Abstract
Background
The genus Onchocerca Diesing, 1841 includes species of medical importance, such as O. volvulus (Leuckart, 1893), which causes river blindness in the tropics. Recently, zoonotic onchocercosis has been reported in humans worldwide. In Japan, O. dewittei japonica Uni, Bain & Takaoka, 2001 from wild boars is a causative agent for this zoonosis. Many filarioid nematodes are infected with Wolbachia endosymbionts which exhibit various evolutionary relationships with their hosts. While investigating the filarial fauna of Borneo, we discovered an undescribed Onchocerca species in the bearded pig Sus barbatus Müller (Cetartiodactyla: Suidae).
Methods
We isolated Onchocerca specimens from bearded pigs and examined their morphology. For comparative material, we collected fresh specimens of O. d. dewittei Bain, Ramachandran, Petter & Mak, 1977 from banded pigs (S. scrofa vittatus Boie) in Peninsular Malaysia. Partial sequences of three different genes (two mitochondrial genes, cox1 and 12S rRNA, and one nuclear ITS region) of these filarioids were analysed. By multi-locus sequence analyses based on six genes (16S rDNA, ftsZ, dnaA, coxA, fbpA and gatB) of Wolbachia, we determined the supergroups in the specimens from bearded pigs and those of O. d. dewittei.
Results
Onchocerca borneensis Uni, Mat Udin & Takaoka n. sp. is described on the basis of morphological characteristics and its genetic divergence from congeners. Molecular characteristics of the new species revealed its close evolutionary relationship with O. d. dewittei. Calculated p-distance for the cox1 gene sequences between O. borneensis n. sp. and O. d. dewittei was 5.9%, while that between O. d. dewittei and O. d. japonica was 7.6%. No intraspecific genetic variation was found for the new species. Wolbachia strains identified in the new species and O. d. dewittei belonged to supergroup C and are closely related.
Conclusions
Our molecular analyses of filarioids from Asian suids indicate that the new species is sister to O. d. dewittei. On the basis of its morphological and molecular characteristics, we propose to elevate O. d. japonica to species level as O. japonica Uni, Bain & Takaoka, 2001. Coevolutionary relationships exist between the Wolbachia strains and their filarial hosts in Borneo and Peninsular Malaysia.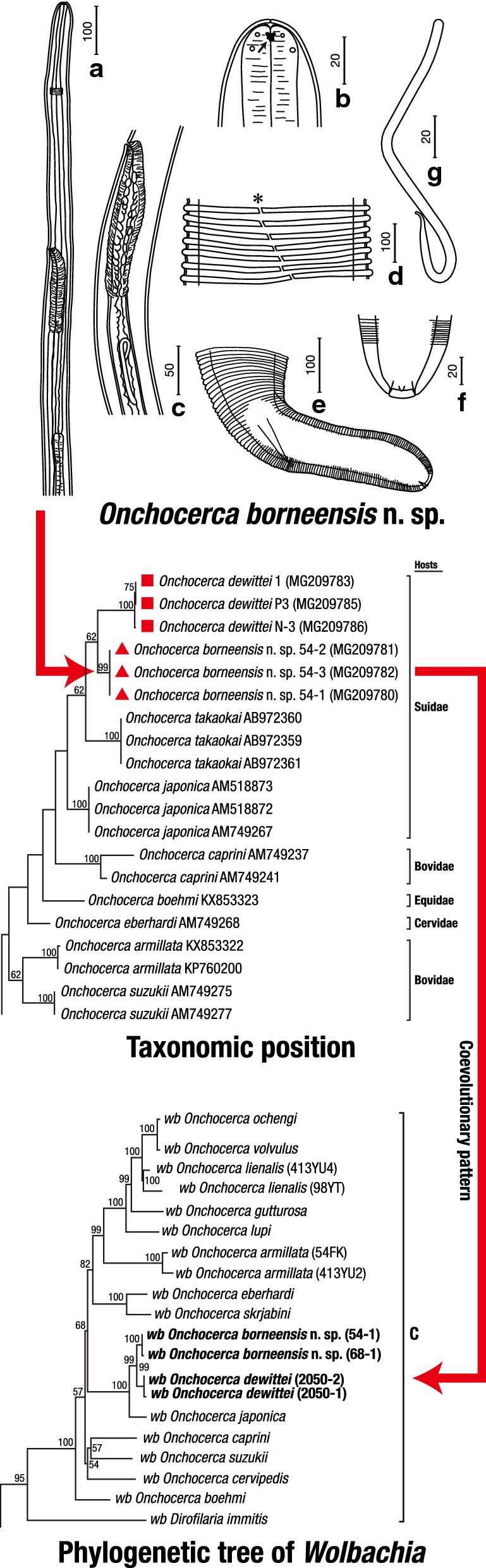
Keywords: Coevolution, Indomalayan realm, Malayfilaria sofiani, Onchocerca dewittei, Onchocerca japonica, Suidae
Background
The genus Onchocerca Diesing, 1841 (Onchocercidae) is well known for its medical importance, with O. volvulus (Leuckart, 1893) causing river blindness or onchocercosis in humans. In 2018, the World Health Organization listed onchocercosis as a major cause of ocular and skin diseases in sub-Saharan Africa and parts of Latin America [1]. Recently, zoonotic infections with Onchocerca spp. have been reported in humans worldwide. Onchocerca lupi Rodonaja, 1967 from wolves, dogs and cats has been reported to cause lesions in the subconjunctival region, skin or spinal cord in humans in the USA, Turkey, Tunisia, Iran and Germany [2–4]. In Japan, zoonotic cases owing to O. dewittei japonica Uni, Bain & Takaoka, 2001 from the Japanese wild boar Sus scrofa leucomystax Temminck (Suidae) have been documented [5–8]. In other zoonotic cases, the causative agents have been identified as O. gutturosa Neumann, 1910 from cattle, O. cervicalis Railliet & Henry, 1910 from horses and O. jakutensis (Gubanov, 1964) from European roe deer [9–11]. In order to assess the zoonotic potential of Onchocerca spp., it is necessary to better define the biodiversity within the genus, and their epidemiology in their domestic or wild hosts throughout their geographic range [12, 13].
To date, 33 species of Onchocerca and one subspecies (O. d. japonica) have been recorded worldwide [6, 14–18]. According to Lefoulon et al. [19], multi-locus sequence analyses suggest a division of members of the Onchocercidae into five clades (ONC1–ONC5), with ONC3 containing three genera, namely Onchocerca, Loxodontofilaria Berghe & Gillain, 1939 and Dirofilaria Railliet & Henry, 1910. Moreover, members of the genus Onchocerca are divided into three clades: O. d. japonica from the Suidae is placed in the first clade, which is the most diverse with regard to the host range [20, 21]. Although 35 species of filarial parasites from 22 genera have previously been recorded in vertebrates in Malaysia [15, 22–24], O. dewittei Bain, Ramachandran, Petter & Mak, 1977 is the only species of Onchocerca reported there. However, the origin, molecular characteristics and evolutionary relationships of O. dewittei remain to be clarified [25–27].
While participating in the Heart of Borneo Scientific Expedition organized by the Department of Forestry, Sarawak in 2016, we found an undescribed Onchocerca species in the bearded pig S. barbatus Müller in Long Banga, Sarawak. To elucidate the diversity and molecular relationships of Onchocerca spp. from suid hosts in Asia, we compared the specimens isolated from Bornean bearded pigs with specimens of O. d. dewittei recently collected from the banded pig S. s. vittatus Boie in Peninsular Malaysia and with those of O. d. japonica previously obtained from wild boars in Japan. Our results indicate that the specimens from Borneo differ from their congeners at the species level but are closely related to O. d. dewittei. In addition, our molecular analyses proved that O. d. dewittei and O. d. japonica are distinct species.
Wolbachia endosymbionts are alpha-proteobacteria widely distributed in arthropods and filarioid nematodes [28–33]. Wolbachia strains infect the female germline, lateral hypodermal chords and intestinal wall cells in many species in the family Onchocercidae [34, 35]. The endosymbionts are essential for the fertility and growth of their filarial hosts and exhibit various evolutionary relationships with them [28, 30, 31, 34, 35]. We examined Wolbachia endosymbionts in the specimens from Bornean bearded pigs and those of O. d. dewittei from banded pigs in Peninsular Malaysia. The Wolbachia supergroups were identified by multi-locus sequence analyses [19, 28]. On the basis of our findings, we discuss the molecular relationships between Wolbachia strains and their Onchocerca hosts in Asia. Furthermore, to compare Wolbachia strains of other filarial nematodes obtained in Malaysia with Wolbachia strains of the present Onchocerca spp., we detected a Wolbachia strain in Malayfilaria sofiani Uni, Mat Udin & Takaoka, 2017 (Onchocercidae), previously isolated from the common treeshrew Tupaia glis Diard & Duvaucel (Mammalia: Scandentia) in Peninsular Malaysia [24]. We discuss the molecular relationships between Wolbachia strains and their onchocercid hosts M. sofiani, Wuchereria bancrofti (Cobbold, 1877) and species of Brugia Buckley, 1958.
Methods
Collection of hosts and parasites
The Heart of Borneo Scientific Expedition organized by the Department of Forestry in Long Banga (3°12′0″N, 115°22′59.99″E), Sarawak, Malaysia, was undertaken between 20 August 2016 and 2 September 2016. During the expedition, the feet (n = 12) of three Bornean bearded pigs (ID nos. WB30, WB54 and WB68), legally captured by local inhabitants, were examined. Filarial parasites obtained from the footpads of the bearded pigs (ID nos. WB54 and WB68) were subjected to subsequent morphological and molecular studies, and pieces of muscle obtained from these two bearded pigs were used for molecular identification of the host species.
Morphological methods
To search for parasites, the footpads were dissected under a stereomicroscope. Isolated adult worms were fixed in 70% ethanol for morphological examination. Worms were cleared in lactophenol (R & M Chemicals, Essex, UK) and drawn under a compound microscope equipped with a camera lucida (Olympus U-DA, Olympus, Tokyo, Japan). The mid-region of a fixed female was embedded in paraffin, and sections were stained with haematoxylin and eosin (HE). Skin snips were taken from the limbs of the infected bearded pigs to examine microfilariae [36]. Thick blood smears were made and stained with 3% Giemsa solution (pH 7.4). Skin snips and blood smears were examined for microfilariae under a compound microscope.
Comparative material examined
Onchocerca dewittei dewittei collected from banded pigs in Peninsular Malaysia
Five banded pigs (ID nos. WP1-WP5) from Peninsular Malaysia were examined for filarial parasites. Because of the absence of a beard, these animals were identified as banded pigs. Three of the five animals (ID nos. WP1, WP4 and WP5) were legally captured by local inhabitants at the Field Studies Center (3°19′29.0″N, 101°45′09.7″E) of the University of Malaya, situated in the primary forest of Ulu Gombak, Selangor, on 27 February 2012, 14 December 2012 and 20 May 2016, respectively. Two of the banded pigs (ID nos. WP2 and WP3) were captured on an oil palm plantation at Sungai Besar (3°42′11.8″N, 101°05′53.2″E), Sabak Bernam District, Selangor, on 7 April 2012 and 14 October 2012, respectively. The two animals were provided by local residents with the permission of the oil palm plantation owner. A piece of muscle of one banded pig (ID no. WP5), captured at Ulu Gombak, was used for molecular identification of the host species. Skin snips were taken from the limbs of the banded pigs to examine microfilariae. Adult worms were obtained from the footpads of the banded pigs and examined as described above. Skin snips and blood smears were examined for microfilariae.
Onchocerca dewittei japonica obtained from the wild boar in Japan
Seven adults (four fragments of females and three fragments of males) of O. d. japonica had been collected from the footpads of two Japanese wild boars (ID nos. OB4 and OB5) in Bungoono (32°58′41.1″N, 131°35′06.2″E), Oita, Kyushu, Japan, on 7 November 2011. Six specimens were fixed in 70% ethanol and examined as described above. One female fragment, fixed in 80% ethanol, was used for the present molecular analysis.
Malayfilaria sofiani isolated from the common treeshrew in Peninsular Malaysia
One fragment of a female of M. sofiani (ID no. KE-2), fixed in 80% ethanol, was used to determine Wolbachia and its supergroup affiliation [24].
Molecular analysis of filarioid nematodes and host animals
The following materials were transferred directly into 80% ethanol and used for molecular analyses: Four Onchocerca females (ID nos. 54-1, 54-2, 54-3 and 68-1) collected from bearded pigs (ID nos. WB54 and WB68) in Borneo; seven fragments of females (ID nos. 1, N2-1, N-3, No3, No4, P-1 and P3) of O. d. dewittei obtained from banded pigs (ID nos. WP1, WP4 and WP5) in Peninsular Malaysia; one fragment of a female of O. d. japonica (ID no. 1; see above) in Japan; microfilariae obtained from the skin snips from a banded pig (ID no. WP5); pieces of muscle from limbs of the bearded pigs (ID nos. WB54, WB68) and a banded pig (WP5).
In order to determine the partial sequences of the mitochondrial cox1 and 12S rRNA genes of Onchocerca specimens, DNA extraction, polymerase chain reaction (PCR) amplification, and sequencing were performed as described previously [7, 24, 37, 38]. Filaria martis Gmelin, 1790 (Filariidae) was selected as the outgroup for the phylogenetic analyses based on the cox1 and 12S rRNA gene sequences. We also cloned the PCR products of the nuclear ITS region into pGEM-T vectors and determined the sequences of the recombinant plasmid [39]. Wuchereria bancrofti and B. malayi (Brug, 1927) were selected as outgroups for the ITS region. In addition, we determined the sequences of the cytochrome b gene (cytb) of bearded pigs in Borneo and a banded pig in Ulu Gombak, Peninsular Malaysia, following the protocol of Watanobe et al. [40].
The newly generated sequences were deposited in the GenBank database. GenBank accession numbers of the new sequences and those of other filarioid nematodes and Suidae used to compare the present specimens to are provided in the figures. We calculated uncorrected p-distances between species of filarial parasites in MEGA7 as an estimate of the accumulated number of nucleotide substitutions per site [41]. Phylogenetic trees of the nucleotide sequences of the cox1 and 12S rRNA genes and the ITS region of Onchocerca spp. and the cytb gene of the host animals were constructed using the maximum-likelihood (ML) method in MEGA7 [41], with 500 bootstrap replicates. The lengths of the sequence datasets used for the analyses were as follows: cox1, 393 bp; 12S rRNA, 304 bp; ITS, 866 bp; and cytb, 741 bp.
Wolbachia detection methods
Immunohistochemical staining
Sections of a female worm of O. d. dewittei were stained with a rabbit polyclonal antiserum raised against the surface protein of Wolbachia from B. pahangi (Buckley & Edeson, 1956) (1:2000 dilution), as described by Kramer et al. [42] and Ferri et al. [34].
Molecular screening
DNA was extracted from two female fragments (ID nos. 54-1 and 68-1) of Onchocerca specimens obtained from Bornean bearded pigs (ID nos. WB54 and WB68, respectively), two fragments of females (ID nos. 2050-1 and 2050-2) of O. d. dewittei obtained from a banded pig (ID no. WP5), and one fragment of a female (ID no. KE-2) of M. sofiani from a common treeshrew in Peninsular Malaysia, using the QIAamp® DNA Mini kit, following the protocol “DNA purification from tissues” recommended by the manufacturer (Qiagen, Courtaboef, France). Wolbachia symbionts were determined by nested PCR screening of the six genes (16S rDNA, ftsZ, dnaA, coxA, fbpA and gatB), as described by Lefoulon et al. [19, 28, 35]. PCR products were purified and sequenced by Eurofins Genomics. Supergroups of Wolbachia were identified as described by Lo et al. [43] and Lefoulon et al. [28].
Phylogenetic analysis
The sequences of the six genes of Wolbachia were aligned with sequences available in GenBank (Additional file 3: Table S2) using MAFFT [44]. For the nucleotide supermatrix, the phylogeny of Wolbachia strains was performed by ML inference using TIM + F + I + G4 with IQ-TREE version 1.5 [45, 46]. The robustness of nodes was assessed with 1000 bootstrap replicates. The length of the supermatrix dataset was 3086 bp.
Results
Family Onchocercidae Leiper, 1911
Subfamily Onchocercinae Leiper, 1911
Onchocerca borneensis Uni, Mat Udin & Takaoka n. sp.
Type-host: Sus barbatus Müller (Cetartiodactyla: Suidae), Bornean bearded pig.
Type-locality: Long Banga (3°12′0″N, 115°22′59.99″E), Ulu Baram, Sarawak, Malaysia.
Type-material: Holotype female (MNHN 103YT) and allotype male (MNHN 104YT) were deposited in the Muséum National d’Histoire Naturelle, Paris, France. Paratypes (7 females: Ob-B54-1–2, Ob-B54-4, Ob-B54-6–7, Ob-54-9, Ob-B68-2; 10 males: Ob-B54-2M1–2, Ob-B54-3M1, and Ob-B68-2-M3–9) were deposited in the Institute of Biological Sciences, University of Malaya, Malaysia. Collection dates: 31.viii.2016 and 1.ix.2016.
Site in host: Adult worms were found in nodular fibrous structures in the adipose tissue of footpads of fore- and hindlimbs.
Prevalence and intensity of infection: Two of three bearded pigs were infected with adult worms: seven females and four males in the bearded pig WB54, and one female and seven males in the bearded pig WB68.
Representative DNA sequences: Sequence data were deposited in the GenBank database as follows: cox1 (MG209780-MG209782), 12S rRNA gene (MG209790-MG209792) and ITS (MG192125-MG192127) for O. borneensis n. sp.; cytb (MG657264-MG657265) for S. barbatus. Accession numbers of Wolbachia sequences are provided in Additional file 3: Table S2.
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [47], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub: F33A99AB-EDB5-40BF-BFBB-34033F4FF1CF. The LSID for the new name Onchocerca borneensis Uni, Mat Udin & Takaoka n. sp. is urn:lsid:zoobank.org:act: 018F7D26-7650-46AF-9330-4826C46CD53B.
Etymology: The specific epithet is derived from Borneo Island, the location where the type-host was collected.
Description
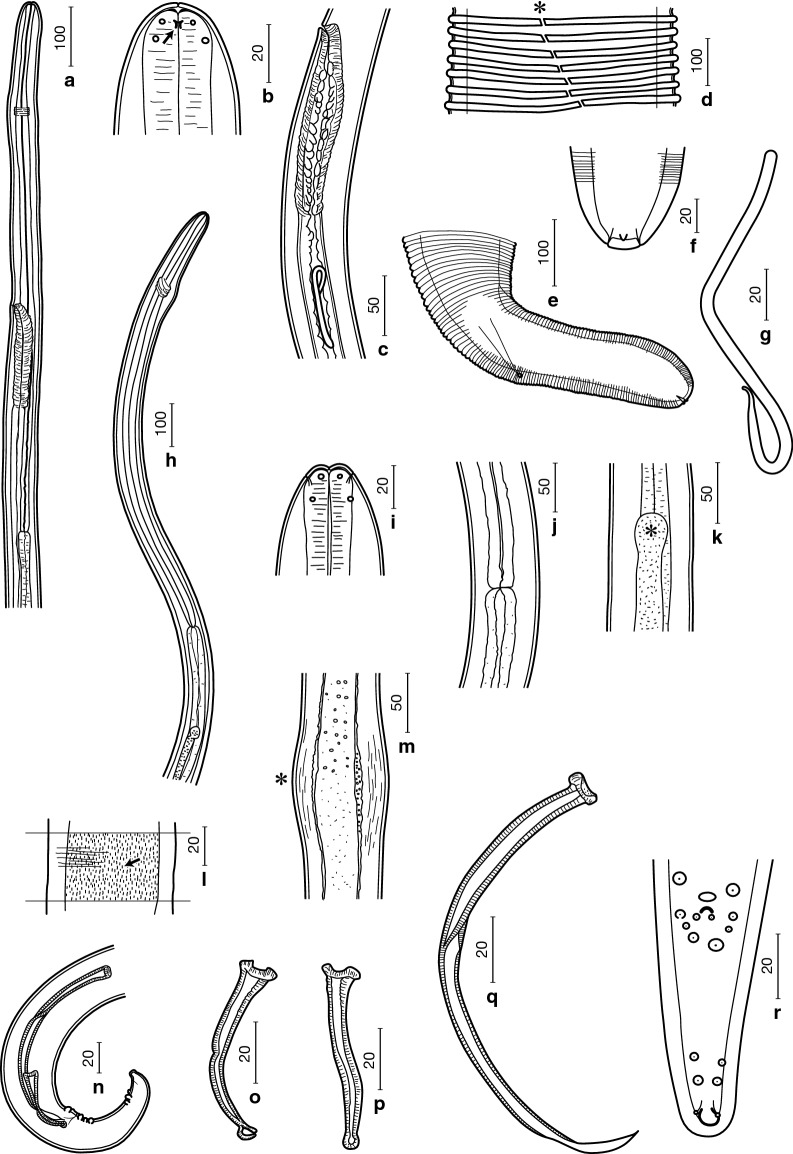
General. [Table 1; Figs. 1, 2]. Body slender, tapering towards both extremities. Anterior extremity very thin, straight, with rounded apex (Fig. 1a). Labial and cephalic papillae arranged in a circle of 4 each (Fig. 1b). Amphids lateral, on level of labial papillae. Mouth opening small; thin sclerotized lamella present between head cuticle and oesophageal apex (Fig. 1b). Oesophagus not divided, anterior portion thin, gradually widening posteriorly. Nerve-ring surrounding oesophagus on level of anterior portion. Slight cervical swelling present at nerve-ring. Deirids and excretory pore not observed. Body cuticle of females with external transverse ridges (Fig. 1d). Caudal papillae in males prominent, grouped near cloaca (Fig. 1r). Microfilaria unsheathed (Fig. 1g).
Table 1.
Comparative morphometric data for Onchocerca borneensis n. sp. and congeners recorded from wild suids
| Species | Onchocerca borneensis n. sp.a | Onchocerca dewittei dewittei Bain, Ramachandran, Petter & Mak, 1977 | Onchocerca dewittei japonica Uni, Bain & Takaoka, 2001 | Onchocerca takaokai Uni, Fukuda & Bain, 2015d | Onchocerca ramachandrini Bain, Wahl & Renz, 1993 |
|---|---|---|---|---|---|
| Reference | Present study | Present study | Present study | [18] | [16] |
| Host | Sus barbatus | Sus scrofa vittatus | Sus scrofa leucomystax | Sus scrofa leucomystax | Phacochoerus africanus |
| Locality | Long Banga, Sarawak, Malaysia | Ulu Gombak, Selangor, Malaysia | Bungoono, Oita, Japan | Oita, Japan | Cameroon |
| Female | (n = 8) | (n = 14) | (n = 3) | Holotype | Holotype |
| Body length (cm) | 21.5 (17.5–21.5) | 30.7b | 16.7–27.3c | – | 12.8 |
| Body width at midbody | 490 (410–590) | 220–320 | 260–430 | 110 | 210 |
| Nerve-ring from anterior end | 193 (170–225) | 195–200 | 207–233 | 190 | 250 |
| Oesophagus length | 975 (900–1288) | 988–1438 | 1020–1120 | 1270 | 1250 |
| Vulva from anterior end | 513 (475–680) | 343–497 | 520–620 | 475 | 650 |
| Distance of ridges at midbody | 17 (10–25) | 40–93 | 235–340 | Ridges absent | Longitudinal crests |
| Ridges (height/width) at midbody | 5 (4–5)/10 (9–10) | 10–11/15–20 | 23–35/75–100 | – | – |
| Cuticle thickness at midbody | 17 (10–36) | 20–40 | 13–33 | 8 | 15–20 |
| Tail length | 258 (183–258) | 143–188 | 100–218 | 130 | 170 |
| Microfilaria | (n = 10) | (n = 10) | (n = 10) | (n = 4) | (n = 11) |
| Body length | 160–188 | 198–245 | 158–203 | 295–329 | 290–325 |
| Body width | 5–6 | 5–8 | 5–6 | 6–9 | 7.0–7.5 |
| Male | (n = 11) | (n = 2) | (n = 3) | – | (n = 2) |
| Body length (mm) | 18 (16–24) | 42–45 | 51–54c | – | 32.3–34 |
| Body width at midbody | 65 (50–110) | 88–93 | 100–113 | – | 65–72 |
| Cuticular crests at midbody | Present | Present | Present | – | None |
| Nerve-ring from anterior end | 225 (163–225) | 213 | 167–247 | – | 175–190 |
| Oesophagus length | 1040 (800–1070) | 905 | 888–1007 | – | 1000–1050 |
| Right spicule length (RS) | 63 (58–85) | 75–88 | 73–77 | – | 78 |
| Left spicule length (LS) | 163 (125–198) | 223–245 | 223–250 | – | 220–240 |
| Spicule length ratio (LS/RS) | 2.6 (2.1–2.8) | 2.8–3.0 | 3.1–3.3 | – | 2.9 |
| Tail length | 70 (63–80) | 53–63 | 62–80 | – | 105–125 |
| Parasitic location of adults | Footpads | Footpads | Footpads | Skin of head, neck and back | Subcutaneous connective tissues of feet |
Fig. 1.
Line drawings of Onchocerca borneensis n. sp. Females (a–f), microfilaria (g) and males (h–r). a Anterior end, left lateral view. b Anterior extremity, lateral view, showing amphid (arrow). c Vagina, left lateral view. d Transverse cuticular ridges at midbody region, showing lateral field (*). e Posterior end, left lateral view. f Posterior extremity, ventral view, showing internal terminal point and two subterminal phasmids. g Microfilaria without sheath. h Anterior end, lateral view. i Anterior extremity, dorsoventral view. j Oesophago-intestinal junction. k Apex of testis (*). l Short longitudinal cuticular crests (arrow) at midbody region. m Body swelling (*). n Posterior end, right lateral view. o Right spicule, lateral view. p Right spicule, dorsoventral view. q Left spicule, lateral view. r Posterior end, ventral view. Scale-bars are in micrometres
Fig. 2.
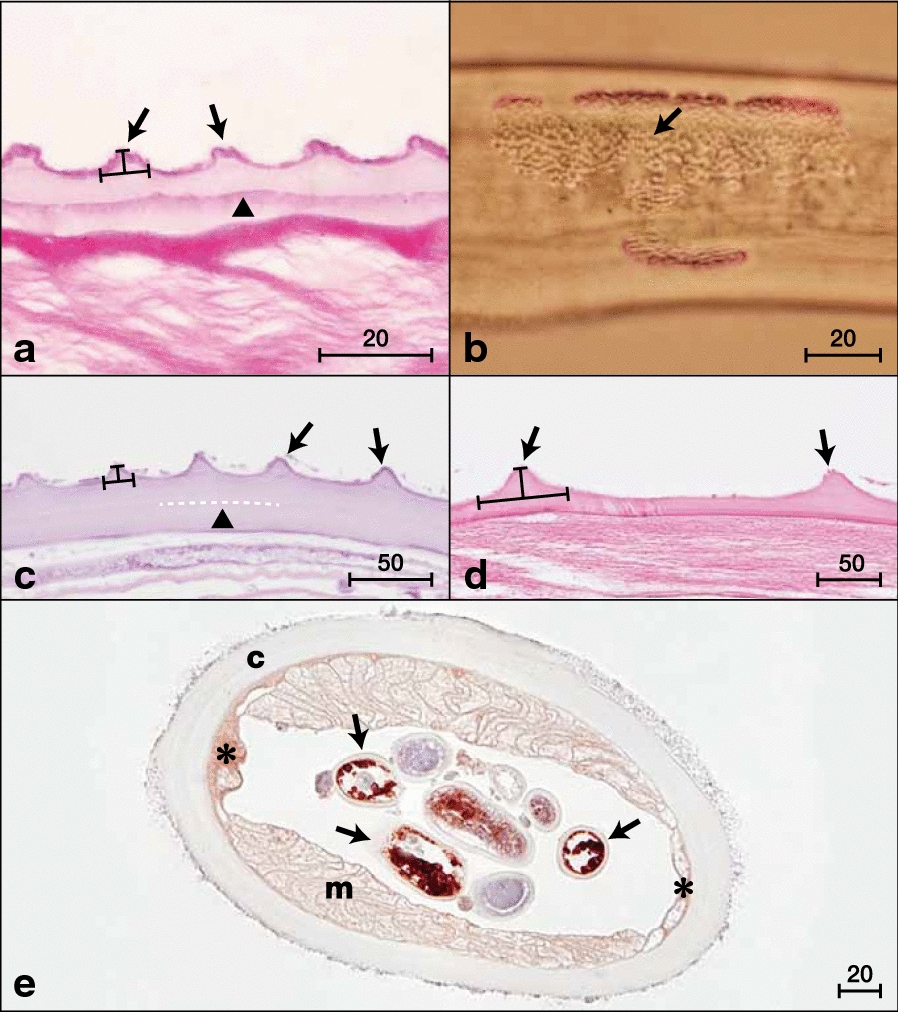
Light micrographs of Onchocerca borneensis n. sp. (a and b), O. dewittei dewittei (c and e) and O. d. japonica (d). a Longitudinal section of female at midbody: transverse cuticular ridges, triangular or trapezoid in shape (arrows; height and width, dark lines) and absence of internal striae at mid-line (arrowhead) (HE staining). b Short longitudinal cuticular crests (arrow) of male at midbody. c Longitudinal section of female at midbody: triangular transverse cuticular ridges (arrows; height and width, dark lines) and absence of internal striae (arrowhead) at mid-line (dotted line) (HE staining). d Longitudinal section of female at midbody: large triangular transverse cuticular ridges (arrows; height and width, dark lines), showing long distance between adjacent ridges (HE staining). e Transverse section of female with Wolbachia immunostaining. Dark red dots in ovaries and uterus (arrows) are Wolbachia. Lateral chords (*), muscles (m) and cuticle (c). Scale-bars are in micrometres
Female. [Based on the holotype, 3 complete worms and 4 fragmented specimens; Table 1, Figs. 1a–f, 2a.] Vulva at mid-level of oesophagus (Fig. 1a). Vagina straight, simple (Fig. 1c). Ovejector straight, parallel to oesophagus, posteriorly directed. Uterus didelphic and opisthodelphic. Body distinctly widened over much of its length (Table 1). Cuticle with fine external transverse ridges at 1.6 mm from anterior extremity, more pronounced posteriorly. Transverse ridges straight, interrupted in lateral fields, without bifurcations (Fig. 1d). In longitudinal sections at midbody, ridges triangular or trapezoid in shape, with rounded or flattened tips. Median layer of cuticle without internal striae (Fig. 2a). Tail bent dorsally (Fig. 1e), tail extremity with internal terminal point and 2 subterminal phasmids (Fig. 1f).
Male. [Based on the allotype and 10 complete specimens; Table 1, Figs. 1h–r, 2b.] Anterior extremity attenuated, rounded (Fig. 1h). Apex of testis 1.6–2.7 mm from anterior extremity, posterior to oesophago-intestinal junction (Fig. 1k). Main part of body with short longitudinal cuticular crests (Figs. 1l, 2b). In anterior half of body, 2 to 4 body swellings with pseudocoelomocytes (Fig. 1m). In one male (16 mm long), three body swellings at 2.5 mm, 4.8 mm and 7.4 mm from anterior extremity. Area rugosa absent. Right spicule with dent in mid-section and knobbed distal end in lateral view (Fig. 1o). Left spicule divided into handle and lamina; lamina with attenuated long membranous extremity (Fig. 1q). Gubernaculum absent. Narrow caudal alae present. Caudal papillae 7 pairs, arranged in 2 groups: 4 paracloacal sublateral pairs and 1 small subventral postcloacal pair; and 2 sublateral pairs in posterior third of tail. One unpaired precloacal ventral papilla present anterior to cloaca. Tail bent ventrally. Posterior extremity with large subcuticular knob and phasmids at its base (Fig. 1r).
Microfilaria. [Based on 10 specimens from uterus of a fixed female; Table 1, Fig. 1g.] Body 160–188 long, 5–6 wide, unsheathed. Anterior extremity rounded, tail extremity attenuated to curled blunt point.
Remarks
The present specimens were assigned to the genus Onchocerca as defined by Anderson & Bain [48]: preoesophageal ring absent; anterior extremity rounded; body not markedly tapering anteriorly and posteriorly in female; body cuticle of females with transverse ridges; and caudal papillae in males prominent, grouped near cloaca.
We thus compared the morphological characteristics of the present specimens with those of the 33 species and a single subspecies (O. d. japonica) currently included in the genus [6, 14, 17, 18, 20, 49]. Females of many Onchocerca spp. possess transverse cuticular ridges and internal cuticular striae [14, 17]. However, O. borneensis n. sp. closely resembles its congeners described from suid hosts, O. d. dewittei in Peninsular Malaysia and O. d. japonica in Japan, in having transverse cuticular ridges but lacking internal striae in females (Figs. 1d, 2a, c, d). The new species, O. d. dewittei and O. d. japonica possess short longitudinal crests in males (Figs. 1l, 2b) [6]. The new species is distinct from O. d. dewittei and O. d. japonica in that females are wider at midbody and have a longer tail, as well as that males are shorter, only reaching half the length of males of the latter two species, and have a shorter left spicule (Table 1). The distance between two adjacent cuticular ridges at midbody in females of O. borneensis n. sp. is one-third that of O. d. dewittei and less than one-tenth that of O. d. japonica. In longitudinal sections of females at midbody, the ridges of O. borneensis n. sp. are half as high and half as wide at base as those of O. d. dewittei (Table 1; Fig. 2a, c). In addition, microfilariae of the new species are shorter than those of O. d. dewittei (Table 1). Furthermore, O. borneensis n. sp. can be differentiated from the other two remaining species of Onchocerca parasitizing suid hosts. These are O. takaokai Uni, Fukuda & Bain, 2015, described from S. s. leucomystax in Japan, whose females lack transverse cuticular ridges [18], and O. ramachandrini Bain, Wahl & Renz, 1993, described from Phacochoerus africanus (Gmelin) [as P. aethiopicus (Pallas)] in Cameroon, whose females lack transverse cuticular ridges but bear discontinuous longitudinal crests, and whose males possess inconspicuous transverse striations without short longitudinal crests at midbody and 11 pairs of caudal papillae [16].
Prevalence, intensity and site in host
Onchocerca borneensis n. sp. was found in two (ID nos. WB54 and WB68) of the three Bornean bearded pigs examined in the Long Banga area. These pigs were identified as S. barbatus by molecular methods (Additional file 2: Figure S1). The third pig (ID no. WB30) was small, probably one year-old. In the bearded pig WB54, one female worm was collected from the footpad of the forelimb, and six females and four males were obtained from nodular fibrous structures in the adipose tissue of two footpads of the hindlimbs. In the bearded pig WB68, one female and seven males were obtained from the footpad of the hindlimb. Many specimens were found in the hindlimbs. Several of the males collected were entangled with a female. We did not find microfilariae in the blood smears or skin snips of the limbs of either of the two infected bearded pigs.
Onchocerca dewittei dewittei from Sus scrofa vittatus in Peninsular Malaysia
Host: Sus scrofa vittatus Boie (Cetartiodactyla: Suidae), banded pig.
Locality: Field Studies Center (3°19′29.0″N, 101°45′09.7″E) of the University of Malaya, situated in the primary forest of Ulu Gombak, Selangor, Peninsular Malaysia.
Voucher material: Specimens of O. d. dewittei were deposited in the Institute of Biological Sciences, University of Malaya, Malaysia (accession numbers: Od-F1–35 and Od-M1–2). Collection dates: 27.ii.2012, 14.xii.2012 and 20.v.2016.
Site in host: Adult worms were found in nodular fibrous structures in the adipose tissue of footpads of fore- and hindlimbs.
Prevalence and intensity of infection: Three of five banded pigs were infected with adult worms: 26 fragments of females and two entire males in WP1; one fragment of a female in WP4; and five fragments of females in WP5.
Representative DNA sequences: Sequence data were deposited in the GenBank database as follows: cox1 (MG209783, MG209785-MG209786); 12S rRNA gene (MG209793-MG209798, MG973148); ITS (MG192128-MG192133, MK045758) for O. d. dewittei; cytb (MG657266) for S. s. vittatus.
Description
Female. [Based on 3 anterior parts, 9 midsections and 2 posterior parts; Table 1, Fig. 2c.] Transverse cuticular ridges spaced 40–93 apart at midbody. In longitudinal sections at midbody, ridges triangular, with pointed tip. Median layer of cuticle without internal striae (Fig. 2c). A single body swelling present at midbody.
Male. [Based on 2 entire males; Table 1.] Apex of testis 2.8 mm from anterior extremity. Two body swellings at 5.3 mm and 14 mm from anterior extremity present in one male (45 mm long). Main part of body with small longitudinal cuticular crests.
Microfilaria. [Based on 10 uterine microfilariae from fixed females and microfilariae from skin snips; Table 1.] Microfilariae from uteri unsheathed, not curled. Microfilariae from skin snips: body usually curled, 175–220 long, 5 wide, identified on the basis of morphological and molecular characteristics (data not shown).
Prevalence, intensity and site in host
Adult worms of O. d. dewittei were found in three (ID nos. WP1, WP4 and WP5) of five banded pigs (S. s. vittatus) in Peninsular Malaysia. The animals were captured in the primary forest of Ulu Gombak. The following entire worms and fragments were detected: three fragments of females in nodular fibrous structures in footpads of the forelimbs, 26 fragments of females and two entire males in footpads of the hindlimbs of WP1; one fragment of a female in the footpad of a forelimb of WP4; and five fragments of females in the footpads of the four limbs of WP5. As deduced from the number of teeth in wild boar, the animal (WP1) was 2.5 years-old. Four and five microfilariae of O. d. dewittei were found in skin snips taken from the limbs of WP1 and WP5, respectively. No microfilariae of O. d. dewittei were found in the skin snips of the remaining banded pigs. No microfilariae were found in the blood smears.
Immunohistochemical staining of Wolbachia in the transverse sections
Wolbachia symbionts were detected in the uteri and ovary but not in the lateral hypodermal chords or in the intestinal wall cells in a transverse section of a female O. d. dewittei (Fig. 2e).
Onchocerca dewittei japonica from Sus scrofa leucomystax in Japan
Host: Sus scrofa leucomystax Temminck (Cetartiodactyla: Suidae), Japanese wild boar.
Locality: Bungoono (32°58′41.1″N, 131°35′06.2″E), Oita, Kyushu, Japan.
Voucher material: Specimens of O. d. japonica were deposited in the Institute for Research Promotion, Oita University, Japan (Accession numbers: Oj-F1–3 and Oj-M1–3). Collection date: 7.xi.2011.
Site in host: Adult worms were obtained from nodular fibrous structures in the footpads of fore- and hindlimbs.
Prevalence and intensity of infection: Both of two Japanese wild boars examined were infected with adult worms: two fragments of females and two fragments of males in OB4; two fragments of females and one fragment of a male in OB5.
Representative DNA sequences: Sequence data for O. d. japonica were deposited in the GenBank database as follows: 12S rRNA gene (MG209799); ITS (MG192124).
Female. [Based on 3 specimens; metrical data in Table 1.] Transverse cuticular ridges spaced 235–340 apart at midbody. In longitudinal sections at midbody, ridges triangular with wide base (Fig. 2d). Median layer of cuticle without internal striae.
Male. [Based on 3 specimens; metrical data in Table 1.] Main part of body with small longitudinal cuticular crests.
Microfilaria. [Based on 10 uterine microfilariae from fixed females.] Body 158–203 long, 5–6 wide.
Morphological differentiation of Onchocerca dewittei dewittei and O. d. japonica
We confirm that the two subspecies differ in the distance between the transverse cuticular ridges as described previously [6]. Microfilariae of O. d. dewittei are slightly longer than those of O. d. japonica as reported by Uni et al. [6]. Moreover, we observed distinct differences in the shape of the ridges in longitudinal sections: the ridges of O. d. dewittei are one-third as high and one-fifth as wide at base as those of O. d. japonica (Table 1; Fig. 2c, d). Furthermore, females of O. d. dewittei are narrower at midbody than those of O. d. japonica (Table 1). The above characteristics can be used to morphologically distinguish O. d. japonica from O. d. dewittei.
Molecular analyses
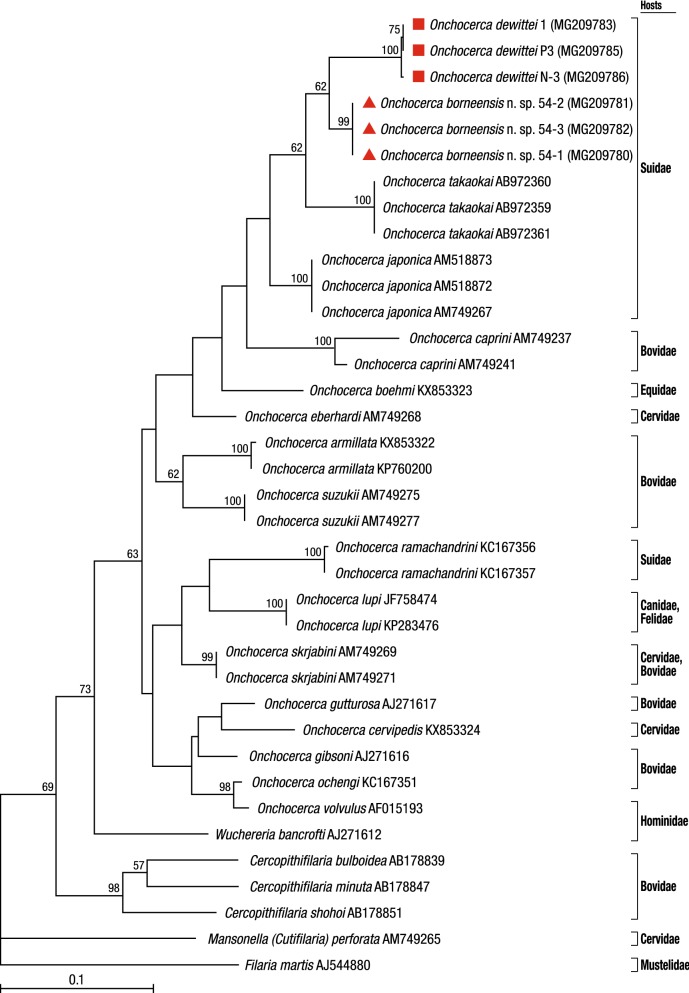
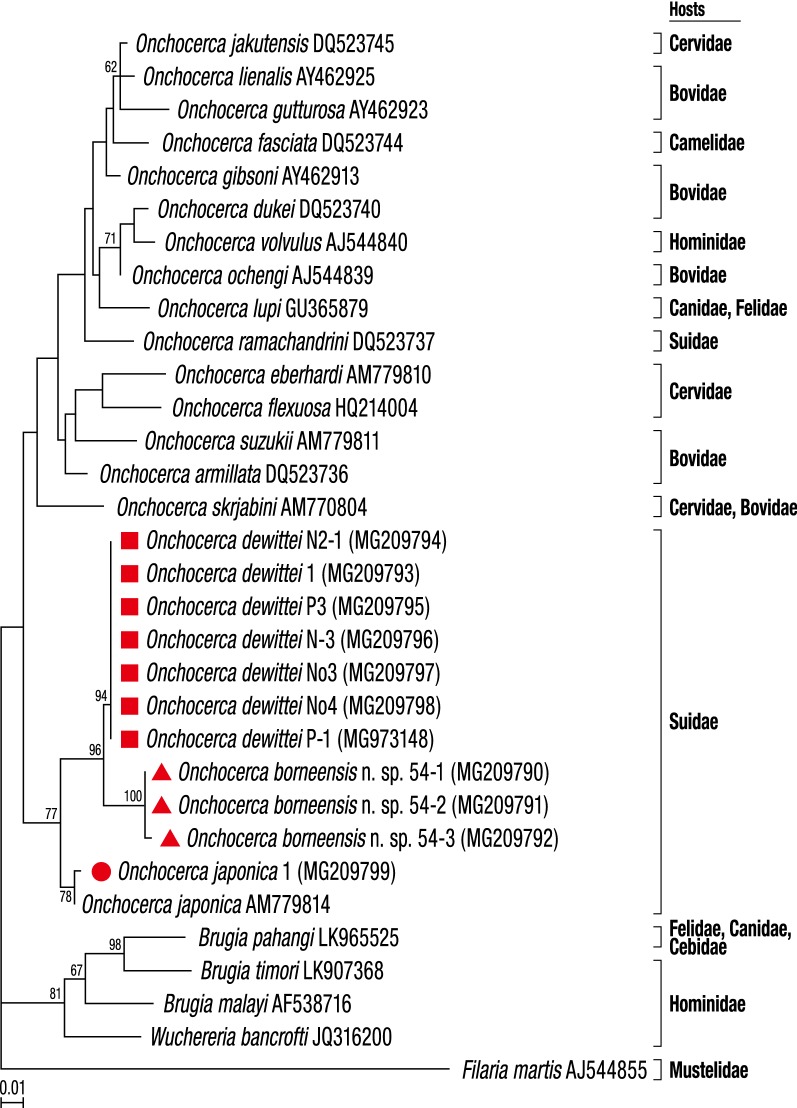
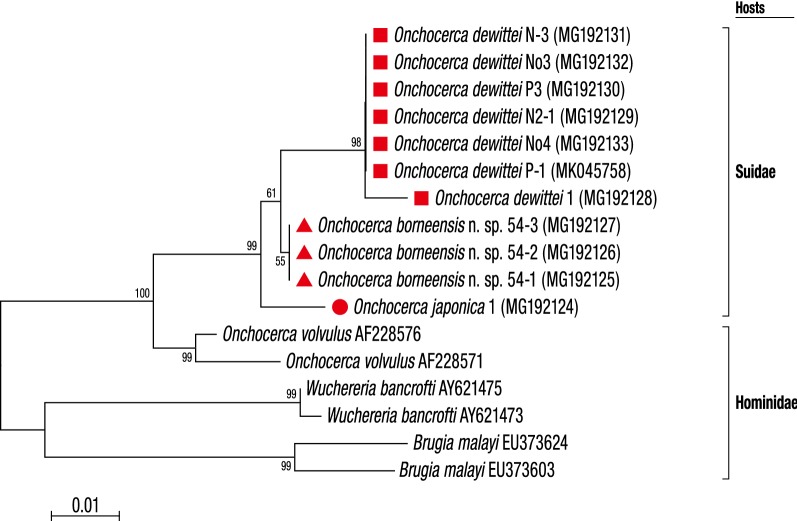
To elucidate the evolutionary relationships between O. borneensis n. sp., O. d. dewittei, O. d. japonica, O. takaokai and O. ramachandrini from suid hosts, we compared their cox1, 12S rDNA and ITS sequences with those of other filarial parasites available on GenBank. Calculated p-distances for the cox1 sequences between O. borneensis n. sp. and its congeners were 5.9% for O. d. dewittei, 6.4% for O. takaokai, 6.9% for O. d. japonica and 11.5–11.7% for O. ramachandrini. The p-distance for the cox1 sequences between O. d. dewittei and O. d. japonica was 7.6% (Additional file 1: Table S1). According to Ferri et al. [50], filarioid nematodes can be considered different species if the genetic distance based on the cox1 sequences is greater than 4.8%. In Onchocerca spp., cox1 interspecific distances are higher than 4.5% and intraspecific distances are lower than 2% [20]. Therefore, the molecular findings corroborate the morphological data, further supporting O. borneensis n. sp. as a species distinct from O. d. dewittei described in Peninsular Malaysia and other congeners. Moreover, on the basis of the morphological differences between O. d. dewittei and O. d. japonica set out above and the high molecular divergence between these two taxa, we propose to elevate O. d. japonica to species level as O. japonica Uni, Bain & Takaoka, 2001. In the phylogenetic trees based on ML inference using sequence data from the two mitochondrial genes, cox1 and 12S rRNA, as well as the nuclear ITS region, O. borneensis n. sp. was placed as a sister species to O. dewittei from Suidae (Figs. 3, 4 and 5).
Fig. 3.
Taxonomic position of Onchocerca borneensis n. sp. and O. dewittei based on cox1 nucleotide sequences. The maximum-likelihood phylogenetic tree was generated under the Tamura-Nei model in MEGA7 with 500 bootstrap replicates. The scale-bar below the diagram indicates the number of inferred changes along each branch. Red triangles and squares indicate sequences generated in this study
Fig. 4.
Taxonomic position of Onchocerca borneensis n. sp., O. dewittei and O. japonica based on 12S rDNA nucleotide sequences. The maximum-likelihood phylogenetic tree was generated under the Tamura-Nei model in MEGA7 with 500 bootstrap replicates. The scale-bar below the diagram indicates the number of inferred changes along each branch. Red triangles, squares and circle indicate sequences generated in this study
Fig. 5.
Taxonomic position of Onchocerca borneensis n. sp., O. dewittei and O. japonica based on ITS nucleotide sequences. The maximum-likelihood phylogenetic tree was generated under the General Time Reversible model in MEGA7 with 500 bootstrap replicates. The scale-bar below the diagram indicates the number of inferred changes along each branch. Red triangles, squares and circle indicate sequences generated in this study
The molecular characteristics of two bearded pigs (WB54 and WB68) in Borneo and a banded pig (WP5) in Peninsular Malaysia are presented in Additional file 2: Figure S1. The two host animals in Borneo were identified as S. barbatus by the present molecular analysis, whereas the host animal at Ulu Gombak in Selangor, Peninsular Malaysia, was morphologically identified as S. s. vittatus. Sequence data for comparison with S. s. vittatus from other studies were not available. The host animal from Peninsular Malaysia and S. barbatus from Borneo occupy vastly different positions in the present phylogenetic tree (Additional file 2: Figure S1).
Wolbachia infection
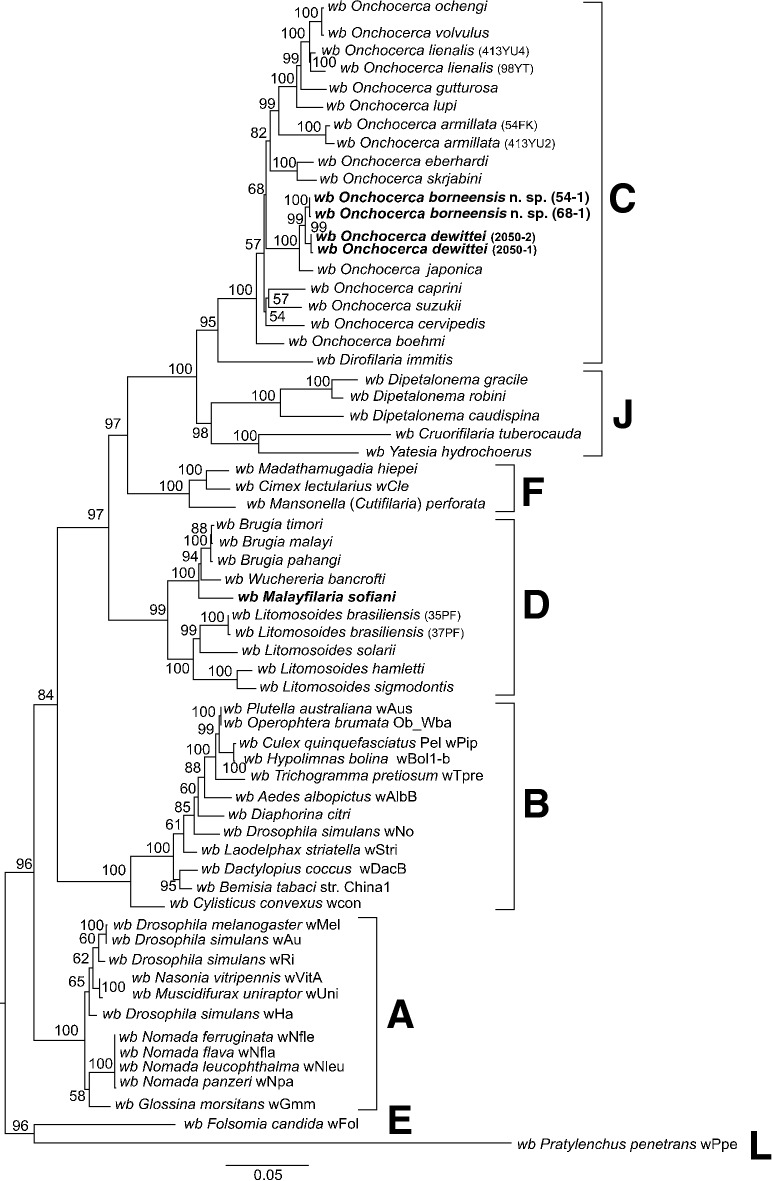
Onchocerca borneensis n. sp. and O. dewittei both harboured Wolbachia strains belonging to supergroup C (Fig. 6). The Wolbachia strains in O. borneensis n. sp. were closely related to the strains in O. dewittei. Malayfilaria sofiani harboured a strain of Wolbachia supergroup D; this strain was placed in a sister position to the clade formed by strains of W. bancrofti and species of Brugia.
Fig. 6.
Phylogenetic tree of Wolbachia based on six markers using maximum-likelihood inference. Analysis based on concatenation of 16S rDNA, ftsZ, dnaA, coxA, fbpA and gatB (3086 bp in total length). The topology was generated using model TIM + F + I + G4 with IQ-TREE. The robustness of nodes was assessed with 1000 bootstrap replicates. The Wolbachia supergroups (A–F, J and L) were identified according to Lefoulon et al. [28] and Lo et al. [43]. The scale-bar indicates the distance in substitutions per nucleotide. Abbreviation: wb, Wolbachia
Discussion
Based on their morphological characteristics, O. borneensis n. sp., O. dewittei and O. japonica constitute a sub-group within the five species of Onchocerca parasitizing hosts of the Suidae. Their predilection sites in the host as well as the hosts themselves (Sus Linnaeus) are equally similar. In contrast, O. ramachandrini, whose females possess longitudinal, not transverse, cuticular crests, is not only morphologically distinct from the above sub-group (Table 1), but its host, P. africanus in the Afrotropics [16], is markedly different from the hosts (Sus) of the four Onchocerca species in Asia. The host genus Phacochoerus F. Cuvier belongs to Phacochoerini, a distinct tribe within the Suinae [51].
Molecular analyses based on cox1, 12S rDNA and ITS sequences indicated that O. borneensis n. sp. and O. dewittei form a monophyletic clade (Figs. 3, 4 and 5). Onchocerca takaokai from Japan was close to the clade in the cox1 tree (Fig. 3) and also O. japonica from Japan was close to the clade in other trees (Figs. 5, 6). Furthermore, the four species (O. dewittei, O. borneensis n. sp., O. takaokai and O. japonica) parasitic in modern pigs (Sus) were closely related in the cox1 tree (Fig. 3) and based on the genetic distances (Additional file 1: Table S1). It is interesting to note that O. borneensis n. sp. and O. dewittei infect S. barbatus and S. s. vittatus, respectively, in the Indomalayan realm, but O. japonica and O. dewittei infect different subspecies of the same host species (S. scrofa Linnaeus) in the Palaearctic realm and the Indomalayan realm, respectively. This fact suggests that the host biogeographical area is a crucial factor for Onchocerca diversification. Conversely, the genetic distance between O. borneensis n. sp. and O. ramachandrini was pronounced and ranged from 11.5–11.7% (Additional file 1: Table S1). Krueger et al. [52] suggested that O. ramachandrini is one of the ancestral lineages in the genus Onchocerca.
Considering the evolutionary history of the Suidae, the ancestor of modern pigs (Sus) has been separated from the ancestor of sub-Saharan suids, including warthogs (Phacochoerus), since the late Miocene (> 10 Ma) [53–55]. During the late Pliocene/early Pleistocene (2.5 Ma), the genus Sus arose in the tropical inland Southeast Asia (ISEA) and subsequently diversified into multiple species. While S. barbatus and several other species are restricted to the ISEA, S. scrofa expanded from Southeast Asia to the whole of Eurasia and North Africa [54–57]. Our molecular analysis identified S. barbatus as host of O. borneensis n. sp. in Borneo (Additional file 2: Figure S1). In Peninsular Malaysia, we morphologically identified S. s. vittatus as host of O. dewittei, on the basis of the absence of a beard. Except for the data generated in the present study, no sequence data for S. s. vittatus are available. Our molecular analysis indicates that the banded pigs from Peninsular Malaysia differ from S. barbatus and S. verrucosus Boie. The results obtained for Sus species in the present study generally resemble the phylogeny of the Suinae proposed by Frantz et al. [54, 55]. We thus speculate that Onchocerca species in the Suidae, in conjunction with the diversification of their hosts, diversified into two lineages: one ancestral lineage leading to O. ramachandrini in the African Suinae, and another ancestral lineage leading to O. borneensis n. sp. and O. dewittei when Sus diversified in the ISEA. With regard to the origin of O. japonica, Lefoulon et al. [20] proposed the transfer of L. caprini Uni & Bain, 2006 to the genus Onchocerca and suggested that O. japonica arose from the ancestral lineage of O. caprini following a host switch from Caprinae to Suidae.
In sections of O. dewittei examined by immunostaining in the present study (Fig. 2e), Wolbachia symbionts were detected in the female germ line, but not in the lateral chords or intestinal wall. In O. japonica, Wolbachia was found in the intra-uterine ova and ovaries in transverse sections [34]. Although not detected in the lateral chords in these sections, a few Wolbachia were observed in the lateral chords when using whole mount fluorescent analysis [34, 58]. It is thus difficult to decide if the absence of Wolbachia in the lateral chords of O. dewittei in the present study reflects a true absence or the limitations of our methodology. Among several bacterial strains identified as Wolbachia supergroups, supergroups C, D, F and J have been detected in the Onchocercidae [28, 31, 34]. These supergroups have been detected in the female germline and the lateral hypodermal chords in many filarioid nematodes by immunostaining and/or whole-mount fluorescent analysis [28, 34, 35].
With regard to the phylogenetic relationships between Wolbachia supergroups and clades (ONC1–ONC5) of Onchocercidae, horizontal transfer events of Wolbachia, secondary losses and local coevolution with host filarioid nematodes are discussed [28, 34, 35]. Our molecular analyses indicate that the Wolbachia strains in O. borneensis n. sp. and O. dewittei form the sister group and are closely related to the strain of O. japonica (Fig. 6). In other words, the molecular relationships of these Wolbachia strains are congruent with the local molecular pattern of their host filarioid nematodes (O. borneensis n. sp. and O. dewittei) from Asian suids (Figs. 3, 4 and 5). Our results support the notion of a coevolutionary pattern between supergroup C strains and species of ONC3, as proposed by Lefoulon et al. [20, 28]. In addition, the Wolbachia strain infecting M. sofiani belongs to supergroup D and is closely related to Wolbachia strains in W. bancrofti and Brugia spp. of ONC5 (Fig. 6). Thus, we also found a coevolutionary pattern between these Wolbachia strains and their respective host filarioid nematodes. Close molecular relationships between M. sofiani, W. bancrofti and Brugia spp. have previously been proposed by Uni et al. [24].
Human infections caused by O. japonica have recently been reported in Japan [7, 8], and the prevalence of O. japonica in S. s. leucomystax was found to be 89% in Oita, Japan [59]. In this study, we found O. borneensis n. sp. from S. barbatus in Borneo. Consequently, there is a potential for zoonotic infection by O. borneensis n. sp. and O. dewittei to occur, given that various species within the genus are known to cause zoonotic infection.
Conclusions
We described O. borneensis n. sp. found in the footpads of bearded pigs (S. barbatus) in Borneo. The new species differs from O. dewittei in banded pigs of Peninsular Malaysia in several morphological characteristics. According to molecular analyses, the cox1 gene sequence of O. borneensis n. sp. differs from that of O. dewittei by 5.9%. Taking into consideration both morphological characteristics and genetic divergence, we conclude that O. borneensis n. sp. is most closely related to O. dewittei from Peninsular Malaysia. Furthermore, because cox1 sequences of O. japonica and O. dewittei differ by 7.6%, we consider that O. japonica is a distinct species, separate from O. dewittei. We detected Wolbachia supergroup C strains in O. borneensis n. sp. and O. dewittei, confirming a coevolutionary pattern between Wolbachia strains and their host Onchocerca spp. from Asian suids.
Supplementary information
Additional file 1: Table S1. Uncorrected p-distances for the cox1 gene sequences between Onchocerca borneensis n. sp., O. dewittei and other known filarial species.
Additional file 2: Figure S1. Taxonomic position of Sus barbatus and S. scrofa vittatus based on cytb nucleotide sequences. The maximum-likelihood phylogenetic tree was generated under the General Time Reversible model in MEGA7 with 500 bootstrap replicates. The scale-bar below the diagram indicates the number of inferred changes along each branch. Red triangles and square indicate sequences generated in this study.
Additional file 3: Table S2. GenBank accession numbers for endosymbiont Wolbachia. Accession numbers in bold (Malayfilaria sofiani, Onchocerca borneensis n. sp. and O. dewittei) represent sequences produced for the present study. Abbreviations: ext., samples from other studies; Ø, no sequences.
Acknowledgements
We are grateful to Professor Emeritus Dr Mohamed Bin Abdul Majid, University of Malaya, and Dr Ahmad Ampeng, Department of Forestry, Sarawak, for conducting the Heart of Borneo Scientific Expedition in Long Banga, Borneo. We thank Professor Dr Ulmar Grafe, Universiti Brunei Darussalam, Mr Gabriel Jau Liran in Long Banga, Mr Muhd Amsyari Morni, Mr Julius William Dee, Mr Qhairil Shyamri Rosli, Ms Norfarhana Mazlan, Ms Nursyafiqah Shazali and Ms Nurmukminah Naharuddin at the University of Malaysia Sarawak, who assisted with sample collection.
Abbreviations
- cox1
cytochrome c oxidase subunit 1 gene
- cytb
cytochrome b gene
- HE
haematoxylin and eosin
- ISEA
inland Southeast Asia
- ITS
internal transcribed spacer
- Ma
mega annum
- ML
maximum-likelihood
- MNHN
Muséum National d’Histoire Naturelle
- CIH
Commonwealth Institute of Helminthology
- PCR
polymerase chain reaction
- wb
Wolbachia
Authors’ contributions
SUni, ASMU and KJ conceived the research and wrote the first draft. SUni, ASMU, KJ, CM, EL, MM, SUga and PLC performed the morphological studies and contributed to the data analysis and interpretation. TA, NB, WS, MF, VLL, CM, EL, AL, YA-LL, HO and SB contributed to the molecular analyses for filarioids and Wolbachia. SUni, ASMU, FAAK, RR, DMB, NAZ, RH, HT and MSA collected samples. All authors read and approved the final manuscript.
Funding
This study was supported by the Ministry of Higher Education, Malaysia (FRGS EP020-2012 to SUni).
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional files. The holotype and allotype of O. borneensis n. sp. were deposited in the MNHN, Paris, France, under accession numbers MNHN 103YT and 104YT, and the paratypes were deposited in the Institute of Biological Sciences, University of Malaya, Malaysia, under accession numbers Ob-B54-1–2, Ob-B54-4, Ob-B54-6–7, Ob-54-9, Ob-B68-2, Ob-B54-2M1–2, Ob-B54-3M1 and Ob-B68-2-M3–9. Specimens of O. dewittei were deposited in the Institute of Biological Sciences, University of Malaya, Malaysia, under accession numbers Od-F1–35 and Od-M1–2. Specimens of O. japonica were deposited in the Institute for Research Promotion, Oita University, Japan, under accession numbers Oj-F1-3 and Oj-M1-3. Sequences were deposited in the GenBank database under the accession numbers: cox1 (MG209780-MG209782), 12S rRNA gene (MG209790-MG209792), ITS (MG192125-MG192127) for O. borneensis n. sp.; cytb (MG657264-MG657265) for S. barbatus; cox1 (MG209783, MG209785-MG209786), 12S rRNA gene (MG209793-MG209798, MG973148), ITS (MG192128-MG192133, MK045758) for O. dewittei; cytb (MG657266) for S. s. vittatus; 12S rRNA gene (MG209799), ITS (MG192124) for O. japonica. Data for Wolbachia are provided in Additional file 3: Table S2.
Ethics approval and consent to participate
Culling of animals and all experimental procedures were carried out in strict accordance with the policy and protocols approved by the Institutional Animal Care and Use Committee, University of Malaya, Kuala Lumpur, Malaysia. The surveys were carried out in accordance with the conservation and control policies of the Department of Wildlife, Malaysia.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shigehiko Uni, Email: unishigehiko@um.edu.my, Email: uni@med.osaka-cu.ac.jp.
Ahmad Syihan Mat Udin, Email: akifsirhan@gmail.com.
Takeshi Agatsuma, Email: agatsuma@kochi-u.ac.jp.
Kerstin Junker, Email: JunkerK@arc.agric.za.
Weerachai Saijuntha, Email: weerachai.s@msu.ac.th.
Naruemon Bunchom, Email: aoy_narumon@hotmail.com.
Masako Fukuda, Email: mfukuda@oita-u.ac.jp.
Coralie Martin, Email: coralie.martin@mnhn.fr.
Emilie Lefoulon, Email: elefoulon@neb.com.
Amandine Labat, Email: alabat@mnhn.fr.
Faisal Ali Anwarali Khan, Email: fanwaral@gmail.com.
Van Lun Low, Email: vanlun_low@um.edu.my.
Phaik Leng Cheah, Email: cheahpl@um.edu.my.
Yvonne Ai-Lian Lim, Email: limailian@um.edu.my.
Rosli Ramli, Email: rosliramli@um.edu.my.
Daicus Martin Belabut, Email: daicus@yahoo.com.
Nur Afiqah Zainuri, Email: afiquahzainuri@gmail.com.
Makoto Matsubayashi, Email: matsubayashi@vet.osakafu-u.ac.jp.
Hasmahzaiti Omar, Email: zaiti_1978@um.edu.my.
Subha Bhassu, Email: subhabhassu@um.edu.my.
Shoji Uga, Email: s-uga@yg.kobe-wu.ac.jp.
Rosli Hashim, Email: roslihashim@um.edu.my.
Hiroyuki Takaoka, Email: takaoka@oita-u.ac.jp.
Mohd Sofian Azirun, Email: sofian@um.edu.my.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-3907-8.
References
- 1.WHO. Onchocerciasis. Geneva: World Health Organization; 2018. https://www.who.int/news-room/fact-sheets/detail/onchocerciasis. Accessed 30 Jan 2019.
- 2.Otranto D, Sakru N, Testini G, Gürlü VP, Yakar K, Lia RP, et al. Case report: first evidence of human zoonotic infection by Onchocerca lupi (Spirurida, Onchocercidae) Am J Trop Med Hyg. 2011;84:55–58. doi: 10.4269/ajtmh.2011.10-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eberhard ML, Ostovar GA, Chundu K, Hobohm D, Feiz-Erfan I, Mathison BA, et al. Case report: zoonotic Onchocerca lupi infection in a 22-month-old child in Arizona: first report in the United States and a review of the literature. Am J Trop Med Hyg. 2013;88:601–605. doi: 10.4269/ajtmh.12-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mowlavi G, Farzbod F, Kheirkhah A, Mobedi I, Bowman DD, Naddaf SR. Human ocular onchocerciasis caused by Onchocerca lupi (Spirurida, Onchocercidae) in Iran. J Helminthol. 2014;88:250–255. doi: 10.1017/S0022149X13000060. [DOI] [PubMed] [Google Scholar]
- 5.Takaoka H, Bain O, Uni S, Korenaga M, Kozek WJ, Shirasaka C, et al. Zoonotic onchocerciasis caused by a parasite from wild boar in Oita, Japan. A comprehensive analysis of morphological characteristics of the worms for its diagnosis. Parasite. 2004;11:285–292. doi: 10.1051/parasite/2004113285. [DOI] [PubMed] [Google Scholar]
- 6.Uni S, Bain O, Takaoka H, Miyashita M, Suzuki Y. Onchocerca dewittei japonica n. subsp., a common parasite from wild boar in Kyushu Island, Japan. Parasite. 2001;8:215–222. doi: 10.1051/parasite/2001083215. [DOI] [PubMed] [Google Scholar]
- 7.Uni S, Fukuda M, Otsuka Y, Hiramatsu N, Yokobayashi K, Takahashi H, et al. New zoonotic cases of Onchocerca dewittei japonica (Nematoda: Onchocercidae) in Honshu, Japan. Parasit Vectors. 2015;8:59. doi: 10.1186/s13071-015-0655-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda M, Uni S, Igari T, Utsumi Y, Otsuka Y, Nakatani J, et al. Human case of Onchocerca dewittei japonica infection in Fukushima, Northeastern Honshu, Japan. Parasitol Int. 2019;72:101943. doi: 10.1016/j.parint.2019.101943. [DOI] [PubMed] [Google Scholar]
- 9.Beaver PC, Horner GS, Bilos JZ. Zoonotic onchocercosis in a resident of Illinois and observations on the identification of Onchocerca species. Am J Trop Med Hyg. 1974;23:595–607. doi: 10.4269/ajtmh.1974.23.595. [DOI] [PubMed] [Google Scholar]
- 10.Burr WE, Jr, Brown MF, Eberhard ML. Zoonotic Onchocerca (Nematoda: Filarioidea) in the cornea of a Colorado resident. Ophthalmology. 1998;105:1494–1497. doi: 10.1016/S0161-6420(98)98035-6. [DOI] [PubMed] [Google Scholar]
- 11.Koehsler M, Soleiman A, Aspöck H, Auer H, Walochnik J. Onchocerca jakutensis filariasis in humans. Emerg Infect Dis. 2007;13:1749–1752. doi: 10.3201/eid1311.070017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colwell DD, Dantas-Torres F, Otranto D. Vector-borne parasitic zoonoses: emerging scenarios and new perspectives. Vet Parasitol. 2011;182:14–21. doi: 10.1016/j.vetpar.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Otranto D, Dantas-Torres F, Brianti E, Traversa D, Petrić D, Genchi C, et al. Vector-borne helminths of dogs and humans in Europe. Parasit Vectors. 2013;6:16. doi: 10.1186/1756-3305-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bain O. Le genre Onchocerca: Hypothèses sur son évolution et clé dichotomique des espèces. Ann Parasitol Hum Comp. 1981;56:503–526. doi: 10.1051/parasite/1981565503. [DOI] [PubMed] [Google Scholar]
- 15.Bain O, Ramachandran CP, Petter F, Mak JW. Description d’Onchocerca dewittei n. sp. (Filarioidea) chez Sus scrofa en Malaisie. Ann Parasitol Hum Comp. 1977;52:471–479. doi: 10.1051/parasite/1977524471. [DOI] [PubMed] [Google Scholar]
- 16.Bain O, Wahl G, Renz A. Onchocerca ramachandrini n. sp. from the warthog in Cameroon. Ann Parasitol Hum Comp. 1993;68:139–143. doi: 10.1051/parasite/1993683139. [DOI] [Google Scholar]
- 17.Uni S, Bain O, Agatsuma T, Harada M, Torii H, Fukuda M, et al. Onchocerca eberhardi n. sp. (Nematoda: Filarioidea) from sika deer in Japan; relationships between species parasitic in cervids and bovids in the Holarctic region. Parasite. 2007;14:199–211. doi: 10.1051/parasite2007143199. [DOI] [PubMed] [Google Scholar]
- 18.Uni S, Fukuda M, Agatsuma T, Bain O, Otsuka Y, Nakatani J, et al. Onchocerca takaokai n. sp. (Nematoda: Filarioidea) in Japanese wild boars (Sus scrofa leucomystax): description and molecular identification of intradermal females. Parasitol Int. 2015;64:493–502. doi: 10.1016/j.parint.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Lefoulon E, Bain O, Bourret J, Junker K, Guerrero R, Cañizales I, et al. Shaking the tree: multi-locus sequence typing usurps current onchocercid (filarial nematode) phylogeny. PloS Negl Trop Dis. 2015;9:e0004233. doi: 10.1371/journal.pntd.0004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefoulon E, Giannelli A, Makepeace BL, Mutafchiev Y, Townson S, Uni S, et al. Whence river blindness? The domestication of mammals and host-parasite co-evolution in the nematode genus Onchocerca. Int J Parasitol. 2017;47:457–470. doi: 10.1016/j.ijpara.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Mirzaei M, Ghahvei Y, Lefoulon E, Lia RP, Otranto D, Martin C, et al. Morphological and molecular characterization of Onchocerca fasciata (Nematoda, Onchocercidae) from dromedary camels (Camelus dromedarius) in Iran. Parasite. 2018;25:50. doi: 10.1051/parasite/2018045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen PKF. Taxonomy of Malaysian filarial parasites. In: Mak JW, editor. Filariasis. Kuala Lumpur: Institute for Medical Research; 1983. pp. 17–35. [Google Scholar]
- 23.Gibbons LM. Keys to the nematode parasites of vertebrates. Wallingford: CAB International; 2010. [Google Scholar]
- 24.Uni S, Mat Udin AS, Agatsuma T, Saijuntha W, Junker K, Ramli R, et al. Morphological and molecular characteristics of Malayfilaria sofiani Uni, Mat Udin & Takaoka n. g., n. sp. (Nematoda: Filarioidea) from the common treeshrew Tupaia glis Diard & Duvaucel (Mammalia: Scandentia) in Peninsular Malaysia. Parasit Vectors. 2017;10:194. doi: 10.1186/s13071-017-2105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chabaud AG, Bain O. The evolutionary expansion of the Spirurida. Int J Parasitol. 1994;24:1179–1201. doi: 10.1016/0020-7519(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 26.Bain O. Evolutionary relationships among filarial nematodes. In: Klei TR, Rajan TV, editors. World class parasites, The Filaria. Dordrecht: Kluwer Academic Publishers; 2002. pp. 21–29. [Google Scholar]
- 27.Morales-Hojas R. Molecular systematics of filarial parasites, with an emphasis on groups of medical and veterinary importance, and its relevance for epidemiology. Inf Genet Evol. 2009;9:748–759. doi: 10.1016/j.meegid.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Lefoulon E, Bain O, Makepeace BL, d’Haese C, Uni S, Martin C, et al. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ. 2016;4:e1840. doi: 10.7717/peerj.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoerauf A, Mand S, Volkmann L, Büttner M, Marfo-Debrekyei Y, Taylor M, et al. Doxycycline in the treatment of human onchocerciasis: kinetics of Wolbachia endobacteria reduction and of inhibition of embryogenesis in female Onchocerca worms. Microbes Infect. 2003;5:261–273. doi: 10.1016/S1286-4579(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 30.Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- 31.Bain O, Casiraghi M, Martin C, Uni S. The Nematoda Filarioidea: critical analysis linking molecular and traditional approaches. Parasite. 2008;15:342–348. doi: 10.1051/parasite/2008153342. [DOI] [PubMed] [Google Scholar]
- 32.Hosokawa T, Koga R, Kikuchi Y, Meng X-Y, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci USA. 2010;107:769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE. 2012;7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferri E, Bain O, Barbuto M, Martin C, Lo N, Uni S, et al. New insights into the evolution of Wolbachia infections in filarial nematodes inferred from a large range of screened species. PLoS ONE. 2011;6:e20843. doi: 10.1371/journal.pone.0020843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefoulon E, Gavotte L, Junker K, Barbuto M, Uni S, Landmann F, et al. A new type F Wolbachia from Splendidofilariinae (Onchocercidae) supports the recent emergence of this supergroup. Int J Parasitol. 2012;42:1025–1036. doi: 10.1016/j.ijpara.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Uni S, Bain O, Takaoka H, Katsumi A, Fujita H, Suzuki Y. Diversification of Cercopithifilaria species (Nematoda: Filarioidea) in Japanese wild ruminants with description of two new species. Parasite. 2002;9:293–304. doi: 10.1051/parasite/2002094293. [DOI] [PubMed] [Google Scholar]
- 37.Casiraghi M, Anderson TJC, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122:93–103. doi: 10.1017/S0031182000007149. [DOI] [PubMed] [Google Scholar]
- 38.Agatsuma T, Iwagami M, Uni S, Takaoka H, Katsumi A, Kimura E, et al. Molecular phylogenetic relationships among seven Japanese species of Cercopithifilaria. Parasitol Int. 2005;54:195–199. doi: 10.1016/j.parint.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Saijuntha W, Tantrawatpan C, Agatsuma T, Wang C, Intapan PM, Maleewong W, et al. Revealing genetic hybridization and DNA recombination of Fasciola hepatica and Fasciola gigantica in nuclear introns of the hybrid Fasciola flukes. Mol Biochem Parasitol. 2018;223:31–36. doi: 10.1016/j.molbiopara.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Watanobe T, Okumura N, Ishiguro N, Nakano M, Matsui A, Sahara M, et al. Genetic relationship and distribution of the Japanese wild boar (Sus scrofa leucomystax) and Ryukyu wild boar (Sus scrofa riukiuanus) analysed by mitochondrial DNA. Mol Ecol. 1999;8:1509–1512. doi: 10.1046/j.1365-294x.1999.00729.x. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kramer LH, Passeri B, Corona S, Simoncini L, Casiraghi M. Immunohistochemical/immunogold detection and distribution of the endosymbiont Wolbachia of Dirofilaria immitis and Brugia pahangi using a polyclonal antiserum raised against WSP (Wolbachia surface protein) Parasitol Res. 2003;89:381–386. doi: 10.1007/s00436-002-0765-6. [DOI] [PubMed] [Google Scholar]
- 43.Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. How many Wolbachia supergroups exist? Mol Biol Evol. 2002;19:341–346. doi: 10.1093/oxfordjournals.molbev.a004087. [DOI] [PubMed] [Google Scholar]
- 44.Katoh K, Toh H. Recent development in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.International Commission on Zoological Nomenclature Amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Zootaxa. 2012;3450:1–7. doi: 10.11646/zootaxa.3450.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson RC, Bain O. Keys to genera of the order Spirurida, Part 3, Diplotriaenoidea, Aproctoidea and Filarioidea. In: Anderson RC, Chabaud AG, Willmott S, editors. CIH keys to the nematode parasites of vertebrates. Farnham Royal: Commonwealth Agricultural Bureaux; 1976. pp. 59–116. [Google Scholar]
- 49.Uni S, Bain O, Agatsuma T, Katsumi A, Baba M, Yanai T, et al. New filarial nematode from Japanese serows (Naemorhedus crispus: Bovidae) close to parasites from elephants. Parasite. 2006;13:193–200. doi: 10.1051/parasite/2006133193. [DOI] [PubMed] [Google Scholar]
- 50.Ferri E, Barbuto M, Bain O, Galimberti A, Uni S, Guerrero RA, et al. Integrated taxonomy: traditional approach and DNA barcoding for the identification of filarioid worms and related parasites (Nematoda) Front Zool. 2009;6:1. doi: 10.1186/1742-9994-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson DE, Reeder DM, editors. Mammal species of the World: a taxonomic and geographic reference. 3. Baltimore: Johns Hopkins University Press; 2005. [Google Scholar]
- 52.Krueger A, Fischer P, Morales-Hojas R. Molecular phylogeny of the filaria genus Onchocerca with special emphasis on Afrotropical human and bovine parasites. Acta Trop. 2007;101:1–14. doi: 10.1016/j.actatropica.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Gongora J, Cuddahee RE, do Nascimento FF, Palgrave CJ, Lowden S, Ho SYW, et al. Rethinking the evolution of extant sub-Saharan African suids (Suidae, Artiodactyla) Zool Scripta. 2011;40:327–335. doi: 10.1111/j.1463-6409.2011.00480.x. [DOI] [Google Scholar]
- 54.Frantz LAF, Schraiber JG, Madsen O, Megens H-J, Bosse M, Paudel Y, et al. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol. 2013;14:R107. doi: 10.1186/gb-2013-14-9-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frantz L, Meijaard E, Gongora J, Haile J, Groenen MAM, Larson G. The evolution of Suidae. Annu Rev Anim Biosci. 2016;4:61–85. doi: 10.1146/annurev-animal-021815-111155. [DOI] [PubMed] [Google Scholar]
- 56.Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 57.Larson G, Cucchi T, Dobney K. Genetic aspects of pig domestication. In: Rothschild MF, Ruvinsky A, editors. The genetics of the pig. 2. Wallingford: CAB International; 2011. pp. 14–37. [Google Scholar]
- 58.Landmann F, Bain O, Martin C, Uni S, Taylor MJ, Sullivan W. Both asymmetric mitotic segregation and cell-to-cell invasion are required for stable germline transmission of Wolbachia in filarial nematodes. Biol Open. 2012;1:536–547. doi: 10.1242/bio.2012737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uni S, Boda T, Daisaku K, Ikura Y, Maruyama H, Hasegawa H, et al. Zoonotic filariasis caused by Onchocerca dewittei japonica in a resident of Hiroshima Prefecture, Honshu, Japan. Parasitol Int. 2010;59:477–480. doi: 10.1016/j.parint.2010.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Uncorrected p-distances for the cox1 gene sequences between Onchocerca borneensis n. sp., O. dewittei and other known filarial species.
Additional file 2: Figure S1. Taxonomic position of Sus barbatus and S. scrofa vittatus based on cytb nucleotide sequences. The maximum-likelihood phylogenetic tree was generated under the General Time Reversible model in MEGA7 with 500 bootstrap replicates. The scale-bar below the diagram indicates the number of inferred changes along each branch. Red triangles and square indicate sequences generated in this study.
Additional file 3: Table S2. GenBank accession numbers for endosymbiont Wolbachia. Accession numbers in bold (Malayfilaria sofiani, Onchocerca borneensis n. sp. and O. dewittei) represent sequences produced for the present study. Abbreviations: ext., samples from other studies; Ø, no sequences.
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its additional files. The holotype and allotype of O. borneensis n. sp. were deposited in the MNHN, Paris, France, under accession numbers MNHN 103YT and 104YT, and the paratypes were deposited in the Institute of Biological Sciences, University of Malaya, Malaysia, under accession numbers Ob-B54-1–2, Ob-B54-4, Ob-B54-6–7, Ob-54-9, Ob-B68-2, Ob-B54-2M1–2, Ob-B54-3M1 and Ob-B68-2-M3–9. Specimens of O. dewittei were deposited in the Institute of Biological Sciences, University of Malaya, Malaysia, under accession numbers Od-F1–35 and Od-M1–2. Specimens of O. japonica were deposited in the Institute for Research Promotion, Oita University, Japan, under accession numbers Oj-F1-3 and Oj-M1-3. Sequences were deposited in the GenBank database under the accession numbers: cox1 (MG209780-MG209782), 12S rRNA gene (MG209790-MG209792), ITS (MG192125-MG192127) for O. borneensis n. sp.; cytb (MG657264-MG657265) for S. barbatus; cox1 (MG209783, MG209785-MG209786), 12S rRNA gene (MG209793-MG209798, MG973148), ITS (MG192128-MG192133, MK045758) for O. dewittei; cytb (MG657266) for S. s. vittatus; 12S rRNA gene (MG209799), ITS (MG192124) for O. japonica. Data for Wolbachia are provided in Additional file 3: Table S2.