables with the analyzed proteomics data for easy access. Furthermore, the two proteomes are available with pre-made graphs for each protein on the webpage https://neuronicheproteome.org. The microarray dataset is accessible at GEO: GPL15692. Custom-written scripts used for motorised stage control, processing of AFM raw data, and the generation and alignment of colormaps can be found at https://github.com/FranzeLab.