Abstract
Systematic Evolution of Ligands by Exponential Enrichment (SELEX) is the iterative process by which nucleic acids that can bind with high affinity and specificity (termed aptamers) to specific protein targets are selected. Using a SELEX protocol adapted for Xeno-Nucleic Acid (XNA) as a suitable substrate for aptamer generation, 2′-fluoroarabinonucleic acid (FANA) was used to select several related aptamers to HIV-1 integrase (IN). IN bound FANA aptamers with equilibrium dissociation constants (KD,app) of ∼50–100 pM in a buffer with 200 mM NaCl and 6 mM MgCl2. Comparisons to published HIV-1 IN RNA and DNA aptamers as well as IN genomic binding partners indicated that FANA aptamers bound more than 2 orders of magnitude more tightly to IN. Using a combination of RNA folding algorithms and covariation analysis, all strong binding aptamers demonstrated a common four-way junction structure, despite significant sequence variation. IN aptamers were selected from the same starting library as FA1, a FANA aptamer that binds with pM affinity to HIV-1 Reverse Transcriptase (RT). It contains a 20-nucleotide 5′ DNA sequence followed by 59 FANA nucleotides. IN-1.1 (one of the selected aptamers) potently inhibited IN activity and intasome formation in vitro. Replacing the FANA nucleotides of IN-1.1 with 2′-fluororibonucleic acid (F-RNA), which has the same chemical formula but with a ribose rather than arabinose sugar conformation, dramatically reduced binding, suggesting that FANA adopts unique structural conformations that promote binding to HIV-1 IN.
Human immunodeficiency virus (HIV-1) integrase (IN), the enzyme responsible for incorporating virus-derived proviral double stranded DNA (dsDNA) into the host cell chromosome, has become an important drug target for HIV therapy.1,2 HIV-1 IN is a 32-kilodalton (kDa) protein with three distinct domains: an N-terminus that coordinates zinc; a catalytic core domain that catalyzes the transfer of viral nucleic acid into the host cell chromosome via a conserved “D,D(35)E” motif commonly found in members of a nucleotidyltransferase superfamily, which includes HIV-1 IN, other retroviral integrases, and bacterial and eukaryotic transposases;3,4 and a C-terminus.5 IN initiates the transfer of dsDNA to the host cell chromosome through two distinct but catalytically symmetrical reactions. Initially, IN mediates cleavage of the penultimate bases from the 3′ ends of the dsDNA, exposing terminal hydroxyl groups for nucleophilic attack on the target host DNA. These newly exposed hydroxyl groups are then coordinated to hydrolyze the phosphodiester backbone of the host cell chromosome that results in covalent attachment of the viral nucleic acid to the host genome.6,7 These reactions are carried out by nucleoprotein complexes called intasomes in which a pair of viral DNA ends are synapsed by a multimer of IN. Intasomes are the target of currently approved drugs for the treatment of AIDS that target DNA integration. The first FDA approved inhibitor to IN was granted approval in 2007 and is an Integrase Strand Transfer Inhibitor (INSTI) known as Raltegravir (RAL).8 Second generation INSTIs have since been approved for HIV treatment including Dolutegravir (DTG), Elvitegravir (EVG), and Bictegravir (BIC). INSTIs have dramatically improved the current regimens for the treatment of HIV and Acquired Immune Deficiency Syndrome (AIDS). However, emergent drug resistance emphasizes the need to further understand the mechanisms underlying viral biochemistry and pathogenesis.9
One approach for studying and potentially inhibiting protein targets is to generate aptamers (short nucleic acid molecules that bind with high affinity and specificity to their target) to the specific protein. Aptamers can serve a myriad of purposes including, among others, replacement of antibodies in flow cytometry, cell-phenotyping, and ELISA assays; biosensors to detect and quantify proteins; and inhibitors for the neutralization of bacterial toxins and other targets.10−24 Currently, only one aptamer has received FDA approval (Macugen) for the treatment of macular degeneration, but several other candidates are in or entering the drug pipeline.16,25 Recently a “primer-template mimicking” aptamer was used to aid in the crystallization and characterization of HIV and hepatitis B virus (HBV) reverse transcriptases (RT), uncovering yet another potential use for aptamers.21,26,27 In the case of HIV-1, aptamers have been successfully used in vitro and display potent antiviral activity with minor cytopathic effects, as well as demonstrating efficacy in animal models.14,27−37 Current DNA aptamers to IN are potent inhibitors of enzyme activity and resemble G-quadruplex structures.1,12,27 In some cases, these aptamers can be targeted to both IN and the RNase H domain of HIV RT, as these proteins are structurally related through their phosphotransferase domains.27,38,39 RNA aptamers containing G-rich sequences have also been raised to IN and exhibited low nanomolar binding KD values.11,40
Aptamers can be raised to specific proteins using the iterative multistep process known as the Systematic Evolution of Ligands by Exponential Enrichment (SELEX).41,42 Successful selection of aptamers relies on several rounds of SELEX and a sufficient degree of sequence diversity (∼1014 or more sequences) to identify specific binding partners. Since its inception, SELEX has been used to select hundreds of aptamers to an array of diverse proteins from several structural and functional classes.
In order to use aptamers as inhibitors in vivo or in assays with cellular lysates, they must be stable in an environment wrought with nucleases that degrade foreign nucleic acid. One interesting approach to this problem is the replacement of conventional nucleic acid substrates with their synthetic equivalents, potentially avoiding detection by the immune system and acquiring enhanced resistance to nucleases. Xeno-Nucleic Acid (XNA) refers to any nucleic acid analogue that differs from natural ones in at least one of the three chemical moieties: sugar, phosphate, or nucleobase.43 XNAs allow the adaptation of chemical or structural configurations distinct from traditional hereditary material.44 Despite chemical and conformational differences that are theorized to explain the nuclease resistance seen by these analogues, XNAs retain the ability to adopt secondary structures and store information similarly to their DNA and RNA counterparts.45−47 Aptamers containing XNA can be made by modifying some of the nucleosides in aptamers produced with conventional nucleic acids (Macugen was produced using this approach48) or by direct selection using XNA nucleotides in a SELEX protocol. Some XNA nucleotides (e.g., 2′-fluororibonucleic acid (F-RNA) pyrimidines) can be incorporated using natural polymerases; however, most require engineered polymerases for incorporation.49
The list of XNA aptamers continues to grow, although it is restricted by the limited commercial availability of XNA triphosphates. Directly selected aptamers include those produced with HNA (1,5-anhydrohexitol nucleic acid), TNA (threose nucleic acid (α-l-threofuranosyl-(3′,2′) nucleic acid), F-RNA/RNA, and FANA among others49−58 (for a review, see ref (54)). The first FANA aptamer was raised to HIV-1 RT and bound with picomolar affinity.55 FANA is structurally and chemically distinct from F-RNA or RNA due to its difference in sugar pucker as an arabinose-based analogue and the presence of an electronegative fluorine atom in the 2′ position in the β conformation in place of a hydroxyl group.59,60 The sugar adopts a C2′/O4′-endo conformation much like DNA, in stark contrast to the C3′-endo conformation seen by both F-RNA and RNA.44,59,61−63
This report builds upon the framework that allowed for the selection of a FANA aptamer to HIV-1 RT and applies it to HIV IN.55 FANA aptamers selected for IN binding are the second set of FANA aptamers selected for binding to a protein (a catalytically active RNA cleaving FANA aptamer has also been described56). These XNA aptamers are structurally and chemically distinct from other DNA and RNA aptamers that bind IN with low nanomolar affinity and display markedly tighter affinity in the low picomolar range.
Materials and Methods
Materials
The 2′-deoxy-2′-fluoro-arabino-nucleotides (faATP, faCTP, faGTP, faUTP) required for FANA synthesis were obtained from Metkinen Chemistry (Kuusisto, Finland). Deoxyribonucleotide triphosphates (dNTPs) were from Roche or United States Biochemical (USB). Enzymes and buffers including Taq polymerase, T4 polynucleotide kinase (PNK), 10X ThermoPol buffer (Mg2+-free), MgSO4, and 10× CutSmart buffer were from New England BioLabs. Radiolabeled ATP (γ-32P) was from PerkinElmer. G-25 spin columns were from Harvard Apparatus. Miniprep DNA preparation kits were purchased from Qiagen. Nitrocellulose filter disks (Protran BA 85, 0.45 μm pore size and 25 mm diameter) were from Whatman. All DNA oligonucleotides were from Integrated DNA Technologies. All other chemicals were from VWR, Fisher Scientific, or Sigma. The HIV-1 IN wild type (INwt) and mutants (F185H/C280S (INF185H/C280S) and Sso7d-IN fusion (INSso7d)) were prepared as described.37,64 Prototype Foamy Virus (PFV) IN protein was prepared as described.65 All IN proteins were stored in aliquots at −80 °C. Thermostable polymerase D4K was prepared as described and stored in aliquots at −80 °C.49
Methods
End-Labeling of Oligonucleotides with T4 PNK
DNA and RNA oligonucleotides were 5′ end-labeled in a 50 μL volume containing 10–50 pmol of the oligonucleotide of interest, 1 × T4 PNK reaction buffer (provided by manufacturer), 10 U of T4 PNK, and 10 μL of (γ-32P) ATP (3000 Ci/mmol, 10 μCi/μL). The labeling reaction was done at 37 °C for 30 min according to the manufacturer’s protocol. The PNK enzyme was heat inactivated by incubating the reaction at 75 °C for 15 min. Excess radiolabeled nucleotides were then removed by centrifugation using a Sephadex G-25 column.
Selection of FANA Aptamers with HIV-1 INF185H/C280S Using Nitrocellulose Filter Binding
The 79 nucleotide FANA random pool starting material for SELEX containing a 40 nucleotide central random region flanked at the 5′ end by 20 nucleotides of DNA (5′-AAAAGGTAGTGCTGAATTCG-3′) and at the 3′ end by 19 nucleotides of fixed FANA sequence (5′-UUCGCUAUCCAGUUGGCCU-3′) was prepared as described previously.55 About 100 pmol of the 5′-32P-labeled FANA starting pool was snap-cooled on ice and then incubated with 10 pmol of HIV-1 IN (INF185H/C280S was used for all experiments unless otherwise indicated) and 10 μg of yeast tRNA competitor in 40 μL of Binding Buffer (20 mM Tris-HCl at pH 7.5, 200 mM NaCl, 6 mM MgCl2, and 1 mM DTT) for 10 min at RT. A separate control was performed identically except that INF185H/C280S was omitted. The control was used to ensure that selection in the presence of INF185H/C280S bound significantly more material than those in its absence. The material was applied to a nitrocellulose filter that was preincubated in a Low Salt Buffer (20 mM Tris-HCl at pH 7.5, 10 mM KCl). A vacuum was applied at a rate of ∼0.25 mL/s to pull the sample through the filter. Filters were washed three times with 1 mL of Low Salt Buffer. Filters were exposed to imaging screens, and the portion of the filter containing radioactive material in selections containing INF185H/C280S was excised. The excised filter slice was mixed with ∼300 μL of 7 M urea and 5 mM EDTA (pH 8) and heated for 5 min at 95 °C. A solution of phenol/chloroform/isoamyl alcohol (25:24:1) saturated with 100 mM Tris-HCl (pH 8; 300 μL) was added, and the material was vortexed then centrifuged in a microfuge to separate phases. FANA sequences were recovered in the aqueous phase. The phenol-chloroform phase was re-extracted a second time by adding 400 μL of water and repeating the centrifugation (selection round 1 only). The recovered material was reverse transcribed to DNA and processed to produce FANA material for round 2 as previously described.55 For subsequent rounds, ∼ 40 pmol of FANA material was used for selection with a 1:10 ratio of IN/FANA. In some rounds, extra FANA was made to perform a control reaction in the absence of INF185H/C280S. After six, eight, and nine rounds, the amount of competitor tRNA was increased from 10 to 20, 40, and 50 μg, respectively, to increase the stringency of the binding conditions. After the second round, some material from PCR 1 was saved as a source to regenerate the selected material from these rounds. Filter binding assays (see below) were used to access the binding affinity to INF185H/C280S of the pools recovered from some of the rounds. The SELEX was stopped after round 10 as no further binding affinity increase was detected between rounds 8 and 10.
Sequence Analysis of FANA Products Recovered from Round 10
PCR products were prepared from FANA sequences recovered from round 10. The PCR material was cloned using a TOPO TA cloning kit from Life Technologies. DNA mini-preps were prepared, and the products were sequenced by Macrogen (Rockville, Maryland). The appropriate DNA oligonucleotide templates for some recovered sequences were synthesized and generation of FANA material was performed as described.55
Determination of Apparent Equilibrium Dissociation Constant (KD,app) Using Nitrocellulose Filter Binding Assay
Standard reactions for KD,app determinations were performed in 1 mL of Binding Buffer with 0.1 mg mL–1 BSA and 2 pM 5′-32P end-labeled aptamer. Increasing amounts of IN (INwt, INF185H/C280S, or INSso7d as indicated) diluted in the above buffer were added in amounts that approximately flanked the KD,app value (estimated from initial experiments) for the aptamer. For aptamers with very low KD,app values (e.g., IN-1.1 (Table 1)), the amount of IN used was typically 0, 4, 8, 16, 31, 63, 125, 250, and 2000 pM. For controls and aptamer constructs with higher KD,app values, the concentrations of IN and aptmer were increased, and the volume of the reaction was decreased to 100 μL (with 20 pM aptamer) or 20 μL (with 100 pM aptamer). After 10 min at RT, the reactions were applied to a 25 mm nitrocellulose disk (0.45 μm pore, Protran BA 85, Whatman) presoaked in filter wash buffer (25 mM Tris-HCl at pH = 7.5, 10 mM KCl). The filter was washed three times under a vacuum with 1 mL of wash buffer at a flow rate of ∼ 0.25 mL/s. Filters were then counted in a scintillation counter. A plot of bound aptamer vs IN concentration was fit to the following equation for ligand binding one-site saturation in SigmaPlot in order to determine the KD,app: y = Bmax(x)/(KD + x) where x is the concentration of IN and y is the amount of bound aptamer. The experiment was performed three times, and the KD,app values in Table 1 are an average of those experiments ± standard deviations. See Figure S1 for a graphical example.
Table 1. Equilibrium Dissociation Constants for Aptamers and Constructs.
| substrate (IN type) | KD,appa |
|---|---|
| P5 (INF185H/C280S) | 53 ± 26 nM |
| HIV-1 TAR (INF185H/C280S) | NDc |
| HIV-1 TAR (INwt) | NDc |
| FANA random pool (INF185H/C280S)b | 4.3 ± 3.2 μM |
| FANA random pool (INSso7d) | 634 ± 355 nM |
| IN-1.1 (INwt) | 103 ± 18 pM |
| IN-1.1 (INF185H/C280S) | 51 ± 18 pM |
| IN-1.1 (INSso7d) | 74 ± 53 pM |
| IN-2.1 (INF185H/C280S) | 44 ± 9 pM |
| IN-3.1 (INF185H/C280S) | 102 ± 42 pM |
| IN-6.1 (INF185H/C280S) | 39 ± 12 pM |
| IN-8.1 (INF185H/C280S) | NDc |
| IN-13.1 (INF185H/C280S) | NDc |
| IN-14.1 (INF185H/C280S) | 2.2 ± 1.1 μM |
| modifications bound to INF185H/C280S | |
| IN-1.1(FANA to F-RNA) | NDc |
| IN-1.1(P1flip) | 89 ± 31 pM |
| IN-1.1(−10) | 56 ± 28 pM |
| IN-2.1(P1flip) | 106 ± 31 pM |
| IN-2.1(altP1) | 8.9 ± 0.8 nM |
| IN-2.1(−10) | 101 ± 67 pM |
| IN-6.1/2.1 | 118 ± 40 pM |
| IN-6.1/2.1(−10) | 146 ± 9 pM |
| IN 1.1 binding to PFV INd | |
| IN-1.1 (PFV IN) | 102 ± 36 nM |
Apparent equilibrium dissociation constants (KD,app) were measured as described in the Materials and Methods.
Starting material for the FANA SELEX experiments.
ND: not determined. The construct bound too weakly to determine a binding constant in reaction with up to 5 μM IN.
PFV IN, Prototype Foamy Virus Integrase.
Competition Binding Assays
Five nanomolar 5′-32P end-labeled IN-1.1 was incubated at room temperature in Binding Buffer with various amounts of excess unlabeled competitor (IN-1.1, P5 aptamer,11 or HIV TAR (1–57),66 at 0-, 1-, 2-, 4-, 8-, or 16-fold excess over radiolabeled IN-1.1). HIV INF185H/C280S was added to a final concentration of 5 nM. The total volume was 20 μL. Incubations were continued for 50 min. Samples were run over a nitrocellulose filter and washed and quantified as described above.
Concerted Integration and Intasome Assembly Assays
INSso7d (2.5 μM) and 1.0 μM FAM-labeled 27-bp viral DNA substrates were preincubated on ice in 20 mM HEPES at pH 7.5, 25% glycerol, 10 mM DTT, 5 mM MgCl2, 4 μM ZnCl2, 300 ng of target plasmid DNA pGEM-9zf, and 100 mM NaCl in a 20 μL reaction volume.64 The reaction was carried out at 37 °C for 1 h. For integration product analysis, the reactions were stopped by the addition of SDS and EDTA to 0.2% and 10 mM, respectively, together with 5 μg of proteinase K. Incubation was continued at 37 °C for a further 1 h. The DNA was then recovered by ethanol precipitation and subjected to electrophoresis in a 1.5% agarose gel in 1 × TBE buffer. Intasome assembly was carried out in the same way except that no target DNA was added, and MgCl2 was substituted with CaCl2. For electrophoretic mobility shift assays of intasomes (EMSA), the reaction was stopped by chilling on ice and the addition of 10 μg/mL heparin. A 2.5 μL aliquot was subjected to electrophoresis on a 3.0% low melting 1 × TBE agarose gel (SeaKem LE agarose) containing 10 μg/mL heparin. DNA was visualized by fluorescence using a Typhoon 8600 fluorescence scanner (GE Healthcare).
Comparative Sequence Analysis and Structure Prediction
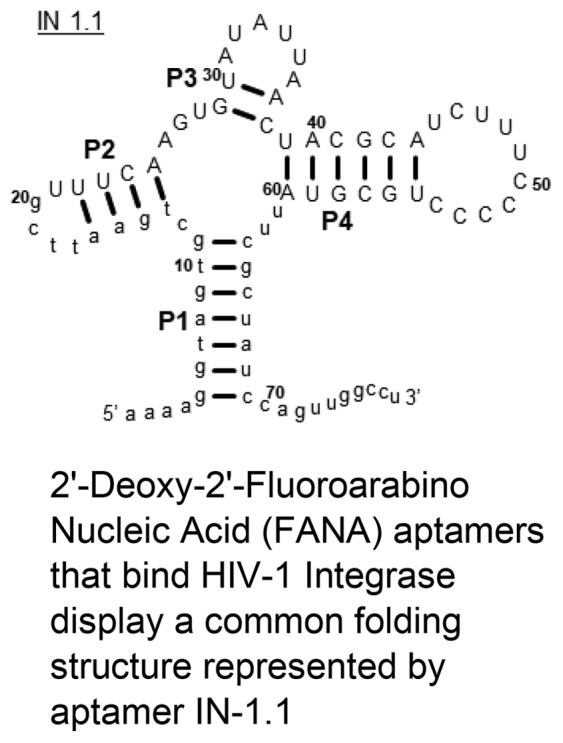
Recovered sequences (26 total) were clustered into 14 separate lineages (shown in Figure 1). We then searched for sequence-level patterns that are common to multiple lineages. We began by identifying all six-nucleotide long sequences within the variable region of the most abundant sequence (lineage 1) and then searched for those 6-mers within the other 13 lineages. Two 6-mers, UCAAGU and AUAUUA, are both present in six of the lineages (lineages 1–6 in Figure 1), and in each instance, the 6-mers are separated by two nucleotides. A very similar sequence pattern was then identified in lineage 7. Next, we took one representative sequence from each of the lineages that contain these 6-mers and performed a sequence alignment using Mafft.67 We then looked for sequence covariation between lineages that might suggest base-pairing or other physical interactions between specific positions in the sequences. From our alignment, we could see covariation between sequences. We generated a secondary structure prediction based on this alignment using RNAalifold,68,69 and it predicts the pairing shown for P3 and P4 (Figure 2). Because the sequences within the 5′ and 3′ constant regions do not vary, we could not use covariation to support structure predictions in these parts of the molecule. We therefore looked at the structure predictions of individual sequences to determine likely base pairing patterns involving the constant regions. We used RNAfold,68,69 to predict secondary structures both with and without folding constraints that force the predicted structure to contain P3 and P4. RNAfold predicted multiple different base pairing patterns for the constant regions (Figure S2). To identify which of these predicted structures is correct, we tested the impact that mutations in the constant regions had on binding. Mutations predicted to disrupt either the P1 or P2 pairing shown in Figure 2 have a pronounced negative impact on binding (Figures S3–S5). Alternatively, mutations that are predicted to maintain the pairing in P1 and P2 have binding comparable to the original aptamer. We therefore used the pairing for P1 and P2 shown in Figure 2 to generate our final structure-based alignment.
Figure 1.

Sequence of the random region from 26 sequences obtained after 10 rounds of selection with INF185H/C280S. Constructs recovered from round 10 of SELEX were PCR amplified and cloned and sequenced as described in the Methods. The recovered sequences are divided into 14 different lineages based on sequence relationships. One sequence (IN-1.1) appeared in the selected pool three times (parentheses), and four sequences differed from it by 1–3 nucleotides (IN-1.2–1.5). IN-2.1 appeared twice, and all other sequences were recovered only one time. For members of the same lineage, an alignment with the first member is shown. Each sequence was flanked at the 5′ end by 20 DNA nucleotides (5′-AAAAGGTAGTGCTGAATTCG-3′) and at the 3′ end by 19 nucleotides of fixed FANA sequence (5′-UUCGCUAUCCAGUUGGCCU-3′).
Figure 2.
Predicted structure and motif alignment of recovered aptamers. The first seven lineages shown in Figure 1 are presented and aligned with the various predicted stem motifs (P1–P4; lower panel). See Methods and Results for a description of how the motifs were predicted. Structures for three of the aptamers are shown (upper panel). The model predicts that all seven lineages form a similar four way junction secondary structure. For lineages with more than one member, only the first member from Figure 1 is shown.
Results
Protocol for FANA Aptamer Selection
Aptamers were selected from a starting pool of approximately 6 × 1013 different sequences using the previously established approach for HIV RT55 with the modifications noted in the Materials and Methods. Note that this approach produces “chimeric” aptamers with a 20-nucleotide fixed DNA sequence at the 5′ end followed by a ∼40-nucleotide region of random FANA nucleotides, then 19 nucleotides of the FANA fixed sequence. With HIV RT, most of the DNA region played no role in the binding of the selected aptamers to RT.55 This was not the case for IN where the DNA region was pivotal for tight binding (see below). Selection was stopped after round 10 of SELEX as there was essentially no difference in the binding of pooled material for rounds 9 and 10, both of which bound at ∼2 nM.
Sequences Recovered from the FANA SELEX
Material from round 10 of the SELEX was cloned, and 26 sequences were recovered (Figure 1, only random region nucleotides are shown, and Clustal was used to compare the sequences from the same lineage). The sequences are arranged in 14 different lineages based on sequence relationships. One sequence appeared three times (IN-1.1) and another twice (IN-2.1). All other sequences appeared only once. IN-1.1 was also closely related to four other sequences (IN-1.2–IN-1.5) and showed strong homology at the 5′ end to IN-2.1. A second cluster of four closely related sequences (IN-3.1–IN-3.4) was also present, although they shared little sequence homology with lineage 1 or 2. Finally, IN-8.1 and IN-8.2 differed by only three nucleotides but were not strongly related to any other sequences. Several sequences (notably lineages 8–14) showed G-rich runs intermittently spaced throughout the sequence. This suggested the possibility of intra- or intermolecular G tetraplex formation, although this was not tested. While relatively little is known about the ability of XNAs to form G quartets, both FANA70 and TNA71 have been shown to form these structures.
For several of the sequences, the KD,app values for binding to IN were calculated using filter binding assays as described in the Methods section. Table 1 shows values for the SELEX starting material and several aptamer constructs. The selection process was performed using INF185H/C280S, a more soluble version of IN that retains full activity.72 For some constructs, values were calculated for this enzyme as well as INwt and INSso7d. The Sso7d version contains a ∼7-kDa N-terminal DNA binding domain corresponding to Sulfolobus solfataricus chromosomal protein Sso7d. This modification improves IN activity and solubility.64 Binding to the starting material was in the high nanomolar to low micromolar range for all IN enzymes tested. IN-1.1, IN-2.1, IN-3.1, and IN 6.1 bound strongly to IN showing KD,app values in the 40–100 pM range with INF185H/C280S. Note that this is well over 1000-fold tighter than binding to the starting material. Interestingly, IN-1.1 and IN-2.1 shared strong homology over the first 16 nucleotides derived from the random region, but less homology over nucleotides 17–40. IN-1.1 and 3.1 were nearly identical to other members of these lineages, and it was assumed that these sequences would bind similarly to IN, although they were not tested. Other sequences from round 10 (Figure 1) that were tested bound poorly. This included the G-rich sequence IN-8.1 (which is nearly identical to IN-8.2), IN-13.1, and IN-14.1. This suggests that their high prevalence in the round 10 SELEX material may be related to another selection parameter such as a higher affinity for nitrocellulose filters or over-representation in the starting material.
FANA IN Aptamers of the Same Sequence Composed of 2′-Fluororibonucleic Acid (F-RNA) Bound INF185H/C280S Weakly and Aptamers Bound Poorly to Protoptype Foamy Virus (PFV) IN
Changing all FANA nucleotides in IN-1.1 to F-RNA (D4K can incorporate both analogs) resulted in a loss in binding (IN-1.1 (FANA to F-RNA); Table 1). In fact, the IN-1.1 F-RNA construct bound so poorly that we were unable to determine the KD,apps in standard assays with up to 5 μM INF185H/C280S. These results illustrated that aptamer binding specificity cannot be mimicked by F-RNA. This is consistent with findings for FANA aptamers selected to HIV RT.55 Since FANA and F-RNA have identical chemical formulas, the complete loss of specific binding may seem counterintuitive. However, as was noted in the introduction, these analogs are structurally divergent with FANA duplexes more closely resembling DNA (the natural substrate of IN) while F-RNA duplexes are more RNA-like.43,58,60−62 IN 1.1 binding to PFV IN was also tested and was much weaker than binding to HIV IN (Table 1). The HIV and PFV IN share only ∼20% sequence homology73 but are predicted to fold similarly.74 This suggests that IN 1.1 has high specificity for HIV IN, just as FA-1 had high specificity for HIV RT.55
Prediction of the Structure of FANA IN Aptamers Was Guided by Covariation
The 26 sequences were clustered into 14 different lineages based on sequence conservation and then analyzed to identify the presence of shared structural motifs. Predicting the secondary structure of these FANA aptamers and other XNAs is complicated by a lack of available programs or enzymatic assays to interrogate XNA folding. Available programs for RNA or DNA can be used to approximate some XNA structures as many XNAs retain canonical bases. However, some XNAs,44 like FANA,63,75 form stronger duplex structures with DNA and RNA than natural nucleic acids, suggesting stronger base-pairing interactions. Although FANA shares properties with DNA (e.g., stimulation of RNase H activity in FANA-RNA hybrids,63,76 susceptibility to DNases,77 and incorporation by some DNA polymerases78), the stability of FANA–RNA hybrids is significantly greater than DNA–RNA and comparable to RNA–RNA duplexes.77 For our structure analysis, we relied on a combination covariation analysis and RNA folding programs to predict the structures of the FANA IN aptamers (see Methods). We identified a potential four-way junction secondary structure for sequences from seven (first seven lineages in Figure 1) of the 14 lineages. Representative aptamers (IN 1.1, 2.1, and 6.1) are shown in Figure 2 along with an alignment illustrating the positions of the common stem structures in the aptamers from the seven different lineages. The presence of P3 and P4 in the functional aptamer structure is supported by covariation between sequences that conform to the predicted four-way junction structure. Because P1 and P2 are either entirely (P1) or in part (P2) derived from invariable sequences at the 5′ and 3′ end of the aptamers, covariation cannot be used to support base pairing of these elements. Additionally, RNA folding software predicts multiple different base-pairing patterns for the invariable region of these sequences. For sequence 1.1, 1.2, 1.4, and 2.1, RNA structure prediction software predicts the pairing shown for P1 and P2, but the other sequences are predicted to form alternative base pairing patterns (Figure S2) that are inconsistent with the structure shown in Figure 2.
To test the proposed structure in Figure 2 and to resolve the ambiguity in predicting base pairs in the invariant region of the sequences, we generated additional aptamer sequences and tested their affinity for IN. Constructs designed to stabilize the pairing of P1 as shown in Figure 2 retain picomolar affinity for IN, but constructs that stabilize competing structures significantly increased the KD,app (Figures S3–S5). The constructs IN 1.1(P1flip) and IN 2.1(P1flip) contain a set of compensatory mutations that maintain P1 pairing for IN 1.1 and IN 2.1 but eliminate the base pairs of the competing alternative structures. IN 1.1(−10) and IN 2.1(−10) lack the last 10 nucleotides at the 3′ end of the aptamers. These last 10 nucleotides are involved in base pairs in the alternative P1 structure, but they are predicted to be single stranded in Figure 2. The shortened constructs therefore eliminated competing base pairs in the alternative structure. All four of these modifications bound IN with affinity that was comparable to the parent constructs (Table 1). Alternatively, IN-2.1(altP1), which contains changes in the invariant regions that are predicted to stabilize an alternative structure, exhibited a dramatic loss in affinity for IN, although it still bound much better than the starting material (Table 1). Several additional constructs (Figure S3) that are predicted to disrupt the secondary structure shown in Figure 2 exhibited even more dramatic losses in binding affinity. Overall, these results show that the structure depicted in Figure 2 likely represents the aptamer structure responsible for high affinity IN binding.
Finally, it was notable that all the stem-loop structures of IN 1.1 and IN 2.1 were highly similar despite the differences in sequence in P3 and P4. In contrast, despite binding strongly (Table 1), IN 6.1 was clearly different with a larger P2 stem and smaller P4 stem and loop (P1 and P3 were structurally invariant in all aptamers). Folded structures for lineages 3, 4, and 5 had less pronounced changes (when compared to IN 1.1 and IN 2.1) in P2 and P4 than IN 6.1. To test for possible interactions between P2 and P4, a construct that contained the P2 stem-loop from IN 6.1 and P4 stem-loop from IN 2.1 was constructed (IN-(6.1/2.1), Figure S3). This construct bound IN F185H/C280S with affinity similar to the parent constructs (Table 1). A version of the construct with the 10 3′ nucleotides deleted (IN-(6.1/2.1)(−10)) was also made and showed similar binding affinity (Table 1). These results suggest that the P2 and P4 stem loops are interchangeable and probably contribute independently to IN binding.
Comparison of FANA IN Aptamers with Other IN Aptamers
DNA and RNA aptamers to HIV IN have been selected by other groups,11,27,38,40 and IN has been shown to bind to various elements in the viral RNA genome.66 A 77 nucleotide RNA aptamer termed “P5” was isolated using SELEX and bound to INF185H/C280S (binding to wild type IN was similar (data not shown)) with a KD of ∼10 nM under conditions with similar ionic strength to those used here (see below).11 IN was also shown to bind to the TAR loop (bases 1–57 of the HIV-1 genome) with a KD of ∼3 nM (measured in a buffer with relatively lower ionic strength).66 Using the same conditions used for FANA aptamer measurements, a KD,app of 53 ± 26 nM was observed for P5 (Table 1). This is somewhat higher than the reported KD but in the same range. Differences may have resulted from inclusion of Mn2+ rather than Mg2+ in the original P5 measurements.11 TAR bound poorly to INF185H/C280S and INwt under the high salt conditions used in our assays. We could not determine a KD value for IN binding to TAR under the conditions used for FANA as the binding was too weak. This may be related to the lower ionic strength as well as the different measurement methods used in the previous work.66
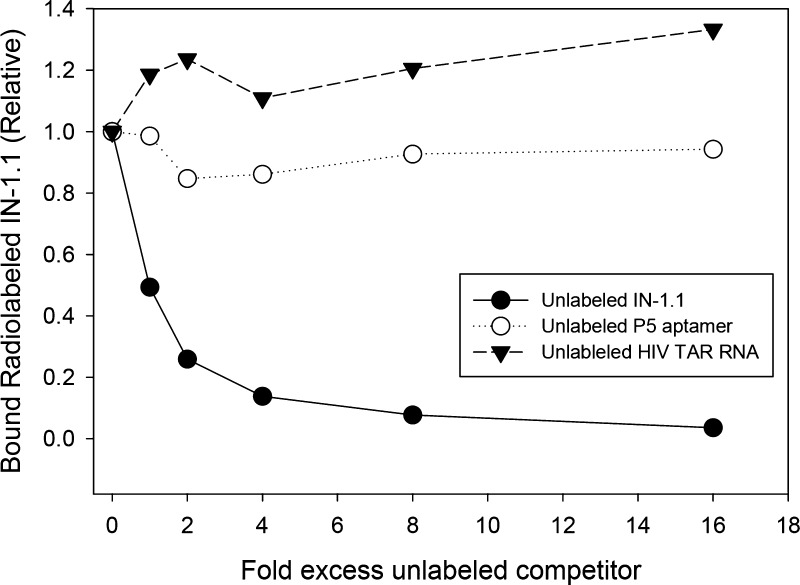
The P5 and TAR RNAs were also compared to IN-1.1 for binding to INF185H/C280S in a competition binding assay (Figure 3). The assay contained 5 nM radiolabeled IN-1.1 and 5 nM INF185H/C280S with an increasing amount of unlabeled competitor. As expected, unlabeled IN-1.1 competed the radiolabeled IN-1.1 off of INF185H/C280S. When equal amounts of labeled and unlabeled IN-1.1 were added, the level of radiolabeled IN-1.1 bound to INF185H/C280S decreased by about 50% as expected, and further decreases were observed as more unlabeled IN-1.1 was added. In contrast, neither P5 nor HIV TAR produced any significant decrease in binding of radiolabeled IN-1.1, even when they were added at 16-fold excess. This is consistent with IN-1.1 binding much more strongly to INF185H/C280S than the other constructs. Alternatively, the constructs may bind to different locations on INF185H/C280S such that they do not affect each other’s binding. However, this possibility was ruled out since radiolabeled P5 was completely displaced from INF185H/C280S with IN-1.1 (data not shown).
Figure 3.
HIV TAR and Aptamer P5 cannot compete with IN-1.1 for binding to IN. 5 nM 5′ 32P labeled IN-1.1 was incubated with 5 nM HIV INF185H/C280S in 20 mM Tris-HCl at pH 7.5, 200 mM NaCl, 6 mM MgCl2, 1 mM DTT, and 20 μL total volume at room temperature. Various amounts of excess unlabeled competitor (IN-1.1, P5 aptamer, or HIV TAR (1–57)) were added (fold excess: 0, 1, 2, 4, 8, 16) and incubated for 50 min. Samples were run over nitrocellulose filters, washed, and counted in a scintillation counter. Values on the y axis are relative to the sample with no competitor added.
The IN-1.1 Aptamer Is a Potent Inhibitor of HIV IN Activity and Intasome Formation
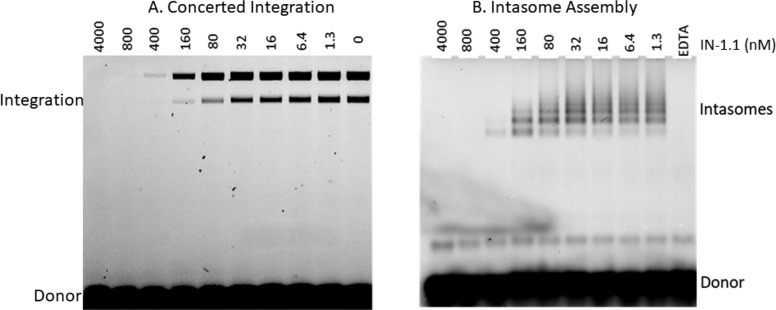
HIV IN, together with viral DNA, forms multiple intasome species, including tetramers and dodecamers that are functional for DNA integration.79 Similar structures with other retroviruses consisting of four or eight and 16 IN subunits have been uncovered.80−82 Both intasome formation and concerted integration activity were monitored in the presence and absence of different concentrations of IN-1.1 using INSso7d (Figure 4A). Concerted integration was inhibited by IN-1.1 with an IC50 of approximately 80 nM with complete inhibition at about 160 nM IN-1.1. This correlated well with the formation of intasomes, which began to decline at ∼80 nM IN-1.1, and no intasome formation was observed past 160 nM IN-1.1 (Figure 4B). Although these values are large compared to the measured KD,app for IN binding to IN-1.1 (Table 1), it is important to note that the concentration of INSso7d in the reactions was 2.5 μM; currently established in vitro integration assays require an IN protein concentration that is orders of magnitude higher than the KD,apps of the aptamer, which precludes measuring the true IC50. Complete inhibition of integration and intasome formation was observed when the ratio of IN-1.1/INSso7d was ∼1:10 in the reaction. This is consistent with concerted integration requiring the formation of the intasome complex, which can apparently be completely abrogated when a relatively small proportion of the total INSso7d is occupied by aptamer.
Figure 4.
(A and B) IN-1.1 aptamer inhibits concerted integration and intasome assembly. (A) Integrase strand transfer assay. 2.5 μM INSso7d and 1 μM 27 bp FAM labeled viral DNA and 300 ng of target plasmid DNA pGEM-9zf substrate were incubated in 20 mM HEPES at pH 7.5, 25% glycerol, 10 mM DTT, 5 mM MgCl2, 4 μM ZnCl2, and 100 mM NaCl in a 20 μL reaction volume. The reaction was carried out at 37 °C for 1 h. The integration products were recovered by ethanol precipitation and subjected to electrophoresis in a 1.5% agarose gel. (B) Intasome assembly was carried out in the same way except that no target DNA was added, and CaCl2 was substituted for MgCl2. Intasome assembly mix was subjected to electrophoresis on a 3.0% low melting agarose gel. DNAs were visualized using a fluorescence scanner.
Discussion
In this report, a previously described method to directly select aptamers to HIV-1 RT containing only FANA nucleotides in the randomized region was employed to generate aptamers to HIV IN. Like the HIV RT aptamers, the IN aptamers bound with a KD,app in the low picomolar range (Table 1). With HIV RT, previous aptamers made with natural RNA and DNA typically bound RT with modestly lower affinities that were in the same range as the FANA aptamers.55 In contrast, previous natural nucleic acid aptamers to IN bind much more weakly than the FANA aptamers produced here.31,45,48,49 Like other XNAs, FANA nucleic acids form unique hybrid structures with binding free energies that are different from natural nucleic acids.44,60,83 Coupled with the 2′ fluorine and arabinose sugar, this may allow FANA to sample unique structural spaces or make distinct interactions with amino acid moieties and domains in proteins. Other XNAs or noncanonical nucleotide analogs would likely have different properties that may make them more or less amenable to specific proteins. The commercial availability of FANA and F-RNA nucleotides has contributed to a more rapid advancement of these XNAs. Whether XNAs or natural nucleic acid will yield better aptamers with respect to binding affinity likely depends on the particular protein target.
In addition to the altered target binding specificity imparted by the chemical and structural diversity noted above, XNA modifications of the phosphate backbone and sugar groups in particular can enhance biological stability (reviewed in refs (84), (85)). This property would be pivotal for potential aptamer drugs or aptamers that would be used in assays with biological material. Macugen, an aptamer that is used for treatment of macular degeneration, was originally selected from a nucleic acid pool containing 2′-fluoropyrimidines. The resulting aptamer bound with low micomolar affinity to the VEGF target protein. Further postselection modification increased the stability of the aptamer without substantially sacrificing binding affinity.48 However, postselection modification can be time-consuming and is hindered by the potential loss of affinity.
The FANA aptamers described here and previously differ from most other modified aptamers in that the selected region is composed solely of FANA nucleotides, a property imparted by the ability of modified or natural polymerases to efficiently and accurately incorporate all four FANA nucleotides.49,50 In contrast, T7 RNA polymerase efficiently incorporates F-RNA pyrimidines, which limits selection, while other RNA polymerases that can more efficiently incorporate F-RNA are being explored.86 During the current experiments, we found that the modified D4K enzyme used for FANA incorporation also incorporates all four F-RNA nucleotides (Table 1, IN-1.1 (FANA to F-RNA)). The fidelity and efficiency of this process was not explored, but it has the potential for allowing the selection of homogeneous F-RNA aptamers. Like the F-RNA aptamers, most other modified aptamers produced thus far are also heterogeneous. In some cases, the various nucleotide analogs are incorporated by natural or modified polymerases, while moieties can also be added to select nucleotides using “click” chemistry or other techniques.84,87−89
The structural models for the IN aptamers were guided by covariation analysis that predicted the presence of two stem structures (P3 and P4 in Figure 2) in the variable region of several of the sequences from round 10, indicating that they adopt the same functional structural motif, despite considerable sequence variation. Interestingly, using RNA free-energy parameters90 resulted in structure predictions that were identical or nearly identical to our final structure prediction for the sequences IN 1.1, IN 1.2, IN 1.4, and IN 2.1 (Figure S2). For these sequences, predicting FANA structures using RNA folding parameters produced results that were consistent with both the covariation analysis and mutational analysis. Alternatively, when DNA free-energy parameters68 were used, none of the sequences were predicted to fold into the structure supported by covariation or mutational analysis. This suggests that RNA folding parameters, as opposed to DNA, are better predictors of FANA secondary structure, but further analysis with other aptamers will be necessary to establish this. Several methods of confirming the secondary structures of nucleic acids cannot be applied to FANA and other XNAs. Development of new methods for analyzing XNA structures should be helpful in future experiments characterizing XNAs and other modified nucleic acids.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.9b00237.
Five figures: an example of a binding affinity analysis and the others pertinent to the covariation data and structural analysis of the integrase aptamers (PDF)
This work was supported by the National Institute of Allergy and Infectious Diseases (R01AI150480 to J.J.D.); Intramural Program of the National Institute of Diabetes and Digestive Diseases of the National Institutes of Health (R.C.); P.H. was supported by the Medical Research Council (program no. MC_U105178804).
The authors declare no competing financial interest.
Supplementary Material
References
- Chou S. H.; Chin K. H.; Wang A. H. (2005) DNA aptamers as potential anti-HIV agents. Trends Biochem. Sci. 30, 231–234. 10.1016/j.tibs.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Quashie P. K.; Sloan R. D.; Wainberg M. A. (2012) Novel therapeutic strategies targeting HIV integrase. BMC Med. 10, 34. 10.1186/1741-7015-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins T. M.; Esposito D.; Engelman A.; Craigie R. (1997) Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16, 6849–6859. 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta K.; Knizewski L.; Wyrwicz L. S.; Rychlewski L.; Ginalski K. (2009) Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37, 7701–7714. 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeschla E. M. (2008) Integrase, LEDGF/p75 and HIV Replication. Cell. Mol. Life Sci. 65, 1403–1424. 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R. (2001) HIV Integrase, a Brief Overview from Chemistry to Therapeutics. J. Biol. Chem. 276, 23213–23216. 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- Craigie R. (2012) The molecular biology of HIV integrase. Future Virol. 7, 679–686. 10.2217/fvl.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow D. E.; Bartlett J. A. (2014) Dolutegravir, the Second-Generation of Integrase Strand Transfer Inhibitors (INSTIs) for the Treatment of HIV. Infect. Dis. T. 3, 83–102. 10.1007/s40121-014-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepik K. J.; Harrigan P. R.; Yip B.; Wang L.; Robbins M. A.; Zhang W. W.; Toy J.; Akagi L.; Lima V. D.; Guillemi S.; Montaner J. S. G.; Barrios R. (2017) Emergent drug resistance with integrase strand transfer inhibitor-based regimens. AIDS 31, 1425–1434. 10.1097/QAD.0000000000001494. [DOI] [PubMed] [Google Scholar]
- Brody E. N.; Gold L. (2000) Aptamers as therapeutic and diagnostic agents. Rev. Mol. Biotechnol. 74, 5–13. 10.1016/S1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- Allen P.; Worland S.; Gold L. (1995) Isolation of High-Affinity RNA Ligands to HIV-1 Integrase from a Random Pool. Virology 209, 327–336. 10.1006/viro.1995.1264. [DOI] [PubMed] [Google Scholar]
- Virgilio A.; Amato T.; Petraccone L.; Esposito F.; Grandi N.; Tramontano E.; Romero R.; Haider S.; Gomez-Monterrey I.; Novellino E.; Mayol L.; Esposito V.; Galeone A. (2018) Improvement of the activity of the anti-HIV-1 integrase aptamer T30175 by introducing a modified thymidine into the loops. Sci. Rep. 8, 7447. 10.1038/s41598-018-25720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James W. (2007) Aptamers in the virologists’ toolkit. J. Gen. Virol. 88, 351–364. 10.1099/vir.0.82442-0. [DOI] [PubMed] [Google Scholar]
- Joshi P. J.; Fisher T. S.; Prasad V. R. (2003) Anti-HIV inhibitors based on nucleic acids: emergence of aptamers as potent antivirals. Curr. Drug Targets: Infect. Disord. 3, 383–400. 10.2174/1568005033481060. [DOI] [PubMed] [Google Scholar]
- Mok W.; Li Y. (2008) Recent progress in nucleic acid aptamer-based biosensors and bioassays. Sensors 8, 7050–7084. 10.3390/s8117050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimjee S. M.; Rusconi C. P.; Sullenger B. A. (2005) Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 56, 555–583. 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Porschewski P.; Grattinger M. A.; Klenzke K.; Erpenbach A.; Blind M. R.; Schafer F. (2006) Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification. J. Biomol. Screening 11, 773–781. 10.1177/1087057106292138. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Blank M.; Schluesener H. J. (2004) Nucleic acid aptamers in human viral disease. Arch. Immunol. Ther. Exp. (Warsz). 52, 307–315. [PubMed] [Google Scholar]
- Dolot R.; Lam C. H.; Sierant M.; Zhao Q.; Liu F.-W.; Nawrot B.; Egli M.; Yang X. (2018) Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 46, 4819. 10.1093/nar/gky268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M.; Scheper T.; Walter J.-G. (2013) Aptamers: versatile probes for flow cytometry. Appl. Microbiol. Biotechnol. 97, 7097–7109. 10.1007/s00253-013-5070-z. [DOI] [PubMed] [Google Scholar]
- Miller M. T.; Tuske S.; Das K.; DeStefano J. J.; Arnold E. (2016) Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer. Protein Sci. 25, 46–55. 10.1002/pro.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedighian H.; Halabian R.; Amani J.; Heiat M.; Taheri R. A.; Imani Fooladi A. A. (2018) Manufacturing of a novel double-function ssDNA aptamer for sensitive diagnosis and efficient neutralization of SEA. Anal. Biochem. 548, 69–77. 10.1016/j.ab.2018.02.017. [DOI] [PubMed] [Google Scholar]
- Todd G. C.; Duchon A.; Inlora J.; Olson E. D.; Musier-Forsyth K.; Ono A. (2017) Inhibition of HIV-1 Gag–membrane interactions by specific RNAs. RNA 23, 395–405. 10.1261/rna.058453.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen A. J.; Kupis-Rozmysłowicz J.; Gigli C.; Schuergers N.; Boghossian A. A. (2018) Xeno Nucleic Acid Nanosensors for Enhanced Stability Against Ion-Induced Perturbations. J. Phys. Chem. Lett. 9, 4336–4343. 10.1021/acs.jpclett.8b01879. [DOI] [PubMed] [Google Scholar]
- Parashar A. (2016) Aptamers in Therapeutics. J. Clin. Diagn. Res. 10, BE01–BE06. 10.7860/JCDR/2016/18712.7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K.; Balzarini J.; Miller M. T.; Maguire A. R.; DeStefano J. J.; Arnold E. (2016) Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet. ACS Chem. Biol. 11, 2158–2164. 10.1021/acschembio.6b00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Soultrait V. R.; Lozach P.-Y.; Altmeyer R.; Tarrago-Litvak L.; Litvak S.; Andréola M. L. (2002) DNA Aptamers Derived from HIV-1 RNase H Inhibitors are Strong Anti-integrase Agents. J. Mol. Biol. 324, 195–203. 10.1016/S0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- Andreola M. L.; Pileur F.; Calmels C.; Ventura M.; Tarrago-Litvak L.; Toulme J. J.; Litvak S. (2001) DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry 40, 10087–10094. 10.1021/bi0108599. [DOI] [PubMed] [Google Scholar]
- Ferguson M. R.; Rojo D. R.; Somasunderam A.; Thiviyanathan V.; Ridley B. D.; Yang X.; Gorenstein D. G. (2006) Delivery of double-stranded DNA thioaptamers into HIV-1 infected cells for antiviral activity. Biochem. Biophys. Res. Commun. 344, 792–797. 10.1016/j.bbrc.2006.03.201. [DOI] [PubMed] [Google Scholar]
- Fisher T. S.; Joshi P.; Prasad V. R. (2005) HIV-1 reverse transcriptase mutations that confer decreased in vitro susceptibility to anti-RT DNA aptamer RT1t49 confer cross resistance to other anti-RT aptamers but not to standard RT inhibitors. AIDS Res. Ther. 2, 8. 10.1186/1742-6405-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotoda H.; Koizumi M.; Koga R.; Kaneko M.; Momota K.; Ohmine T.; Furukawa H.; Agatsuma T.; Nishigaki T.; Sone J.; Tsutsumi S.; Kosaka T.; Abe K.; Kimura S.; Shimada K. (1998) Biologically active oligodeoxyribonucleotides. 5. 5′-End-substituted d(TGGGAG) possesses anti-human immunodeficiency virus type 1 activity by forming a G-quadruplex structure. J. Med. Chem. 41, 3655–3663. 10.1021/jm970658w. [DOI] [PubMed] [Google Scholar]
- Kolb G.; Reigadas S.; Castanotto D.; Faure A.; Ventura M.; Rossi J. J.; Toulme J. J. (2006) Endogenous expression of an anti-TAR aptamer reduces HIV-1 replication. RNA Biol. 3, 150–156. 10.4161/rna.3.4.3811. [DOI] [PubMed] [Google Scholar]
- Matzen K.; Elzaouk L.; Matskevich A. A.; Nitzsche A.; Heinrich J.; Moelling K. (2007) RNase H-mediated retrovirus destruction in vivo triggered by oligodeoxynucleotides. Nat. Biotechnol. 25, 669–674. 10.1038/nbt1311. [DOI] [PubMed] [Google Scholar]
- Moelling K.; Abels S.; Jendis J.; Matskevich A.; Heinrich J. (2006) Silencing of HIV by hairpin-loop-structured DNA oligonucleotide. FEBS Lett. 580, 3545–3550. 10.1016/j.febslet.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Neff C. P.; Zhou J.; Remling L.; Kuruvilla J.; Zhang J.; Li H.; Smith D. D.; Swiderski P.; Rossi J. J.; Akkina R. (2011) An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci. Transl. Med. 3, 66ra6. 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasunderam A.; Ferguson M. R.; Rojo D. R.; Thiviyanathan V.; Li X.; O’Brien W. A.; Gorenstein D. G. (2005) Combinatorial selection, inhibition, and antiviral activity of DNA thioaptamers targeting the RNase H domain of HIV-1 reverse transcriptase. Biochemistry 44, 10388–10395. 10.1021/bi0507074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V. M.; Martin M. E.; Fernandez G.; Garcia-Sacristan A. (2016) Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses. Pharmaceuticals 9, 78. 10.3390/ph9040078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley S.; Boroda S.; Musier-Forsyth K.; Kankia B. I. (2011) HIV-integrase aptamer folds into a parallel quadruplex: A thermodynamic study. Biophys. Chem. 155, 82–88. 10.1016/j.bpc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Métifiot M.; Leon O.; Tarrago-Litvak L.; Litvak S.; Andréola M.-L. (2005) Targeting HIV-1 integrase with aptamers selected against the purified RNase H domain of HIV-1 RT. Biochimie 87, 911–919. 10.1016/j.biochi.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Zhang Y.; Ye G.; Yang Z.; Zhang L.; Zhang L. (2008) In vitro selection of G-rich RNA aptamers that target HIV-1 integrase. Sci. China, Ser. B: Chem. 51, 401. 10.1007/s11426-008-0056-x. [DOI] [Google Scholar]
- Ellington A. D.; Szostak J. W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Herdewijn P.; Marliere P. (2009) Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodiversity 6, 791–808. 10.1002/cbdv.200900083. [DOI] [PubMed] [Google Scholar]
- Anosova I.; Kowal E. A.; Dunn M. R.; Chaput J. C.; Van Horn W. D.; Egli M. (2016) The structural diversity of artificial genetic polymers. Nucleic Acids Res. 44, 1007–1021. 10.1093/nar/gkv1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro V. B.; Holliger P. (2012) The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr. Opin. Chem. Biol. 16, 245–252. 10.1016/j.cbpa.2012.05.198. [DOI] [PubMed] [Google Scholar]
- Schmidt M. (2010) Xenobiology: A new form of life as the ultimate biosafety tool. BioEssays 32, 322–331. 10.1002/bies.200900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele F. R.; Gold L. (2012) The sweet allure of XNA. Nat. Biotechnol. 30, 624. 10.1038/nbt.2298. [DOI] [PubMed] [Google Scholar]
- Ruckman J.; Green L. S.; Beeson J.; Waugh S.; Gillette W. L.; Henninger D. D.; Claesson-Welsh L.; Janjic N. (1998) 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 273, 20556–20567. 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- Pinheiro V. B.; Taylor A. I.; Cozens C.; Abramov M.; Renders M.; Zhang S.; Chaput J. C.; Wengel J.; Peak-Chew S. Y.; McLaughlin S. H.; Herdewijn P.; Holliger P. (2012) Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344. 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel A. E.; Chen Z.; Ayele T. M.; Heemstra J. M. (2018) In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 46, 8057–8068. 10.1093/nar/gky667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H.; Liao J.-Y.; Jimenez R. M.; Wang Y.; Bala S.; McCloskey C.; Switzer C.; Chaput J. C. (2018) Synthesis and Evolution of a Threose Nucleic Acid Aptamer Bearing 7-Deaza-7-Substituted Guanosine Residues. J. Am. Chem. Soc. 140, 5706–5713. 10.1021/jacs.7b13031. [DOI] [PubMed] [Google Scholar]
- Dunn M. R.; Chaput J. C. (2014) An In Vitro Selection Protocol for Threose Nucleic Acid (TNA) Using DNA Display. Curr. Protoc. Nucleic Acid Chem. 57, 9.8.1. 10.1002/0471142700.nc0908s57. [DOI] [PubMed] [Google Scholar]
- Yu H.; Zhang S.; Chaput J. C. (2012) Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 4, 183–187. 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- Lipi F.; Chen S.; Chakravarthy M.; Rakesh S.; Veedu R. N. (2016) In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 13, 1232–1245. 10.1080/15476286.2016.1236173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves Ferreira-Bravo I.; Cozens C.; Holliger P.; DeStefano J. J. (2015) Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 43, 9587–9599. 10.1093/nar/gkv1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Ngor A. K.; Nikoomanzar A.; Chaput J. C. (2018) Evolution of a General RNA-Cleaving FANA Enzyme. Nat. Commun. 9, 5067. 10.1038/s41467-018-07611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A.; Kirby L.; Sayer N.; Wellesley R.; Disterer P.; Sylvester I.; Gill A.; Hope J.; James W.; Tahiri-Alaoui A. (2003) Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J. Biol. Chem. 278, 39697–39705. 10.1074/jbc.M305297200. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Swiderski P.; Li H.; Zhang J.; Neff C. P.; Akkina R.; Rossi J. J. (2009) Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 37, 3094–3109. 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. K.; Damha M. J. (2008) 2′F-Arabinonucleic acid (2′F-ANA) - history, properties, and new frontiers. Can. J. Chem. 86, 641–656. 10.1139/v08-049. [DOI] [Google Scholar]
- Wilds C. J.; Damha M. J. (2000) 2′-Deoxy-2′-fluoro-beta-D-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 28, 3625–3635. 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Pintado N.; Deleavey G. F.; Portella G.; Campos-Olivas R.; Orozco M.; Damha M. J.; Gonzalez C. (2013) Backbone FC□H···O Hydrogen Bonds in 2′F-Substituted Nucleic Acids. Angew. Chem., Int. Ed. 52, 12065–12068. 10.1002/anie.201305710. [DOI] [PubMed] [Google Scholar]
- Trempe J.-F.; Wilds C. J.; Denisov A. Y.; Pon R. T.; Damha M. J.; Gehring K. (2001) NMR Solution Structure of an Oligonucleotide Hairpin with a 2‘F-ANA/RNA Stem: Implications for RNase H Specificity toward DNA/RNA Hybrid Duplexes. J. Am. Chem. Soc. 123, 4896–4903. 10.1021/ja003859p. [DOI] [PubMed] [Google Scholar]
- Li F.; Sarkhel S.; Wilds C. J.; Wawrzak Z.; Prakash T. P.; Manoharan M.; Egli M. (2006) 2′-Fluoroarabino- and arabinonucleic acid show different conformations, resulting in deviating RNA affinities and processing of their heteroduplexes with RNA by RNase H. Biochemistry 45, 4141–4152. 10.1021/bi052322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Jurado K. A.; Lin S.; Engelman A.; Craigie R. (2014) Engineered hyperactive integrase for concerted HIV-1 DNA integration. PLoS One 9, e105078. 10.1371/journal.pone.0105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z.; Lapkouski M.; Yang W.; Craigie R. (2012) Assembly of prototype foamy virus strand transfer complexes on product DNA bypassing catalysis of integration. Protein Sci. 21, 1849–1857. 10.1002/pro.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessl J. J.; Kutluay S. B.; Townsend D.; Rebensburg S.; Slaughter A.; Larue R. C.; Shkriabai N.; Bakouche N.; Fuchs J. R.; Bieniasz P. D.; Kvaratskhelia M. (2016) HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis. Cell 166, 1257–1268. 10.1016/j.cell.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]; e1212.
- Katoh K.; Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R.; Lorenz R.; Bernhart S. H.; Neubock R.; Hofacker I. L. (2008) The Vienna RNA websuite. Nucleic Acids Res. 36, W70–74. 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R.; Bernhart S. H.; Honer Zu Siederdissen C.; Tafer H.; Flamm C.; Stadler P. F.; Hofacker I. L. (2011) ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26. 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietard J.; Abou Assi H.; Gómez-Pinto I.; González C.; Somoza M. M.; Damha M. J. (2017) Mapping the affinity landscape of Thrombin-binding aptamers on 2′F-ANA/DNA chimeric G-Quadruplex microarrays. Nucleic Acids Res. 45, 1619–1632. 10.1093/nar/gkw1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J. Y.; Anosova I.; Bala S.; Van Horn W. D.; Chaput J. C. (2017) A parallel stranded G-quadruplex composed of threose nucleic acid (TNA). Biopolymers 107, e22999. 10.1002/bip.22999. [DOI] [PubMed] [Google Scholar]
- Jenkins T. M.; Engelman A.; Ghirlando R.; Craigie R. (1996) A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271, 7712–7718. 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- Quashie P. K.; Han Y. S.; Hassounah S.; Mesplede T.; Wainberg M. A. (2015) Structural Studies of the HIV-1 Integrase Protein: Compound Screening and Characterization of a DNA-Binding Inhibitor. PLoS One 10, e0128310. 10.1371/journal.pone.0128310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. C.; Metifiot M.; Ferris A.; Pommier Y.; Hughes S. H. (2013) A homology model of HIV-1 integrase and analysis of mutations designed to test the model. J. Mol. Biol. 425, 2133–2146. 10.1016/j.jmb.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. K.; Martin-Pintado N.; Gomez-Pinto I.; Schwartzentruber J.; Portella G.; Orozco M.; Gonzalez C.; Damha M. J. (2010) Differential stability of 2′F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 38, 2498–2511. 10.1093/nar/gkp1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damha M. J.; Wilds C. J.; Noronha A.; Brukner I.; Borkow G.; Arion D.; Parniak M. A. (1998) Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are subsrates of ribonuclease H. J. Am. Chem. Soc. 120, 12976–12977. 10.1021/ja982325+. [DOI] [Google Scholar]
- Watts J. K.; Katolik A.; Viladoms J.; Damha M. J. (2009) Studies on the hydrolytic stability of 2′-fluoroarabinonucleic acid (2′F-ANA). Org. Biomol. Chem. 7, 1904–1910. 10.1039/b900443b. [DOI] [PubMed] [Google Scholar]
- Peng C. G.; Damha M. J. (2007) Polymerase-directed synthesis of 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acids. J. Am. Chem. Soc. 129, 5310–5311. 10.1021/ja069100g. [DOI] [PubMed] [Google Scholar]
- Passos D. O.; Li M.; Yang R.; Rebensburg S. V.; Ghirlando R.; Jeon Y.; Shkriabai N.; Kvaratskhelia M.; Craigie R.; Lyumkis D. (2017) Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355, 89–92. 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballandras-Colas A.; Brown M.; Cook N. J.; Dewdney T. G.; Demeler B.; Cherepanov P.; Lyumkis D.; Engelman A. N. (2016) Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature 530, 358–361. 10.1038/nature16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawenhoff J.; Engelman A. N. (2017) Retroviral integrase protein and intasome nucleoprotein complex structures. World J. Biol. Chem. 8, 32–44. 10.4331/wjbc.v8.i1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S.; Gupta S. S.; Valkov E.; Engelman A.; Cherepanov P. (2010) Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236. 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B.; Wengel J. (2004) LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 43, 13233–13241. 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- Antipova O. M.; Zavyalova E. G.; Golovin A. V.; Pavlova G. V.; Kopylov A. M.; Reshetnikov R. V. (2018) Advances in the Application of Modified Nucleotides in SELEX Technology. Biochemistry (Moscow) 83, 1161–1172. 10.1134/S0006297918100024. [DOI] [PubMed] [Google Scholar]
- Keefe A. D.; Cload S. T. (2008) SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 12, 448–456. 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Zhu B.; Hernandez A.; Tan M.; Wollenhaupt J.; Tabor S.; Richardson C. C. (2015) Synthesis of 2′-Fluoro RNA by Syn5 RNA polymerase. Nucleic Acids Res. 43, e94–e94. 10.1093/nar/gkv367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlisberger P.; Hollenstein M. (2018) Aptamer chemistry. Adv. Drug Delivery Rev. 134, 3–21. 10.1016/j.addr.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Dellafiore M. A.; Montserrat J. M.; Iribarren A. M. (2016) Modified Nucleoside Triphosphates for In-vitro Selection Techniques. Front. Chem. 4, 18. 10.3389/fchem.2016.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer F.; Tolle F.; Rosenthal M.; Brandle G. M.; Ewers J.; Mayer G. (2018) Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat. Protoc. 13, 1153–1180. 10.1038/nprot.2018.023. [DOI] [PubMed] [Google Scholar]
- Mathews D. H.; Disney M. D.; Childs J. L.; Schroeder S. J.; Zuker M.; Turner D. H. (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 101, 7287–7292. 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.