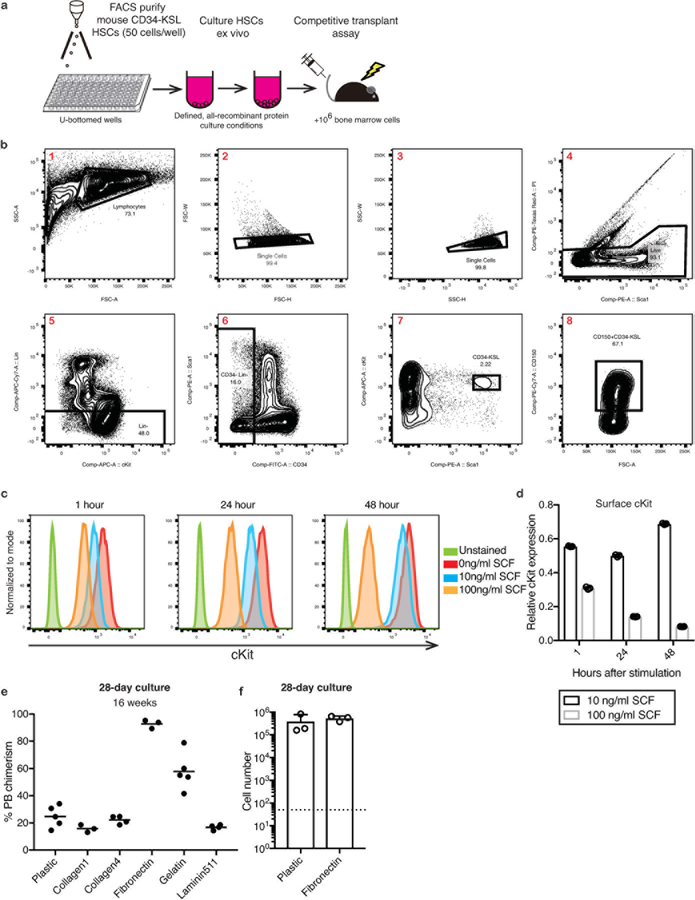
Multipotent self-renewing hematopoietic stem cells (HSCs) regenerate the adult blood system following transplantation1, a curative therapy for numerous diseases such as immunodeficiencies and leukemias2. While significant effort has been applied to identify HSC maintenance factors through characterization of the in vivo bone marrow (BM) HSC microenvironment or niche3–5, stable ex vivo HSC expansion has been unattainable6,7. Here, we describe the development of a defined, albumin-free culture system that supports long-term ex vivo expansion of functional HSCs. Through systematic optimization, we have found that high thrombopoietin (TPO) synergizes with low stem cell factor (SCF) and fibronectin to sustain HSC self-renewal. Furthermore, we identified polyvinyl alcohol (PVA) as a functionally-superior, Good Manufacturing Practice (GMP)-compatible replacement for serum albumin, which has long been a major source of biological contaminants in HSC cultures8. These conditions afford between 236- and 899-fold expansion of functional HSCs over 1 month, although analysis of clonally-derived cultures suggests significant heterogeneity in ex vivo HSC self-renewal capacity. Using this system, HSC cultures derived from just 50 cells robustly engrafted in recipients without the normal requirement for toxic pre-conditioning (e.g. radiation), suggesting new approaches for HSC transplantation. These findings therefore have important implications both for basic HSC research and clinical hematology.
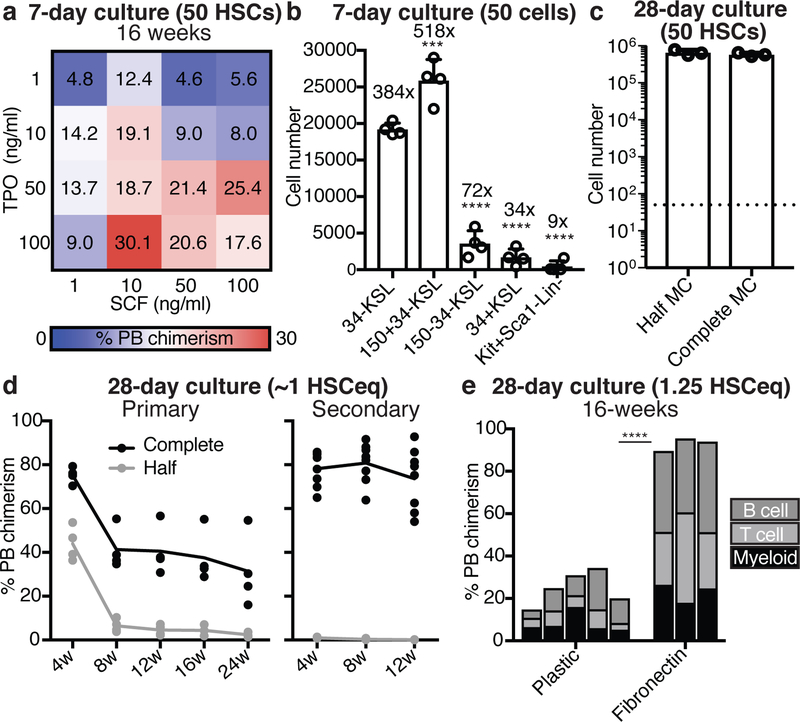
To optimize HSC cultures, we initially titrated TPO against SCF in 7-day CD34-cKit+Sca1+Lin- (CD34-KSL) HSC cultures (Extended Data Figure 1a,b), and determined the consequence by competitive transplantation into lethally-irradiated recipient mice against 1×106 BM competitor cells. Highest 16-week peripheral blood (PB) chimerism (~30%) was observed with 100 ng/ml TPO and 10 ng/ml SCF (Figure 1a), perhaps due to the increased cKit internalization at higher SCF concentrations causing loss of SCF-sensitivity (Extended Data Figure 1c,d).
Figure 1: High TPO synergizes with low SCF and fibronectin to enhance HSC expansion.
(a) Mean 16-week donor PB chimerism from 50 CD34-KSL HSCs following a 7-day culture in mouse TPO (1–100 ng/ml) and mouse SCF (1–100 ng/ml), as described in Extended Data Figure 1a. Competitive transplantation against 1×106 BM competitors.
(b) Cell number derived from 50 CD34-KSL, 50 CD150+CD34-KSL, 50 CD150-CD34-KSL CD34+KSL, or 50 cKit+Sca1-Lin- BM cells after 7-day culture in 100 ng/ml TPO and 10 ng/ml SCF. Statistical significance calculated using ANOVA. *** denotes p=0.004 and **** denotes p<0.0001. Mean ± s.d. of 4 independent cultures.
(c) 28-day growth of 50 CD34-KSL HSCs in 100 ng/ml TPO and 10 ng/ml SCF, and with half or complete media changes (MC) every 3-days. Mean ± s.d. of 4 independent cultures.
(d) Donor PB chimerism in recipient mice from 1×104 HSC-derived cells (~1 starting HSC equivalent; ~1 HSCeq) following a 28-day culture (started from 50 CD34-KSL), as described in (c). Competitive transplantation against 1×106 BM competitors. Donor PB chimerism at 4–24-week in primary recipients (left) and at 4–12 weeks in secondary recipients (right).
(e) 16-week donor PB chimerism from 1×104 HSC-derived cells (1.25 HSCeq) following a 28-day culture (started from 50 CD34-KSL) on plastic (n=5) or fibronectin (n=3) plates cultured in 100 ng/ml TPO and 10 ng/ml SCF with complete media changes. Competitive transplantation against 1×106 BM competitors. Each column represents an individual mouse. Statistical significance calculated using an unpaired two-tailed t-test. **** denotes p<0.0001.
The 100 ng/ml TPO and 10 ng/ml SCF condition preferentially induced proliferation from BM-derived CD150+CD34-KSL HSCs, rather than BM-derived CD34+KSL hematopoietic progenitor cells (HPCs) (Figure 1b). We therefore determined whether longer-term ex vivo HSC expansion was possible by attempting 1-month cultures. As 50 starting HSCs expanded by ~13,000-fold during culture (Figure 1c), we transplanted 1×104 cells per recipient, approximately 1/50th of the culture or ~1 starting HSC equivalent (termed ~1 HSCeq). Using half-media changes, we only detected short-term reconstitution (Figure 1d). However, by performing complete media changes on the HSC cultures, we achieved similar cellular expansion but also sustained long-term HSC activity from ~1 HSCeq (1×104 cells) (Figure 1c,d).
Given the need for complete media changes during the culture, we hypothesized that HSC-plate attachment may help to retain HSCs during media changes. Of the 5 plate-coatings tested, fibronectin improved 16-week PB chimerism the most (Extended Data Figure 1e). Although HSC proliferation was similar on fibronectin (Extended Data Figure 1f), 1×104 cells (1.25 HSCeq) from day-28 fibronectin cultures gave almost 100% PB chimerism at 16-weeks (Figure 1e). This was consistent with recent suggestions that fibronectin is a BM niche factor9 and fibronectin signaling improves HSC maintenance10,11.
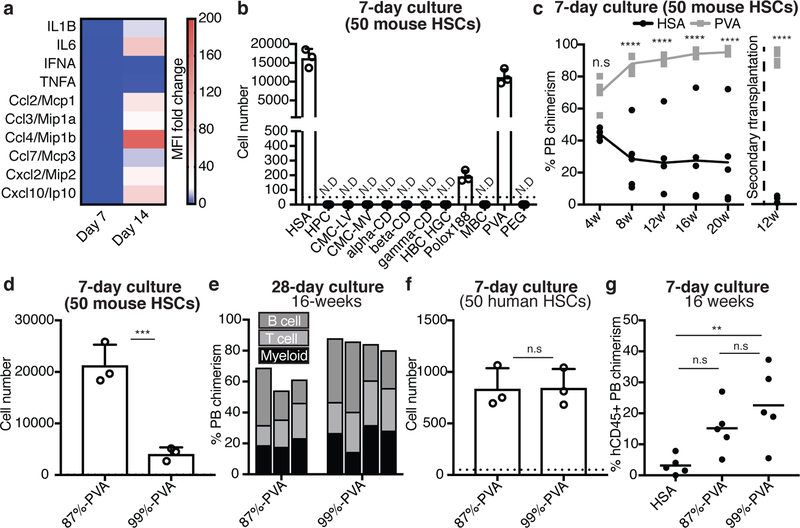
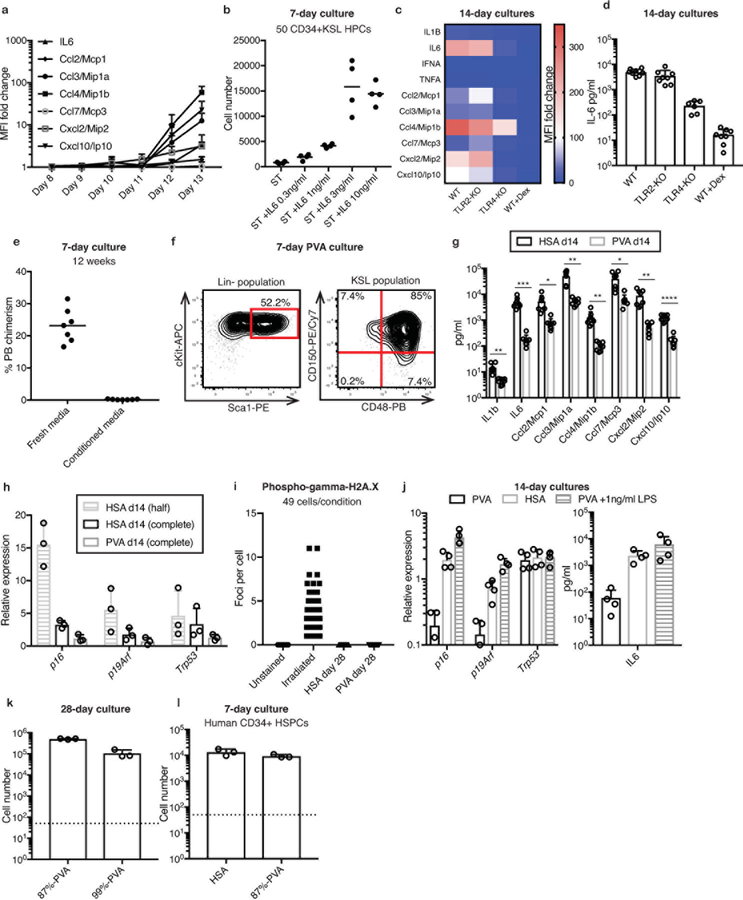
Similar to human hematopoietic stem/progenitor cell (HSPC) cultures12, several cytokines and chemokines (e.g. IL-6 and Ccl2–4) were abundant in day-14 cultures (Figures 2a, Extended Data Figure 2a) and suggested mechanisms of HPC contamination (just 3 ng/ml IL-6 enhanced ex vivo CD34+KSL HPC proliferation; Extended Data Figure 2b). The secretion profile also suggested activation of an innate immune response13. Consistent with this idea, cytokine secretion was reduced from TLR4−/− HSCs or by addition of dexamethasone (Extended Data Figure 2c,d). Conditioned media also induced loss of HSC activity, suggesting differentiation-inducing factors were soluble (Extended Data Figure 2e).
Figure 2: Polyvinyl alcohol can replace serum albumin for ex vivo HSC expansion.
(a) Heatmap displaying the fold-change in cytokine immunoassay mean fluorescence intensity (MFI) using conditioned media from day 7 and 14 HSC cultures. Mean of 4 independent cultures with fold-change relative to unconditioned media.
(b) Cellular expansion of 50 CD34-KSL HSCs after a seven-day culture in serum albumin-free conditions, supplemented with various potential chemically-defined serum albumin replacements (see Methods section for details). Recombinant human serum albumin (HSA) used as a positive control. Mean ± s.d. of 3 independent cultures. N.D. denotes not detected.
(c) Mean donor PB chimerism in primary recipients (n=5 per group) and secondary recipients (n=4 per group) from 50 CD34-KSL HSCs following a 7-day culture in HSA-based or PVA-based media, supplemented with 100 ng/ml TPO and 10 ng/ml SCF in U-bottom plates. Competitive transplantation against 1×106 BM competitors in primary recipients. Statistical significance calculated using ANOVA. **** denotes p<0.0001, n.s. denotes not statistically significant.
(d) 7-day expansion of 50 CD150+CD34-KSL HSCs in media containing 87%-hydrolyzed PVA (87%-PVA) or >99%-hydrolyzed PVA (99%-PVA). Mean ± s.d. of 3 independent cultures. Statistical significance calculated using t-test. *** denotes p=0.0021.
(e) 16-week donor PB chimerism from 1×104 cells from 28-day in 87%-PVA (n=3) and 99%-PVA (n=4) cultures (see Extended Data Figure 2k for 28-day cell counts). Each column represents an individual mouse.
(f) 7-day expansion of 50 human umbilical cord blood-derived CD34+CD38-CD90+CD49f+ HSCs in HSA- or PVA-based cultures supplemented with 10 ng/ml human SCF and 100 ng/ml human TPO. Mean ± s.d. of 3 independent cultures.
(g) Mean 16-week human CD45+ PB chimerism within sub-lethally irradiated NOG mice (n=5 per group) following transplantation with 7-day cultures derived from 2×103 CD34+ cells. Statistical significance calculated using ANOVA. ** denotes p=0.0098. n.s. denotes not statistically significant.
Our HSC cultures used yeast-derived recombinant human serum albumin (HSA). We hypothesized that recombinant protein contaminants might be responsible for the inflammatory phenotype14,15 and sought HSA replacements. Serum albumin could have several possible functions in HSC cultures, including as a “carrier-molecule” or as a source of amino acids. As HSCs fail to grow in low amino acid medias containing albumin16,17, we focused on replacing the carrier-molecule function. Of the 11 chemically-synthesized potential replacements screened (see Methods for chemical list), only polyvinyl alcohol (PVA) afforded HSC survival, growth, and phenotypic HSPC maintenance (Figure 2b, Extended Data Figure 2f). Additionally, PVA-cultured HSCs outperformed HSA-cultured HSCs in competitive transplantation assays (Figure 2c). We also observed significantly lower concentrations of secreted factors in PVA-cultures (Extended Data Figure 2g). Senescence-associated gene expression18,19 (p16Ink4a, p19Arf, and Trp53) was also reduced in PVA-cultures and accumulation of gamma-histone 2A.X phosphorylation20 was not detected (Extended Data Figure 2h,i). Consistent with the role of TLR4 in the HSA phenotype, addition of TLR4-agonist lipopolysaccharide to PVA-cultures caused similar induction of p16 and p19, as well as IL-6 secretion (Extended Data Figure 2j).
While PVA has been used for culturing embryonic cell types21,22, its mechanistic role is still poorly understood. To investigate the properties of PVA that allowed it to act as an albumin replacement, we compared PVA hydrolysis states. Our initial screen used 87%-hydrolyzed PVA (87%-PVA), an amphiphilic polymer containing acetate and alcohol domains. By contrast, >99%-hydrolyzed PVA (99%-PVA) lacks acetate domains. HSCs survived in media containing either PVA, but proliferation was 5-fold less in 99%-PVA cultures (Figure 2d, Extended Data Figure 2k). However, competitive transplantation of 1×104 cells from day-28 87%-PVA and 99%-PVA cultures demonstrated that both PVA-types supported HSCs ex vivo (Figure 2e).
As an inexpensive but GMP-compatible albumin-replacement, PVA may also have important implications for human HSC expansion. As proof-of-concept, we confirmed that PVA can replace serum albumin in human umbilical cord blood-derived CD34+ HSPC cultures (Extended Data Figure 2l). However, human CD34+CD38-CD90+CD49f+ HSCs proliferated similarly in 87%-PVA and 99%-PVA (Figure 2f) suggesting that unlike mouse, human HSC proliferation was not sensitive to amphiphilic PVA. Both PVA-types could maintain functional HSC activity ex vivo (Figure 2g).
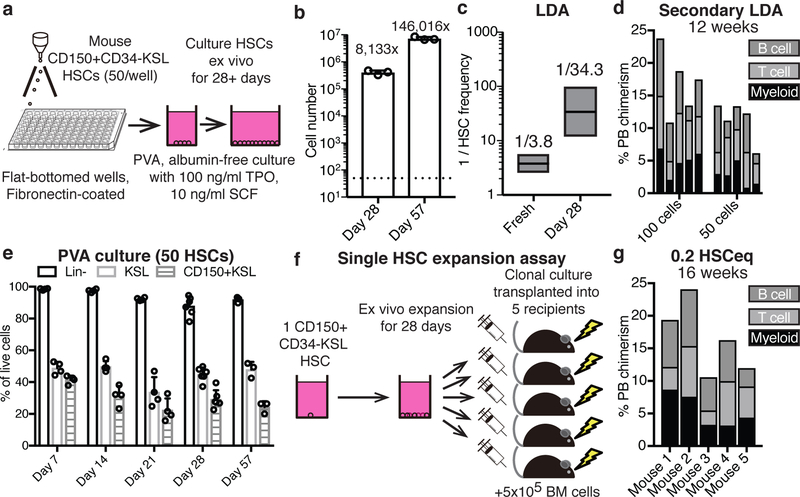
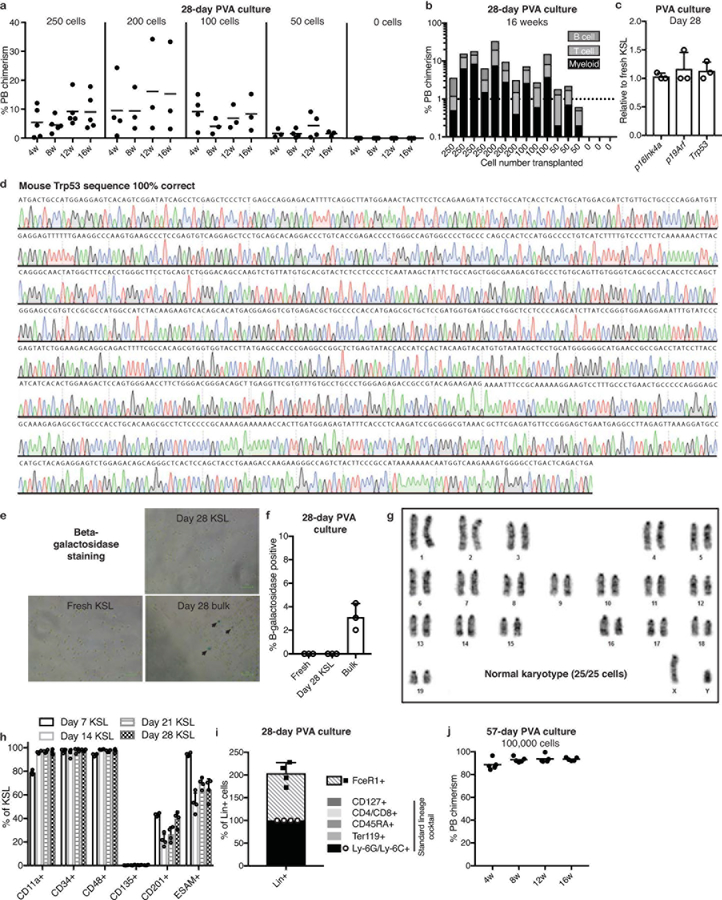
From the results above, we defined optimal mouse HSC culture conditions as 100 ng/ml TPO, 10 ng/ml SCF, and 87%-PVA on fibronectin (Figure 3a). In these conditions, 50 CD150+CD34-KSL HSCs expanded ~8,000-fold over 28-days (Figure 3b). In limiting dilution assays (LDAs) of 28-day cultures (transplanted against 2×105 BM competitors), just 50 cell aliquots displayed >1% multilineage PB chimerism at 16-weeks in 2 out of 3 recipients (Extended Data Figures 3a,b). Using Extreme Limiting Dilution Analysis23, we calculated HSC frequency at 1:34.3 cells, equivalent to 1.2×104 functional HSCs in the day-28 culture (Figure 3c). This compares to the 1:3.8 frequency of functional HSCs within the freshly-isolated CD150+CD34-KSL population (based on previous transplantation data24). We therefore estimate expansion of functional HSCs at between 236-fold (assuming all 50 starting cells were functional HSCs) and 899-fold (assuming 1:3.8 starting cells [~13 cells] were functional HSCs).
Figure 3: Long-term ex vivo expansion of functional HSCs.
(a) Schematic of the optimized mouse HSC expansion culture: 50 CD150+CD34-KSL HSCs were sorted into flat-bottom fibronectin-coated plate wells containing albumin-free F12 media supplemented with 1 mg/ml PVA, 100 ng/ml TPO, and 10 ng/ml SCF.
(b) Mean number of live cells after culturing 50 CD150+CD34-KSL HSCs for 28 days (n=6) or 57 days (n=3) on fibronectin-coated plates in PVA-based culture media. Error bars denote s.d.
(c) Box plots representing limiting dilution analysis of fresh CD150+CD34-KSL (total of 138 mice; published previously24,25) and 28-day HSC cultures (total of 16 mice; see Extended Data Figure 3a) calculated with ELDA software23, using a positive cutoff of >1% multilineage PB chimerism at 16-weeks. Box plots denote calculated mean, and upper and lower limits.
(d) 12-week donor PB chimerism in secondary recipient mice (n=5 per group), from 100 and 50 donor cells from day-28 PVA cultures (using primary recipients in Extended Data Figure 3b). BM from three primary recipients was pooled and 1×106 cells transplanted into secondary recipients. Each column represents an individual mouse.
(e) Mean percentage of phenotypic Lineage-, KSL and CD150+KSL cells during cultures, as described in (a), at day 7 (n=4), day 14 (n=4), day 21 (n=4), day 28 (n=6), and day 57 (n=3). Error bars denote s.d.
(f) Schematic of the single HSC expansion assay: single CD150+CD34-KSL HSCs were expanded for 28-days and then transplanted into five lethally-irradiated recipient mice against 5×105 BM competitors. Single HSCs expanded into ~5×105 cells meaning each recipient received ~1×105 cells (0.2 HSCeq). 10 single HSC-derived cultures were transplanted.
(g) 16-week donor PB chimerism from 1/5th of a 28-day culture derived from a single CD150+CD34-KSL HSC (n=5), as describe in (f). Each column represents an individual mouse. Representative data for 3 independent single HSC cultures (out of 10 transplanted).
Secondary transplantation was performed by pooling BM from the primary LDA recipients (3 survived to 16-weeks): all secondary recipients of 106 BM cells from 100-cell and 50-cell primary transplants displayed donor PB chimerism at 12-weeks (Figure 3d). As these were pooled secondary transplants, we can only conclude that at least 1 in 150 day-28 cells were serially-transplantable long-term HSCs. We therefore estimate expansion of serially-engraftable HSCs at a minimum of between 54-fold (assuming all starting cells were functional HSCs) and 204-fold (assuming only 1:3.8 of starting cells [~13 cells] were functional HSCs).
Consistent with the high functional activity at day-28, senescence markers were not increased and Trp53 remained unmutated (Extended Data Figure 3c,d). The KSL population also remained negative for senescence-associated beta-galactosidase (Extended Data Figure 3e,f). Additionally, engrafting HSCs were karyotypically normal (Extended Data Figure 3g). Ex vivo phenotypic KSL populations remained fairly stable during culture (Figure 3e, Extended Data Figure 3h) and most cells in the cultures remained lineage-negative (based on a Ly-6G/Ly-6C/Ter119/CD45RA/CD4/CD8/CD127 antibody lineage cocktail; Figure 3e). The lineage cocktail-positive cells were Ly-6G/Ly-6C+ although additional analysis identified FceR1+ cells at similar frequencies (Extended Data Figure 3i). HSC cultures could also be continued longer-term: by culture day-57, the 50 starting HSCs had generated ~7.3×106 cells while retaining a stable phenotypic KSL population and functional HSC activity (Figures 3b,e; Extended Data Figure 3j).
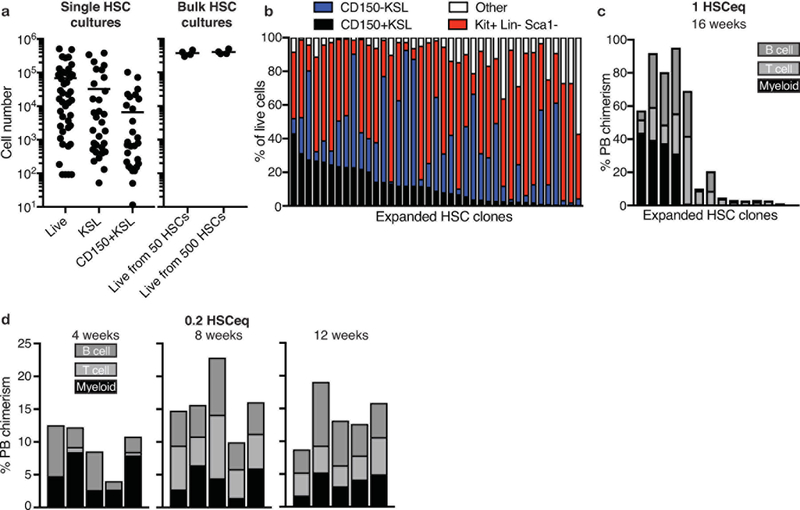
Although mouse BM HSCs can be highly-enriched based on surface-marker expression, purified CD150+CD34-KSL cells exhibit significant functional heterogeneity in single cell transplantation assays24,25. Consistent with this, significant variability in the expansion of single HSCs was observed, with some cells only generating <100 cells while others expanded to ~5×105 cells (although >90% generated colonies; Extended Data Figure 4a). The most proliferative clones generated similar cell numbers as cultures from 50 or 500 HSCs (Extended Data Figure 4a), suggesting surface area limited growth. Phenotypic heterogeneity was also observed in clonally-derived cultures; some clones retained ~90% KSL (Extended Data Figure 4b) while others almost exclusively generated cKit+Sca1-Lin- cells. Transplantation of clonal cultures resulted in high-level multipotent activity in 4 out of 14 recipients (Extended Data Figure 4c). We also detected robust (~15%) PB chimerism when clonally-derived cultures were divided and transplanted into 5 recipients against 5×105 BM competitors (for 3 out of 10 single HSC cultures tested; Figure 3f,g). These experiments confirmed bona fide HSC self-renewal ex vivo, but suggested that self-renewal capacity is unevenly distributed within the phenotypic HSC compartment.
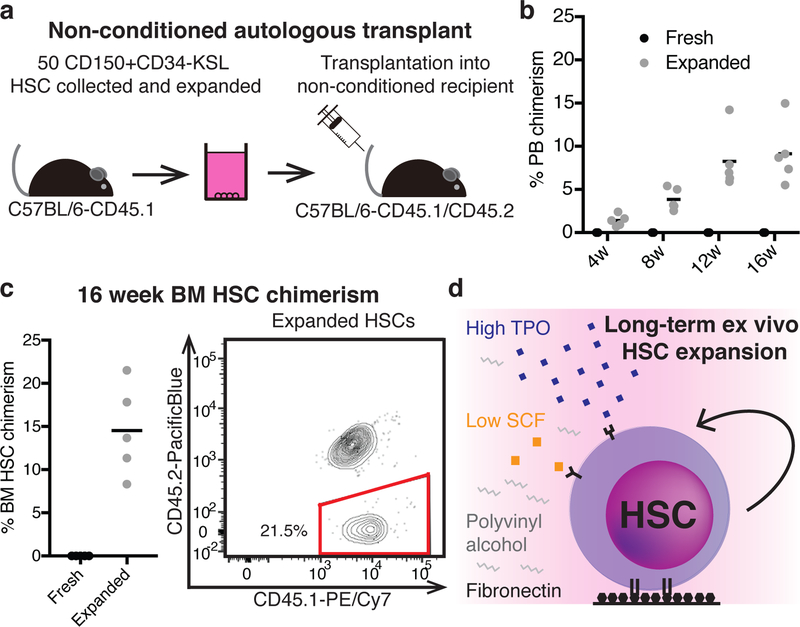
Radiation-based BM conditioning is normally required to make space for donor HSCs in HSC transplantation. Donor engraftment in non-conditioned recipients is possible, but not normally feasible, because very large numbers of HSCs are required for the transplant26,27. By expanding 50 HSCs for 28-days before transplantation, we could achieve long-term donor PB and BM HSC chimerism in non-conditioned immunocompetent mice (Figure 4a-c). This approach could also be used to engraft immunodeficient NOD/SCID mice, a model of congenital immunodeficiency; multilineage PB chimerism was observed in all recipients with donor lymphoid B and T cells detected long-term (Extended Data Figure 5a-c).
Figure 4: Ex vivo expanded HSCs engraft in non-conditioned recipients.
(a) Schematic of non-conditioned autologous transplantation: 50 CD150+CD34-KSL cells from C57BL/6-CD45.1 mice were expanded for 28-days before being transplanted into non-conditioned C57BL/6-CD45.1/CD45.2 recipients.
(b) Mean 4–16 week donor PB chimerism from 50 fresh HSCs (n=5) or a 28-day culture derived from 50 HSCs (50 HSCeq; n=5), transplanted as described in (a).
(c) Mean 16-week donor BM CD34-KSL HSC chimerism (n=5) for the assay described in (a) (left) and an example flow cytometric plot displaying CD45.1 and CD45.2 expression within the BM CD34-KSL compartment of recipient mice (right).
(d) Graphical summary of the optimized conditions for expansion of functional mouse HSCs.
In summary, we have developed an albumin-free ex vivo culture condition that expands functional mouse HSCs (Figure 4d) with broad applications in HSC research. Although we have been able to isolate HSCs at high purity for over 20 years1, stable ex vivo expansion of functional HSCs has remained elusive. Our results suggest that poor optimization of existing culture constituents combined with media supplement impurities have been a major barrier to ex vivo HSC expansion.
Methods:
Data Reporting.
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to outcome assessment.
Mice.
C57BL/6-CD45.2 and C57BL/6-CD45.1 (PepboyJ) mice were purchased from Japan SLC, Sankyo-Lab Service, Jackson Laboratories (000664, 002014), or bred in-house. For congenic transplantation experiments, 8–12 week-old male mice were used as donors and 8–12 week-old female mice were used as recipients. TLR2-knockout mice, TLR4-knockout mice, and NOD/Scid (NOD.Cg-Prkdcscid) mice were purchased from Jackson Laboratories (004650, 007227, 005557, and 001303, respectively). NG (NOD.Cg-Prkdcscid Il-2rγnull/SzJ) mice were purchased from In Vivo Science Inc. All mice were housed in a specific pathogen-free (SPF) condition with free access to food and water. All animal protocols were approved by the Animal Care and Use Committee of the Institute of Medical Science University of Tokyo, the Animal Care and Use Committee of RIKEN Tsukuba Branch, and/or the Administrative Panel on Laboratory Animal Care at Stanford University.
Cell collection by fluorescent-activated cell sorting (FACS).
Mouse BM cells were isolated from the tibia, femur, and pelvis, stained with APC-cKit antibody and cKit-positive cells enriched using anti-APC magnetic beads and LS columns (Miltenyi Biotec). cKit-enriched cells were then stained with a lineage antibody cocktail (biotinylated-CD4, -CD8, -CD45RA/B220, -TER119, -Gr1, and -CD127), before being stained with anti-CD34, anti-cKit, anti-Sca1, and streptavidin-APC/eFluor 780 (as detailed in Extended Data Table 1) for 90 minutes. Where indicated, cells were also stained with anti-CD150. Cell populations were then purified using a FACS AriaII (BD) by direct sorting into wells containing media using PI as a live/dead stain (Extended Data Figure 1b).
Serum albumin-based mouse cell cultures.
Serum albumin-based cultures were performed using F12 media (Life Technologies), 1% insulin-transferrin-selenium-ethanolamine (ITSX; Life Technologies), 1% penicillin/streptomycin/ glutamine (P/S/G; Life Technologies), 10 mM HEPES (Life Technologies), 0.1% recombinant human serum albumin (HSA; Albumin Biosciences), at 37°C with 5% CO2. Cultures were supplemented with recombinant mouse SCF and recombinant mouse TPO (Peprotech), as indicated. All long-term cultures used 10 ng/ml SCF and 100 ng/ml TPO, with media changes made every 3 days after the first 5 days by manually removing conditioned media by pipetting and replacing fresh media as indicated. In 96-well plate wells containing 200 μl of media, this involved gentle removal of 190–200 μl of the conditioned media using a pipette to avoid disturbing cells that were lightly adherent to the well bottom, and then gently pipetting in 200 μl of pre-warmed and freshly-prepared media down the side of the well to minimize disturbing the cell layer. Any cells removed from the well in the conditioned media were discarded. Long-term cultures were performed using flat-bottomed plates, tissue culture-treated and/or coated with fibronectin (Corning; 354409), Collagen1 (Corning; 354407), Collagen4 (Corning; 354429), Gelatin (Sigma; G2500), or Laminin511 (iMatrix; 892018). Where indicated, various concentrations of recombinant mouse IL-6 (Peprotech) were added to cultures.
Serum albumin-free mouse cell cultures.
HSCs were cultured in media composed of F12 media, 1% ITSX, 1%, 10 mM HEPES, 1% P/S/G, 100 ng/ml mouse TPO, 10 ng/ml mouse SCF and 0.1% of one the following chemicals (all from Sigma): hydroxypropyl cellulose (HPC; 435007), low-viscosity carboxymethylcellulose sodium salt (CMC-LV; C5678), medium viscosity carboxymethylcellulose sodium salt (CMC-MV; 21902), alpha-cyclodextrin (alpha-CD; C4642), beta-cyclodextrin (beta-CD; C4767), gamma-cyclodextrin (gamma-CD; C4930), 2-hydroxypropyl-betal—cylodextrin (HBC; H107), 2-hydroxypropyl-gamma-cycldextrin (HGC; H125), methyl-beta-cylodextrin (MBC; C4555), poloxamer 188 (polox188; P5556), or polyvinyl alcohol (PVA; P8136, 363081, or 363146), at 37°C with 5% CO2. For long-term cultures in PVA-based cultures, complete media changes were made every 2–3 days after the first 5–6 days, as described above for albumin-based cultures. Long-term cultures were split 1:3 at ~90% confluency. Where indicated, lipopolysaccharide (LPS; Sigma L2762) or IL-6 (Peprotech) were added to cultures.
Analysis of cultured cells.
Following ex vivo culture, cells were counted (using a hemocytometer, a CYTORECON cytometer, or a Nucleocounter NC-3000). For flow cytometric analysis, cells were stained with a lineage cocktail (biotinylated-CD4, -CD8, -CD45RA/B220, -TER119, -Gr-1, and -CD127) and then with antibodies detailed in Extended Data Table 1 for 30–90 minutes. Following a wash step, flow cytometric analysis was performed using a FACS AriaII (BD), LSRFortessa (BD), or FACS Canto (BD) using PI as a live/dead stain.
Competitive transplantation assay.
Culture HSCs from C57BL/6-CD45.1 mice were transplanted alongside 1×106 whole BM competitor cells C57BL/6-CD45.1/CD45.1 (F1) mice into C57BL/6-CD45.2 mice following lethal-dose irradiation (9.5 Gy). Donor chimerism was tracked by collecting peripheral blood (PB) cells and staining with anti-CD45.1, anti-CD45.2, anti-CD11b, anti-Ly-6G/Ly-6C, anti-CD45RA (B220), anti-CD4, anti-CD8 antibodies (detailed in Extended Data Table 1) for 30 minutes. Following a wash step, cells were analyzed by flow cytometry (as above) using PI as a live/dead stain. Secondary BM transplantation assay were performed by transferring 1×106 BM cells from the primary recipient mice into lethally-irradiated C57BL/6-CD45.2 mice, with donor chimerism analyzed as above.
Limiting dilution assays.
For limiting dilution assays, set numbers of cultured C57BL/6-CD45.1 cells were aliquoted by FACS after culture and transplanted into lethally-irradiated C57BL/6-CD45.2 recipient mice together with 2×105 F1 BM competitor cells. Donor chimerism was analyzed as above. Limiting dilution analysis was performed using ELDA software23, based on a 1% PB multilineage chimerism (at least 0.2% myeloid donor chimerism) as the threshold for positive engraftment. To calculate fresh HSC frequency, the same criteria were applied to a total of 138 transplantation assays from freshly-isolated CD150+CD34-KSL cells that we published previously24,25. Where indicated, secondary transplantation assays were performed as described above.
Non-conditioned transplantation assays.
CD150+CD34-KSL cells were purified from C57BL/6-CD45.1 or C57BL/6-CD45.2 mice and expanded, as described above. Freshly-isolated or expanded bulk cell cultures were then transplanted into non-irradiated C57BL/6-CD45.1/CD45.2 recipient mice or NOD/SCID (CD45.1) recipient mice, split into three doses over consecutive days.
Cytokine immunoassays.
Conditioned media was collected at day-7 (before any media change) or day-14 (following a media change at day 10), or as indicated in the figure legends. Mouse 39-plex kits (eBiosciences) were used according to the manufacturer’s recommendations with modifications described below. Briefly, beads were added to a 96 well plate and washed in a Biotek ELx405 washer. Samples were added to the plate containing the mixed antibody-linked beads and incubated at room temperature for one hour followed by overnight incubation at 4°C with shaking. Cold and Room temperature incubation steps were performed on an orbital shaker at 500–600 rpm. Following the overnight incubation plates were washed in a Biotek ELx405 washer and then biotinylated detection antibody added for 60 minutes at room temperature with shaking. Plate was washed as above and streptavidin-PE was added. After incubation for 30 mins at room temperature wash was performed and reading buffer was added to the wells. Plates were read using a Luminex Flex3D instrument with a lower bound of 50 beads per sample per cytokine. Custom Control beads (Assay CHEX) by Radix Biosolutions were added to all wells.
Human cell cultures and xenograft assays.
Human umbilical cord blood-derived CD34+ cells (purchased from Lonza or Stem Cell Technologies) or FACS-purified CD34+CD38-CD90+CD49f+ cells (following staining with APC-CD34 [Biolegend; 560940], PE-CD38 [BD; 347687], FITC-CD90 [Biolegend; 328113], and APC/Cy7-CD49f [Biolegend; 313611]) were cultured in IMDM (Life Technologies) containing 0.1% HSA or 0.1% PVA (Sigma P8136, 363081, or 363146), supplemented with 1% ITSX, 1% P/S/G, 10 mM HEPES. For proliferation assays, 50 cells were seeded per well and supplemented with 10 ng/ml human SCF and 100 ng/ml human TPO (Peprotech). During the cultures, media was refreshed every three days and counted at day-7. For xenograft assays, 2×103 cells were expanded for 7-days before being injected intravenously into sub-lethally-irradiated (1.5 Gy) NOG mice. Human cell chimerism in the PB was calculated at 16-weeks post-transplantation using PE/Cy7-CD45.1 (Biolegend; 110730) and V450-hCD45 antibodies (BD; 560367).
Senescence analysis.
Total KSL cells RNA was extracted using the RNeasy Mini Kit (QIAGEN) and reverse transcribed using SuperScript III First-Strand Synthesis System and Oligo (dT) primers (Invitrogen). qPCR was performed on a Thermal Cycler Dice Real Time System (Takara) using SsoAdvancedTM Universal SYBR Green Supermix and the following primer sets: p16Ink4a (GAACTCTTTCGGTCGTACCC and CGAATCTGCACCGTAGTTGA), p19Arf (GGGTTTTCTTGGTGAAGTTCG and TTGCCCATCATCATCACCT), and Trp53 (CAGTCTACTTCCCGCCATAA and GTCTCAGCCCTGAAGTCATAAG). The reaction conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec, and 72°C for 20 sec. Gene expression of genes was normalized relative to Gapdh expression (using primer set: CGACTTCAACAGCAACTCCCACTCTTCC and TGGGTGGTCCAGGGTTTCTTACTCCTT). Senescence beta-galactosidase staining (Cell Signaling, kit 9860S) was performed according to the manufacturer’s instructions following cell attachment to poly-L-lysine-coated plates. For Sanger sequencing, Trp53 cDNA was PCR amplified (using primer set: CATCCTGGCTGTAGGTAGCG ACCCTATGAGGGCCCAAGAT) and sequenced using nested sequencing primers: AAAAGTCTGCCTGTCTTCCAG; TGATGGCCTGGCTCCTCC; CACGTACTCTCCTCCCCTCA; and CTTCTGTACGGCGGTCTCTC. Sanger sequencing was outsourced to FASMAC Co. Ltd, and visualized and aligned using SnapGene software.
Phospho-protein fluorescence immunostaining.
Fluorescent immunostaining was performed by attaching, fixing, and staining cells on poly-l-lysine-coated glass slides (Matsunami Glass, Osaka, Japan), with imaging and quantification performed with a Cellomics ArrayScan VTI HCS Reader (ThermoScientific), using methods described previously20,28.
Karyotyping analysis.
Karyotyping was performed on CD45.1+ BM cells FACS-purified from primary recipients of 28-day PVA cultured HSCs at 16 weeks post-transplantation, and performed by Nihon Gene Research Laboratories Inc. (Japan).
Statistical analysis.
One-way and two-way ANOVA tests and unpaired two-tailed t-tests, were performed as indicated in the figures using Prism 7 software.
Extended Data
Extended Data Figure 1: Optimizing conditions for long-term HSC culture.
(a) Schematic of the standard HSC culture assay: C57BL/6-CD45.1 BM CD34-cKit+Sca1+Lineage- (CD34-KSL) HSCs were sorted (50 cells/well) into U-bottom 96-well plate wells, see (b) for sorting scheme. HSC growth can be observed during culture by counting or flow cytometry, with media changes made every three days (after initial seven days in culture). After 7–28 days, functional HSC activity was determined using competitive transplantation into irradiated C57BL/6-CD45.2 mice, against 1×106 BM competitor cells from C57BL/6-CD45.1/CD45.2 (F1) mice. Donor chimerism within peripheral blood (PB) myeloid, T cell, and B cell lineages was determined after 4–16 weeks or longer. Where indicated, secondary transplantation assays were performed by transplanting 106 BM cells from primary recipients into irradiated C57BL/6-CD45.2 mice.
(b) FACS gating strategy for sorting CD34-KSL cells (gates 1–7) and CD150+CD34-KSL cells (gates 1–8) from cKit-enriched mouse BM. Representative of at least 5 experiments.
(c) Flow cytometric histograms for cell surface cKit staining of HSCs following stimulating with 100 ng/ml TPO and either 0, 10, or 100 ng/ml SCF for 1, 24, and 48 hours. Representative of 3 independent cultures.
(d) Mean florescence intensity (MFI) of cKit antibody staining on HSCs cultured in 100 ng/ml TPO supplemented with 10 ng/ml or 100 ng/ml SCF, analyzed after 1–72 hours in culture, relative to cultures containing 100 ng/ml TPO without SCF. Mean of 3 independent cultures. Error bars denote s.d.
(e) Mean 16-week donor PB chimerism from 1×104 HSC-derived cells following a 28-day culture on plastic (n=5), Collagen1 (n=3), Collagen4 (n=4), Fibronectin (n=3), Gelatin (n=5), or Laminin511 (n=4) culture plates (cultured in 100 ng/ml TPO and 10 ng/ml SCF with complete media changes). Competitive transplantation against 1×106 BM competitors.
(f) Number of live cells after culturing 50 CD34-KSL HSCs for 28 days on plastic (tissue culture-treated) plates or fibronectin-coated plates. Mean of 3 independent cultures. Error bars denote s.d.
Extended Data Figure 2: Identification of PVA-based HSC culture conditions.
(a) Fold-change in mean fluorescence intensity (MFI) from cytokine immunoassays performed on HSA-based HSC cultures between day 8 and 13. Media changes performed at day 7 and day 10. Mean of 4 independent cultures with fold-change relative to unconditioned media. Error bars denote s.d.
(b) Mean 7-day expansion of 50 CD34+KSL HPCs in 100 ng/ml TPO and 10 ng/ml SCF with or without addition of 0.3 ng/ml-10 ng/ml mouse IL-6 (n=4).
(c) Heatmap displaying the MFI fold change from cytokine immunoassays using conditioned media from day-14 HSC cultures. CD34-KSL HSCs were isolated from C57BL/6 WT, TLR2-KO or TLR4-KO mice and cultured in HSA-based cultures. Dexamethasone (+Dex) at 50nM was added where indicated. Mean of 4 independent cultures with fold-change relative to unconditioned media.
(d) Concentration of IL-6 observed in 14-day HSA-based cultures of WT HSCs (n=8), TLR2-KO HSCs (n=8), TLR4-KO HSCs (n=6), or WT HSCs +Dex (n=8). Error bars denote s.d.
(e) Mean 12-week donor PB chimerism from 7-day cultured HSCs, in fresh media (n=7) or in media composed of 50% media collected from a 12-day HSC culture and 50% fresh media (termed conditioned media; n=7). Competitive transplantation against 1×106 BM competitors.
(f) Example flow cytometry plots displaying cKit and Sca1 expression on the Lin- progeny (left), and CD150 and CD48 expression in the KSL population (right) after a seven-day polyvinyl alcohol (PVA)-based HSC culture. Representative of 4 independent cultures.
(g) Concentration of various cytokines in day-14 conditioned media from HSA- or PVA-based CD34-KSL HSC cultures. Mean of 8 independent cultures. Error bars denote s.d. Statistical significance was calculated using t-tests. *, **, ***, and **** denote p<0.05, p<0.01, p<0.001, and p<0.0001, respectively.
(h) Relative expression of p16Ink4a, p19Arf, and p53 in KSL cells collected from 14-day cultures (HSA-based cultures with half-media changes, HSA-based cultures with complete media changes, and PVA-based cultures with complete media changes), relative to expression in freshly-isolated KSL cells. Mean of 3 independent cultures, with gene expression normalized to Gapdh expression. Error bars denote s.d.
(i) Number of phospho-gamma histone 2AX (H2AX) nuclear foci in day-28 KSL cells from HSA-based or PVA-based HSC cultures. Irradiated cells were included as a positive control. 49 cells quantified per condition.
(j) Relative expression of p16Ink4a, p19Arf, and p53 in KSL cells collected from 14-day cultures (left): HSA-based cultures, PVA-based cultures, and PVA-based cultures supplemented with 1 ng/ml LPS. Mean of technical quadruplets, with gene expression normalized to Gapdh expression. Concentration of IL-6 observed in these culture conditions (right). Mean of 4 independent cultures. Error bars denote s.d.
(k) 28-day expansion of 50 CD150+CD34-KSL HSCs in media containing 87% hydrolyzed PVA (87%-PVA) or >99% hydrolyzed PVA (99%-PVA). 1×104 day 28 cells represents ~1 HSCeq for 87%-PVA and ~5 HSCeq for 99%-PVA. Mean of 3 independent cultures. Error bars denote s.d.
(l) 7-day expansion of 50 human CB CD34+ cells in HSA- or PVA-based cultures supplemented with 10 ng/ml human SCF and 100 ng/ml human TPO. Mean of 3 independent cultures. Error bars denote s.d.
Extended Data Figure 3: Characterization of long-term PVA-based HSC cultures.
(a) Mean 4–16-week donor PB chimerism from 28-day PVA-based (CD150+CD34-KSL) HSC cultures using 100 ng/ml TPO and 10 ng/ml SCF in fibronectin-coated wells with complete media changes. Indicated cell numbers transplanted against 2×105 BM cells. Data from two independent transplantation experiments.
(b) 16-week multilineage donor PB chimerism for each individual mouse in (a).
(c) Expression of p16Ink4a, p19Arf, and p53 in 28-day PVA-cultured KSL cells, relative to expression in freshly-isolated KSL cells. Mean of 3 independent cultures, with gene expression normalized to Gapdh expression. Error bars denote s.d.
(d) Sanger sequencing trace of p53 cDNA amplified from KSL cells collected from 28-day PVA-based HSC cultures (n=1).
(e) Representative images of beta-galactosidase activity staining of freshly-isolated KSL, KSL isolated from 28-day PVA-based cultures, and bulk 28-day PVA-based cultures. Representative of two biological replicates.
(f) Percentage of beta-galactosidase-positive cells in conditions described in (e). Mean of technical triplicates (50–100 cells counted per replicate). Error bars denote s.d.; N.D. denotes not detected.
(g) Karyotype of CD45.1+ BM-repopulating progeny of day-28 expanded functional HSCs in PVA-based media at 16-weeks post-transplantation. All chromosomes analyzed were normal in 25 out of 25 cells analyzed (performed by Nihon Gene Research Laboratories Inc.).
(h) Frequency of CD11a+, CD34+, CD48+, CD135+, CD201+, and ESAM+ cells within the phenotypic KSL population during ex vivo HSC culture (derived from 50 CD150+CD34-KSL). Mean of 4 independent cultures. Error bars denote s.d.
(i) Composition of the lineage marker-positive compartment of day-28 HSC cultures. The lineage antibody cocktail used in this study comprised of CD4, CD8, CD45RA, Ter119, Ly-6C/Ly-6G, and CD127. A non-overlapping FceR1+ cell population was also identified within the culture, and is quantitated relative to the lineage antibody-cocktail positive population. Mean of 4 independent cultures. Error bars denoting s.d.
(j) Mean 4–16 week donor PB chimerism from 1×105 cells from a 57-day PVA-based HSC culture using fibronectin-coated plates and supplemented with 100 ng/ml TPO and 10 ng/ml SCF (n=5). Competitive transplantation against 1×106 BM competitors.
Extended Data Figure 4: Characterization of clonally-derived HSC expansion cultures.
(a) Mean number of live cells, KSL cells, and CD150+KSL cells derived from single CD150+CD34-KSL HSCs after 28-days culture (n=48; left), and mean number of live cells from bulk (50 and 500) CD150+CD34-KSL HSC cultures after 28-days (n=4; right).
(b) Proportion of phenotypic cell types that constituent day-28 cultures derived from single CD150+CD34-KSL HSCs. Only cultures with >10,000 cells analyzed (39 wells of 84 wells analyzed).
(c) 16-week donor PB chimerism of 28-day expanded single CD150+CD34-KSL HSC cultures transplanted into lethally-irradiated recipients against 2×105 BM cells. Each column represents an individual mouse.
(d) 4–12-week donor PB chimerism from 1/5th of a 28-day culture derived from a single CD150+CD34-KSL HSC, as describe in Figure 3f,g. Each column represents an individual mouse. Representative data for 3 independent single HSC cultures (out of 10 transplanted).
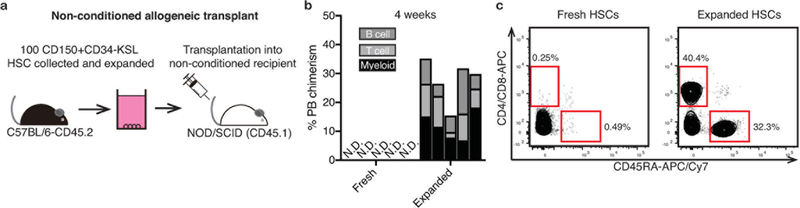
Extended Data Figure 5: Nonconditioned transplantation into immunodeficient recipients.
(a) Schematic of nonconditioned allogeneic transplantation: 100 CD150+CD34-KSL cells from C57BL/6-CD45.2 mice were expanded for 28-days before being transplanted into non-conditioned immunodeficient NOD/SCID recipients.
(b) 4-week donor PB chimerism of 100 fresh HSCs (n=5) or a 28-day HSC culture derived from 100 HSCs (n=5), transplanted as described in (a). Each column represents an individual mouse. N.D. denotes not detected.
(c) Example flow cytometric plots displaying T cell (CD4/CD8) and B cell (CD45RA, also known as B220) PB lineages within non-conditioned NOD/SCID mice at 16-weeks post-transplantation (representative of 5 mice), as described in (a).
Extended Data Table 1: List of antibodies.
Table of anti-mouse antibodies used in this study including Supplier and Identifier.
| Antibody | Source | Identifier |
|---|---|---|
| Biotin anti-CD4 | eBioscience | Cat# 13-0041-85 |
| Biotin anti-CD8 | eBioscience | Cat# 13-0081-86 |
| Biotin anti-CD45RA/B220 | eBioscience | Cat# 36-0452-85 |
| Biotin anti-TER-119 | eBioscience | Cat# 13-5921-85 |
| Biotin anti-Ly-6G/Ly-6C (RB6-8C5) | eBioscience | Cat# 13-5931-85 |
| Biotin anti-CD 127 (A7R34) | eBioscience | Cat# 13-1271-85 |
| APC anti-c-Kit (2B8) | eBioscience | Cat# 17-1171-83 |
| PE-Cy7 anti-c-Kit (2B8) | Biolegend | Cat# 105814 |
| FITC anti-CD34 (RAM34) | eBioscience | Cat# 11-0341-85 |
| PE-Cy7 anti-CD 150 (TC15-12F12.2) | BioLegend | Cat# 115914 |
| PE anti-Ly-6A/E (Sca-1) (D7) | BioLegend | Cat# 122508 |
| PE anti-Ly-6A/E (Sca-1) (D7) | eBioscience | Cat# 12-5981-83 |
| FITC anti-Ly-6A/E (Sca-1) (D7) | Biolegend | Cat# 108105 |
| Streptavidin-APC/ePluor 780 | eBioscience | Cat# 47-4317-82 |
| PE-Cy7 anti-CD45.1 | BioLegend | Cat# 110730 |
| BrilliantViolet421 anti-CD45.2 (104) | BioLegend | Cat# 109832 |
| ePluor450 anti-CD45.2 (104) | eBioscience | Cat# 48-0454-82 |
| FITC anti-Ly-6G/Ly-6C (RB6-8C5) | eBioscience | Cat# 11-5931-85 |
| FITC anti-CD lib (Ml/70) | eBioscience | Cat# 11-0112-41 |
| PE anti-Ly-6G/Ly-6C (RB6-8C5) | eBioscience | Cat# 12-5931-82 |
| PE anti-CDllb (Ml/70) | eBioscience | Cat# 12-0112-82 |
| APC-ePluor780 anti CD45R (RA3-6B2) | eBioscience | Cat# 47-0452-82 |
| APC anti-CD4 (RM4-5) | eBioscience | Cat# 17-0042-83 |
| APC anti-CD8 (53-6.7) | eBioscience | Cat# 17-0081-83 |
| Pacific Blue anti-Terll9 (TER119) | eBioscience | Cat# 48-5921-82 |
| PE-Cy7 anti-Ly-6G/Ly-6C (RB6-8C5) | eBioscience | Cat# 25-5931-82 |
| FITC anti-CD 127 (A7R34) | eBioscience | Cat# 11-1271-85 |
| PE anti-FceRl (MAR-1) | eBioscience | Cat# 12-5898-81 |
| BrilliantViolet421 anti-CD135 (A2F10) | Biolegend | Cat# 135353 |
| PE anti-CDlla (M17/4) | eBioscience | Cat# 12-0111-081 |
| APC anti-CD201 (eBiol560) | eBiosciences | Cat# 17-2012-82 |
| Pacific Blue anti-CD48 (BCM1) | Biolegend | Cat# 103418 |
| APC anti-ESAM (1G8/ESAM) | Biolegend | Cat# 136207 |
Supplementary Material
Acknowledgements:
We thank S. Takaki, Y. Ishii, H. Hasegawa, M. Hayashi, Stanford Human Immune Monitoring Center for technical support, and J. Bhadury for advice. This research was funded by JSPS KAKENHI Grant-in-Aid for Scientific Research (JP18H05095; JP17H05086), Japan Agency for Medical Research and Development (JP18bm0404025), CIRM (LA1_C12–06917; DISC1–10555), the NIH (R01DK116944; R01HL147124) and the Ludwig Foundation. ACW was funded by Bloodwise (15050), the Leukemia and Lymphoma Society (3385–19), and the JSPS. KML was supported by the NIH Director’s Early Independence Award (DP5OD024558), Siebel Stem Cell Institute, Baxter Foundation, and The Anthony DiGenova Endowed Faculty Scholar.
Footnotes
Conflict-of-interest: H.N. is a co-founder and shareholder of ReproCELL. Inc.
Data Availability:
All graphed datasets can be found in the Supplementary Source Data Files. Additional data files will be made available upon reasonable request from the corresponding authors.
Supplementary Information: 5 Extended Data Figures and 1 Extended Data Table available in the online version of the paper.
References:
- 1.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 1996;273(5272):242–245. [DOI] [PubMed] [Google Scholar]
- 2.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med 2006;354(17):1813–1826. [DOI] [PubMed] [Google Scholar]
- 3.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature 2014;505(7483):327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood 2015;125(17):2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamazaki S, Ema H, Karlsson G, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 2011;147(5):1146–1158. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S, Geiger H. HSC Niche Biology and HSC Expansion Ex Vivo. Trends Mol Med 2017;23(9):799–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaves CJ. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood 2015;125(17):2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ieyasu A, Ishida R, Kimura T, et al. An All-Recombinant Protein-Based Culture System Specifically Identifies Hematopoietic Stem Cell Maintenance Factors. Stem Cell Reports 2017;8(3):500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutu DL, Kokkaliaris KD, Kunz L, Schroeder T. Three-dimensional map of nonhematopoietic bone and bone-marrow cells and molecules. Nat Biotechnol 2017;35(12):1202–1210. [DOI] [PubMed] [Google Scholar]
- 10.Gekas C, Graf T. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood 2013;121(22):4463–4472. [DOI] [PubMed] [Google Scholar]
- 11.Umemoto T, Yamato M, Ishihara J, et al. Integrin-αvβ3 regulates thrombopoietin-mediated maintenance of hematopoietic stem cells. Blood 2012;119(1):83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csaszar E, Kirouac DC, Yu M, et al. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell 2012;10(2):218–229. [DOI] [PubMed] [Google Scholar]
- 13.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol 2014;5:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netea MG, Van der Graaf C, Van der Meer JW, Kullberg BJ. Recognition of fungal pathogens by Toll-like receptors. Eur J Clin Microbiol Infect Dis 2004;23(9):672–676. [DOI] [PubMed] [Google Scholar]
- 15.Loures FV, Pina A, Felonato M, Araújo EF, Leite KR, Calich VL. Toll-like receptor 4 signaling leads to severe fungal infection associated with enhanced proinflammatory immunity and impaired expansion of regulatory T cells. Infect Immun 2010;78(3):1078–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson AC, Morita M, Nakauchi H, Yamazaki S. Branched-chain amino acid depletion conditions bone marrow for hematopoietic stem cell transplantation avoiding amino acid imbalance-associated toxicity. Exp Hematol 2018;63:12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taya Y, Ota Y, Wilkinson AC, et al. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science 2016;354(6316):1152–1155. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of Cellular Senescence. Trends Cell Biol 2018;28(6):436–453. [DOI] [PubMed] [Google Scholar]
- 19.de Haan G, Lazare SS. Aging of hematopoietic stem cells. Blood 2018;131(5):479–487. [DOI] [PubMed] [Google Scholar]
- 20.Flach J, Bakker ST, Mohrin M, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 2014;512(7513):198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kane MT, Bavister BD. Protein-free culture medium containing polyvinylalcohol, vitamins, and amino acids supports development of eight-cell hamster embryos to hatching blastocysts. J Exp Zool 1988;247(2):183–187. [DOI] [PubMed] [Google Scholar]
- 22.Wiles MV, Johansson BM. Embryonic stem cell development in a chemically defined medium. Exp Cell Res 1999;247(1):241–248. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347(1–2):70–78. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 2013;154(5):1112–1126. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto R, Wilkinson AC, Ooehara J, et al. Large-Scale Clonal Analysis Resolves Aging of the Mouse Hematopoietic Stem Cell Compartment. Cell Stem Cell 2018;22(4):600–607.e604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharya D, Rossi DJ, Bryder D, Weissman IL. Purified hematopoietic stem cell engraftment of rare niches corrects severe lymphoid deficiencies without host conditioning. J Exp Med 2006;203(1):73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimoto M, Sugiyama T, Nagasawa T. Numerous niches for hematopoietic stem cells remain empty during homeostasis. Blood 2017;129(15):2124–2131. [DOI] [PubMed] [Google Scholar]
- 28.Seita J, Ema H, Ooehara J, et al. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc Natl Acad Sci U S A 2007;104(7):2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.