Abstract
Global climate change (GCC) increasingly threatens biodiversity through the loss of species, and the transformation of entire ecosystems. Many species are challenged by the pace of GCC because they might not be able to respond fast enough to changing biotic and abiotic conditions. Species can respond either by shifting their range, or by persisting in their local habitat. If populations persist, they can tolerate climatic changes through phenotypic plasticity, or genetically adapt to changing conditions depending on their genetic variability and census population size to allow for de novo mutations. Otherwise, populations will experience demographic collapses and species may go extinct. Current approaches to predicting species responses to GCC begin to combine ecological and evolutionary information for species distribution modelling. Including an evolutionary dimension will substantially improve species distribution projections which have not accounted for key processes such as dispersal, adaptive genetic change, demography, or species interactions. However, eco‐evolutionary models require new data and methods for the estimation of a species' adaptive potential, which have so far only been available for a small number of model species. To represent global biodiversity, we need to devise large‐scale data collection strategies to define the ecology and evolutionary potential of a broad range of species, especially of keystone species of ecosystems. We also need standardized and replicable modelling approaches that integrate these new data to account for eco‐evolutionary processes when predicting the impact of GCC on species' survival. Here, we discuss different genomic approaches that can be used to investigate and predict species responses to GCC. This can serve as guidance for researchers looking for the appropriate experimental setup for their particular system. We furthermore highlight future directions for moving forward in the field and allocating available resources more effectively, to implement mitigation measures before species go extinct and ecosystems lose important functions.
Keywords: Biodiversity loss, eco‐evolutionary dynamics, genomic quantitative genetics, models
Impact Summary.
Global climate change (GCC) will lead to severe environmental changes and many species will lose their habitats. According to the recent Global Assessment Report on Biodiversity and Ecosystem Services (IPBES 2019), 5% of all species are at risk of extinction from 2°C global warming alone. To escape demographic decline or extinction, species have three different strategies to respond to changing environmental conditions: (1) range shift to track their ecological niche, (2) phenotypic plasticity to tolerate environmental change, and (3) genetic evolution to adapt to new local conditions. The ability to disperse depends on species specific characteristics, for example, birds can fly, whereas trees are sessile and, thus, have to cope with their local conditions. The ability of a species to genetically adapt to changing conditions depends on species‐specific characteristics such as genetic variability and population size. If we want to counteract species extinction by conservation strategies, we first need to be able to predict how different species will respond to GCC. A key element for accurate predictions is the potential for dispersal and/or evolutionary response of as many species as possible, and particularly of so‐called keystone species which are of major importance for their respective ecosystem.
Here, we argue that it is possible to use genomic data to understand a species’ evolutionary potential and then use this to improve prediction models that can reliably predict how species will respond to changing climate conditions across their distribution area. A lot has already been learned about the genomic footprints of adaptation to climate in different organisms and we highlight some important research strategies. Similarly, there have been great advances in prediction modelling. So‐called eco‐evolutionary models are most promising for successfully integrating ecological and evolutionary information. However, these models depend on large amounts of ecological and genomics (i.e., evolutionary) data. Scientists will therefore have to establish large consortia and engage with local communities to collect the required data before the consequences of global climate change are irreversible.
A Global Evolutionary Challenge
Global climate change (GCC) is proceeding at an unprecedented rate and has major ecological consequences. Changes in temperature regimes or precipitation patterns strongly impact local environmental conditions of a species’ habitat (Walther et al. 2002; Root et al. 2003; Parmesan 2006). Consequently, the overall question is whether and how species can cope with the accelerated rate of environmental change. There are following four general and mutually nonexclusive strategies on how species and their populations may respond to changes of local environmental conditions (Fig. 1): (1) shifting their distribution range (niche tracking), (2) persisting in their local habitat because they are phenotypically plastic enough to tolerate environmental changes, (3) persisting in their local habitat by genetic adaptation to new conditions (niche evolution), and (4) persisting their ecological niche but experience demographic decline or even go extinct.
Figure 1.
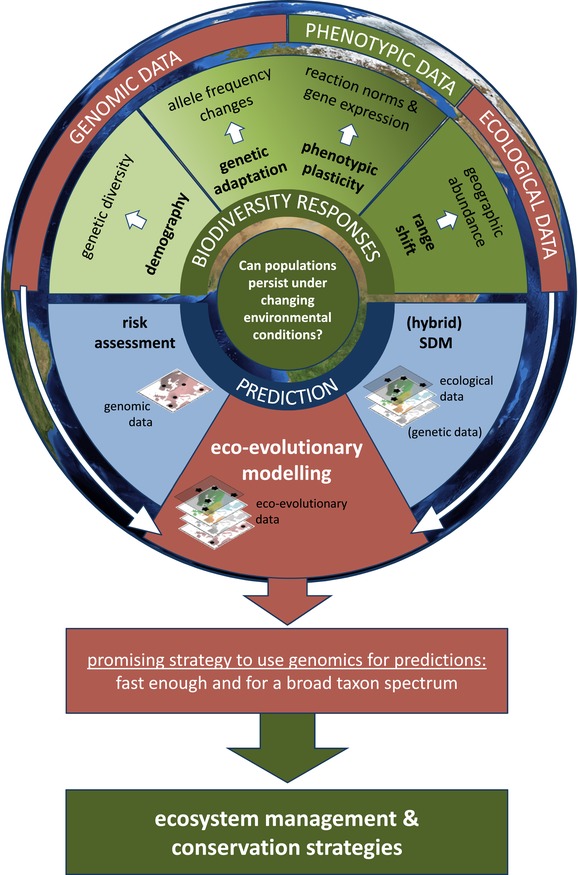
Graphical outline of how biodiversity can respond to changing environmental conditions under GCC, how to investigate these mechanisms, and how different types of empirical data can be used for predicting biodiversity responses to climate change (read from centre to top, then from centre to bottom). Sections in red highlight the connection of genomic and ecological data as basis for eco‐evolutionary modelling as most promising strategy to generate predictions for a relevant proportion of biodiversity fast enough to meet the accelerating pace of GCC. For a final implementation of this strategy, there is further demand for the development of tools to reliably estimate fitness from cohort/time‐series data. Predictions of how biodiversity responds to GCC are fundamental to urgently needed strategies for ecosystem management and conservation in order to counter‐act the imminent loss of biodiversity, ecosystem functioning, and ecosystem services.
While GCC proceeds and ecological consequences are intensifying, conservation and management strategies to cope with changes in global biodiversity are lagging behind, because few studies investigate the response of species to GCC quantitatively and evolutionarily. We argue that genomic data can be used to infer the evolutionary potential for future adaptations of a relevant proportion of biodiversity and in due time. We outline research developments and advances in the field of GCC genomics, as well as in the field of predictive species distribution modelling. Furthermore, we encourage the establishment of international consortia, also involving society at large, to meet challenges in data collection and analysis. It is our objective to motivate future GCC research, to encourage the integration of GCC genomics with predictive modelling, and to generate comprehensive eco‐evolutionary data in the context of climate change to improve predictions of species’ potential to adapt to GCC.
Investigating Biodiversity Responses to Climate Change
Identifying general features of biodiversity responses to climate change is a crucial but challenging mission (Fitzpatrick and Edelsparre 2018). Unravelling the underpinnings of evolutionary responses and, especially, of mechanisms of adaptation, can deliver the missing information, that is, “evolution,” for integrative models to improve the prediction of species responses to GCC.
NICHE TRACKING
Due to the gradual variation of climate factors across geographic space, climatic changes can shift climatic conditions along gradients, excluding mountain tops, ocean currents, or terrestrial regions isolated by geographic barriers. If species are mobile enough or at least possess the ability to disperse their offspring across the borders of their local habitat (e.g., seed or larval dispersal), they have the potential to track their ecological niche by shifting their distribution range along the climate gradient. Evidence for such range shifts comes from studies investigating invasive species (Dukes and Mooney 1999; Fridley 2012; Wolkovich et al. 2013), shifts in breeding and overwintering ranges of migratory birds (Strode 2003; Rolshausen et al. 2009; Both et al. 2010; Zurell et al. 2018), as well as geographical range shifts in insects (Hill et al. 1999; Hickling et al. 2005; Crozier and Dwyer 2006) and plants (Walther et al. 2002; Thuiller et al. 2005; Kelly and Goulden 2008; Lenoir et al. 2008; Chen et al. 2011).
Generally, such investigations rely on multiple time points of occurrence/abundance data of populations across distribution ranges and might be extended by information of phenotypic differences among populations. However, catching, marking, and tracking a large number of individuals often involve logistical complications. Genetic tools provide a great variation of potential solutions to measuring dispersal, because dispersal leads to gene flow (reviewed in Broquet and Petit 2009).
NICHE PERSISTENCE BY PHENOTYPIC PLASTICITY
As long as environmental changes do not exceed the physiological limits of an organism, individuals can persist by plastic responses. Plastic reprogramming of the unchanged genomic basis allows individuals to respond to environmental changes (Aubin‐Horth and Renn 2009). These responses can be investigated via acclimation and climate associated life‐history experiments with populations from different climate regimes. The aim is to obtain reaction norms, which give information on the degree of phenotypic plasticity versus genetic adaptation, and to be able to make predictions on the climate niche breadth of individual species as well as vulnerability in respect to GCC (Fangue et al. 2006; Calosi et al. 2008; Andrew et al. 2013; Foray et al. 2014; Gaitán‐Espitia et al. 2017). Investigations targeting plastic phenotypes have shed light on the molecular basis of cold and heat stress (Gleason and Burton 2015; Zhang et al. 2015; Xiao et al. 2016; Chou et al. 2018), desiccation resistance (Mizrahi et al. 2015; Menzel et al. 2018), and thermal tolerance (Mock et al. 2017; Gunderson et al. 2018; Hermann et al. 2018).
Since phenotypic plasticity, by definition, is the ability of one genotype to generate multiple phenotypes, genomic data need to be supplemented with experiments and other omics approaches such as transcriptomics, epigenomics, or proteomics (Aubin‐Horth and Renn 2009).
NICHE EVOLUTION BY GENETIC ADAPTATION
Changes in environmental conditions are expressed as changes in the local selection regime acting on populations. Such altered selection regimes will eventually drive genetic adaptation. Evidence for genetic adaptation is manifold, including climate‐driven genetic changes in phenology and phenotype such as changes in the timing of vegetation development (Willis et al. 2008; Anderson et al. 2012; van Asch et al. 2013), reproduction (Bradshaw and Holzapfel 2001; Franks et al. 2007), seasonal range shifts (Pulido and Berthold 2003), body size (Daufresne et al. 2009; Gardner et al. 2011), and the strength of competition (Mboup et al. 2012; Bocedi et al. 2013).
With regard to long‐term physiological costs and fitness benefits, genetic adaptation is the most relevant scenario of species’ responses to climate change. Genomic data provides insight in the genomic basis underlying evolutionary adaptation and, thus, contribute most to understand mechanisms of genetic adaptation and to solve the problem of delivering clear evidence (Gienapp et al. 2008; Merilä 2012). To investigate the evolution of genetic adaptation under changing environmental conditions, knowledge of the initial genetic state, that is, the ancestral state, as well as the adapted/evolved state to the new conditions is necessary. To this end, the ancestral state is either approximated by correlation of environmental and genetic heterogeneity (space‐for‐time approach), or by directly following the evolutionary trajectory in time‐series data (time‐for‐time approach). Space‐for‐time approaches can inform on the amount of standing genetic variation present and on the genetic changes necessary to adapt from the current (initial) condition to a potential future condition. Time‐for‐time approaches inform on the likelihood of the change from the known initial condition.
Space‐for‐time approach
Studying the correlation between genomic and environmental variation across populations along climatic gradients, provides a first approximation of how much genetic change might be expected given future climate change projections (Rellstab et al. 2015; Li et al. 2017). Environmental association analyses (EAAs) provide a correlative insight in genotype‐environment interactions (Coop et al. 2010). EAAs are especially powerful when combined with traditional population genomic approaches to detect highly differentiated outlier loci.
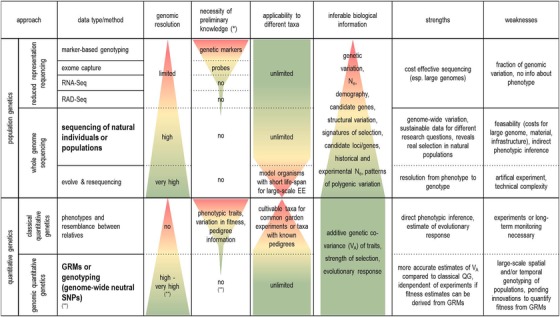
Most genomic studies of climate adaptation to date are based on reduced‐representation sequencing of natural populations, that is, sequencing parts of an organism's DNA by RAD‐Seq, RNA‐Seq, targeted sequencing (e.g., exome capture), SNP‐chips, or the sequencing of previously known candidate genes (Hoban et al. 2016). The resulting genetic polymorphism data provide insight into the genetic variation of populations and can be used to identify signatures of selection and patterns of local adaptation (Savolainen et al. 2013). There is a growing list of studies that identify important candidate genes or loci involved in climate adaptation of nonmodel species (e.g., Jaramillo‐Correa et al. 2015; Pluess et al. 2016; Roschanski et al. 2016; Rellstab et al. 2017; Housset et al. 2018, Ahrens et al. 2019). Reduced‐representation sequencing is especially useful for landscape genomics studies with species having large genome sizes. For example in two distantly related conifer species (with genome sizes above 20 Gb), analysis using exome capture data of most of the coding region provided evidence of convergent adaptation to climate (Yeaman et al. 2016). However, studies relying on reduced‐representation sequencing approaches by definition only evaluate a small to moderate proportion of the genome and will, therefore, likely miss many signals of local adaptation (Lowry et al. 2017, but also see Catchen et al. 2017 or Benjelloun et al. 2019). There are additional limitations, such as some reduced‐representation approaches depend on previous knowledge of candidate loci, regions, or genes (e.g., SNP‐chips) and generally the link to fitness‐relevant phenotypic traits cannot be tested (Table 1).
Table 1.
Experimental approaches to assess evolutionary responses to climate change using different sets of biological data. The column data type/method comprises different sequencing techniques and experimental setups. Genomic resolution outlines the detail of genomic information at which the genomic footprint of climate adaptation can be investigated in a population genetics or quantitative genetics context. Inferable biological information lists parameters that can be estimated applying the respective approach (cumulative across approaches with the complete parameter‐set inferable with the lower approach). Colour bars in the background visualise to what extend genomic information can be assessed for a broad range of taxa and contribute valuable information to be used in eco‐evolutionary prediction models. The two approaches with the best compromise of suitability are printed in bold
|
(*) Does not include genomic resources, that is, reference genome with/without annotation.
(**) Ideally WGS data of cohorts or time‐series data that allow estimating relatedness as well as fitness from genomic relatedness matrices (GRMs), otherwise preliminary knowledge of phenotypic traits will be necessary and applicability will be restricted to taxa that are suitable for trait measuring.
In contrast, whole genome sequencing (WGS) delivers data on the entire genome of individuals or pooled individuals from a population (Pool‐Seq). When a reference genome is available, WGS provides information on the spatial pattern of variation along chromosomes. Genome‐wide information can be used to infer population history based on genome‐wide patterns of neutral genetic diversity, for the estimation of genome‐wide signatures of selection, or the analysis of variation in chromosome structure, or the genome‐wide landscape of differentiation and recombination (Table 1; Lexer and Stölting 2012; Hoban et al. 2016).
If EAAs are combined with WGS data, a comprehensive set of loci is used for correlation to environmental variables and thereby, depending on population structure and the strength of differentiation along the environmental gradient, minor effect loci can ideally also be uncovered (de Villemereuil et al. 2014). This approach requires WGS data of natural populations that should be distributed along environmental gradients. This is in principle applicable to all organisms with a small to medium genome size, a reference genome, even of suboptimal quality, and populations spanning a sufficiently steep environmental gradient. EAA do not require prior knowledge of specific phenotypic traits; they are, therefore, less labour intensive and will become affordable even for intermediate size genomes due to decreasing costs of sequencing technologies.
Conceptually, genome‐wide association studies (GWAS) are similar to EAA with the major difference that GWAS require both genomic information obtained from individuals and phenotypic measurements on certain traits (continuous or categorial; Balding 2006) on the same set of individuals. Because GWAS identifies associations between genomic regions and phenotypic traits, it is a powerful approach to uncover the genomic underpinnings of quantitative traits (such as phenotypic responses to most climate factors).
Investigations applying EEA or GWAS have provided important insights into genetic processes underlying climate adaptation. For example, the Drosophila and Arabidopsis models have a long tradition in the study of climate adaptation, with extensive knowledge of phenotypic traits and many well‐studied candidate polymorphisms along clines on different continents (Atwell et al. 2011; Fournier‐Level et al. 2011; Fabian et al. 2012; reviewed in Adrion et al. 2015). These studies have provided evidence for rapid and stable adaptive oscillations of allele frequencies over seasonal time scales (Bergland et al. 2014), the importance of inversion polymorphisms (Kapun et al. 2014, 2016), polygenic adaptation (Exposito‐Alonso et al. 2018b), migrants with beneficial alleles but also novel mutations and adaptive introgression (Hancock et al. 2011; Flood and Hancock 2017) in climate adaptation.
Sequencing entire genomes is important, since climate adaptation might not be restricted to single genomic loci but might depend on larger genomic regions including structural variants such as inversions. With decreasing sequencing costs and improved genome assembly and analysis algorithms, it is now feasible to perform WGS studies also in nonmodel species such as lichens (lichen‐forming fungus Lasallia pustulata, Dal Grande et al. 2017), insects (Anopheles gambiae, Cheng et al. 2012; Apis mellifera, Chen et al. 2016; Chironomus riparius, Waldvogel et al. 2018), molluscs (Crassostrea gigas, Zhang et al. 2012), and vertebrates (domesticated sheep, Yang et al. 2016).
Time‐for‐time approach
While the space‐for‐time approach will only deliver approximations and might be confounded by covariation, the time‐for‐ time approach offers the possibility to track evolutionary changes directly. To this end, evolve and re‐sequence (E&R) studies typically combine WGS of individuals or populations with experimental evolution (Kofler and Schlötterer 2013; Long et al. 2015). We can experimentally follow evolutionary trajectories of individuals during an environmental perturbation. Temporal genome analyses allow us to disentangle even weakly selected loci from genomic background noise. Given that the organism is experimentally amenable (e.g., sufficient number of individuals, appropriate number of generations to allow for sufficient recombination), E&R can deliver the highest level of genomic resolution of all currently used approaches. With such data, we can test whether populations carry sufficient standing genetic variation to adapt to climate change, or we can identify the genomic architecture of climate adaptation (e.g., polygenic patterns vs. selective sweeps at a few loci with large effect).
Again, the Drosophila model has pioneered E&R studies, providing important insights into the evolution of climate adaptation: evolution of Drosophila melanogaster populations under hot climate conditions resulted in genetic adaptation and was not due to phenotypic plasticity (Orozco‐Terwengel et al. 2012; Tobler et al. 2014); adaptation seemed to be polygenic and with different genomic footprints underlying similar adaptive phenotypic changes among replicates (Barghi et al. 2019). With genome‐wide scans in evolving Drosophila simulans populations, it was moreover possible to identify a central metabolic switch as key factor for thermal adaptation (Mallard et al. 2018). Large‐scale and long‐term experimental evolution to search for genomic innovation is broadly applicable in bacteria and an E&R study with Escherichia coli populations revealed detailed insights in adaptation to temperature: during the evolution to different temperature regimes, genomic signatures of adaptation were found to be highly specific with 17% overlap in mutated genes in strains that evolved under the same regime (Deatherage et al. 2017). Parallel evolutionary responses to the same environments are often observed at the level of genes, operons, and functional complexes, but less so at the nucleotide level (Tenaillon et al. 2012, 2016). Modes of adaptation of polygenic traits can either lead to convergence or genomic redundancy among individual genomes. In the latter case, it will be difficult to derive general pathways to adaptation. Exploring the likelihood for convergence or redundancy will, thus, pose a challenge to population genomicists and bear high potential for analytical innovation in the field.
E&R experiments have certain limitations. They remain artificial and cannot reflect ecological reality, for instance, they neglect complex selection regimes, community effects, and different migration scenarios. Furthermore, they require an extensive experimental set‐up (Kofler and Schlötterer 2013; Schlötterer et al. 2015), and come with many technical complications (e.g., large variation between replicates, increased mutation rates, and unconscious selection bias). Thus, E&R studies are hardly feasible for most multicellular organisms with long generation times. Furthermore, many organisms are partially asexual or selfing, and will, therefore, show limited amounts of recombination over the course of an E&R experiment.
Instead of performing complex E&R experiments under laboratory conditions with all their limitations, there is also naturally occurring data that allow the study of evolutionary change at the genome level over evolutionary timescales. This includes remains of zooplankton in dateable sediments (e.g., Brede et al. 2009; Rellstab et al. 2011; Orsini et al. 2012), water flea individuals resurrected from resting stages in biological archives (e.g., Decaestecker et al. 2007), historical samples from archives (e.g., archived otholits Therkildsen et al. 2013b), herbarium specimens (Lang et al. 2019), or trees of different ages inferred by dendrochronological methods (e.g., Jump et al. 2006; Elleouet and Aitken 2018). Especially, resurrection experiments offer the possibility to span longer evolutionary timescales when dormant propagules (e.g., ephippia, seeds) from past times are retrieved from nature and compared to descendant individuals from the same localities in a common garden (Franks et al. 2018). These approaches have been mainly used in the pregenomic era (see Therkildsen et al. 2019), but could be easily analysed with WGS (Bálint et al. 2018). Herbarium specimens of Arabidopsis thaliana, for instance, have been used to study the emergence of de novo mutations through time (Exposito‐Alonso et al. 2018a).
However, time‐for‐time analyses, either based on E&R experiments or on time‐series data collected from natural populations, can only be applied to a small proportion of species and are, thus, less relevant for the broad‐scale assessment of biodiversity responses to climate change.
EXTINCTION RISK DUE TO DEMOGRAPHIC DECLINE
The potential of a species to genetically adapt to changed environmental conditions depends on the level of genetic diversity of a population in a respective habitat, the extent of migration and gene flow between populations, and the rate of genetic innovation (e.g., by de novo mutations). If incoming gene flow and dispersal are limited, rapid environmental changes can lead to dramatic demographic shifts (especially in small populations) and only genetic adaptation can restore positive population growth and allow for the so‐called evolutionary rescue of the population (Carlson et al. 2014). Adaptation from standing genetic variation is particularly important under rapid environmental change (as due to GCC) because evolution can act on alleles that already segregate at higher frequencies in the population (speed of selection depending on the effective population size) and which can carry an adaptive value under new environmental conditions (Tigano and Friesen 2016).
Genomic data can provide information on within‐species genetic variation, a species’ demography, and effective population size (Ne). These population genomic parameters can be estimated with high confidence and serve as proxies or estimators of the vulnerability of a population or species to succumb to changing climate conditions (Fitzpatrick and Keller 2015; Rellstab et al. 2016; Bay et al. 2018; Exposito‐Alonso et al. 2018b)—a major step toward predicting evolutionary responses.
Prediction Models of Species Responses to Global Climate Change
PREDICTING RANGE SHIFTS WITH AND WITHOUT EVOLUTION
Most studies forecasting biodiversity responses to climate change currently use statistical and functional species distribution models (hereafter SDMs; broadly including niche, envelope, and bioclimatic models). In brief, they correlate geographic location data of a species’ occurrences and experimentally gained functional models with a comprehensive set of climatic variables (Pearman et al. 2008; Elith and Leathwick 2009). This ecological modelling approach yields a coarse estimation of current or future Grinellian niche dimensions of a species, depending on the statistical association of species occurrences and environmental data. However, these models generally consider species as uniform and static with regards to climate‐relevant traits, and disregard intraspecific variation, local adaptation, and genetic potential for rapid evolutionary change (Jay et al. 2012).
Evolutionary models rely on quantitative genetics theory to estimate trait heritability. Such models have been used in animal and plant breeding, as well as in predicting responses to selection of nondomestic species including responses to changing climate conditions (e.g., Alberto et al. 2013; Gonzalez et al. 2013). It was possible, for instance, to reveal a lack of adaptive potential in a threatened New Zealand bird species by combining quantitative genetics with long‐term phenotypic data and fitness proxies (de Villemereuil et al. 2019). It has further been shown that SDMs lacking quantitative genetics can vastly underestimate species range dynamics, for example, range expansions in Aedes aegypti, the mosquito that transmits dengue virus (Kearney et al. 2009).
To improve predictions of climate responses, quantitative genetics should be an integral part of modelling frameworks. However, obtaining pedigrees and documentation of phenotypic key traits is time consuming and only possible in a handful of species. These species must be amenable to controlled crosses, common garden experiments, and/or long‐term individual monitoring (e.g., birds, large mammals). Moreover, fitness traits have to be identified, and ideally experimentally confirmed (Shaw 2019).
The integration of ecological and evolutionary models can potentially lead to more realistic predictions of species' persistence under GCC (Hoffmann and Sgrò 2011; Urban et al. 2016; Benito Garzón et al. 2019). Today, several models incorporate genetic or evolutionary information within SDMs (e.g., hybrid SDMs; Dormann et al. 2012). For example, genomic hybrid SDMs use geographical covariation of SNP frequencies with environmental variation, basically subdividing the species’ distribution into genetic clusters associated with climatic conditions (Fitzpatrick and Keller 2015; Exposito‐Alonso et al. 2018b; Razgour et al. 2018, 2019; Lowry et al. 2019). By building one SDM per SNP and using joint projections of the respective SNP niches into future climates (Exposito‐Alonso et al. 2018b) or by calculating genetic distances based on many SNPs along environmental gradients (Fitzpatrick and Keller 2015; Rellstab et al. 2016; Bay et al. 2018), these approaches attempt to assess mismatches between current allelic compositions and predicted future local conditions, assuming that local populations are currently adapted to their environment. Such gene‐environment mismatches are sometimes called genomic vulnerability, genetic offset, or risk of maladaptation (see above and Rellstab et al. 2016; Bay et al. 2018).
However, hybrid SDMs remain a static association of genetic clusters with environmental variation. Genomic vulnerability as such, only refers to the mismatch of current allele frequencies with potential future climatic conditions. It does not incorporate predictions of shifts in allele frequencies caused by selection or gene flow among populations. When explicitly taken into account, migration and selection may decrease the mismatch of current genomic compositions and future abiotic conditions, and lead to a prediction of lower genomic vulnerability (Exposito‐Alonso et al. 2018b). Furthermore, measures of genomic vulnerability cannot directly predict whether a species will adapt or succumb to extinction, as it does not include information on evolutionary rates, adaptive phenotypic changes, or future population growth rates (i.e., absolute fitness). This is caused by the general lack of estimates of effect size of the genetic variants associated with environmental variables (but see Taylor et al. 2019). Hybrid SDMs are therefore unable to model local population density as a function of the degree of local adaptation. Nevertheless, genomic data can provide the demographic information needed to make more accurate predictions. In particular, spatial patterns of genetic diversity can provide estimates of migration rates and effective population sizes required to parametrize process‐based models (see below). Moreover, when coupled with past climatic information, models of temporal change in population density can be constructed with coalescent‐based simulations. By associating past changes in population density with past changes in climatic conditions, demographic models can then be validated by hindcasting (see Brown et al. 2016; Prates et al. 2016).
PREDICTING ECO‐EVOLUTIONARY RESPONSES OF BIODIVERSITY TO CLIMATE CHANGE
Current hybrid SDMs do not fully integrate eco‐evolutionary dynamics, but rather a subset of the processes involved. For instance, some approaches incorporate migration and demography, but not evolution (e.g., Dullinger et al. 2012), or dispersal and evolutionary trait dynamics, but not demography (Bush et al. 2016), or climate‐driven selection of genetic variants, but not demography and migration (Exposito‐Alonso et al. 2019). To predict species’ range shifts, other approaches couple mechanistic, process‐based, physiological, or phenological models with information on evolutionary adaptation (Kearney et al. 2009; Oddou‐Muratorio and Davi 2014) or phenological plasticity (Wilczek et al. 2010; Duputié et al. 2015). Only recently, Cotto et al. (2017) have developed a full, individual‐based eco‐evolutionary model (EEM) of adaptation to climatic changes that integrates quantitative genetics, thus, disregarding specific loci. EEMs are process‐based in which they take into account life‐history traits and genetic characteristics of a species, and simulate population dynamics and evolution on a spatial grid for a given scenario of environmental change. This approach requires extensive data on the evolutionary potential of a species (genetic covariance of phenotypic traits, strength of selection), its spatial occurrence and ecological characteristics (e.g., dispersal kernels, vital rates). EEMs are, thus, bound to be limited to a handful of species, but can deliver valuable information on species’ extinction risks in specific geographical areas. Given appropriate computational resources, and adequate approximations, individual‐based (e.g., Guillaume and Rougemont 2006) or population‐based (e.g., Dullinger et al. 2012; Bush et al. 2016) simulation approaches offer an exciting avenue to build EEMs for forecasting species’ responses to climate change. Nevertheless, the genetic information these models require is in the form of additive genetic variance or heritability of quantitative traits, or, even better, genetic covariation of traits with fitness. Thus, there is a disconnect with the previous genetic/genomic hybrid SDM approach because information on allele frequencies across environmental space is directly transformed to changes across time without translating it into adaptive genetic variation of traits, and temporal demographic feedbacks, as EEMs do. One avenue to connect these two approaches is to utilize genomic SDMs to define parameters across space needed for EEMs.
The predictive potential of EEMs can be leveraged by a better integration with ecological genomics. The discipline of ecological genomics combines classical ecological research with population genomic approaches in order to study the genetic basis underlying responses of organisms to variation in their natural environment (Ungerer et al. 2008). Genomic data can provide EEMs with estimates of quantitative genetics parameters from natural populations sampled across environmental gradients (Gienapp et al. 2017). Traditionally, relatedness between individuals has been estimated from pedigrees or from controlled crosses in common gardens. Although not yet widely applied, so called “genomic quantitative genetics” (gQG) relies on estimates of genetic relatedness between individuals that are based on genome‐wide polymorphisms (see Speed and Balding 2015 for an overview; see Gienapp et al. 2019 for an example in birds). Phenotypic trait covariation between individuals with known levels of relatedness is then used to estimate the additive genetic covariation of a set of quantitative traits. The response of a population to a shift in local trait optima (e.g., caused by climate change) can then be predicted from the additive genetic covariance between traits and fitness, given that the strength of selection is known (Etterson and Shaw 2001). Even though gQG can help extending the quantitative genetics approach of EEMs to any wild species, the need for phenotypic data and reliable fitness estimates across environments remains. For some species where cohort or time‐series data are available, knowledge of the genetic relatedness among individuals through time can help identify the number of surviving offspring per reproducing adult (Truffaut et al. 2017). Estimates of reproductive success can then be related to phenotypes in order to estimate the strength of selection within populations; or it can directly deliver an estimate of the species’ evolvability from the genetic additive variation in fitness. This will still require large sampling efforts. Further developments of the gQG approach are, thus, necessary to identify the best sampling strategies to allow for its application in wild species. Nevertheless, gQG has the potential to yield estimates of the evolutionary potential of wild species for which classical breeding plans or pedigree information are not accessible. When quantitative genetic parameters are not available, the EEM approach has the advantage to allow for sensitivity analyses on eco‐evolutionary parameter values and deliver process‐based estimates of extinction probabilities. Doing so will allow deploying EEM approaches to a broader range of species in natural contexts.
Challenges and Outlook
The severe impact of GCC on biodiversity necessitates the development of predictive models that can help to take timely actions (Urban 2015). Current approaches are limited by either technical issues (e.g., statistical methods that integrate genomic and environmental information) or data acquisition challenges (e.g., data availability). Here, we have highlighted some recent technical advances that integrate genetic information for ecological projections, and identified major challenges in the area of data acquisition for a wide range of organisms (see above and Fitzpatrick and Keller 2015; Bush et al. 2016; Rellstab et al. 2016; Cotto et al. 2017; Exposito‐Alonso et al. 2018b).
Realistic eco‐evolutionary prediction models of species distributions require large amounts of data, magnitudes higher than what is currently available. Thus, several logistical challenges have to be addressed to build and expand databases for environmental data, species abundance data, and genomic data on a global scale (Fig. 2). Environmental data, and especially climate data, are already available in public databases with sufficient resolution for large parts of the globe (e.g., WorldClim2, Fick and Hijmans 2017; or CHELSA, Karger et al. 2017). Species’ distribution records are publicly available for an increasing number of species (GBIF.org). Future research could also draw from satellite and remote sensing data to build an analytical platform for near‐term prediction of habitat change. Automatic platforms that use SDMs could combine the abundance data with environmental and land use systems (worldclim.org, CORINE, or USGS Land Cover) to produce current geographic distributions (GFBIO.org, IUCN) and predict changes in geographic distribution boundaries.
Figure 2.
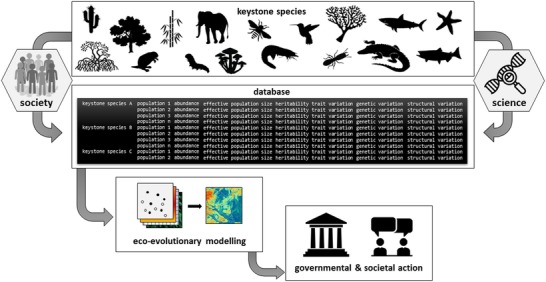
Outline of our proposed mode of action to collect, store, and analyse eco‐evolutionary data as a joint venture of citizens and scientists. A comprehensive database can then support eco‐evolutionary modelling to predict biodiversity responses to GCC. These predictions will provide government and society with educated recommendations in order to take significant actions for conservation.
Combined with corresponding phenotypic variation or geographical and ecological information, genomic data can help estimating some of the evolutionary quantities relevant for eco‐evolutionary forecasting. Genomic data is constantly accumulating in public databases, such as NCBI and EMBL‐EBI, however there is a strong imbalance of sequencing model versus nonmodel species and resequencing of laboratory versus natural populations. Databases will grow even faster as soon as portable devices allow ecologists to sequence directly in the field (Michael et al. 2018), as is routinely done for trait, photographic, or environmental data. Even though genome sequencing is improving in terms of cost and portability (https://www.genome.gov/sequencingcostsdata/), so far sequencing effort is still focusing on an unrepresentative fraction of biodiversity, that is, mainly few (model) species. In the future, however, genetic monitoring for conservation requires a systematic approach that encompasses relevant biodiversity (Reside et al. 2018). To address this challenge, sequencing effort may prioritise keystone species, which might inform about ecosystem‐wide evolutionary responses (Fig. 1). Keystone species act as major ecosystem hubs, for example, the most predominant tree species in a forest, and their decline or major range shifts would most likely generate cascading effects that affect the whole ecosystem (Mills et al. 1993; Valls et al. 2015). Studying networks of species interactions can help to identify keystone species (Bascompte et al. 2006; Tylianakis et al. 2008). Another set of high‐priority species should be those flagged as being threatened in the red list of the International Union for Conservation of Nature (https://www.iucnredlist.org/), as arguably those are undergoing early impacts.
The overall mission will be to comprehensively collect occurrence and abundance data of the prioritised set of species and finally sample populations across the species’ distribution range. In order to realize this task on a short‐term time scale, the involvement of citizens bears high potential. From plant ecology experiments (NutNet, DryNet), to the worldwide “watch” on any species (iNaturalist or iSpot), citizen science‐based platforms open the door for global‐scale scientific databases. Citizens can even be involved in sampling efforts, when integrated in well‐organised scientific projects that deal with distribution of required sampling material and the compliance with international legislation concerning biological samples. On different scales, such citizen science projects have already proven to be highly successful, as for example, the EcoAction program to survey the health of coral reefs around the world (reefcheck.org, Done et al. 2017) or the “big wasp survey” to sample wasp populations across the United Kingdom (bigwaspsurvey.org, Sumner et al. 2019).
Genomic sequencing of the collected samples can be realised by scientific consortia that integrate genome sequencing centres, as there already exists, for example, the Earth Biogenome Project (earthbiogenome.org, Lewin et al. 2018) and the Darwin Tree of Life UK project (sanger.ac.uk/news/view/genetic‐code‐66000‐uk‐species‐be‐sequenced). The ultimate aim would be to establish platforms that integrate different databases (climate data, species abundance data, phenotypic data, genomic data, etc.; Fig. 2) with the necessary analysis pipelines, to produce publicly available and easily interpretable species vulnerability projections. A successful example that has pioneered large‐scale community‐based data collection comes from human‐pathogenic organisms as Ebola, Flu, or Zika, where researchers have shared significant amounts of data during critical outbreaks and analyses were generated and published in almost real time (http://nextstrain.org). New online technologies facilitate forming consortia of citizens and scientists that can coordinate, process, and share data in safe and reproducible ways. We think that these new ways of collecting and processing data are promising avenues for improving our understanding of the limits of species tolerances and adaptation and might, thus, help to parametrize the next generation of species response models (Swan et al. 2010; Özdemir et al. 2013; Grossman 2019).
As soon as projections on the responses of biodiversity to climate change have reached nationwide, continental or even global scales, these results gain in importance to become integrated in political decision making. Publicly available databases will sensitise citizens for the effects of GCC on biodiversity and, thus, strengthen the need for political awareness, especially on local scale. Results can furthermore be integrated in reports of intergovernmental panels like IPCC and/or IPBES (ipcc.ch, ipbes.net) in order to use already established and acknowledged channels to policymakers.
Conclusion
GCC poses severe risks to biodiversity and latest extrapolations alert an extinction risk due to climate change for more than 5% of all species from 2°C global warming alone (IPBES 2019). We, thus, urgently need to better understand and accurately predict how species respond to changing environmental conditions in order to inform policymakers and implement conservation strategies. We need to incorporate ecological as well as evolutionary parameters in our models to account for a species’ potential to compensate changing environmental conditions by either range shift or genetic evolution for adaptation. A species’ evolutionary potential for adaptation can be estimated from genomic data. Hybrid SDMs, which attempt to at least partially incorporate eco‐evolutionary dynamics, are becoming more frequent and deliver informed estimates of a species' vulnerability. EEM that incorporate gQG approaches can yield more realistic predictions and be applied to any wild species. Such models require extensive amounts of data and especially the required genomic data could be generated rapidly enough (i.e., without too much experimental effort) and for a representative large group of taxa (e.g., keystone species). We, here propose a roadmap of how science and society can work together to facilitate sampling, estimating of fitness parameters, and genome sequencing for a broad range of species to meet the urgent need of action in face of the accelerating speed of GCC.
Associate Editor: Z. Gompert
AUTHOR CONTRIBUTIONS
All authors together conceived the study, AMW drafted and streamlined the manuscript, all authors contributed to the writing and revision of the manuscript.
ACKNOWLEDGMENTS
This commentary is a joint effort of the invited speakers of the symposium "Genomic basis of climate adaptation" which was held on January 17–19, 2018 at the Senckenberg Biodiversity and Climate Research Centre in Frankfurt, Germany. The symposium was funded by the Senckenberg Gesellschaft für Naturforschung and the DFG (PF‐390/12‐1). The authors declare no conflicts of interest.
DATA ARCHIVING
This is a commentary article; data not applicable.
LITERATURE CITED
- Adrion, J. R. , Hahn M. W., and Cooper B. S.. 2015. Revisiting classic clines in Drosophila melanogaster in the age of genomics . Trends Genet. 31:434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens, C. W. , Byrne M., and Rymer P. D.. 2019. Standing genomic variation within coding and regulatory regions contributes to the adaptive capacity to climate in a foundation tree species. Mol. Ecol. 28:2502–2516. [DOI] [PubMed] [Google Scholar]
- Alberto, F. J. , Aitken S. N., Alía R., González‐Martínez S. C., Hänninen H., Kremer A., et al. 2013. Potential for evolutionary responses to climate change—evidence from tree populations. Glob. Chang. Biol. 19:1645–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. T. , Inouye D. W., McKinney A. M., Colautti R. I., and Mitchell‐Olds T.. 2012. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc. R. Soc. B Biol. Sci. 279:3843–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew, N. R. , Hart R. A., Jung M. P., Hemmings Z., and Terblanche J. S.. 2013. Can temperate insects take the heat? A case study of the physiological and behavioural responses in a common ant, Iridomyrmex purpureus (Formicidae), with potential climate change. J. Insect Physiol. 59:870–880. [DOI] [PubMed] [Google Scholar]
- Atwell, S. , Huang Y. S., Vilhjálmsson B. J., Willems G., Horton M., Li Y., et al. 2011. Genome‐wide asscociaion study of 107 phenotype in a common set of Arabidopsis thalia inbred lines. Nature 465:627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin‐Horth, N. , and Renn S. C. P.. 2009. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol. Ecol. 18:3763–3780. [DOI] [PubMed] [Google Scholar]
- Balding, D. J. 2006. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 7:781–791. [DOI] [PubMed] [Google Scholar]
- Bálint, M. , Pfenninger M., Grossart H. P., Taberlet P., Vellend M., Leibold M. A., et al. 2018. Environmental DNA time series in ecology. Trends Ecol. Evol. 33:945–957. [DOI] [PubMed] [Google Scholar]
- Barghi, N. , Tobler R., Nolte V., Jakšić A. M., Mallard F., Otte K. A., et al. 2019. Genetic redundancy fuels polygenic adaptation in Drosophila . 10.1371/journal.pbio.3000128 [DOI] [PMC free article] [PubMed]
- Bascompte, J. , Jordano P., and Olesen J. M.. 2006. Asymmetric coevolutionary networks facilitate biodiversity maintenance. Science 312:431–433. [DOI] [PubMed] [Google Scholar]
- Bay, R. A. , Harrigan R. J., Le Underwood V., Gibbs H. L., Smith T. B., and Ruegg K.. 2018. Genomic signals of selection predict climate‐driven population declines in a migratory bird. Science 359:83–86. [DOI] [PubMed] [Google Scholar]
- Benito Garzón, M. , Robson T. M., and Hampe A.. 2019. ΔTraitSDMs: species distribution models that account for local adaptation and phenotypic plasticity. New Phytol. 222:1757–1765. [DOI] [PubMed] [Google Scholar]
- Benjelloun, B. , Boyer F., Streeter I., Zamani W., Engelen S., Alberti A., et al. 2019. An evaluation of sequencing coverage and genotyping strategies to assess neutral and adaptive diversity. Mol. Ecol. Resour. 00:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland, A. O. , Behrman E. L., O'Brien K. R., Schmidt P. S., and Petrov D. A.. 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila . PLoS Genet. 10:e1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocedi, G. , Atkins K. E., Liao J., Henry R. C., Travis J. M. J., and Hellmann J. J.. 2013. Effects of local adaptation and interspecific competition on species’ responses to climate change. Ann. N. Y. Acad. Sci. 1297:83‐97. [DOI] [PubMed] [Google Scholar]
- Both, C. , Van Turnhout C. A. M., Bijlsma R. G., Siepel H., Van Strien A. J., and Foppen R. P. B.. 2010. Avian population consequences of climate change are most severe for long‐distance migrants in seasonal habitats. Proc. R. Soc. B Biol. Sci. 277:1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, W. E. , and Holzapfel C. M.. 2001. Genetic shift in photoperiodic response correlated with global warming. Proc. Natl. Acad. Sci. 98:14509–14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brede, N. , Sandrock C., Straile D., Spaak P., Jankowski T., Streit B., et al. 2009. The impact of human‐made ecological changes on the genetic architecture of Daphnia species. Proc. Natl. Acad. Sci. 106:4758–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquet, T. , and Petit E. J.. 2009. Molecular estimation of dispersal for ecology and population genetics. Annu. Rev. Ecol. Evol. Syst. 40:193–216. [Google Scholar]
- Brown, J. L. , Weber J. J., Alvarado‐Serrano D. F., Hickerson M. J., Franks S. J., and Carnaval A. C.. 2016. Predicting the genetic consequences of future climate change: the power of coupling spatial demography, the coalescent, and historical landscape changes. Am. J. Bot. 103:153–163. [DOI] [PubMed] [Google Scholar]
- Bush, A. , Mokany K., Catullo R., Hoffmann A., Kellermann V., Sgrò C., et al. 2016. Incorporating evolutionary adaptation in species distribution modelling reduces projected vulnerability to climate change. Ecol. Lett. 19:1468–1478. [DOI] [PubMed] [Google Scholar]
- Calosi, P. , Bilton D. T., and Spicer J. I.. 2008. Thermal tolerance, acclimatory capacity and vulnerability to global climate change. Biol. Lett 4:99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, S. M. , Cunningham C. J., and Westley P. A. H.. 2014. Evolutionary rescue in a changing world. Trends Ecol. Evol. 29:521–530. [DOI] [PubMed] [Google Scholar]
- Catchen, J. M. , Hohenlohe P. A., Bernatchez L., Funk W. C., Andrews K. R., and Allendorf F.. 2017. Unbroken: RADseq remains a powerful tool for understanding the genetics of adaptation in natural populations. Mol. Ecol. Resour. 17:362–365. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Hill J. K., Ohlemüller R., Roy D. B., and Thomas C. B.. 2011. Rapid range shifts of species associated with high levels of climate warming. Science 333:1024–1026. [DOI] [PubMed] [Google Scholar]
- Cheng, C. , White B. J., Kamdem C., Mockaitis K., Costantini C., Hahn M. W., et al. 2012. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population‐resequencing approach. Genetics 190:1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Liu Z., Pan Q., Chen X., Wang H., Guo H., et al. 2016. Genomic analyses reveal demographic history and temperate adaptation of the newly discovered honey bee subspecies Apis mellifera sinisxinyuan n. ssp. Mol. Biol. Evol. 33:1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, H. , Pathmasiri W., Deese‐spruill J., Sumner S. J., Jima D. D., Funk D. H., et al. 2018. The good, the bad, and the lethal: gene expression and metabolomics reveal physiological mechanisms underlying chronic thermal effects in mayfly larvae (Neocloeon triangulifer). Front. Ecol. Evol. 6:1–11. [Google Scholar]
- Coop, G. , Witonsky D., Di Rienzo A., and Pritchard J. K.. 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto, O. , Wessely J., Georges D., Klonner G., Schmid M., Dullinger S., et al. 2017. A dynamic eco‐evolutionary model predicts slow response of alpine plants to climate warming. Nat. Commun 8:15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier, L. , and Dwyer G.. 2006. Combining population‐dynamic and ecophysiological models to predict climate‐induced insect range shifts. Am. Nat. 167:853–866. [DOI] [PubMed] [Google Scholar]
- Dal Grande, F. , Sharma R., Meiser A., Rolshausen G., Büdel B., Mishra B., et al. 2017. Adaptive differentiation coincides with local bioclimatic conditions along an elevational cline in populations of a lichen‐forming fungus. BMC Evol. Biol. 17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daufresne, M. , Lengfellner K., and Sommer U.. 2009. Global warming benefits the small in acquatic ecostystems. PNAS 106:12788–12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villemereuil, P. , Frichot E., Bazin E., Francois O., and Gaggiotti O. E.. 2014. Genome scan methods against more complex models: when and how much should we trust them? Mol. Ecol. 23:2006–2019. [DOI] [PubMed] [Google Scholar]
- de Villemereuil, P. , Rutschmann A., Lee K. D., Ewen J. G., Brekke P., and Santure A. W.. 2019. Little adaptive potential in a threatened passerine bird. Curr. Biol. 29:889‐894. [DOI] [PubMed] [Google Scholar]
- Deatherage, D. E. , Kepner J. L., Bennett A. F., Lenski R. E., and Barrick J. E.. 2017. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc. Natl. Acad. Sci. 114:E1904–E1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker, E. , Gaba S., Raeymaekers J. A. M., Stoks R., Van Kerckhoven L., Ebert D., et al. 2007. Host‐parasite “Red Queen” dynamics archived in pond sediment. Nature 450:870–873. [DOI] [PubMed] [Google Scholar]
- Done, T. , Roelfsema C., Harvey A., Schuller L., Hill J., Schläppy M. L., et al. 2017. Reliability and utility of citizen science reef monitoring data collected by Reef Check Australia, 2002–2015. Mar. Pollut. Bull. 117:148–155. [DOI] [PubMed] [Google Scholar]
- Dormann, C. F. , Schymanski S. J., Cabral J., Chuine I., Graham C., Hartig F., et al. 2012. Correlation and process in species distribution models: bridging a dichotomy. J. Biogeogr. 39:2119–2131. [Google Scholar]
- Dukes, J. S. , and Mooney H. A.. 1999. Does global change increase the success of biological invaders? Trends Ecol. Evol. 14:135–139. [DOI] [PubMed] [Google Scholar]
- Dullinger, S. , Willner W., Plutzar C., Englisch T., Schratt‐Ehrendorfer L., Moser D., et al. 2012. Post‐glacial migration lag restricts range filling of plants in the European Alps. Glob. Ecol. Biogeogr. 21:829–840. [Google Scholar]
- Duputié, A. , Rutschmann A., Ronce O., and Chuine I.. 2015. Phenological plasticity will not help all species adapt to climate change. Glob. Chang. Biol. 21:3062–3073. [DOI] [PubMed] [Google Scholar]
- Elith, J. , and Leathwick J. R.. 2009. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40:677–697. [Google Scholar]
- Elleouet, J. S. , and Aitken S. N.. 2018. The interplay between demography and neutral evolution at the expansion front of a widespread conifer, Picea sitchensis. bioRxiv:1‐25. 10.1101/327742 [DOI] [Google Scholar]
- Etterson, J. R. , and Shaw R. G.. 2001. Constraint to adaptive evolution in response to global warming. Science 294:151–154. [DOI] [PubMed] [Google Scholar]
- Exposito‐Alonso, M. , Becker C., Schuenemann V. J., Reiter E., Setzer C., Slovak R., et al. 2018a. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 14:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito‐Alonso, M. , Vasseur F., Ding W., Wang G., Burbano H. A. A., and Weigel D.. 2018b. Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana . Nat. Ecol. Evol. 2:352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito‐Alonso, M. , 500 Genomes Field Experiment Team , Burbano H. A., Bossdorf O., Nielsen R., and Weigel D.. 2019. Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature 573:126–129. [DOI] [PubMed] [Google Scholar]
- Fabian, D. K. , Kapun M., Nolte V., Kofler R., Schmidt P. S., Schlötterer C., et al. 2012. Genome‐wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21:4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangue, N. A. , Hofmeister M., and Schulte P. M.. 2006. Intraspecific variation in thermal tolerance and heat shock protein gene expression in common killifish, Fundulus heteroclitus . J. Exp. Biol. 209:2859–2872. [DOI] [PubMed] [Google Scholar]
- Fick, S. E. , and Hijmans R. J.. 2017. WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37:4302–4315. [Google Scholar]
- Fitzpatrick, M. C. , and Keller S. R.. 2015. Ecological genomics meets community‐level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 18:1–16. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, M. J. , and Edelsparre A. H.. 2018. The genomics of climate change. Science 359:29–30. [DOI] [PubMed] [Google Scholar]
- Flood, P. J. , and Hancock A. M.. 2017. The genomic basis of adaptation in plants. Curr. Opin. Plant Biol. 36:88–94. [DOI] [PubMed] [Google Scholar]
- Foray, V. , Desouhant E., and Gibert P.. 2014. The impact of thermal fluctuations on reaction norms in specialist and generalist parasitic wasps. Funct. Ecol. 28:411–423. [Google Scholar]
- Fournier‐Level, A. , Korte A., Cooper M. D., Nordborg M., Schmitt J., and Wilczek A. M.. 2011. A map of local adaptation in Arabidopsis thaliana . Science 334:86–89. [DOI] [PubMed] [Google Scholar]
- Franks, S. J. , Sim S., and Weis A. E.. 2007. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. USA 104:1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, S. J. , Hamann E., and Weis A. E.. 2018. Using the resurrection approach to understand contemporary evolution in changing environments. Evol. Appl. 11:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley, J. D. 2012. Extended leaf phenology and the autumn niche in deciduous forest invasions. Nature 485:359–362. [DOI] [PubMed] [Google Scholar]
- Gaitán‐Espitia, J. D. , Bacigalupe L. D., Opitz T., Lagos N. A., Osores S., and Lardies M. A.. 2017. Exploring physiological plasticity and local thermal adaptation in an intertidal crab along a latitudinal cline. J. Therm. Biol. 68:14–20. [DOI] [PubMed] [Google Scholar]
- Gardner, J. L. , Peters A., Kearney M. R., Joseph L., and Heinsohn R.. 2011. Declining body size: a third universal response to warming? Trends Ecol. Evol. 26:285–291. [DOI] [PubMed] [Google Scholar]
- Gienapp, P. , Teplitsky C., Alho J. S., Mills J. A., and Merilä J.. 2008. Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol. 17:167–178. [DOI] [PubMed] [Google Scholar]
- Gienapp, P. , Fior S., Guillaume F., Lasky J. R., Sork V. L., and Csilléry K.. 2017. Genomic quantitative genetics to study evolution in the wild. Trends Ecol. Evol. 32:1–12. [DOI] [PubMed] [Google Scholar]
- Gienapp, P. , Calus M. P. L., Laine V. N., and Visser M. E.. 2019. Genomic selection on breeding time in a wild bird population. Evol. Lett. 3:142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, L. U. , and Burton R. S.. 2015. RNA‐seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma funebralis . Mol. Ecol. 24:610–627. [DOI] [PubMed] [Google Scholar]
- Gonzalez, A. , Ronce O., Ferriere R., and Hochberg M. E.. 2013. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Philos. Trans. R. Soc. B Biol. Sci. 368:20120404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, R. L. 2019. Data Lakes, Clouds, and commons: a review of platforms for analyzing and sharing genomic data. Trends Genet. 35:223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume, F. , and Rougemont J.. 2006. Nemo: an evolutionary and population genetics programming framework. Bioinformatics 22:2556–2557. [DOI] [PubMed] [Google Scholar]
- Gunderson, A. R. , Mahler D. L., and Leal M.. 2018. Thermal niche evolution across replicated Anolis lizard adaptive radiations. Proc. R. Soc. B Biol. Sci. 285:20172241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, A. M. , Brachi B., Faure N., Horton M. W., Jarymowycz L. B., Sperone F. G., et al. 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334:83–86. [DOI] [PubMed] [Google Scholar]
- Herrmann, M. , Ravindran S. P., Schwenk K., and Cordellier M.. 2018. Population transcriptomics in Daphnia: the role of thermal selection. Mol. Ecol. 27:387–402. [DOI] [PubMed] [Google Scholar]
- Hickling, R. , Roy D. B., Hill J. K., and Thomas C. D.. 2005. A northward shift of range margins in British Odonata. Glob. Chang. Biol. 11:502–506. [Google Scholar]
- Hill, J. K. , Thomas C. D., and Huntley B.. 1999. Climate and habitat availability determine 20th century changes in a butterfly's range margin. Proc. Royal Soc. London Ser. B Biolog. Sci. 266:1197–1206. [Google Scholar]
- Hoban, S. , Kelley J. L., Lotterhos K. E., Antolin M. F., Bradburd G., Lowry D. B., et al. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am. Nat. 188:379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A. A. , and Sgrò C. M.. 2011. Climate change and evolutionary adaptation. Nature 470:479–485. [DOI] [PubMed] [Google Scholar]
- Housset, J. M. , Nadeau S., Isabel N., Depardieu C., Duchesne I., Lenz P., et al. 2018. Tree rings provide a new class of phenotypes for genetic associations that foster insights into adaptation of conifers to climate change. New Phytol. 218:630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPBES . 2019. Summary for policymakers of the global assessment report on biodiversity and ecosystem services of the Intergovernmental Science‐Policy Platform on Biodiversity and Ecosystem Services In: IPBES secretariat. Díaz S., Settele J., Brondizio E. S., Ngo H. T., Guèze M., Agard J., Arneth A., et al. (eds.). IPBES, Bonn, Germany, pp. 45. [Google Scholar]
- Jaramillo‐Correa, J.‐P. , Rodríguez‐Quilón I., Grivet D., Lepoittevin C., Sebastiani F., Heuertz M., et al. 2015. Molecular proxies for climate maladaptation in a long‐lived tree (Pinus pinaster Aiton, Pinaceae). Genetics 199:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay, F. , Manel S., Alvarez N., Durand E. Y., Thuiller W., Holderegger R., et al. 2012. Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol. Ecol. 21:2354–2368. [DOI] [PubMed] [Google Scholar]
- Jump, A. S. , Hunt J. M., and Pen̈uelas J.. 2006. Rapid climate change‐related growth decline at the southern range edge of Fagus sylvatica . Glob. Chang. Biol. 12:2163–2174. [Google Scholar]
- Kapun, M. , Van Schalkwyk H., McAllister B., Flatt T., and Schlötterer C.. 2014. Inference of chromosomal inversion dynamics from Pool‐Seq data in natural and laboratory populations of Drosophila melanogaster . Mol. Ecol. 23:1813–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun, M. , Fabian D. K., Goudet J., and Flatt T.. 2016. Genomic Evidence for adaptive inversion clines in Drosophila melanogaster . Mol Biol Evol. 33:1317–1336. [DOI] [PubMed] [Google Scholar]
- Karger, D. N. , Conrad O., Böhner J., Kawohl T., Kreft H., Soria‐Auza R. W., et al. 2017. Climatologies at high resolution for the earth's land surface areas. Sci. Data 4:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, M. , Porter W. P., Williams C., Ritchie S., and Hoffmann A. A.. 2009. Integrating biophysical models and evolutionary theory to predict climatic impacts on species’ ranges: the dengue mosquito Aedes aegypti in Australia. Funct. Ecol. 23:528–538. [Google Scholar]
- Kelly, A. E. , and Goulden M. L.. 2008. Rapid shifts in plant distribution with recent climate change. PNAS 105:11823–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler, R. , and Schlötterer C.. 2013. A guide for the design of evolve and resequencing studies. Mol. Biol. Evol. 31:474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, P. L. M. , Willems F. M., Scheepens J. F., Burbano H. A., and Bossdorf O.. 2019. Using herbaria to study global environmental change. New Phytol. 221:110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir, J. , Gégout J. C., Marquet P. A., de Ruffray P., and Brisse H.. 2008. A significant upward shift in plant species optimum elevation during the 20th century. Science 320:1768‐1771. [DOI] [PubMed] [Google Scholar]
- Lewin, H. A. , Robinson G. E., Kress W. J., Baker W. J., Coddington J., Crandall K. A., et al. 2018. Earth BioGenome Project: sequencing life for the future of life. Proc. Natl. Acad. Sci. USA 115:4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer, C. , and Stölting K. N.. 2012. Whole genome sequencing (WGS) meets biogeography and shows that genomic selection in forest trees is feasible. New Phytol. 196:652–654. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang X.‐X., Mao R.‐L., Yang J., Miao C.‐Y., Li Z., et al. 2017. Ten years of landscape genomics: challenges and opportunities. Front. Plant Sci. 8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. , Liti G., Luptak A., and Tenaillon O.. 2015. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Genet. 16:567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, D. B. , Hoban S., Kelley J. L., Lotterhos K. E., Reed L. K., Antolin M. F., et al. 2017. Breaking RAD: an evaluation of the utility of restriction site‐associated DNA sequencing for genome scans of adaptation. Mol. Ecol. Resour. 17:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, D. B. , Lovell J. T., Zhang L., Bonnette J., Fay P. A., Mitchell R. B., et al. 2019. QTL × environment interactions underlie adaptive divergence in switchgrass across a large latitudinal gradient. Proc. Natl. Acad. Sci. 116:12933‐12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard, F. , Nolte V., Tobler R., Kapun M., and Schlötterer C.. 2018. A simple genetic basis of adaptation to a novel thermal environment results in complex metabolic rewiring in Drosophila . Genome Biol. 19:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboup, M. , Bahri B., Leconte M., De Vallavieille‐Pope C., Kaltz O., and Enjalbert J.. 2012. Genetic structure and local adaptation of European wheat yellow rust populations: the role of temperature‐specific adaptation. Evol. Appl. 5:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel, F. , Zumbusch M., and Feldmeyer B.. 2018. How ants acclimate: impact of climatic conditions on the cuticular hydrocarbon profile. Funct. Ecol. 32:657–666. [Google Scholar]
- Merilä, J. 2012. Evolution in response to climate change: in pursuit of the missing evidence. BioEssays 34:811–818. [DOI] [PubMed] [Google Scholar]
- Michael, T. P. , Motley S. T., Sandoval J. P., Lanz C., Weigel D., Jupe F., et al. 2018. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, L. S. , Soulé M. E., Doak D. F., and Soule M. E.. 1993. The keystone‐species concept in ecology and conservation. Bioscience 43:219–224. [Google Scholar]
- Mizrahi, T. , Goldenberg S., Heller J., and Arad Z.. 2015. Natural variation in resistance to desiccation and heat shock protein expression in the land snail Theba pisana along a climatic gradient. Physiol. Biochem. Zool. 88:66–80. [DOI] [PubMed] [Google Scholar]
- Mock, T. , Otillar R. P., Strauss J., McMullan M., Paajanen P., Schmutz J., et al. 2017. Evolutionary genomics of the cold‐adapted diatom Fragilariopsis cylindrus . Nature 541:536–540. [DOI] [PubMed] [Google Scholar]
- Oddou‐Muratorio, S. , and Davi H.. 2014. Simulating local adaptation to climate of forest trees with a Physio‐Demo‐Genetics model. Evol. Appl. 7:453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco‐Terwengel, P. , Kapun M., Nolte V., Kofler R., Flatt T., and Schlötterer C.. 2012. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Mol. Ecol. 21:4931–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini, L. , Spanier K. I., and De Meester L.. 2012. Genomic signature of natural and anthropogenic stress in wild populations of the waterflea Daphnia magna: validation in space, time and experimental evolution. Mol. Ecol. 21:2160–2175. [DOI] [PubMed] [Google Scholar]
- Özdemir, V. , Badr K. F., Dove E. S., Endrenyi L., Geraci C. J., Hotez P. J., et al. 2013. Crowd‐funded micro‐grants for genomics and “big data”: an actionable idea connecting small (artisan) science, infrastructure science, and citizen philanthropy. Omi. A J. Integr. Biol. 17:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan, C. 2006. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37:637–669. [Google Scholar]
- Pearman, P. B. , Guisan A., Broennimann O., and Randin C. F.. 2008. Niche dynamics in space and time. Trends Ecol. Evol. 23:149–158. [DOI] [PubMed] [Google Scholar]
- Pluess, A. R. , Frank A., Heiri C., Lalagüe H., Vendramin G. G., and Oddou‐Muratorio S.. 2016. Genome‐environment association study suggests local adaptation to climate at the regional scale in Fagus sylvatica . New Phytol. 210:589–601. [DOI] [PubMed] [Google Scholar]
- Prates, I. , Xue A. T., Brown J. L., Alvarado‐Serrano D. F., Rodrigues M. T., Hickerson M. J., et al. 2016. Inferring responses to climate dynamics from historical demography in neotropical forest lizards. Proc. Natl. Acad. Sci. USA 113:7978–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido, F. , and Berthold P.. 2003. Quantitative genetic analysis of migratory behavior Pp. 53–77 in Berthold P., Gwinner E., Sonnenschein E., eds. Avian migration. Springer, Berlin, New York. [Google Scholar]
- Razgour, O. , Taggart J. B., Manel S., Juste J., Ibáñez C., Rebelo H., et al. 2018. An integrated framework to identify wildlife populations under threat from climate change. Mol. Ecol. Resour. 18:18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razgour, O. , Forester B., Taggart J. B., Bekaert M., Juste J., Ibáñez C., et al. 2019. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc. Natl. Acad. Sci. 116:10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellstab, C. , Keller B., Girardclos S., Anselmetti F. S., and Spaaka P.. 2011. Anthropogenic eutrophication shapes the past and present taxonomic composition of hybridizing Daphnia in unproductive lakes. Limnol. Oceanogr. 56:292–302. [Google Scholar]
- Rellstab, C. , Gugerli F., Eckert A. J., Hancock A. M., and Holderegger R.. 2015. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 24:4348–4370. [DOI] [PubMed] [Google Scholar]
- Rellstab, C. , Zoller S., Walthert L., Lesur I., Pluess A. R., Graf R., et al. 2016. Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Mol. Ecol. 25:5907–5924. [DOI] [PubMed] [Google Scholar]
- Rellstab, C. , Fischer M. C., Zoller S., Graf R., Tedder A., Shimizu K. K., et al. 2017. Local adaptation (mostly) remains local: reassessing environmental associations of climate‐related candidate SNPs in Arabidopsis halleri . Heredity (Edinb) 118:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reside, A. E. , Butt N., and Adams V. M.. 2018. Adapting systematic conservation planning for climate change. Biodivers. Conserv. 27:1–29. [Google Scholar]
- Rolshausen, G. , Segelbacher G., Hobson K. A., and Schaefer H. M.. 2009. Contemporary evolution of eeproductive isolation and phenotypic divergence in sympatry along a migratory divide. Curr. Biol. 19:2097–2101. [DOI] [PubMed] [Google Scholar]
- Root, T. L. , Price J. T., Hall K. R., and Schneider S. H.. 2003. Fingerprints of global warming on wild animals and plants. 421:57–60. [DOI] [PubMed] [Google Scholar]
- Roschanski, A. M. , Csilléry K., Liepelt S., Oddou‐Muratorio S., Ziegenhagen B., Huard F., et al. 2016. Evidence of divergent selection for drought and cold tolerance at landscape and local scales in Abies alba Mill. in the French Mediterranean Alps. Mol. Ecol. 25:776–794. [DOI] [PubMed] [Google Scholar]
- Savolainen, O. , Lascoux M., Merilä J., and Merila J.. 2013. Ecological genomics of local adaptation. Nat. Rev. Genet. 14:807–820. [DOI] [PubMed] [Google Scholar]
- Schlötterer, C. , Kofler R., Versace E., Tobler R., and Franssen S. U.. 2015. Combining experimental evolution with next‐generation sequencing: a powerful tool to study adaptation from standing genetic variation. Heredity (Edinb) 114:431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, R. G. 2019. From the past to the future: considering the value and limits of evolutionary prediction. Am. Nat. 193. [DOI] [PubMed] [Google Scholar]
- Speed, D. , and Balding D. J.. 2015. Relatedness in the post‐genomic era: is it still useful? Nat. Rev. Genet. 16:33–44. [DOI] [PubMed] [Google Scholar]
- Strode, P. K. 2003. Implications of climate change for North American wood warblers (Parulidae). Glob. Chang. Biol. 9:1137–1144. [Google Scholar]
- Sumner, S. , Bevan P., Hart A. G., and Isaac N. J. B.. 2019. Mapping species distributions in 2 weeks using citizen science. Insect Conserv. Divers 12:382–388. [Google Scholar]
- Swan, M. , Hathaway K., Hogg C., McCauley R., and Vollrath A.. 2010. Citizen science genomics as a model for crowdsourced preventive medicine research. J. Particip. Med 2:e20. [Google Scholar]
- Taylor, M. A. , Wilczek A. M., Roe J. L., Welch S. M., Runcie D. E., Cooper M. D., et al. 2019. Large‐effect flowering time mutations reveal conditionally adaptive paths through fitness landscapes in Arabidopsis thaliana . Proc. Natl. Acad. Sci. 116:17890–17899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, O. , Rodriguez‐Verdugo A., Gaut R. L., McDonald P., Bennett A. F., Long A. D., et al. 2012. The molecular diversity of adaptive convergence. Science 335:457–461. [DOI] [PubMed] [Google Scholar]
- Tenaillon, O. , Barrick J. E., Ribeck N., Deatherage D. E., Blanchard J. L., Dasgupta A., et al. 2016. Tempo and mode of genome evolution in a 50,000‐generation experiment. Nature 536:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therkildsen, N. O. , Hemmer‐Hansen J., Hedeholm R. B., Wisz M. S., Pampoulie C., Meldrup D., et al. 2013b. Spatiotemporal SNP analysis reveals pronounced biocomplexity at the northern range margin of Atlantic cod Gadus morhua . Evol. Appl. 6:690–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therkildsen, N. O. , Wilder A. P., Conover D. O., Munch S. B., Baumann H., and Palumbi S. R.. 2019. Contrasting genomic shifts underlie parallel phenotypic evolution in response to fishing. Science 365:487–490. [DOI] [PubMed] [Google Scholar]
- Thuiller, W. , Lavorel S., Araujo M. B., Sykes M. T., and Prentice I. C.. 2005. Climate change threats to plant diversity in Europe. PNAS 102:8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano, A. , and Friesen V. L.. 2016. Genomics of local adaptation with gene flow. Mol. Ecol. 25:2144–2164. [DOI] [PubMed] [Google Scholar]
- Tobler, R. , Franssen S. U., Kofler R., Orozco‐Terwengel P., Nolte V., Hermisson J., et al. 2014. Massive habitat‐specific genomic response in D. melanogaster populations during experimental evolution in hot and cold environments. Mol. Biol. Evol. 31:364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truffaut, L. , Ducousso A., Badeau V., Ehrenmann F., Chancerel E., Kremer A., et al. 2017. Fine‐scale species distribution changes in a mixed oak stand over two successive generations. New Phytol 215:126–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylianakis, J. M. , Didham R. K., Bascompte J., and Wardle D. A.. 2008. Global change and species interactions in terrestrial ecosystems. Ecol. Lett. 11:1351–1363. [DOI] [PubMed] [Google Scholar]
- Ungerer, M. C. , Johnson L. C., and Herman M. A.. 2008. Ecological genomics: understanding gene and genome function in the natural environment. Heredity 100:178–183. [DOI] [PubMed] [Google Scholar]
- Urban, M. C. 2015. Accelerating extinction risk from climate change. Science 348:571–573. [DOI] [PubMed] [Google Scholar]
- Urban, M. C. , Bocedi G., Hendry A. P., Mihoub J.‐B., Peer G., Singer A., et al. 2016. Improving the forecast for biodiversity under climate change. Science 353:aad8466. [DOI] [PubMed] [Google Scholar]
- Valls, A. , Coll M., Christensen V., and Ellison A. M.. 2015. Keystone species: toward an operational concept for marine biodiversity conservation. Ecol. Monogr. 85:29–47. [Google Scholar]
- Van Asch, M. , Salis L., Holleman L. J. M., Van Lith B., and Visser M. E.. 2013. Evolutionary response of the egg hatching date of a herbivorous insect under climate change. Nat. Clim. Chang. 3:244–248. [Google Scholar]
- Waldvogel, A. , Wieser A., Schell T., Patel S., Schmidt H., Hankeln T., et al. 2018. The genomic footprint of climate adaptation in Chironomus riparius . Mol. Ecol. 27:1439–1456. [DOI] [PubMed] [Google Scholar]
- Walther, G.‐R. , Post E., Convey P., Menzel A., Parmesan C., Beebee T. J. C., et al. 2002. Ecological response to recent climate change. Nature 416:389–395. [DOI] [PubMed] [Google Scholar]
- Wilczek, A. M. , Burghardt L. T., Cobb A. R., Cooper M. D., Welch S. M., and Schmitt J.. 2010. Genetic and physiological bases for phenological responses to current and predicted climates. Philos. Trans. R. Soc. B Biol. Sci. 365:3129–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, C. G. , Ruhfel B., Primack R. B., Miller‐Rushing A. J., and Davis C. C.. 2008. Phylogenetic patterns of species loss in Thoreau's woods are driven by climate change. PNAS 105:17029–17033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkovich, E. M. , Jonathan Davies T., Schaefer H., Cleland E. E., Cook B. I., Travers S. E., et al. 2013. Temperature‐dependent shifts in phenology contribute to the success of exotic species with climate change. Am. J. Bot. 100:1407–1421. [DOI] [PubMed] [Google Scholar]
- Xiao, R. , Wang L., Cao Y., and Zhang G.. 2016. Transcriptome response to temperature stress in the wolf spider Pardosa pseudoannulata (Araneae: Lycosidae). Ecol. Evol. 6:3540–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Li W. R., Lv F. H., He S. G., Tian S. L., Peng W. F., et al. 2016. Whole‐genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol. Biol. Evol. 33:2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman, S. , Hodgins K. A., Lotterhos K. E., Suren H., Nadeau S., Degner J. C., et al. 2016. Convergent local adaptation to climate in distantly related conifers. Science 353:1431–1433. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , Fang X., Guo X., Li L., Luo R., Xu F., et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Wu H., Xie J., Jiang R., Deng C., and Pang H.. 2015. Transcriptome responses to heat and cold‐stress in ladybirds (Cryptolaemus montrouzieri Mulasnt) analyzed by deep‐sequencing. Biol. Res. 48:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurell, D. , Graham C. H., Gallien L., Thuiller W., and Zimmermann N. E.. 2018. Long‐distance migratory birds threatened by multiple independent risks from global change. Nat. Clim. Chang. 8:992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]


